- 1Department of Cell Biology and Neuroscience, Rutgers University, Piscataway, NJ, United States
- 2Graduate Program in Cellular and Molecular Pharmacology, Rutgers University, Piscataway, NJ, United States
- 3Department of Pharmacology, Anhui Medical University, Hefei, China
- 4Department of Obstetrics and Gynecology, The Affiliated Suzhou Hospital of Nanjing Medical University, Suzhou, China
- 5Engineering Research Center of the Modern Technology for Eel Industry, Ministry of Education of the People's Republic of China, Fisheries College of Jimei University, Xiamen, China
- 6Member, Rutgers Cancer Institute of New Jersey, New Brunswick, NJ, United States
The tumor necrosis factor receptor (TNF-R)-associated factor (TRAF) family of cytoplasmic adaptor proteins regulate the signal transduction pathways of a variety of receptors, including the TNF-R superfamily, Toll-like receptors (TLRs), NOD-like receptors (NLRs), RIG-I-like receptors (RLRs), and cytokine receptors. TRAF-dependent signaling pathways participate in a diverse array of important cellular processes, including the survival, proliferation, differentiation, and activation of different cell types. Many of these TRAF-dependent signaling pathways have been implicated in cancer pathogenesis. Here we analyze the current evidence of genetic alterations of TRAF molecules available from The Cancer Genome Atlas (TCGA) and the Catalog of Somatic Mutations in Cancer (COSMIC) as well as the published literature, including copy number variations and mutation landscape of TRAFs in various human cancers. Such analyses reveal that both gain- and loss-of-function genetic alterations of different TRAF proteins are commonly present in a number of human cancers. These include pancreatic cancer, meningioma, breast cancer, prostate cancer, lung cancer, liver cancer, head and neck cancer, stomach cancer, colon cancer, bladder cancer, uterine cancer, melanoma, sarcoma, and B cell malignancies, among others. Furthermore, we summarize the key in vivo and in vitro evidence that demonstrates the causal roles of genetic alterations of TRAF proteins in tumorigenesis within different cell types and organs. Taken together, the information presented in this review provides a rationale for the development of therapeutic strategies to manipulate TRAF proteins or TRAF-dependent signaling pathways in different human cancers by precision medicine.
Introduction
The tumor necrosis factor receptor (TNF-R)-associated factor (TRAF 1–7) family of cytoplasmic adaptor proteins regulates the signal transduction pathways of a variety of receptors, including the TNF-R superfamily, Toll-like receptors (TLRs), NOD-like receptors (NLRs), RIG-I-like receptors (RLRs), and cytokine receptors (1–4). TRAF proteins function as both adaptor proteins and E3 ubiquitin ligases to regulate receptor signaling, leading to the activation of canonical and noncanonical nuclear factor-κBs (NF-κB1 and NF-κB2), mitogen-activated protein kinases (MAPKs: ERK1/2, JNK1/2, and p38), or interferon-regulatory factors (IRFs: IRF3, IRF5, and IRF7) (1–4). The TRAF-dependent signaling pathways participate in a diverse array of important cellular processes, including the survival, proliferation, differentiation, activation, and stress responses of different cell types (1–4). Many of these TRAF-dependent signaling pathways have been implicated in cancer pathogenesis.
With the rapid progress made in next-generation deep sequencing technology and the tremendous efforts put forth on whole genome/exome/transcriptome sequencing and copy number variation (CNV) analyses of cancers at the post-genome era, it has become increasingly clear that genetic alterations of TRAF proteins are commonly present in various human cancers. Here we analyze the current evidence of genetic alterations of TRAF molecules available from the Cancer Genome Atlas (TCGA) (5) and the Catalog of Somatic Mutations in Cancer (COSMIC) (6) as well as the published literature, including the landscape of genetic alterations and the map of recurrent mutations in TRAF molecules in different types of human cancers. Moreover, we summarize the key in vivo and in vitro evidence that demonstrates the causal roles of genetic alterations of TRAF proteins in tumorigenesis within different cell types and organs. Collectively, the information presented in this review identifies TRAF proteins and TRAF-dependent signaling pathways as important therapeutic targets in specific human cancers.
