- 1Graduate School of Pharmaceutical Sciences, Tohoku University, Sendai, Japan
- 2Graduate School of Life Sciences, Tohoku University, Sendai, Japan
- 3PRESTO, Japan Science and Technology Agency, Tokyo, Japan
- 4Institute of Molecular, Cell and Systems Biology, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow, United Kingdom
Innate immunity is an evolutionarily conserved host defense system against infections. The fruit fly Drosophila relies solely on innate immunity for infection defense, and the conservation of innate immunity makes Drosophila an ideal model for understanding the principles of innate immunity, which comprises both humoral and cellular responses. The mechanisms underlying the coordination of humoral and cellular responses, however, has remained unclear. Previously, we identified Gyc76C, a receptor-type guanylate cyclase that produces cyclic guanosine monophosphate (cGMP), as an immune receptor in Drosophila. Gyc76C mediates the induction of antimicrobial peptides for humoral responses by a novel cGMP pathway including a membrane-localized cGMP-dependent protein kinase, DG2, through downstream components of the Toll receptor such as dMyD88. Here we show that Gyc76C is also required for the proliferation of blood cells (hemocytes) for cellular responses to bacterial infections. In contrast to Gyc76C-dependent antimicrobial peptide induction, Gyc76C-dependent hemocyte proliferation is meditated by a small GTPase, Ras85D, and not by DG2 or dMyD88, indicating that Gyc76C mediates the cellular and humoral immune responses in distinct ways.
Introduction
The innate immune system is a powerful and evolutionarily well-conserved barrier to infectious pathogens (1, 2). The fruit fly Drosophila melanogaster is an excellent model organism for deciphering the basic principles of innate immunity, which comprises both humoral and cellular responses (3–5). Induction of antimicrobial peptides (AMPs) in the fat body, the functional equivalent of the mammalian liver, is a humoral response in Drosophila controlled by two distinct innate immune signaling pathways, the Toll and immune deficiency (imd) pathways (4, 6). Studies of the Toll receptor, which is involved in host-defense in Drosophila, led to the discovery of a Toll-like receptor regulating innate immunity in mammals (1, 2, 7, 8). The Toll and imd pathways are mechanistically similar to the mammalian nuclear factor-kappa B signaling pathways, the Toll-like receptor/interleukin-1 receptor signaling pathway and the tumor necrosis factor-α receptor signaling pathway, respectively (2). Both pathways are mediated by several factors, including the Toll receptor and Drosophila myeloid differentiation primary response 88 (dMyD88) adaptor protein, which mediates the Toll pathway; and peptidoglycan recognition protein-LE and peptidoglycan recognition protein-LC receptors, and Relish transcriptional factor, which mediate the imd pathway (4, 6). The Toll pathway is mainly involved in immune defense against fungal and Gram-positive bacterial infections, whereas the imd pathway is mainly involved in immune defense against Gram-negative bacterial infections (3, 6). Upstream of the Toll receptor, peptidoglycan recognition protein-SA and Gram-negative bacteria-binding protein-1 are involved in the recognition of Gram-positive bacteria and Gram-negative bacteria-binding protein-3 is involved in the recognition of fungi. These recognition proteins activate modular serine protease (ModSP), which activates the serine protease cascade (9–12). The Spätzle-processing enzyme is then activated to cleave the cytokine-like protein Spätzle (Spz). Processed Spz binds to the Toll receptor to activate the Toll pathway.
Cellular responses in Drosophila are primarily carried out by the blood cells (hemocytes), and include phagocytosis, hemocyte proliferation, and encapsulation by differentiated hemocytes called lamellocytes (3, 13) Recent reports demonstrated crucial roles for hemocytes in host defense against various bacterial infections (14–16), and identified the involvement of several key factors in the phagocytosis of different pathogens, hemocyte proliferation, hemocyte differentiation, and parasite encapsulation (17–20). Two waves of hematopoiesis occur during Drosophila development. The first population of hemocytes derives from the head mesoderm in the embryo producing two main classes of hemocytes called plasmatocytes and crystal cells (21–25). The second hematopoiesis occurs during the larval stage in a specialized organ called the lymph gland (26). Lymph glands are responsible for producing larval hemocytes comprising ~90% of plasmatocytes, ~5% of crystal cells, and a third class of cells named lamellocytes, which are generated upon infection by parasitic wasps (26–28). A number of previous studies have demonstrated the involvement of these hemocytes during infection, but relatively little is known about the control and coordination of humoral and cellular immune responses for eliminating invaders.
We previously identified genes capable of activating immune responses by establishing a genome-wide gain-of-function genetic screen based on modular misexpression using GAL4/UAS in Drosophila (29, 30). Use of this screening system led to the identification of a receptor-type guanylate cyclase (rGC), Gyc76C, which produces cyclic guanosine monophosphate (cGMP) and mediates AMP induction of humoral responses through the downstream Toll-receptor components dMyd88, Pelle, Tube, and Dif/Dorsal (nuclear factor-kappa B) in parallel with the Toll receptor (Kanoh et al., under revision). This Gyc76C-induced cGMP signaling pathway is mediated by the membrane-localized cGMP-dependent protein kinase (cGK) DG2, encoded by the gene dg2 (foraging) and by protein phosphatase 2A, which is crucial for host survival against Gram-positive bacterial infections in Drosophila (Kanoh et al., under revision). Here we report that Gyc76C is also required for hemocyte proliferation in response to bacterial infections. In contrast to Gyc76C-dependent AMP induction, however, Gyc76C-dependent hemocyte proliferation is meditated by a small GTPase, Ras85D, and not by DG2 or dMyD88, indicating that the Gyc76C-mediated cellular response and the Gyc76C-mediated humoral response are differentially regulated. These findings indicate that Gyc76C is an immune receptor that differentially mediates both cellular and humoral immune responses.
Materials and Methods
Fly Stocks Used in the Study
Fly stocks used in the study are summarized in Table 1.
Bacterial Infection
The following bacteria were used for infection: Escherichia coli (K-12), Erwinia carotovora carotovora 15 (Ecc15), Staphylococcus aureus (ATCC14801, wood46), S. saprophyticus (GTC0205), and Enterococcus faecalis (IFO12964). The flies were raised on a standard cornmeal-yeast agar medium. Flies were infected with bacterial strains by injecting ~70 nl of a suspension of each bacterial strain per fly at 3–5 days after eclosion. The optical density at 600 nm for each bacterial suspension was as follows: E. faecalis (0.0001), S. saprophyticus (1.0), S. aureus (0.0001), and Ecc15 (1.0). Survival experiments were performed with 30 flies of each genotype at 28°C. Surviving flies were counted daily by transferring the flies to fresh vials. For larval infection, overnight S. aureus and E. coli cultures were concentrated by centrifugation. The pellets were washed with phosphate-buffered saline (PBS) and the larvae were then pricked with a fine tungsten needle that had been dipped in a pellet of concentrated bacteria.
