- 1Department of Pediatrics, Morsani College of Medicine, University of South Florida, Tampa, FL, United States
- 2Department of Molecular Genetics, Children's Research Institute, Johns Hopkins All Children's Hospital, St. Petersburg, FL, United States
The gut microbiome of animals consists of diverse microorganisms that include both prokaryotes and eukaryotes. Complex interactions occur among these inhabitants, as well as with the immune system of the host, and profoundly influence the overall health of both the host and its microbial symbionts. Despite the enormous importance for the host to regulate its gut microbiome, the extent to which animals generate immune-related molecules with the capacity to directly influence polymicrobial interactions remains unclear. The urochordate, Ciona robusta, is a model organism that has been adapted to experimental studies of host/microbiome interactions. Ciona variable-region containing chitin-binding proteins (VCBPs) are innate immune effectors, composed of immunoglobulin (Ig) variable regions and a chitin-binding domain (CBD) and are expressed in high abundance in the gut. It was previously shown that VCBP-C binds bacteria and influences both phagocytosis by granular amoebocytes and biofilm formation via its Ig domains. We show here that the CBD of VCBP-C independently recognizes chitin molecules present in the cell walls, sporangia (spore-forming bodies), and spores of a diverse set of filamentous fungi isolated from the gut of Ciona. To our knowledge, this is the first description of a secreted Ig-containing immune molecule with the capacity to directly promote transkingdom interactions through simultaneous binding by independent structural domains and could have broad implications in modulating the establishment, succession, and homeostasis of gut microbiomes.
Introduction
The gut microbiota consists of diverse communities of microorganisms, including: bacteria, archaea, viruses, fungi, and other microbial eukaryotes. Investigations utilizing diverse model systems have defined an active role of the microbiota in both normal host physiology (1) and disease (2, 3). Bacteria are the main components of the microbiota, having been shown to comprise >99% of the digestive tract microbial cohort (4). Bacteria are the best understood constituent of the microbiota, often serving diverse roles in host physiology (5). In recent years, the fungal component, or “mycobiota,” is becoming increasingly better recognized (6–8). It is estimated to constitute 0.01–0.1% of the microbial community in humans (4, 8, 9); however, the fungal contribution to the microbiome could be under-represent by the paucity of fungal genome reference sequences (8, 10). The gut mycobiome is not stable over time (7, 11, 12) and the low abundance and diversity of its composition are influenced by different factors, such as diet, gender, age, and geographical location (13, 14).
A recent report suggests that fungi do not efficiently colonize the digestive tract, rather their presence in the gut may be linked to both diet and the oral flora (15). However, a number of studies demonstrate that the presence of commensal fungi could be important for immune homeostasis. Fungal dysbiosis is associated with several disease etiologies (10, 16, 17). Interspecific interactions among fungi and fungi-bacteria and/or fungi/host can significantly impact health and disease (16). The gut mycobiome has been linked to different gastrointestinal pathologies, including: inflammatory bowel disease (IBD) (18, 19), irritable bowel syndrome (20, 21), peptic ulcers (22), antibiotic associated diarrhea (23) as well as graft-vs.-host disease (24, 25), and to crosstalk with the brain (26).
Although there is a dearth of knowledge on if and how fungi interact with host immune system components under normal “commensal” or healthy conditions or during times of microbial dysbiosis, much is known about how diverse innate immune receptors interact with fungal pathogens (10, 27). Innate immune cells, which have a phylogenetic history extending well-beyond the vertebrates, are equipped with a range of pattern recognition receptors (PRR) to sense and interact with different fungal pathogen-associated molecular patterns (PAMPs), as components of fungal cell wall, including β-glucan, mannans, mannoproteins, chitin, and also fungal-derived nucleic acids (10, 27). Notably, most of these PRRs are localized on the cell surface or in the cytoplasm of blood cells (17, 28) and the binding to their ligands shape antifungal immune responses, activating various signaling cascades that result in fungal internalization via phagocytosis, cytokine production, and/or production of reactive nitrogen and oxygen species (29, 30). Whereas, it has been shown that certain immune effectors influence the growth of bacteria in ways that often promote stable biofilms and/or shape barrier defenses (31–35), equivalent responses have yet to be documented for any fungal species.
We previously have described the variable region containing chitin binding proteins (VCBPs)-encoding gene families in two protochordate model systems (36–38). In C. robusta, four members of the VCBP gene family (VCBP-A, -B, -C, and -D) have been described (38). VCBP A-C consist of two immunoglobulin (Ig)-like variable domains at the N-terminus and a chitin binding domain (CBD) at the C-terminus (39). Unique temporal-spatial expression patterns among VCBP genes during morphogenesis, suggest a role in gut development that also may impact colonization dynamics (40). VCBP-C is the most extensively studied (34, 38). The Ig domains of VCBPs bind bacteria and promote phagocytosis or shape biofilm formation (34, 38); a physiological role for the CBD is less clear. We show here that the CBD of VCBP-C binds chitin molecules present on the cell walls of fungi isolated from the Ciona gut. VCBP-C may well-serve an integral role in promoting transkingdom, bacterial-host-fungal interactions within the gut microbiome, a heretofore-unrecognized function for an immunoglobulin-related molecule.
Materials and Methods
Isolation of Fungi From the Ciona robusta Gut
Upon arrival, wild-harvested animals were allowed to acclimate in 0.22 μm-filtered artificial seawater (ASW) for 3–4 h. All steps to isolate fungi were carried out under a laminar hood, in aseptic conditions, using sterile materials. Briefly, after sterilization of the Ciona tunic with 70% ethanol and washing with sterile ASW (41), gut was excised surgically (aseptically) from five animals and disrupted using a Dounce homogenizer to liberate bacteria and fungi from the mucosal surface of the organ. Host tissue was separated by processing through a 40 μm filter at 1200 × g for 10 min. Gut microorganisms were collected by pelleting at 500 × g or 10 min, washing and re-suspending in 1 ml ASW. An aliquot of this suspension (300 μl) was plated onto Yeast Peptone Dextrose (YPD) Agar plates (2% BactoPeptone, 1% Yeast extract, 2% dextrose, and 2% Agar) dissolved in ASW with Penicillin/Streptomycin (200 U/ml and 0.2 mg/ml, respectively; Fisher BP295950) antibiotics. Fungi were grown for 7–10 days before clonal growth was established and maintained by replica-streaking on YPD plates without antibiotics. Each fungal isolate was then inoculated in 5 ml YPD liquid medium, grown for 1 week in an orbital shaker at 20°C and then mixed with glycerol (10% final concentration) for long-term storage at −80°C.
Fungal species were identified by sequencing (Sanger) across the interspersed spacer (ITS) region of the 18s rRNA gene. Briefly, using Dneasy PowerSoil kit (Qiagen, Cat#12888), DNA extraction was performed on fungi grown in liquid medium for 5–7 days; fungal-specific18s rRNA amplicons were generated with standard ITS primers [ITS1 and ITS4, (42)]. Sequence data were deposited in GenBank and accession numbers are reported in Supplementary Table 1.