TRAF1
Landscape of Genetic Alterations
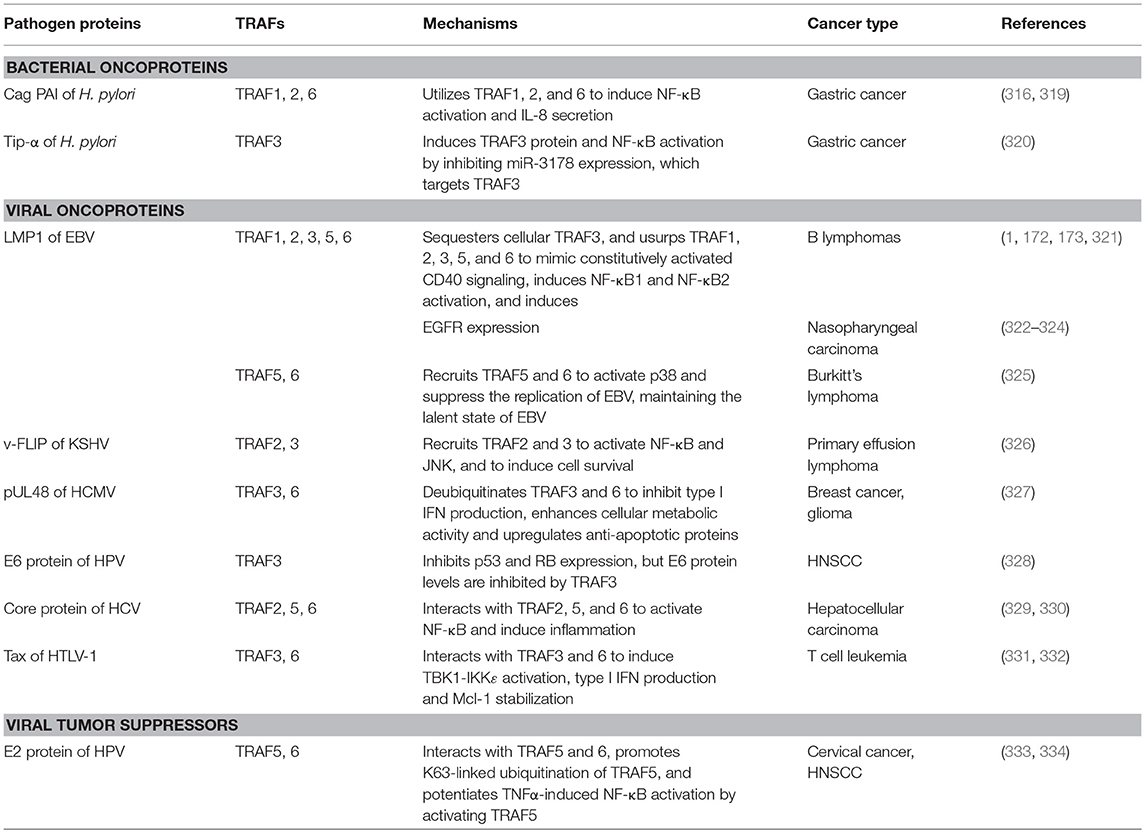
According to the TCGA and COSMIC datasets of sample size n > 100, the frequency of genetic alterations of TRAF1 is generally <4% in human cancers (Figure 1A). The eight human cancers with relatively higher genetic alterations of TRAF1 are pancreatic cancer (3.7%) (7), skin cutaneous melanoma (2.9%) (TCGA, PanCancer Atlas), esophageal cancer (2.8%) (TCGA, PanCancer Atlas), stomach cancer (2.7%) (8), sarcoma (2.4%) (9), ovarian cancer (2.3%) (TCGA, Provisional), lung cancer (2.3%) (10), and prostate cancer (2%) (TCGA, Provisional). The most common genetic alterations of TRAF1 are gene amplification (copy gain) and mutation. Deep deletion (copy loss) is less common but also detected in several types of human cancers (Figure 1). Truncation is rare for TRAF1 in human cancers.