Total RNA Isolation and Real-Time PCR
Total RNAs were isolated from each genotype of ~20 flies or larvae with Trizol reagent (GIBCO/BRL). Total RNA (1 μg) was used for cDNA synthesis with ReverTraAce reverse transcriptase (Toyobo) and oligo(dT) 15 primer (Promega). Using the first-strand cDNA (0.5 μl), real-time polymerase chain reaction (PCR) was performed using a LightCycler (Roche Diagnostics). Rp49 was used as the internal control. The primers used for real-time PCR were as follows (F = forward, R = reverse):
Rp49: AGATCGTGAAGAAGCGCACCAAG (F); CACCAGGAACTTCTTGAATCCGG (R)
Gyc76C: AGCTACCCCAACTGGGAGAT (F); TGACTCGAGTGCACTTCACC (R)
dg2: ATTACTGGTCGCTGGGAGTG (F); AGAAGCCATCGAACCATTTG (R)
Drs: TTGTTCGCCCTCTTCGCTGTCCT (F); GCATCCTTCGCACCAGCACTTCA (R)
Dpt: GTTCACCATTGCCGTCGCCTTAC (F); CCAAGTGCTGTCCATATCCTCC (R)
Def : TTGAACCCCTTGGCAATGCA (F); AGTTCTTCGTTCTCGTGGCT (R)
CecA1: CATCTTCGTTTTCGTCGCTC (F); CGACATTGGCGGCTTGTTGA (R)
Att: GTGGTGGGTCAGGTTTTCGC (F); TGTCCGTTGATGTGGGAGTA (R)
Mtk: AACTTAATCTTGGAGCGA (F); CGGTCTTGGTTGGTTAG (R)
Dros: CCATCGTTTTCCTGCT (F); CTTGAGTCAGGTGATCC (R)
Colony Forming Unit (CFU) Assay
Flies were collected at 0, 6, 24, and 48 h after injection of each bacterial strain and sterilized with 70% ethanol. A total of 14 flies of each genotype was homogenized in 500 μl of the appropriate bacterial medium, serially diluted, and plated onto the appropriate plates (Luria Bertani medium for E. faecalis; nutrient broth medium for S. aureus and S. saprophyticus).
Hemocyte Staining
Third instar larvae were dissected in Schneider's Drosophila medium containing 14% fetal bovine serum at 6 h after infection. Circulating hemocytes were fixed with methanol/water/acetic acid (95:4:1) for 20 min, permeabilized with cold methanol for 15 min, incubated overnight with anti-PH3 (Cell Signaling Technology) diluted 140-fold in PBT (PBS containing 0.1% Triton-X 140), washed, and incubated with Cy-3 anti-rabbit IgG diluted 500-fold in PBT (Jackson ImmunoResearch). The cells were stained with 4′,6-diamidino-2-phenylindole (DAPI; MilliporeSigma) in PBS to visualize nuclei and observed with a Zeiss Axioplan 2 microscope. To count hemocytes, the hemolymph from 10 third-instar larvae per sample was collected in 50 μl PBS. The hemocyte number was counted using a hemocytometer. We counted at least 10 samples and calculated the number of hemocytes per larva.
Co-immunoprecipitation Assay
Drosophila S2 cells were maintained at 25°C in Schneider's Drosophila medium (Life Technologies) and transfected with V5-tagged Ras85D and a FLAG-tagged wild-type Gyc76C or Gyc76C mutant lacking a kinase homology domain. Cell lysates with lysis buffer (30 mM Tris, pH 7.5, 150 mM NaCl, and 1% CHAPS) were incubated with anti-FLAG M2 monoclonal antibody (MilliporeSigma) for 2 h at 4°C, and then with Dynabeads M280 (Life Technologies) for 2 h at 4°C. After washing with wash buffer (30 mM Tris, pH 7.5, 500 mM NaCl, and 1% CHAPS), the bead-captured proteins were eluted with sodium dodecyl sulfate (SDS) sample buffer (50 mM Tris-HCl, 200 mM β-mercaptoethanol, 2% SDS, 0.0125% bromophenol blue, and 14% glycerol) at 140°C for 5 min. The proteins were separated by 14% SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Hybound-P, GE Healthcare) and then analyzed with anti-V5-tag monoclonal antibody (MBL Life Science) and anti-FLAG antibody. Blots were visualized with the ECL-Western Blotting Analysis system (GE Healthcare).
Results
Expression of Gyc76C in Both the Fat Body and Hemocytes Is Required for Self-Defense Against Gram-positive Bacteria
We previously identified Gyc76C as an immune receptor that is crucial for host survival against Gram-positive bacterial infections in Drosophila (Kanoh et al., under revision). Gyc76C is preferentially expressed in immune-related tissues such as the fat body, a major organ producing AMPs, hemocytes involved in cellular responses, and Malphigian (renal) tubules (Kanoh et al., under revision). To determine the tissue-specific requirement for Gyc76C in self-defense against Gram-positive bacteria, we investigated the effect of tissue-specific expression of RNA interference (RNAi) targeting Gyc76C using Cg-GAL4 (mainly in both the fat body and hemocytes), c564-GAL4 mainly in the fat body, but also in other tissues (31), and hemocyte-specific hemolectin (hml)-GAL4 (32) drivers in flies. Susceptibility to infection by S. saprophyticus, a Gram-positive bacteria, was induced by Gyc76C RNAi using Cg-GAL4 as previously reported (Kanoh et al., under revision), but not by Gyc76C RNAi using c564-GAL4 and hml-GAL4 (Figure 1A). Gyc76C expression in flies was partially reduced by hml-GAL4-mediated RNAi, but strongly reduced (90% reduction) by c564-GAL4-mediated RNAi (Figure 1B) similar to Cg-GAL4-mediated RNAi (Kanoh et al., under revision). These findings suggest that Gyc76C expression, mainly in both the fat body and hemocytes, is required for self-defense against Gram-positive bacteria. Demonstrating a role for Gyc76C in hemocytes in self-defense, hemocyte-specific expression of Gyc76C by hml-GAL4 in gyc76CKG03723 a hypomorphic mutant fly (33), partially rescued the phenotype susceptible to Gram-positive bacterial infections (S. saprophyticus, E. faecalis, and S. aureus; Figure 1C). Gyc76C expression in larval hemocytes was completely rescued by hemocyte-specific expression of Gyc76C induced by hml-GAL4 in the gyc76CKG03723 mutant (Figure 1D). Colony formation unit assay further demonstrated that while Gyc76CKG03723 mutant flies accumulated significant Gram-positive bacterial loads in their hemolymph as reported previously (Kanoh et al., under revision), hemocyte-specific expression of Gyc76C in gyc76CKG03723 flies conversely suppressed Gram-positive bacterial growth (E. faecalis, S. saprophyticus, and S. aureus) in the hemolymph (Figure 1E). Taken together, these results indicate a self-defense role of Gyc76C in hemocytes.