Immunofluorescence of Cultured Fungal Isolates Cryo-Sectioned and Labeled With Fc-CBD-C Probe
Each fungal isolate was grown on a YPD agar plate for about 5 days and fixed with 4% paraformaldehyde in PBS overnight at 4°C. Plates were washed three times for 10 min in PBS and small pieces of colonies were embedded and frozen on dry ice in OCT freezing media (Fisher Scientific), and stored at −80°C. Thin sections (7 μm) were collected on poly-L lysine (MP Biomedicals, #0210269125)-coated slides. Immunofluorescence staining was performed using a recombinant, customized, probe possessing a N-terminal human Fc domain fused to the VCBP-C CBD domain, and named the Fc-CBD-C probe. The specificity of this probe for chitin, and the general staining procedures, were previously validated and described in Dishaw et al (34). Briefly, slides were washed twice with PBS and incubated first for 10 min with animal-free blocking solution (Vector Laboratories #SP-5030) diluted in PBS and then incubated with the Fc-CBD-C probe diluted 1:10 in blocking solution for 2 h at room temperature. Control staining experiments were run in parallel using only a human Fc probe. After three PBS washes, slides were incubated with goat anti-human Fc Alexa Fluor 488 antibody (Invitrogen, #H10120) diluted 1:1000 in PBS for 45 min and washed again twice with PBS. Cellular nuclei were stained with Hoechst (Invitrogen, #H3570) diluted 1:2000 in PBS for 20 min. After a PBS wash for 5 min, slides were mounted with Aquamount (Fisher #143905) and viewed using fluorescence microscopy. Overexposed images of control experiments were acquired to verify lack of specific signal compared to those from the experimental slides (fungi incubated with Fc-CBD-C probe), and to verify the presence/absence of diffuse background signal in the controls.
Immunofluorescence Detection of Whole Mount Fungi With the Fc-CBD-C Probe
Fungal strains were grown in YPD liquid medium and orbital shaking at 20°C for 7 days, following which fungal bodies were fixed in 4% paraformaldehyde in PBS overnight at 4°C. Samples were washed several times in PBS and processed for whole mount immunofluorescence staining with the Fc-CBD-C probe as described above. Controls were run in parallel and images acquired as described in the previous section Immunofluorescence of cultured fungal isolates cryo-sectioned and labeled with Fc- CBD-C probe.
Isolation of Fungal Spores for Immunofluorescence and VCBP-C Binding
Spores were isolated from fungal cultures grown for 7 days in 10 ml YPD liquid medium with orbital shaking at 20°C. Briefly, the culture was filtered using twice folded sterile gauze and centrifuged at 2200 × g for 15 min. Spores were re-suspended in 1 ml YPD medium and, either were fixed for immunofluorescence staining with the Fc-CBD-C probe or incubated with VCBP-C recombinant protein, as describe below.
For the CBD-C binding, spores were fixed by adding formamide (4% final concentration) to 400 μl of re-suspended spores, followed by incubation for 30 min with gentle agitation. Spores were pelleted at 2600 × g for 10 min, re-suspended in PBS and plated on poly-L-lysine coated circle glass coverslips (see above). The spore suspension was incubated for 15 min to allow attachment to the coverslip, then the immobilized spores were incubated overnight at 4°C with the Fc-CBD-C probe and BSA 0.5% in PBS was used as blocking solution before detection with secondary antibody.
In order to characterize the biding of VCBP-C, 300 μl of spores were incubated for 2 h with gentle agitation, either with 50 μg/ml VCBP-C recombinant protein (prepared as described in (34), or the same volume of 10 mM Tris pH 8/50 mM NaCl solution as a control. Spores were collected by centrifugation at 2600 × g for 10 min to remove unbound VCBP-C protein, re-suspended in 500 μl PBS, fixed and plated on poly-L lysine coated coverslips as described above. Binding of VCBP protein to spores was detected by immunofluorescence using an anti-VCBP-C antibody (34, 38, 40). Chitin molecules on spore cell walls also were stained with wheat germ agglutinin (WGA) Texas-Red conjugate (Invitrogen, # W7024) diluted 1:200 in PBS and incubated for 20 min. WGA solution was removed with two PBS washes and then nuclei were stained with Hoechst and mounted as described previously.
Results
Chitin-Binding Domain of VCBP-C Recognizes Chitin Molecules in Fungal Cell Wall
In order to define the Ciona gut mycobiome, fungi were recovered from the gut of C. robusta using culture-based isolation methods. The isolates recovered in this study include eight different Penicillium spp. and five different Trichoderma spp., identified by ITS sequencing. Other filamentous fungal species that were recovered include: Acremonium sp., Acrostalagmus sp., Arthrinium sp., Mucor sp., Phaeosphaeria sp., and Phoma sp. It is essential to recognize that the approaches used facilitated the isolation and identification of only those fungal species that could be cultured in the conditions defined in this study; hypothetically, these may represent only a small proportion of the Ciona gut mycobiota.
All of the isolates were cryo-sectioned and stained with the Fc-CBD-C probe. Chitin fibers were localized in the cell wall of the fungal hyphae (Figures 1, 2), whereas control experiments performed using only the human Fc-probe do not show any fluorescent signal (Supplementary Figure 1). In almost all the fungal isolates, CBD-C binds the whole cell wall (Figures 1G,O,S,W, 2); however, in Acremonium sp. (Figure 1C) and Arthrinium sp. (Figure 1K), binding is localized predominantly along the edges of the cell wall. The observed staining patterns do not demonstrate if the CBD is detecting chitin in the inner or outer layer of the cell wall, an important distinction in characterizing the overall function of VCBP molecules. In order to resolve this distinction, whole mount staining with the Fc-CBD-C probe was carried out with Mucor sp., Penicillium spp., and Trichoderma sp. grown in liquid medium. In Mucor sp., the CBD-C mainly binds the spores that are produced in great abundance, but it does not bind the fungal hyphae (Figures 3B,E). In the two Penicillium spp. and in the Trichoderma sp., the CBD-C staining is highly regionalized along fungal hyphae (Figures 3K,N,Q,T,W). In one of the Penicillium sp., spores are bound by CBD-C, as observed in Mucor sp. (Figure 3H). Control experiments using the human Fc-probe show no specific binding to the whole fungal bodies (Supplementary Figure S2).