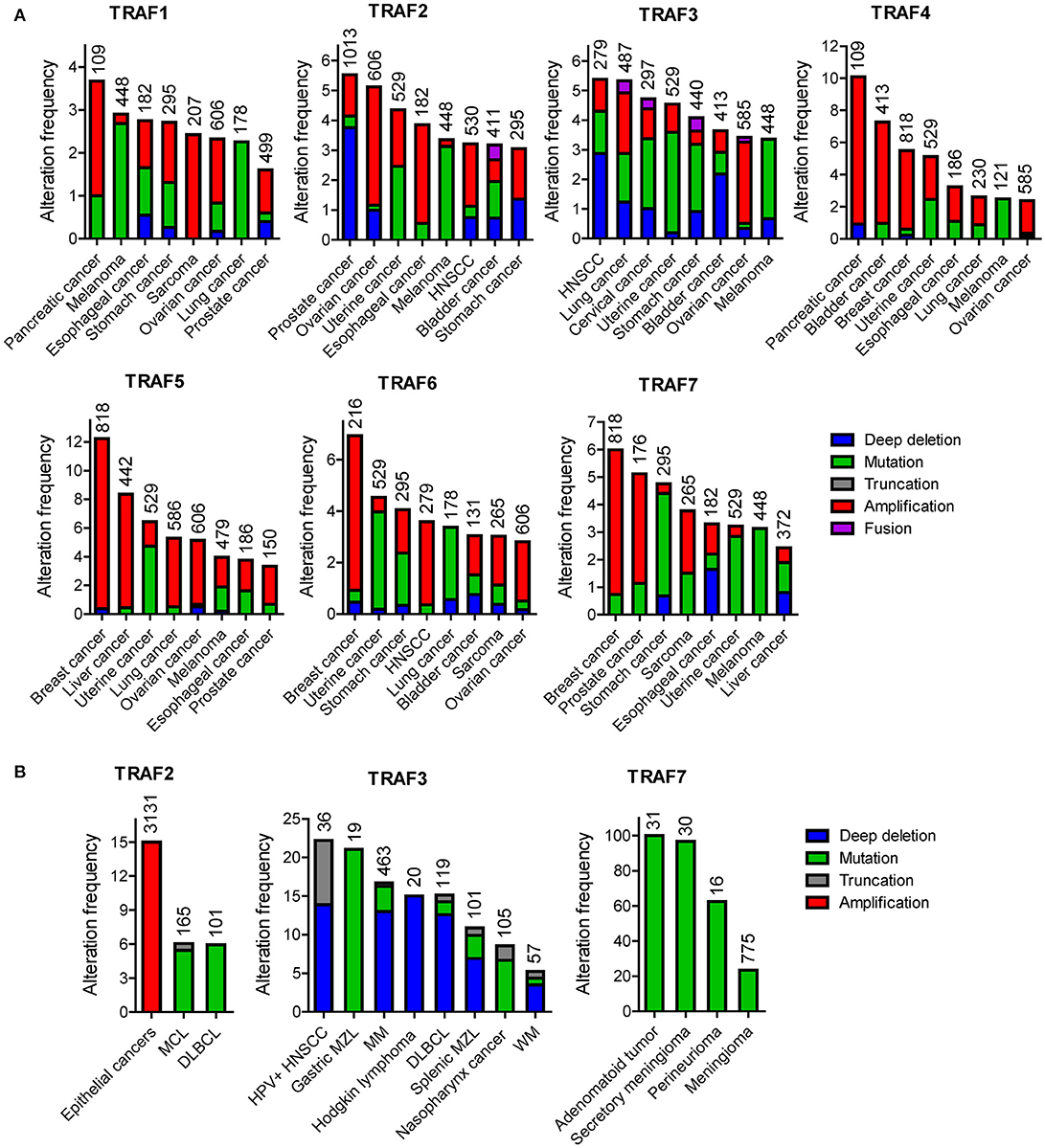
Figure 1. Landscape of genetic alterations of the TRAF family in human cancers. (A) Representative results retrieved from TCGA. For each TRAF gene, eight cancer types that exhibit relatively higher frequency of genetic alterations were selected and datasets with relatively larger sample size (n > 100) are shown. (B) Frequent genetic alterations recognized in the published literature. Genetic alterations shown include deep deletion (copy number loss), mutation (missense mutation, frameshift insertion or deletion, and in frame insertion or deletion), truncation (nonsense mutation), amplification (copy number gain), and fusion. The sample size of each dataset is indicated on top of each bar in the graphs.