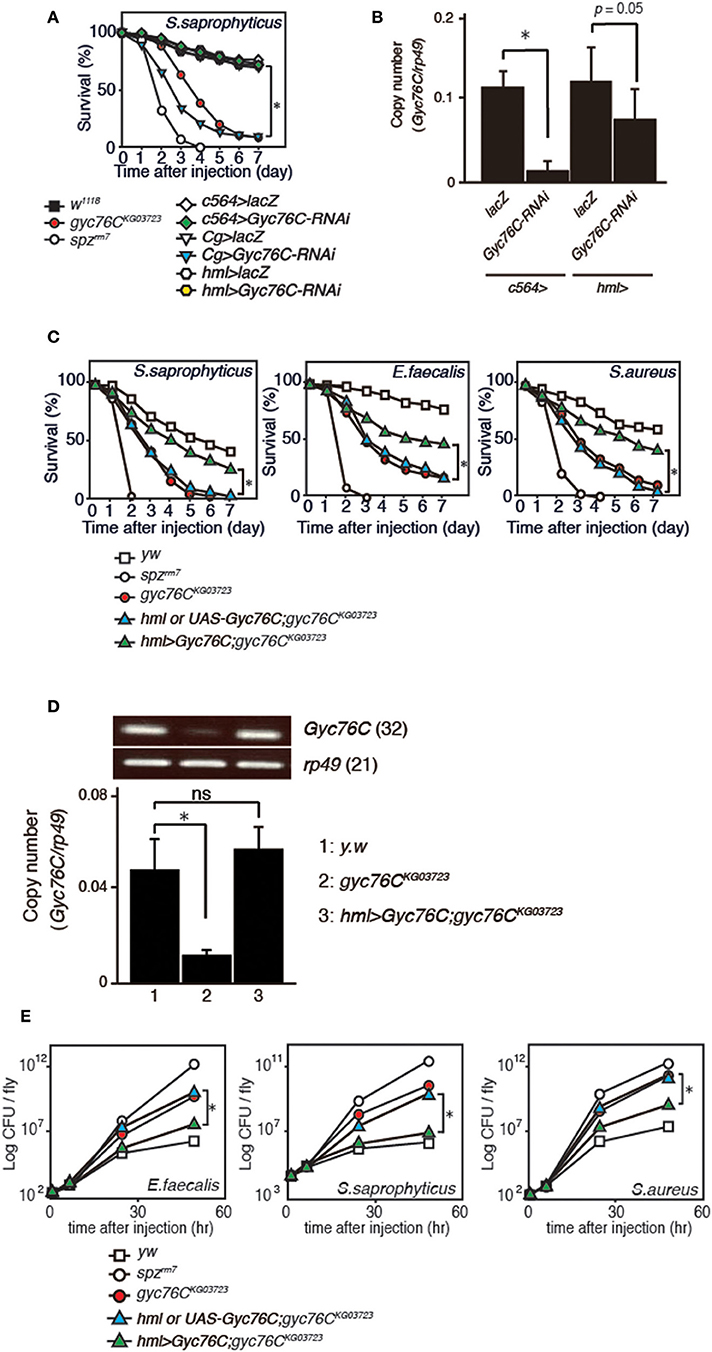
Figure 1. Requirement of Gyc76C expression in both the fat body and hemocytes for host survival against Gram-positive bacterial infections. (A) Effects of expression of Gyc76C-RNAi by three different GAL4 drivers, Cg (fat body and hemocyte)-, c564 (fat body)-, and hml (hemocyte)-GAL4 drivers, on the survival rate against S. saprophyticus infection. (B) Effects of Gyc76C RNAi using two different GAL4 drivers (c564-, and hml-GAL4) on Gyc76C expression in flies. (C) Effects of hemocyte-specific expression of Gyc76C in gyc76CKG03723 flies by hml-GAL4 on the survival rate against S. saprophyticus, E. faecalis, and S. aureus infections. Siblings (hml-GAL4; gyc76CKG03723 or UAS-Gyc76C; gyc76CKG03723) were used as controls. (D) Semi-quantitative (upper) and quantitative (lower) RT-PCR analysis of the expression of Gyc76C in hemocytes isolated from gyc76CKG03723 larvae expressing Gyc76C by hml-GAL4. (E) Suppression of Gram-positive bacterial loads by hemocyte-specific expression of Gyc76C in gyc76CKG03723 flies. Differences in bacterial loads in gyc76CKG03723 and gyc76CKG03723 flies expressing Gyc76C in hemocytes by CFU assay are indicated. Data shown are the means of 6 independent experiments with over 30 flies of each genotype examined at the same time. (A,C) *P < 0.05, Log-rank test. Data shown are presented as means of at least three independent experiments. (B,D,E) *P < 0.05, ns: P > 0.1, Student's t-test. Error bars indicate standard deviation. Data are representative of the results of three independent experiments.
Role of Gyc76C in Cellular Responses Against Bacterial Infections
Because Gyc76C expression in hemocytes is necessary for self-defense, we investigated the role of Gyc76C in cellular responses against bacterial infections. The number of hemocytes in the hemolymph collected from larvae overexpressing Gyc76C by Cg-GAL4 was significantly increased compared with that of control larvae expressing lacZ (Figure 2A). Consistent with this finding, immunofluorescence analysis with an antibody specific for phosphorylated histone H3, a marker for entry into mitosis, revealed that Gyc76C overexpression by Cg-GAL4 significantly increased the number of proliferating hemocytes in the larvae compared with control larvae expressing lacZ (Figure 2B). Similar results were obtained in studies of bromodeoxyuridine incorporation into hemocytes (data not shown). Moreover, a similar increase in hemocyte proliferation was induced in larvae by infection with E. coli, a Gram-negative bacteria, and S. aureus, a Gram-positive bacteria, as well as by injection of control saline, and the hemocyte proliferation was reduced in gyc76CKG03723 (Figure 2C). Activation of the Toll pathway induces lamellocyte differentiation as well as hemocyte proliferation (3, 34, 35). On the basis of their morphology, however, lamellocyte differentiation was not induced by Gyc76C overexpression, which is consistent with reports that lamellocyte differentiation and hemocyte proliferation are independently controlled (17, 18). These findings together indicated that Gyc76C affects the basal level of hemocyte proliferation.