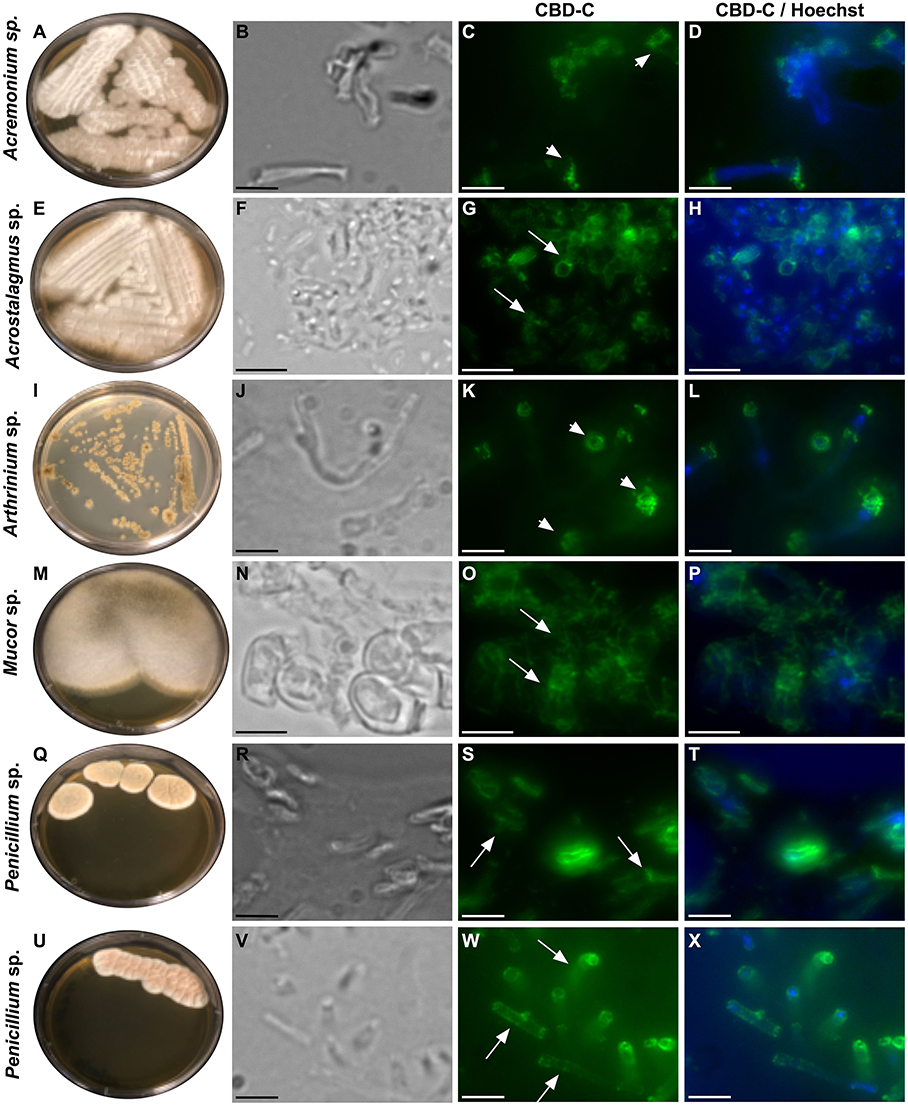
Figure 1. Immunofluorescence with the Fc-CBD-C probe on fungal sections. Several species of fungi, including: Acremonium sp., Acrostalagmus sp., Arthrinium sp., Mucor sp., and Penicillium spp., were isolated from Ciona gut and grown on YPD agar plates (A,E,I,M,Q,U). Immunofluorescence staining, using the Fc-CBD-C probe and detected with a fluorescent secondary antibody (Alexa Fluor 488), on OCT-embedded and cryo-sectioned fungal isolates, indicates that chitin molecules are distributed in the whole cell wall of most of the fungi analyzed (G,O,S,W, arrows). In two fungal species, the Fc-CBD-C probe detects chitin localized in specific regions of the fungal cell wall (C,K, arrowhead). (B,F,J,N,R,V) are brightfield images of fungal sections; (D,H,L,P,T,X) represents merged images of the Fc-CBD-C probe (green) and nuclear staining with Hoechst (blue). Scale bars: (B–D,J–L,R-T,V–X), 5 μm; (F–H, N–P) 10 μm.
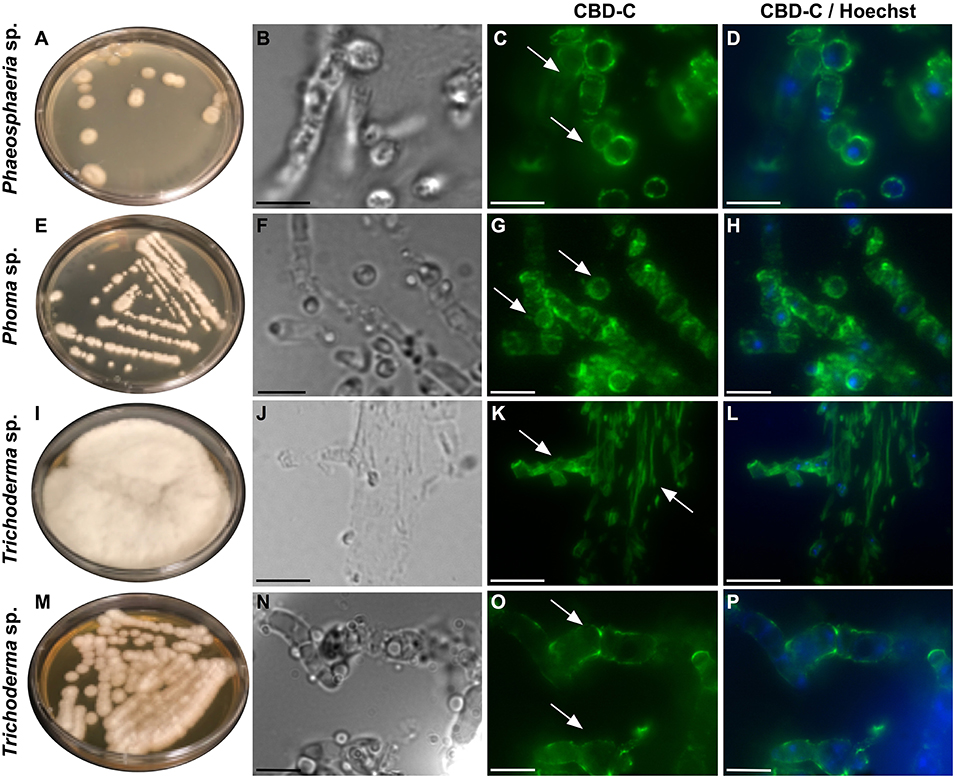
Figure 2. Immunofluorescence with the Fc-CBD-C probe on fungal sections. Fungal species, including Phaeosphaeria sp., Phoma sp., and Trichoderma spp., were isolated from the Ciona gut and grown on YPD agar plates (A,E,I,M). Immunofluorescent staining, using the Fc-CBD-C probe, on OCT-embedded and cryo-sectioned fungal isolates, indicates that chitin molecules are distributed in the whole cell walls of these fungi (C,G,K,O, arrows). (B,F,J,N) are brightfield images of fungal sections; (D,H,L,P) represent merged images of the Fc-CBD-C probe (green) and nuclear staining with Hoechst (blue). Scale bars: (B–D,J–L), 10 μm; (F–H,N–P), 5 μm.
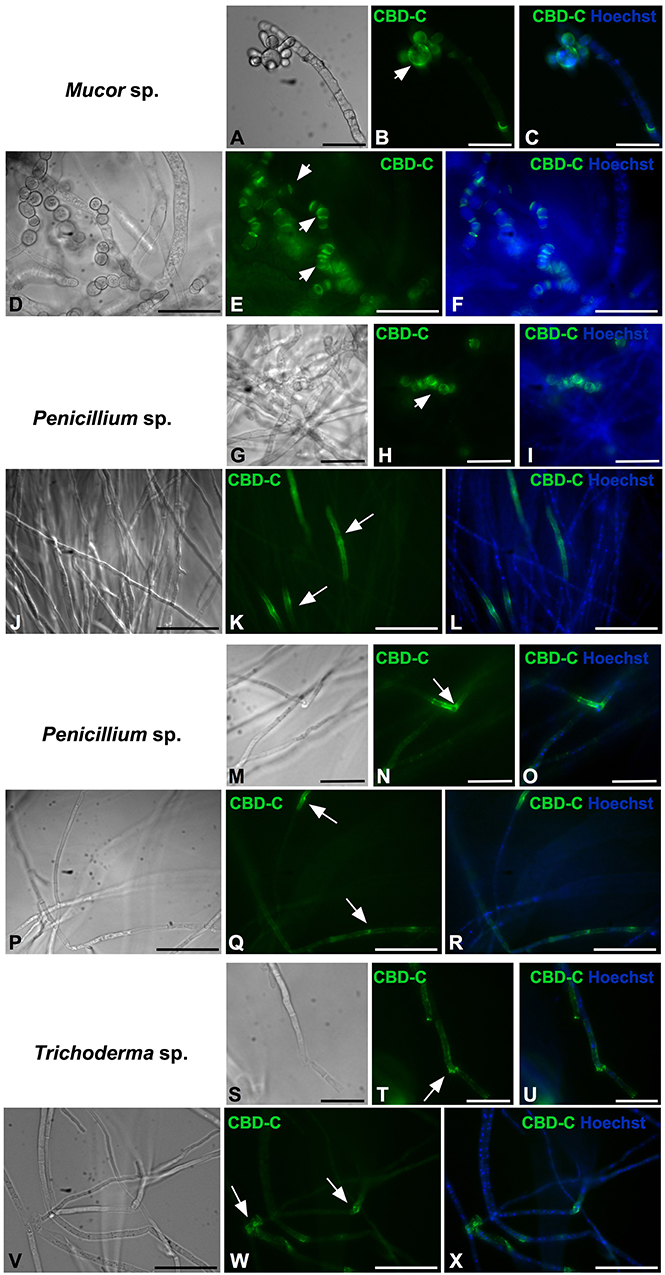
Figure 3. Immunofluorescence with the Fc-CBD-C probe on whole fungi grown in liquid medium. Fungal species, Mucor sp. (A–F), Penicillium spp. (G–R), and Trichoderma sp. (S–X), were grown in liquid medium and characterized by immunofluorescent staining with the Fc-CBD-C probe. In Mucor sp., CBD-C binds fungal spores (B,E, arrowheads); in Penicillium spp., CBD-C recognizes chitin in specific regions of the hyphae (K,N,Q, arrows) and in some cases fungal spores (H, arrowhead); in Trichoderma sp., chitin fibers are apparent in specific regions of the hyphae (T,W, arrows). (A,D,G,J,M,P,S,V) represent brightfield images; (C,F,I,L,O,R,U,X) represents merged images of the Fc-CBD-C probe (green) and nuclear staining with Hoechst (blue). Scale bars: (A–C,D–F,J–L,P–R,V–X), 50 μm; (G–I,M–O,S–U), 25 μm.
To further investigate the binding of CBD-C to fungal spores, Acremonium sp. Acrostalagmus sp., Mucor sp., and Penicillium spp., were grown in liquid medium and spores were isolated. The Fc-CBD-C probe binds the surface of spores recovered from all of these fungal species (Figure 4). In Acremonium sp. Acrostalagmus sp., and Mucor sp., CBD-C exhibits regionalized binding on the spore surface (Figures 4B,E,H). In the Penicillium spp., CBD-C binds almost the entire spore wall; however, in some spores, chitin binding is localized to discrete regions (Figures 4K,N). One explanation for the observed differences in localization of chitin is that it may reflect different stages of the spore cell cycle. No staining was observed in control experiments (Supplementary Figure 3).
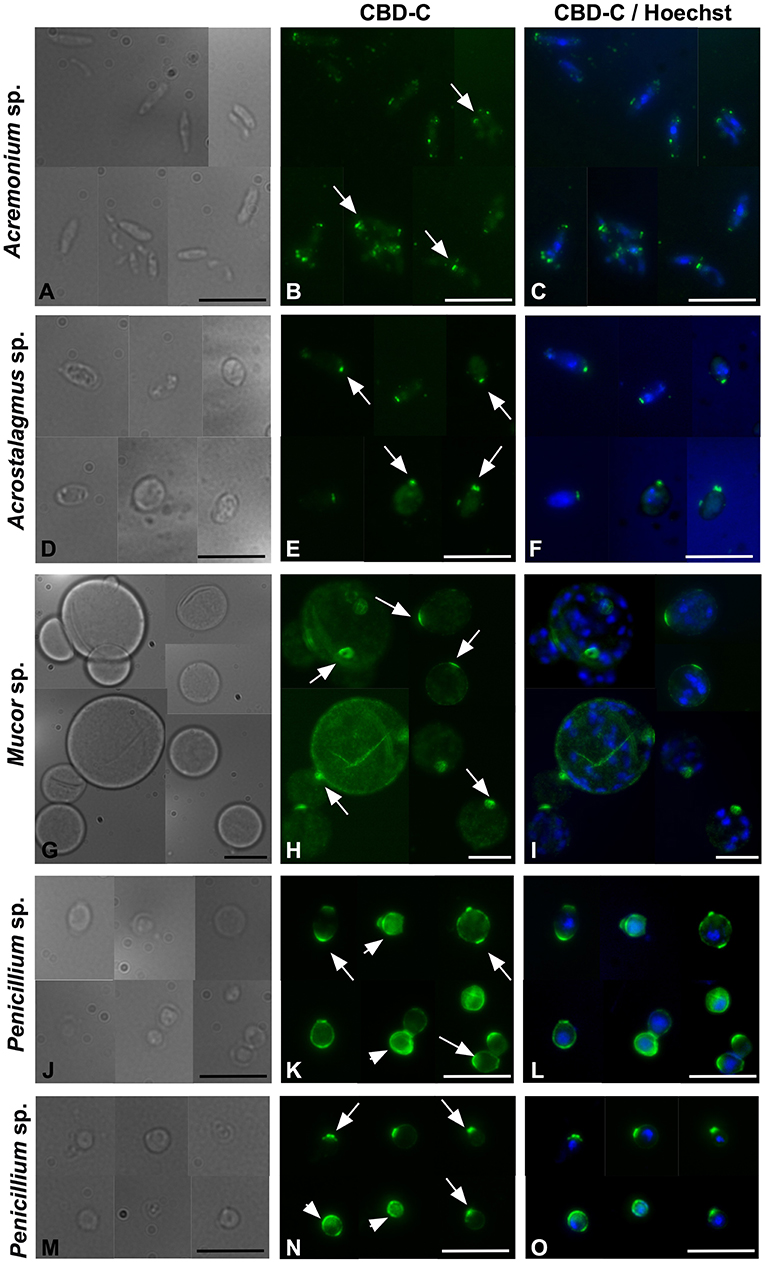
Figure 4. Immunofluorescence with the Fc-CBD-C probe on fungal spores. In Acremonium sp., Acrostalagmus sp., and Mucor sp. the CBD-C probe detects chitin fibers in regionalized areas on the surface of spores isolated from liquid cultures (B,E,H, arrows). In Penicililum spp., CBD-C binds almost the entire spore wall (K,N, arrowheads) in most spores. In some spores, chitin is instead detected in localized spots (K,N, arrow). (A,D,G,J,M) represents brightfield images; (C,F,I,L,O) represent merged images of the Fc-CBD-C probe (green) and nuclear staining with Hoechst (blue). Scale bars: 10 μm.
Collectively, these results demonstrate that the CBD of VCBP-C binds chitin that is localized mainly on the surface of fungal spores. In parallel studies, the dye-conjugated plant lectin, wheat germ agglutinin (WGA), was used to confirm some of these observations; specifically, WGA recognizes chitin fibers localized only on fungal surfaces and not in the inner cell walls (43, 44); hence, this lectin was utilized, as described in the following section, to verify the presence of chitin on fungal surfaces and to confirm the degree of overlap with signals detected by the CBD of VCBP-C. Moreover, chitin molecules that are recognized by CBD-C were not found to be exposed throughout the entirety of the cell walls of fungi, rather are localized to specific regions, e.g., distinct segments of hyphae.