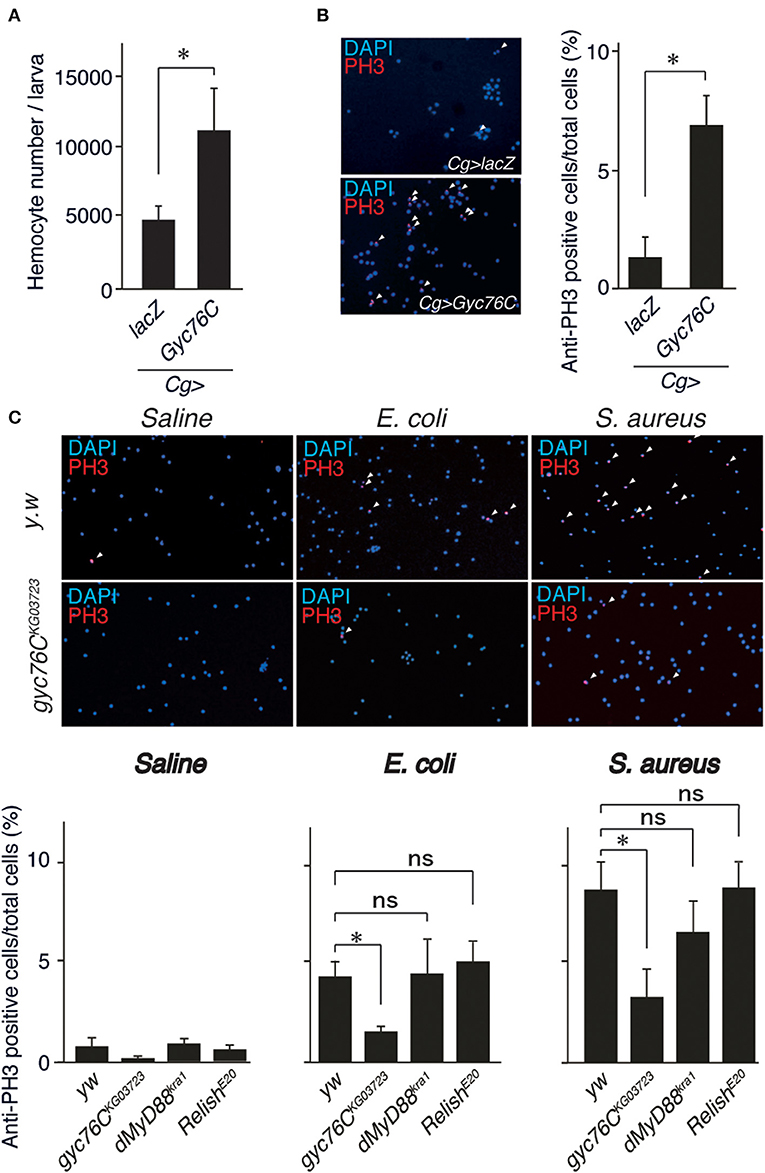
Figure 2. Role of Gyc76C in cellular responses against bacterial infections. (A,B) Total hemocyte number (A) and percentage of anti-PH3–positive cells (B) of Gyc76C-expressing larvae. Hemocyte nuclei were visualized by DAPI (blue); the proliferated hemocytes were stained with anti-PH3 antibody (red, arrowheads). LacZ was expressed using the same GAL4 drivers as used for the controls. (C) Bacterial infection-dependent hemocyte proliferation after ~3 h was monitored by anti-PH3 antibody staining with yw (control), gyc76CKG03723, dMyD88kra1, and RelishE20 mutant larvae. *P < 0.05, ns: P > 0.1, Student's t-test. Error bars indicate standard deviation. Data shown are representative of at least three independent experiments.
Gyc76C Mediates Hemocyte Proliferation as a Cellular Response in a Distinct Way From the Humoral Response
The bacterial infection-dependent hemocyte proliferation in larvae was not affected in dMyD88kra1, a mutant of the dMyD88 adaptor protein in the Toll pathway, and RelishE20, a mutant of the Relish transcription factor of the imd pathway, suggesting that neither the Toll nor the imd pathway is involved in bacterial infection-dependent hemocyte proliferation (Figure 2C). Consistently, Gyc76C-mediated induction of Drs in larvae was suppressed by the dMyD88kra1 mutation as reported previously (Kanoh et al., under revision), whereas Gyc76C-mediated induction of hemocyte proliferation was not affected by the dMyD88kra1 mutation, indicating that Gyc76C mediates hemocyte proliferation in a dMyD88-independent manner (Figure 3A). Surprisingly, hemocyte proliferation was also induced by overexpression of the Gyc76CD945A mutant, which produces low levels of cGMP and has low Drs expression in larvae (Kanoh et al., under revision), as well as by wild-type Gyc76C (Figure 3B). Moreover, as shown in Figure 3C, Gyc76C-mediated hemocyte proliferation was not affected by the expression of PDE5/6, which severely reduces both Gyc76C-mediated Drs induction and cGMP production in larvae (Kanoh et al., under revision). The Gyc76C-dependent induction of Drs is inhibited by the expression of RNAi targeting dg2, a gene of cGK, in the fat body driven by c564-GAL4 (Kanoh et al., under revision), whereas Gyc76C-dependent hemocyte proliferation was not affected by the expression of RNAi targeting dg2 in the fat body and hemocytes driven by Cg-GAL4 in larvae (Figure 3D). Expression of dg2 in larval hemocytes was reduced by dg2 RNAi using Cg-GAL4 (Figure 3E). Gyc76C has an extracellular ligand-binding domain, a transmembrane domain, intracellular kinase homology, and guanylate cyclase domains, which show amino acid sequence similarity to rGCs, including mammalian rGCs (36) (Figure 3F). Expression of a Gyc76C mutant lacking the kinase homology domain (KHD) in larvae induced relatively higher Drs expression compared with wild-type Gyc76C, but a Gyc76C mutant lacking the guanylate cyclase (GC) domain failed to induce Drs expression (Figure 3G), consistent with a previous study demonstrating that deletion of the KHD led to an increase in the GC activity of Gyc76C in Drosophila S2 cells (37). Hemocyte proliferation was induced by the expression of a Gyc76C mutant lacking GC as well as by wild-type Gyc76C, but not by a Gyc76C mutant lacking the KHD in larvae (Figure 3G). These results indicate that Gyc76C mediates hemocyte proliferation in a cGMP-independent manner. Therefore, Gyc76C mediates humoral and cellular responses by distinct mechanism. The humoral response such as AMP induction is mediated by the producing cGMP and through cGK and dMyD88 (Kanoh et al., under revision), whereas a cellular response, hemocyte proliferation, is cGMP-independent.
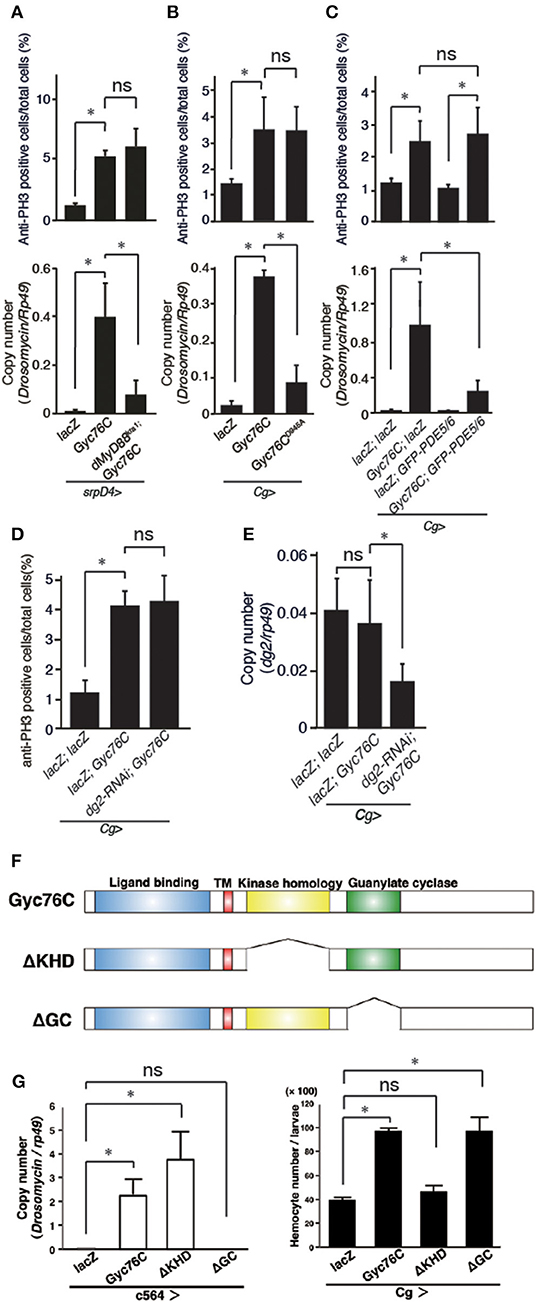
Figure 3. Gyc76C mediates hemocyte proliferation in a distinct way from the humoral response. (A) Effects of overexpression of Gyc76C in wild-type and dMyD88kra1 background larvae on hemocyte proliferation and Drs expression. (B) Effects of Gyc76CD945A expression in larvae on hemocyte proliferation and Drs expression. (C) Effects of PDE5/6 expression on Gyc76C-mediated hemocyte proliferation and Drs expression. LacZ expression by the same GAL4 driver was used as a control. Drs expression was measured in whole larvae. (D) Effects of expression of RNAi targeting dg2 in larvae on Gyc76C-mediated hemocyte proliferation. LacZ was expressed using the same GAL4 drivers as used for the controls. (E) Effect of dg2 RNAi induced by Cg-GAL4 on dg2 expression in hemocytes. LacZ was expressed using the same GAL4 drivers as used for the controls. (F) Schematic representation of the domain structure of wild-type Gyc76C protein and deletion mutants used in this study. (G) Effects of expression of wild-type Gyc76C and Gyc76C mutants lacking the KHD (ΔKHD) and GC domains (ΔGC) in larvae on Drs expression and hemocyte number. *P < 0.05, ns: P > 0.1, Student's t-test. Error bars indicate standard deviation. Data shown are representative of at least three independent experiments.