VCBP-C Binds Chitin Exposed on the Surface of Fungal Spores
In order to determine if the full-length VCBP-C exhibits the same binding pattern to fungi that is exhibited by its CBD (in the form of the Fc-CBD probe), Acremonium sp. Acrostalagmus sp., Mucor sp., and Penicillium sp., were grown in liquid medium, processed for isolation of spores and incubated with recombinant VCBP-C protein. Intact VCBP-C exhibits the same binding pattern to the cell wall surface of these species (Figure 5) that was observed with the CBD probe (Figure 4), whereas no staining is observed in control samples (Supplementary Figure S3). Specifically, in Acremonium sp. Acrostalagmus sp., and Mucor sp., VCBP-C recognizes chitin localized in specific regions of the spore surface (Figures 5B,F,J); in Penicillium sp., the staining either surrounds the entire spore or is detected in specific regions (Figure 5N). WGA lectin was also used to stain the same isolates of spores that were incubated with the VCBP-C protein (Figures 5C,G,K,O) and the fluorescent signals co-localize (Figures 5D,H,L,P), further confirming the specificity of VCBP-C binding to chitin molecules that are localized on fungal surface.
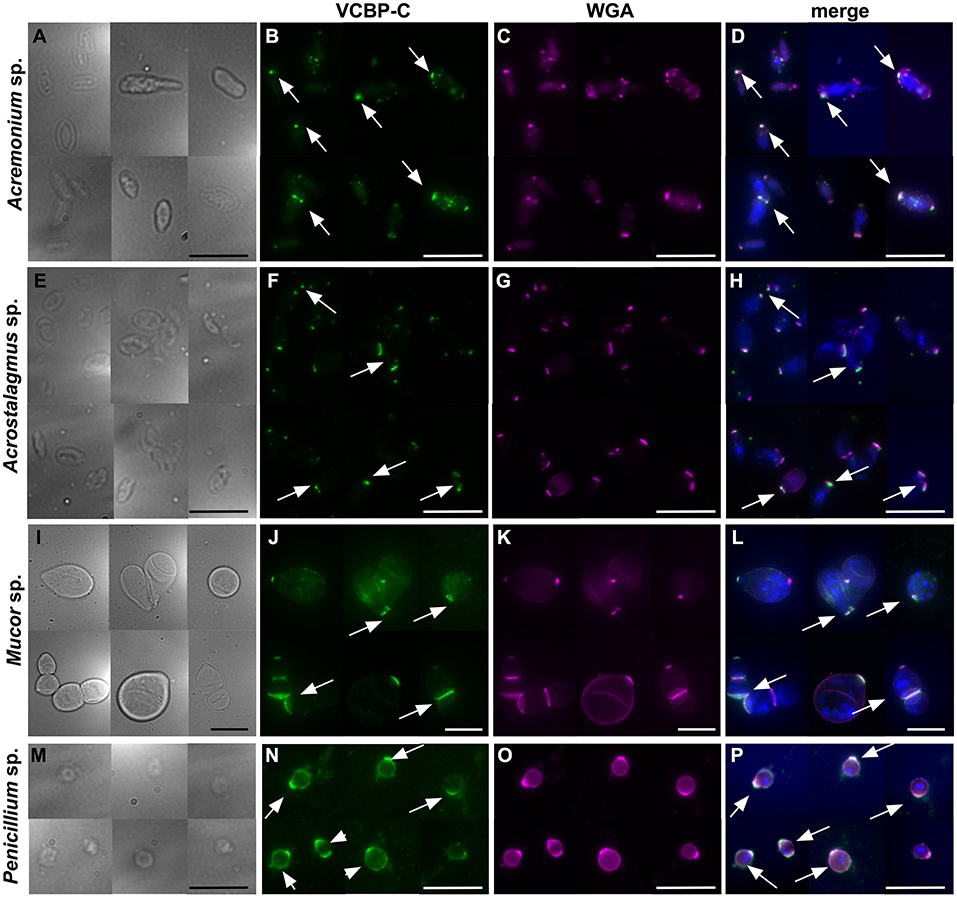
Figure 5. Immunofluorescence on fungal spores incubated with VCBP-C protein and WGA. VCBP-C protein incubated with spores isolated from liquid cultures of Acremonium sp., Acrostalagmus sp. and Mucor sp. binds chitin localized in specific regions of the spore surface (B,F,J, arrow). In Penicillium sp., VCBP-C recognizes chitin surrounding the entire spore (N, arrowheads) or in some cases, localized to specific spots (N, arrow). WGA staining (C,G,K,O) confirms the presence of chitin fibers on fungal spores. WGA co-localizes with VCBP-C staining in almost all of the spores, as observed in the merged images (D,H,L,P, arrows). (A,E,I,M) represent brightfield images; (D,H,L,P) represent merged images: VCBP-C in green, WGA in magenta and nuclear staining with Hoechst in blue. Scale bars: 10 μm.
Discussion
Ciona robusta is a model organism that in recent years has been adapted to studies of host-microbe interactions within the gut (45, 46), and has introduced an entirely unique, non-vertebrate model system to complement studies performed in mouse and zebrafish models (46). Disparate populations of Ciona share a core microbiome of distinct bacterial taxa (47). More recently we have shown that the gut virome of Ciona is temporally stable and is dominated by prophages (48). The isolation of diverse microorganisms from the Ciona gut, including bacterial strains [(34) Dishaw et al., unpublished data], different phages (49, 50) and, as described here, fungal species, is a critical first step in allowing us to investigate the underlying mechanisms that govern the bacterial, viral, and fungal relationships that define homeostasis in the gut of an organism relying solely on innate immunity (51). We have shown previously that epithelial barriers and associated innate functions are phylogenetically ancient and have evolved different mechanisms to maintain dialogue with adherent microbiota (46). However, such relationships appear to have been preserved and are remarkably insightful in gaining a better appreciation of the nature and dynamics of prokaryotic-eukaryotic relationships.
Here, we show that a VCBP, through its CBD, binds chitin localized in the cell wall of filamentous fungi isolated from the gut of Ciona. In most fungi, i.e., Candida yeasts, Saccharomyces cerevisiae, Pneumocystis spp., Aspergillus conidium, Aspergillus fumigatus hyphae, Cryptococcus, Histoplasma capsulatum, and Blastomyces dermatitidis (52), chitin polymers, together with β-1,3- and β-1,6 glucans, are localized within the inner layer of the fungal cell wall where they form a basket-like scaffold around the cell (52–54). The integrity of this scaffold is important for the fungal cell to ensure plasticity, allowing turgor-driven cell expansion as well as robustness and to prevent bursting of the cell (52). Although VCBP-C recognizes chitin within the fungal cell wall, the binding patterns to the whole fungal body indicate that at least some of the chitin is exposed on the fungal cell surface. Binding of the CBD-C probe to Mucor sp. spores and some regions of the Penicillium spp. and Thricoderma sp. hyphae (Figure 3), suggest that these regions could be undergoing cell wall reorganization for production of spores or for hyphal growth in the intercalary compartments. Although the hyphal extension in filamentous fungi normally is confined to the extreme apex, there are exceptions where this process is observed in the intercalary regions of fungal hyphae (55). In yeast cells, bud scars, which are located on the surface of the mother cell and mark the site of cytokinesis and septation from daughter cell, have fewer outer cell wall layers, thereby exposing the inner chitin-glucan to the external environment (56). VCBP-C recognizes chitin fibers on spores isolated from fungal species, such as Acremonium sp., Acrostalagmus sp., Mucor sp., and Penicillium spp. (Figures 4, 5). Different patterns of localization, reminiscent of bud scars are evident. Collectively, the findings are consistent with VCBP-C binding fungal spores or those structures that could originate spores, which in Ciona could be ingested through filtration of water and reside in the gut.