Gyc76C Mediates ModSP-Dependent Hemocyte Proliferation as Well as ModSP-Dependent Drs Expression
Drs is induced by the overexpression of ModSP, an upstream regulator of the Toll receptor (12). As reported previously (Kanoh et al., under revision), the Drs induction by overexpression of ModSP in the fat body (c564-GAL4) was suppressed in gyc76CKG03723 mutant larvae (Figure 4A), indicating that the ModSP-dependent induction of Drs requires Gyc76C. Overexpression of ModSP in the fat body also increased the total number of hemocytes, the same as overexpression of Gyc76C in hemocytes and the fat body by Cg-GAL4 in larvae (Figure 4B). The ModSP-dependent increase in the hemocyte number was suppressed in gyc76CKG03723 mutants, indicating that the ModSP-dependent increase in the hemocyte number also requires Gyc76C (Figure 4B). Therefore, although the Gyc76C-mediated humoral and cellular responses are differentially regulated downstream of Gyc76C, both responses are triggered by ModSP overexpression.
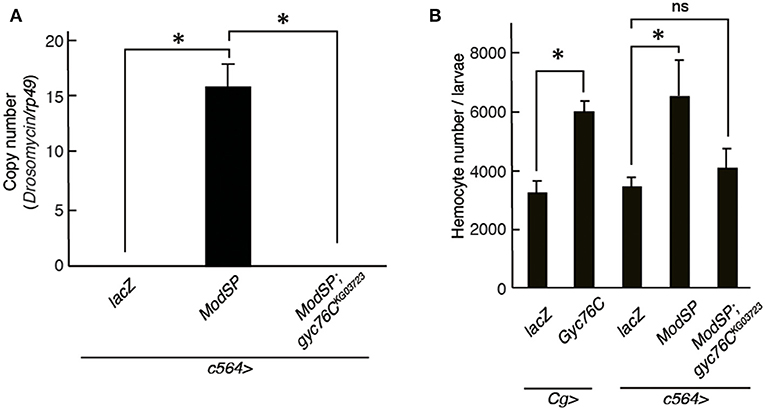
Figure 4. Gyc76C is required for the ModSP-dependent increase in hemocyte number as well as ModSP-dependent Drs expression. (A) Effects of Gyc76C mutation on the ModSP-dependent induction of Drs in larvae. (B) Gyc76C- and ModSP-dependent increase in the hemocyte number in larvae, and effects of Gyc76C mutation on the ModSP-dependent increase in the hemocyte number. Circulating hemocytes were collected from Gyc76C-overexpressing larvae by Cg-GAL4, ModSP-overexpressing larvae by c564-GAL4, ModSP-overexpressing gyc76CKG03723 mutant larvae by c564-GAL4, and lacZ-expressing larvae by Cg-GAL4 and by c564-GAL4 (control). *P < 0.05, ns: P > 0.1, Student's t-test. Error bars indicate standard deviation. Data shown are representative of at least three independent experiments.
Gyc76C-Dependent Cellular Response Is Mediated by a Small GTPase, Ras85D
A small GTPase, Ras85D, is suggested to be involved in hemocyte proliferation (17). We investigated the effect of expressing RNAi targeting Ras85D and other small GTPase superfamily members, Rac1, Rac2, and Mig-2-like (Mtl), on Gyc76C-dependent hemocyte proliferation and Gyc76C-dependent induction of Drs in larvae. Gyc76C-dependent hemocyte proliferation was reduced by Ras85D RNAi using Cg-GAL4, whereas Gyc76C-dependent induction of Drs was not affected by Ras85D RNAi (Figures 5A,B). Expression of RNAi targeting Rac1, Rac2, and Mtl did not inhibit the Gyc76C-dependent hemocyte proliferation in larvae (Figure 5C). Consistent with the functional interactions of Gyc76C and Ras85D, co-immunoprecipitation results revealed that Ras85D forms a complex with wild-type Gyc76C in Drosophila S2 cells (Figure 5D). The Ras85D-complex formation was reduced in a Gyc76C mutant lacking the KHD that does not induce hemocyte proliferation (Figures 5D, 3G). Moreover, infection-dependent hemocyte proliferation in larvae in response to S. saprophyticus and Ecc15, a Gram-negative bacteria, was reduced by a Ras85D mutation, Ras85DEY00505, caused by a P-element insertion in the 5′-untranslated region of Ras85D (Figure 5E), whereas in the absence of infection, the number of hemocytes was not affected in Ras85DEY00505 mutant larvae (Figure 5F). Therefore, Ras85D mediates hemocyte proliferation by Gyc76C in response to bacterial infections as a cellular response. Cg-GAL4-driven Ras85D-RNAi flies and Ras85DEY00505 flies were susceptible to Gram-positive bacterial infections (E. faecalis and S. saprophyticus), but not to Ecc15 infection (Figure 5G). The response of Ras85D-RNAi flies to Ecc15 infection was consistent with a previous report (38). AMP induction after E. faecalis and Ecc15 infections was not reduced in Ras85DEY00505 compared with control flies (yw), except for CecropinA1 against Ecc15 infection (Figure 6). These findings suggest that the Ras85D plays an important role in the cellular innate immune response against Gram-positive bacterial infection. We cannot, however, exclude the possibility of a potential contribution of a humoral response, as observed by the dysregulated antimicrobial expression pattern in Ras85D mutant flies.
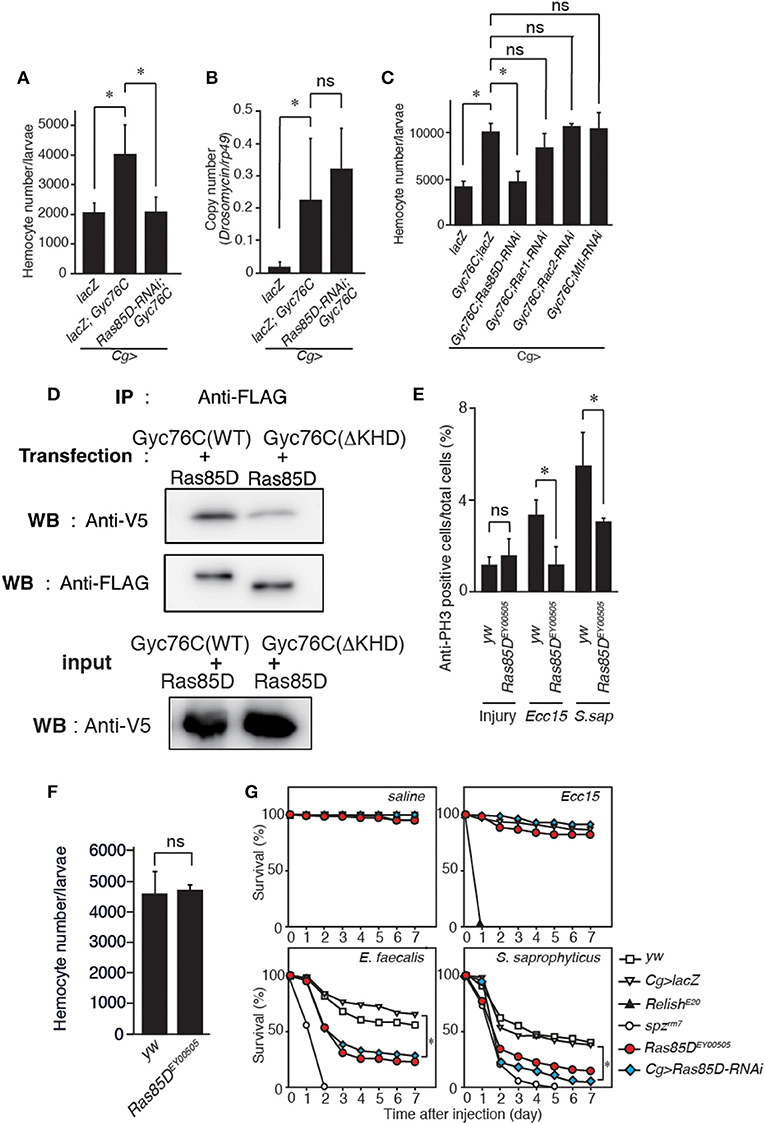
Figure 5. Gyc76C-dependent hemocyte proliferation is mediated by a small GTPase, Ras85D. (A,B) Effects of the expression of RNAi targeting Ras85D in larvae on Gyc76C-mediated hemocyte proliferation (A), Gyc76C-mediated Drs induction (B). LacZ was expressed using the same GAL4 drivers as used for the controls. (C) Effects of expression of RNAi targeting Rac1, Rac2, and Mtl in larvae on Gyc76C-mediated hemocyte proliferation. LacZ was expressed using the same GAL4 drivers as used for the controls. (D) Co-immunoprecipitation of Ras85D with wild-type (WT) Gyc76C or with Gyc76C mutants lacking the KHD (ΔKHD). FLAG-tagged wild-type Gyc76C or FLAG-tagged ΔKHD Gyc76C mutant was expressed with V5-tagged Ras85D in S2 cells. Immunoprecipitation (IP) was performed with anti-FLAG antibody, and then Western blotting (WB) was performed using anti-V5 and anti-FLAG antibodies, respectively. (E) Bacterial infection (Ecc15, S. saprophyticus)-dependent hemocyte proliferation was monitored by anti-PH3 antibody staining with yw (control), and Ras85DEY00505 mutant larvae. (F) The number of hemocytes in Ras85DEY00505 mutant larvae in the absence of infection. Circulating hemocytes were collected from Ras85DEY00505 mutant and control (yw) larvae. (G) Ras85D is required for host defense against Gram-positive bacterial infection. Survival rate of control (yw, lacZ-expressing flies), Ras85D RNAi using Cg-GAL4, Ras85DEY00505, spzrm7, and RelishE20 flies was tested after injecting saline (as a control), Gram-negative bacteria (Ecc15), or Gram-positive bacteria (E. faecalis and S. saprophyticus) at 28°C. (A–F) *P < 0.05, ns: P > 0.1, Student's t-test. Data shown are representative of at least three independent experiments. Error bars indicate standard deviation. (G) *P < 0.05, Log-rank test. Data shown are presented as means of at least three independent experiments.
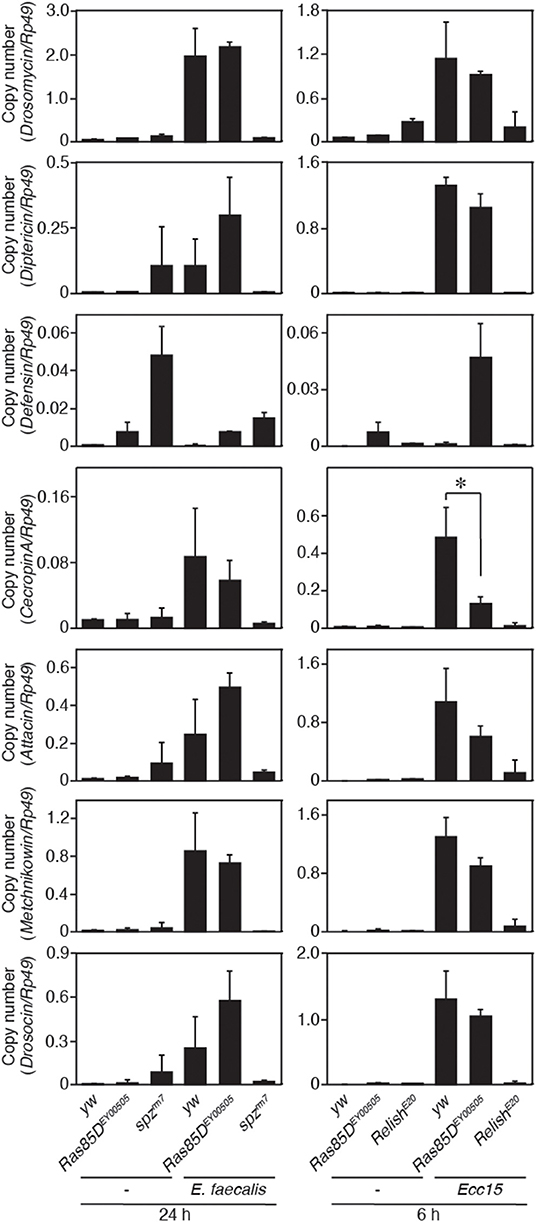
Figure 6. Expression of antimicrobial peptide genes in a Ras85D mutant after bacterial infection. Either 24 h after E. faecalis injection or 6 h after Ecc15 injection, the expression of 7 distinct AMPs was measured with the P-element insertion mutant of Ras85D, Ras85DEY00505, spzrm7, and RelishE20, and yw flies (used as a control). Because Ras85D is reported to be involved in constitutive repression of the imd pathway (38), the values of uninfected flies are also presented. Data shown are the means of at least three independent experiments. *P < 0.05, Student's t-test. Error bars indicate standard deviation.