In vertebrates, chitin is recognized by the murine mannose receptor, Nod2, and TLR9 (57). Although chitin is localized within the inner layers of fungal cell walls and typically is not exposed to innate immune cells, it is recognized as an important immune-reactive polysaccharide (17). Ultrapurified chitin from C. albicans blocks the recognition of this yeast by human peripheral blood mononuclear cells (58). Chitin purified from Aspergillus induces the production of IL-1Ra, an anti-inflammatory cytokine and also stimulates proinflammatory mechanisms (i.e., IL-1β production) when it is present in combination with costimulatory PAMPs (59). The immune response to chitin molecules is complicated by the finding that different particle sizes are recognized by a different combination of PRRs, activating distinct signaling pathways that consequently induce the secretion of diverse cytokines (60, 61). Recently, a role for chitin also has been suggested in trained immunity (62), a form of immunological memory exhibited by innate immunity (63). Whereas, the downstream effects of VCBP-C and chitin interactions, specifically on living fungal species, may involve the shaping of activation-inhibition of developmental pathways in fungal cells, a role for VCBPs in regulating immune responses that shape fungal colonization of the gastrointestinal tract also may be possible. Future studies may help to decipher mechanisms used by the host recognition of fungi within the gut that may have phylogenetic significance.
The capacity of VCBP-C to bind both bacteria and fungi, suggests that the Ciona gut may represent unique challenges for establishing and maintaining homeostasis in this high flow rate, filter feeder. Through interactions involving the IgV domains, VCBP-C binding enhances bacterial phagocytosis by Ciona immunocytes (38) and modulates biofilm formation among bacteria isolated from the Ciona gut (34). The studies reported here indicate that CBD-C binds fungi by recognizing cell surface-exposed chitin molecules. Furthermore, the location of chitin detected by the CBD probe likely reflects physiologic or developmental states of the fungi. It is attractive to consider that the recognition of both bacteria and fungi may in some way facilitate protection against pathogens or support the colonization of gut symbionts; however, the dynamics likely are complicated. Specifically, in Ciona, the endogenous expression of chitin-rich mucus coating the gut epithelium has been shown previously to capture and bind VCBPs via the CBD (34). Thus, VCBPs can be free in the lumen as well as tethered to the chitin-rich mucus. Furthermore, the production of chitinases by some colonizing bacteria could release surface-trapped VCBPs, thereby facilitating recognition and interactions with chitin-rich fungal structures.
Although this may represent an expansive interpretation of the relationship of VCBPs to bacterial and fungal components of the gut, it is well-recognized that fungal and bacterial communities in the gut influence each other, through both positive and negative associations (7, 16, 19, 64). A transkingdom association between potentially pathogenic bacteria, as E. coli and Serratia marcescens, and yeast as C. tropicalis has been described in Crohn's disease (65). Fungal-bacterial interactions also have been demonstrated in an animal model where Saccharomyces boulardii interacts with and alters commensal bacteria, alleviating acute liver failure (66). Hence, bacteria and fungi may regulate each other in the commensal community using different mechanisms, that only now are being recognized and may offer novel targets for manipulating the microbiome (64). The novelty of the studies reported here is that this process may be modulated by the host and mediated by a bifunctional immune receptor.
The VCBP gene family is unique to protochordates (36–38), and thus the relationships that we propose may be unique among molecular mediators. However, the VCBPs may share some functional similarities to other classes of immune molecules found in vertebrates, such as IgA, that often exert their effects via immune inclusion-exclusion mechanisms (34, 40, 46, 67, 68). Most studies focus on the colonization of microbiomes by bacterial communities; however, in regards to colonization by fungi, there have only been a few studies demonstrating a protective and/or regulatory role for secreted immune mediators such as IgA against, for example, systemic candidiasis (69–71). Here, the unique binding of VCBP-C to both bacteria and fungi may influence the positive and negative crosstalk among resident microorganisms and consequently influence the dynamics of co-colonization and the establishment and maintenance of the gut microbiota, providing a first example of transkingdom interactions modulated by secreted immune mediators.
Ethics Statement
The research described herein was performed on C. robusta a marine invertebrate collected in Mission Bay near San Diego, CA (M-REP, Carlsbad, CA, USA), in locations that are not privately-owned nor protected in any way. Ciona is considered an invasive species and is not regulated or protected by environmental agencies in the United States. The collection service contracted in this study maintains current permits and licenses for collection and distribution of marine invertebrates to academic institutions. Handling of live animals was in accordance with the guidelines of our academic institutions. Animals were recovered and brought to the laboratory alive and maintained in clean water with aeration. In accordance with general animal protocols, the least number of animals required per experiment were utilized. Animal waste products were disposed of in accordance with USF Health and Safety Guidelines.
Data Access
The sequences reported in this manuscript have been deposited in the Genbank database with the following accession numbers: MK431096, MK424129, MK424120, MK423932, MK424124, MK424125, MK424126, MK424132, MK424131, MK424130, as reported more in detail in the Supplementary Table 1.
Authors Contributions
AL designed, performed, and analyzed experiments and wrote/edited the manuscript. JC and GL edited the manuscript. LD conceived the original idea and wrote/edited the manuscript.