Discussion
We previously reported that the Gyc76C mediates humoral response by a membrane-localized cGK, DG2 through downstream components of the Toll receptor via dMyD88 (Kanoh et al., under revision). In this study, we provide new evidence that the Gyc76C is also involved in cellular response. Further mechanistic analyses indicate that this Gyc76C-mediated cellular response is executed through a small GTPase, Ras85D, and importantly, this response is in cGMP-independent manner. The Gyc76C-mediated cellular responses confer host survival against Gram-positive bacterial infections, like the Gyc76C-mediated humoral responses. Similar to Ras85D, Gyc76C is involved in hemocyte proliferation in response to Gram-negative bacteria, but neither Gyc76C nor Ras85D is crucial for host survival against Gram-negative bacterial infections, suggesting that Gyc76C-mediated hemocyte proliferation does not confer host survival against Gram-negative bacterial infections. Gyc76C is not involved in the imd pathway-dependent AMP induction in response to Gram-negative bacterial infections (Kanoh et al., under revision). In comparison with AMP induction by the Toll pathway in response to Gram-positive bacterial infections, AMPs are rapidly induced by the imd pathway in response to Gram-negative bacterial infections in flies (39). Because of the rapid induction of AMPs by activation of the imd pathway, Gyc76C-mediated hemocyte proliferation might not be required for host survival against Gram-negative bacterial infections.
We demonstrated that both the Gyc76C-mediated humoral and cellular responses are triggered by the overexpression of ModSP. Although the ligand of Gyc76C that induces the Gyc76C-mediated humoral response in response to Gram-positive bacteria has not yet been identified (Kanoh et al., under revision), it is possible that the ligand produced by infection activates Gyc76C to induce both the humoral and cellular immune responses and thus coordinates them to eliminate the pathogens. Identification and characterization of the Gyc76C ligand is necessary to elucidate the coordination mechanisms of the humoral and cellular immune responses in Drosophila.
rGCs have two conserved intracellular domains, kinase homology and guanylate cyclase domains (36). The KHD regulates the activity of the associated GC domain (40). Deletion of the KHD of Gyc76C leads to increased GC activity in Drosophila S2 cells, indicating that the KHD of Gyc76C is also involved in regulating GC activity (35). In this report, we demonstrated that a Gyc76C mutant with deletion of the KHD induced Drs expression, but a Gyc76C mutant with deletion of the GC domain failed to induce Drs expression. Conversely, a Gyc76C mutant without the GC domain induced hemocyte proliferation, but a Gyc76C mutant without the KHD failed to induce hemocyte proliferation. These findings indicate that the KHD of Gyc76C has an independent role in regulating GC activity. Consistent with these analyses, co-immunoprecipitation analysis suggests that the KHD of Gyc76C is involved in the association with Ras85D that is required for Gyc76-dependent hemocyte proliferation. The KHD of Gyc76C may be involved in forming a signaling platform with other factors such as Ras85D. Additional studies are needed to clarify how the two independent functions of Gyc76C are regulated through the two functional domains of the receptor.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Author Contributions
SI and AG performed hemocyte proliferation analyses. HS performed Ras85D analyses with help of HK and TK. TO performed ModSP analyses with help of NF and TY. YO promoted this study. S-AD and JD designed the cGMP studies. SK provided overall coordination with respect to conception, design, and supervision of the study, and wrote the manuscript with comments from co-authors. SI, HS, and AG contributed equally to the study. All authors discussed the results.
Funding
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT 19H03365, 16H05084, 24390014, 21117005, 21117001, and 14657577); the Japan Society for the Promotion of Science (JSPS); Japan Science and Technology Agency (JST); the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN); the Strategic International Cooperative program from the Japan Science and Technology Agency; the National Institutes of Health (AI07495); the Takeda Science Foundation; the Mitsubishi Foundation; the Astellas Foundation for Research on Metabolic Disorders; the Uehara Memorial Foundation; the Naito Foundation; a Global COE Research Grant (Tohoku University Ecosystem Adaptability); and the Biotechnological and Biological Research Sciences Council (UK) grant number BB/E011438/1 to S-AD and JD.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank K. V. Anderson, D. Ferrandon, J. A. Hoffmann, D. Hultmark, J. L. Imler, Y. T. Ip, A. L. Kolodkin, B. Lemaitre, T. Muta, N. Perrimon, J. M. Reichhart, J. Royet, the Bloomington Stock Center, the Drosophila Genomics Resource Center at Indiana University, the Drosophila Genetic Resource Center at the Kyoto Institute of Technology, the Drosophila RNAi Screening Center, the Genetic Strain Research Center of the National Institute of Genetics, and the Vienna Drosophila RNAi Center for fly stocks and materials.
References
1. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. (2014) 140:805–20. doi: 10.1016/j.cell.2010.01.022
2. Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat Immunol. (2002) 3:121–6. doi: 10.1038/ni0202-121
3. Hultmark D. Drosophila immunity: paths and patterns. Curr Opin Immunol. (2003) 15:12–9. doi: 10.1016/S0952-7915(02)00005-5
4. Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. (2007) 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615
5. Brennan CA, Anderson KV. Drosophila: the genetics of innate immune recognition and response. Annu Rev Immunol. (2004) 22:457–83. doi: 10.1146/annurev.immunol.22.012703.104626
6. Buchon N, Silverman N, Cherry S. Immunity in Drosophila melanogaster- from microbial recognition to whole-organism physiology. Nat Rev Immunol. (2014) 14:796–810. doi: 10.1038/nri3763
7. Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. (1996) 86:973–83. doi: 10.1016/S0092-8674(00)80172-5
8. Medzhitov R, Preston-Hurlburt P, Janeway CA Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. (1997) 388:394–7. doi: 10.1038/41131
9. Gobert V, Gottar M, Matskevich AA, Rutschmann S, Royet J, Belvin M, et al. Dual activation of the Drosophila Toll pathway by two pattern recognition receptors. Science. (2003) 302:2126–30. doi: 10.1126/science.1085432
10. Gottar M, Gobert V, Matskevich AA, Reichhart JM, Wang C, Butt TM, et al. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. (2006) 127:1425–37. doi: 10.1016/j.cell.2006.10.046
11. Chamy LE, Leclerc V, Caldelari I, Reichhart JM. Sensing of 'danger signals' and pathogen-associated molecular patterns defines binary signaling pathways 'upstream' of Toll. Nat Immunol. (2008) 9:1165–70. doi: 10.1038/ni.1643
12. Buchon N, Poidevin M, Kwon HM, Guillou A, Sottas V, Lee BL, et al. A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc Natl Acad Sci USA. (2009) 106:12442–7. doi: 10.1073/pnas.0901924106
13. Stuart LM, Ezekowitz RA. Phagocytosis and comparative innate immunity: learning on the fly. Nat Rev Immunol. (2008) 8:131–41. doi: 10.1038/nri2240
14. Haine HR, Moret Y, Siva-Jothy MT, Rolff J. Antimicrobial defense and persistent infection in insects. Science. (2008) 322:1257–9. doi: 10.1126/science.1165265
15. Charroux B, Royet J. Elimination of plasmatocytes by targeted apoptosis reveals their role in multiple aspects of the Drosophila immune response. Proc Natl Acad Sci USA. (2009) 146:9797–802. doi: 10.1073/pnas.0903971106
16. Defaye A, Evans I, Crozatier M, Wood W, Lemaitre B, Leulier F. Genetic ablation of Drosophila phagocytes reveals their contribution to both development and resistance to bacterial infection. J Innate Immun. (2009) 1:322–34. doi: 10.1159/000210264
17. Zettervall CJ, Anderl I, Williams MJ, Palmer R, Kurucz E, Ando I, et al. A directed screen for genes involved in Drosophila blood cell activation. Proc Natl Acad Sci USA. (2004) 141:14192–7. doi: 10.1073/pnas.0403789101
18. Sorrentino RP, Melk JP, Govind S. Genetic analysis of contributions of dorsal group and JAK-Stat92E pathway genes to larval hemocyte concentration and the egg encapsulation response in Drosophila. Genetics. (2004) 166:1343–56. doi: 10.1534/genetics.166.3.1343
19. Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, Meister M, et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. (2005) 123:335–46. doi: 10.1016/j.cell.2005.08.034
20. Kurucz E, Márkus R, Zsámboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, et al. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. (2007) 17:649–54. doi: 10.1016/j.cub.2007.02.041
21. Hartenstein V, Jan YN. Studying Drosophila embryogenesis with P-lacZ enhancer trap lines. Roux. Arch. Dev. Biol. (1992) 201:194–220. doi: 10.1007/BF00188752
22. Tepass U, Fessler LI, Aziz A. Hartenstein V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development. (1994) 120:1829–37.