Funding
This work was supported in part by grants from the National Science Foundation (IOS1456301), Johns Hopkins All Children's Hospital Foundation Research Grant and a USF College of Medicine Internal Award (LD) and the National Institute of Health AI23338 (GL).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00369/full#supplementary-material
References
1. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. (2012) 148:1258–70. doi: 10.1016/j.cell.2012.01.035
2. Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. (2003) 361:512–9. doi: 10.1016/S0140-6736(03)12489-0
3. Zhang YJ, Li S, Gan RY, Zhou T, Xu DP, Li HB. Impacts of gut bacteria on human health and diseases. Int J Mol Sci. (2015) 16:7493–519. doi: 10.3390/ijms16047493
4. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. (2010) 464:59–65. doi: 10.1038/nature08821
5. Yadav M, Verma MK, Chauhan NS. A review of metabolic potential of human gut microbiome in human nutrition. Arch Microbiol. (2018) 200:203–17. doi: 10.1007/s00203-017-1459-x
6. Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. (2012) 336:1314–7. doi: 10.1126/science.1221789
7. Dollive S, Chen YY, Grunberg S, Bittinger K, Hoffmann C, Vandivier L, et al. Fungi of the murine gut: episodic variation and proliferation during antibiotic treatment. PLoS ONE. (2013) 8:e71806. doi: 10.1371/journal.pone.0071806
8. Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. (2017) 5:153. doi: 10.1186/s40168-017-0373-4
9. Huffnagle GB, Noverr MC. The emerging world of the fungal microbiome. Trends Microbiol. (2013) 21:334–41. doi: 10.1016/j.tim.2013.04.002
10. Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. (2014) 14:405–16. doi: 10.1038/nri3684
11. Hallen-Adams HE, Kachman SD, Kim J, Legge RM, Martinez I. Fungi inhabiting the healthy human gastrointestinal tract: a diverse and dynamic community. Fungal Ecol. (2015) 15:9–17. doi: 10.1016/j.funeco.2015.01.006
12. Hallen-Adams HE, Suhr MJ. Fungi in the healthy human gastrointestinal tract. Virulence. (2017) 8:352–8. doi: 10.1080/21505594.2016.1247140
13. Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS ONE. (2013) 8:e66019. doi: 10.1371/journal.pone.0066019
14. Strati F, Di Paola M, Stefanini I, Albanese D, Rizzetto L, Lionetti P, et al. Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front Microbiol. (2016) 7:1227. doi: 10.3389/fmicb.2016.01227
15. Auchtung TA, Fofanova TY, Stewart CJ, Nash AK, Wong MC, Gesell JR, et al. Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere. (2018) 3:e00092–18. doi: 10.1128/mSphere.00092-18
16. Mukherjee PK, Sendid B, Hoarau G, Colombel JF, Poulain D, Ghannoum MA. Mycobiota in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol. (2015) 12:77–87. doi: 10.1038/nrgastro.2014.188
17. Wheeler ML, Limon JJ, Underhill DM. Immunity to commensal fungi: detente and disease. Annu Rev Pathol. (2017) 12:359–85. doi: 10.1146/annurev-pathol-052016-100342
18. Ott SJ, Kuhbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol. (2008) 43:831–41. doi: 10.1080/00365520801935434
19. Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe. (2016) 19:865–73. doi: 10.1016/j.chom.2016.05.003
20. Santelmann H, Howard JM. Yeast metabolic products, yeast antigens and yeasts as possible triggers for irritable bowel syndrome. Eur J Gastroenterol Hepatol. (2005) 17:21–6. doi: 10.1097/00042737-200501000-00005
21. Botschuijver S, Roeselers G, Levin E, Jonkers DM, Welting O, Heinsbroek SEM, et al. Intestinal fungal dysbiosis is associated with visceral hypersensitivity in patients with irritable bowel syndrome and rats. Gastroenterology. (2017) 153:1026–39. doi: 10.1053/j.gastro.2017.06.004
22. Ramaswamy K, Correa M, Koshy A. Non-healing gastric ulcer associated with Candida infection. Indian J Med Microbiol. (2007) 25:57–8. doi: 10.4103/0255-0857.31064
23. Krause R, Reisinger EC. Candida and antibiotic-associated diarrhoea. Clin Microbiol Infect. (2005) 11:1–2. doi: 10.1111/j.1469-0691.2004.00978.x
24. Marr KA, Seidel K, Slavin MA, Bowden RA, Schoch HG, Flowers ME, et al. Prolonged fluconazole prophylaxis is associated with persistent protection against candidiasis-related death in allogeneic marrow transplant recipients: long-term follow-up of a randomized, placebo-controlled trial. Blood. (2000) 96:2055–61.
25. van der Velden WJ, Netea MG, de Haan AF, Huls GA, Donnelly JP, Blijlevens NM. Role of the mycobiome in human acute graft-versus-host disease. Biol Blood Marrow Transpl. (2013) 19:329–32. doi: 10.1016/j.bbmt.2012.11.008
26. Enaud R, Vandenborght LE, Coron N, Bazin T, Prevel R, Schaeverbeke T, et al. The mycobiome: a neglected component in the microbiota-gut-brain axis. Microorganisms. (2018). 6:E22. doi: 10.3390/microorganisms6010022
27. Iliev ID, Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev Immunol. (2017) 17:635–46. doi: 10.1038/nri.2017.55
28. Patin EC, Thompson A, Orr SJ. Pattern recognition receptors in fungal immunity. Semin Cell Dev Biol. (2018) 17:30541–4. doi: 10.1016/j.semcdb.2018.03.003
29. Bryant CE, Orr S, Ferguson B, Symmons MF, Boyle JP, Monie TP. International Union of Basic and Clinical Pharmacology. XCVI Pattern recognition receptors in health and disease. Pharmacol Rev. (2015) 67:462–504. doi: 10.1124/pr.114.009928
30. Netea MG, Joosten LA, van der Meer JW, Kullberg BJ, van de Veerdonk FL. Immune defence against Candida fungal infections. Nat Rev Immunol. (2015) 15:630–42. doi: 10.1038/nri3897
31. Bollinger RR, Everett ML, Palestrant D, Love SD, Lin SS, Parker W. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology. (2003) 109:580–7. doi: 10.1046/j.1365-2567.2003.01700.x
32. Bollinger RR, Everett ML, Wahl SD, Lee YH, Orndorff PE, Parker W. Secretory IgA and mucin-mediated biofilm formation by environmental strains of Escherichia coli: role of type 1 pili. Mol Immunol. (2006) 43:378–87. doi: 10.1016/j.molimm.2005.02.013
33. Rogier EW, Frantz AL, Bruno ME, Kaetzel CS. Secretory IgA is concentrated in the outer layer of colonic mucus along with gut bacteria. Pathogens. (2014) 3:390–403. doi: 10.3390/pathogens3020390
34. Dishaw LJ, Leigh B, Cannon JP, Liberti A, Mueller MG, Skapura DP, et al. Gut immunity in a protochordate involves a secreted immunoglobulin-type mediator binding host chitin and bacteria. Nat Commun. (2016) 7:10617. doi: 10.1038/ncomms10617
35. Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. (2018) 360:795–800. doi: 10.1126/science.aaq0926
36. Cannon JP, Haire RN, Litman GW. Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat Immunol. (2002) 3:1200–7. doi: 10.1038/ni849
37. Cannon JP, Haire RN, Schnitker N, Mueller MG, Litman GW. Individual protochordates have unique immune-type receptor repertoires. Curr Biol. (2004) 14:R465–6. doi: 10.1016/j.cub.2004.06.009
38. Dishaw LJ, Giacomelli S, Melillo D, Zucchetti I, Haire RN, Natale L, et al. A role for variable region-containing chitin-binding proteins (VCBPs) in host gut-bacteria interactions. Proc Natl Acad Sci USA. (2011) 108:16747–52. doi: 10.1073/pnas.1109687108
39. Hernandez Prada JA, Haire RN, Allaire M, Jakoncic J, Stojanoff V, Cannon JP, et al. Ancient evolutionary origin of diversified variable regions demonstrated by crystal structures of an immune-type receptor in amphioxus. Nat Immunol. (2006) 7:875–82. doi: 10.1038/ni1359
40. Liberti A, Melillo D, Zucchetti I, Natale L, Dishaw LJ, Litman GW, et al. Expression of Ciona intestinalis variable region-containing chitin-binding proteins during development of the gastrointestinal tract and their role in host-microbe interactions. PLoS ONE. (2014) 9:e94984. doi: 10.1371/journal.pone.0094984
41. Leigh BA, Liberti A, Dishaw LJ. Generation of germ-free ciona intestinalis for studies of gut-microbe interactions. Front Microbiol. (2016) 7:2092. doi: 10.3389/fmicb.2016.02092
42. White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols in A Guide to Methods and Application. New York, NY: Academic Press Inc. (1990). p. 315–22.
43. Sherrington SL, Sorsby E, Mahtey N, Kumwenda P, Lenardon MD, Brown I, et al. Adaptation of Candida albicans to environmental pH induces cell wall remodelling and enhances innate immune recognition. PLoS Pathog. (2017) 13:e1006403. doi: 10.1371/journal.ppat.1006403
44. Esher SK, Ost KS, Kohlbrenner MA, Pianalto KM, Telzrow CL, Campuzano A, et al. Defects in intracellular trafficking of fungal cell wall synthases lead to aberrant host immune recognition. PLoS Pathog. (2018) 14:e1007126. doi: 10.1371/journal.ppat.1007126
45. Dishaw LJ, Flores-Torres JA, Mueller MG, Karrer CR, Skapura DP, Melillo D, et al. A Basal chordate model for studies of gut microbial immune interactions. Front Immunol. (2012) 3:96. doi: 10.3389/fimmu.2012.00096
46. Dishaw LJ, Cannon JP, Litman GW, Parker W. Immune-directed support of rich microbial communities in the gut has ancient roots. Dev Comp Immunol. (2014a) 47:36–51. doi: 10.1016/j.dci.2014.06.011
47. Dishaw LJ, Flores-Torres J, Lax S, Gemayel K, Leigh B, Melillo D, et al. The gut of geographically disparate Ciona intestinalis harbors a core microbiota. PLoS ONE. (2014b) 9:e93386. doi: 10.1371/journal.pone.0093386
48. Leigh BA, Djurhuus A, Breitbart M, Dishaw LJ. The gut virome of the protochordate model organism, Ciona intestinalis subtype A. Virus Res. (2018) 244:137–46. doi: 10.1016/j.virusres.2017.11.015
49. Leigh B, Karrer C, Cannon JP, Breitbart M, Dishaw LJ. Isolation and characterization of a shewanella phage-host system from the gut of the tunicate, ciona intestinalis. Viruses. (2017) 9:E60. doi: 10.3390/v9030060
50. Leigh BA, Breitbart M, Oksanen HM, Bamford DH, Dishaw LJ. Genome sequence of PM2-Like phage Cr39582, induced from a Pseudoalteromonas sp. isolated from the gut of ciona robusta. Genome Announc. (2018) 6:e00368–18. doi: 10.1128/genomeA.00368-18
51. Azumi K, De Santis R, De Tomaso A, Rigoutsos I, Yoshizaki F, Pinto MR, et al. Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: “waiting for Godot”. Immunogenetics. (2003) 55:570–81. doi: 10.1007/s00251-003-0606-5
52. Gow NAR, Latge JP, Munro CA. The fungal cell wall: structure, biosynthesis, and function. Microbiol Spectr. (2017) 5:1–25. doi: 10.1128/microbiolspec.FUNK-0035-2016
53. Nguyen TH, Fleet GH, Rogers PL. Composition of the cell walls of several yeast species. Appl Microbiol Biotechnol. (1998) 50:206–12.
54. Latge JP. The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol. (2007) 66:279–90. doi: 10.1111/j.1365-2958.2007.05872.x
55. Voisey CR. Intercalary growth in hyphae of filamentous fungi. Fungal Biol Rev. (2010) 24:123–31. doi: 10.1016/j.fbr.2010.12.001
56. Gantner BN, Simmons RM, Underhill DM. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. (2005) 24:1277–86. doi: 10.1038/sj.emboj.7600594
57. Wagener J, Malireddi RK, Lenardon MD, Koberle M, Vautier S, MacCallum DM, et al. Fungal chitin dampens inflammation through IL-10 induction mediated by NOD2 and TLR9 activation. PLoS Pathog. (2014) 10:e1004050. doi: 10.1371/journal.ppat.1004050
58. Mora-Montes HM, Netea MG, Ferwerda G, Lenardon MD, Brown GD, Mistry AR, et al. Recognition and blocking of innate immunity cells by Candida albicans chitin. Infect Immun. (2011) 79:1961–70. doi: 10.1128/IAI.01282-10
59. Becker KL, Aimanianda V, Wang X, Gresnigt MS, Ammerdorffer A, Jacobs CW, et al. Aspergillus cell wall chitin induces anti- and proinflammatory cytokines in human PBMCs via the Fc-gamma Receptor/Syk/PI3K Pathway. MBio. (2016) 7:e01823–15. doi: 10.1128/mBio.01823-15
60. Lee CG, Da Silva CA, Lee JY, Hartl D, Elias JA. Chitin regulation of immune responses: an old molecule with new roles. Curr Opin Immunol. (2008) 20:684–9. doi: 10.1016/j.coi.2008.10.002
61. Da Silva CA, Chalouni C, Williams A, Hartl D, Lee CG, Elias JA. Chitin is a size-dependent regulator of macrophage TNF and IL-10 production. J Immunol. (2009) 182:3573–82. doi: 10.4049/jimmunol.0802113
62. Rizzetto L, Ifrim DC, Moretti S, Tocci N, Cheng SC, Quintin J, et al. Fungal chitin induces trained immunity in human monocytes during cross-talk of the host with Saccharomyces cerevisiae. J Biol Chem. (2016) 291:7961–72. doi: 10.1074/jbc.M115.699645
63. Netea MG, Quintin J, van der Meer JW. Trained immunity: a memory for innate host defense. Cell Host Microbe. (2011) 9:355–61. doi: 10.1016/j.chom.2011.04.006
64. Limon JJ, Skalski JH, Underhill DM. Commensal fungi in health and disease. Cell Host Microbe. (2017) 22:156–65. doi: 10.1016/j.chom.2017.07.002
65. Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn's disease. MBio. (2016) 7:e01250–16. doi: 10.1128/mBio.01250-16
66. Yu L, Zhao XK, Cheng ML, Yang GZ, Wang B, Liu HJ, et al. Saccharomyces boulardii administration changes gut microbiota and attenuates D-galactosamine-induced liver injury. Sci Rep. (2017) 7:1359. doi: 10.1038/s41598-017-01271-9
67. Liberti A, Leigh B, De Santis R, Pinto MR, Cannon JP, Dishaw LJ, et al. An immune effector system in the protochordate gut sheds light on fundamental aspects of vertebrate immunity. Results Probl Cell Differ. (2015) 57:159–73. doi: 10.1007/978-3-319-20819-0_7
68. Liberti A, Zucchetti I, Melillo D, Skapura D, Shibata Y, De Santis R, et al. Chitin protects the gut epithelial barrier in a protochordate model of DSS-induced colitis. Biol Open. (2018) 7:bio029355. doi: 10.1242/bio.029355
69. Wagner RD, Vazquez-Torres A, Jones-Carson J, Warner T, Balish E. B cell knockout mice are resistant to mucosal and systemic candidiasis of endogenous origin but susceptible to experimental systemic candidiasis. J Infect Dis. (1996) 174:589–97.
70. Casadevall A, Feldmesser M, Pirofski LA. Induced humoral immunity and vaccination against major human fungal pathogens. Curr Opin Microbiol. (2002) 5:386–91. doi: 10.1016/S1369-5274(02)00337-5
Keywords: VCBP-C, Ciona, fungal-immune interaction, mycobiota, innate immunity, gut immunity, transkingdom interactions, host-microorganism interactions
Citation: Liberti A, Cannon JP, Litman GW and Dishaw LJ (2019) A Soluble Immune Effector Binds Both Fungi and Bacteria via Separate Functional Domains. Front. Immunol. 10:369. doi: 10.3389/fimmu.2019.00369
Received: 08 September 2018; Accepted: 13 February 2019;
Published: 06 March 2019.
Edited by:
Uday Kishore, Brunel University London, United KingdomReviewed by:
Jonathan P. Rast, Emory University School of Medicine, United StatesDan Anthony Mitchell, University of Warwick, United Kingdom
Copyright © 2019 Liberti, Cannon, Litman and Dishaw. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Assunta Liberti, libertia@mail.usf.edu; assusy.liberti@gmail.com
Larry J. Dishaw, ldishaw@health.usf.edu
†Present Address: Assunta Liberti, Biology and Evolution of Marine Organisms (BEOM), Stazione Zoologica Anton Dohrn, Naples, Italy
 Assunta Liberti
Assunta Liberti John P. Cannon1
John P. Cannon1 Larry J. Dishaw
Larry J. Dishaw