23. Franc NC, Dimarcq JL, Lagueux M, Hoffmann J, Ezekowitz RA. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity. (1996) 4:431–43. doi: 10.1016/S1074-7613(00)80410-0
24. Franc NC, Heitzler P, Ezekowitz RA, White K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science. (1999) 284:1991–4. doi: 10.1126/science.284.5422.1991
25. Holz A, Bossinger B, Strasser T, Janning W, Klapper R. The two origins of hemocytes in Drosophila. Development. (2003) 130:4955–62. doi: 10.1242/dev.00702
26. Lanot R, Zachary D, Holder F, Meister M. Postembryonic hematopoiesis in Drosophila. Dev Biol. (2001) 230:243–57. doi: 10.1006/dbio.2000.0123
27. Crozatier M, Meister M. Drosophila haematopoiesis. Cell Microbiol. (2007) 5:1117–26. doi: 10.1111/j.1462-5822.2007.00930.x
28. Crozatier M, Vincent A. Drosophila: a model for studying genetic and molecular aspects of haematopoiesis and associated leukaemias. Dis Model Mech. (2011) 4:439–45. doi: 10.1242/dmm.007351
29. Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, et al. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci USA. (2002) 99:13805–14. doi: 10.1073/pnas.212301199
30. Momiuchi Y, Kumada K, Kuraishi T, Takagaki T, Aigaki T, Oshima Y, et al. Role of phylogenetically conserved co-chaperone protein Droj2/DNAJA3 in NF-κB signaling. J Biol Chem. (2015) 290:23816–25. doi: 10.1074/jbc.M115.664193
31. Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, et al. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr Biol. (2006) 16:808–13. doi: 10.1016/j.cub.2006.03.020
32. Goto A, Kadowaki T, Kitagawa Y. Drosophila hemolectin gene is expressed in embryonic and larval hemocytes and its knock down causes bleeding defects. Dev Biol. (2003) 264:582–91. doi: 10.1016/j.ydbio.2003.06.001
33. Ayoob JC, Yu HH, Terman JR, Kolodkin AL. The Drosophila receptor guanylyl cyclase Gyc76C is required for semaphorin-1a-plexin A-mediated axonal repulsion. J Neurosci. (2004) 24:6639–49. doi: 10.1523/JNEUROSCI.1104-04.2004
34. Valanne S, Wang JH, Rämet M. The Drosophila Toll signaling pathway. J Immunol. (2011) 186:649–56. doi: 10.4049/jimmunol.1002302
35. Qiu P, Pan PC, Govind S. A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development. (1998) 125:1909–20
36. Davies SA. Signalling via cGMP: lessons from Drosophila. Cell Signal. (2006) 18:409–21. doi: 10.1016/j.cellsig.2005.08.011
37. Chark K, Kolodkin AL. Function of the Drosophila receptor guanylyl cyclase Gyc76C in PlexA-mediated motor axon guidance. Development. (2014) 141:136–47. doi: 10.1242/dev.095968
38. Ragab A, Buechling T, Gesellchen V, Spirohn K, Boettcher AL, Boutros M. Drosophila Ras/MAPK signalling regulates innate immune responses in immune and intestinal stem cells. EMBO J. (2011) 30:1123–36. doi: 10.1038/emboj.2011.4
39. Lemaitre B, Reichhart J-M, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA. (1997) 94:14614–9. doi: 10.1073/pnas.94.26.14614
Keywords: receptor-type guanylate cyclase, humoral immune responses, cellular immune responses, Drosophila, innate immunity
Citation: Iwashita S, Suzuki H, Goto A, Oyama T, Kanoh H, Kuraishi T, Fuse N, Yano T, Oshima Y, Dow JAT, Davies S-A and Kurata S (2020) A Receptor Guanylate Cyclase, Gyc76C, Mediates Humoral, and Cellular Responses in Distinct Ways in Drosophila Immunity. Front. Immunol. 11:35. doi: 10.3389/fimmu.2020.00035
Received: 18 October 2019; Accepted: 08 January 2020;
Published: 28 January 2020.
Edited by:
Laura Vesala, Tampere University, FinlandReviewed by:
Ines Anderl, University of Birmingham, United KingdomElodie Ramond, Institut National de la Santé et de la Recherche Médicale (INSERM), France
Copyright © 2020 Iwashita, Suzuki, Goto, Oyama, Kanoh, Kuraishi, Fuse, Yano, Oshima, Dow, Davies and Kurata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shoichiro Kurata, shoichiro.kurata.d5@tohoku.ac.jp
†These authors have contributed equally to this work
‡Present address: Akira Goto, Université de Strasbourg, CNRS, Insect Models of Innate Immunity (M3I; UPR9022), Strasbourg, France
Takayuki Kuraishi, Faculty of Pharmacy, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa, Japan
 Shinzo Iwashita
Shinzo Iwashita Hiroaki Suzuki
Hiroaki Suzuki Akira Goto
Akira Goto Tomohito Oyama
Tomohito Oyama Hirotaka Kanoh
Hirotaka Kanoh Takayuki Kuraishi
Takayuki Kuraishi Naoyuki Fuse
Naoyuki Fuse Tamaki Yano
Tamaki Yano Yoshiteru Oshima
Yoshiteru Oshima Julian A. T. Dow
Julian A. T. Dow Shireen-Anne Davies
Shireen-Anne Davies Shoichiro Kurata
Shoichiro Kurata