Overview and Map of Recurrent Mutations
To date, there are 139 different mutations of the TRAF1 gene detected in human cancers, comprising 80% (111/139) mutations that alter the protein sequence of TRAF1 and 20% (28/139) coding silent mutations (Table 1). In the TRAF family, TRAF1 has the lowest count of recurrent mutations. Only 29% (32/111) of the coding-altering mutations of TRAF1 are recurrent and have been detected in at least two patients with various cancers. Almost all the recurrent mutations of TRAF1 are missense mutations (94%, 30/32) except one nonsense mutation (truncation) and one fusion (Table 1 and Figure 2). These recurrent mutations occurred across 24 different amino acids that are distributed in all the major domains of the TRAF1 protein (Figure 3). Interestingly, missense mutations of two specific amino acids are detected in more than three patients: R70C or H in the linker between the Zinc finger and the coiled-coil domain, and M182I of the coiled-coil (also known as TRAF-N) domain of the TRAF1 protein (Figure 3). The R70 mutations are detected in 4 patients with stomach, colon, and colorectal cancers (TCGA) (11–13). M182I is documented in 4 patients with melanoma and chronic lymphocytic leukemia (CLL) (14, 15). The functional significance of R70C/H and M182I mutations of TRAF1 remains to be determined.
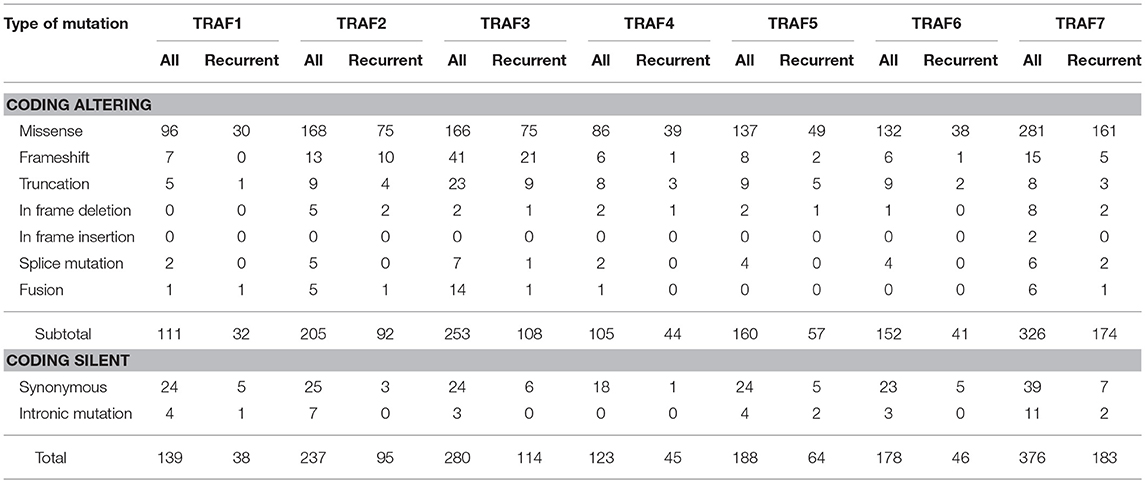
Table 1. Summary of the number of different types of mutations of TRAF proteins detected in human cancers.
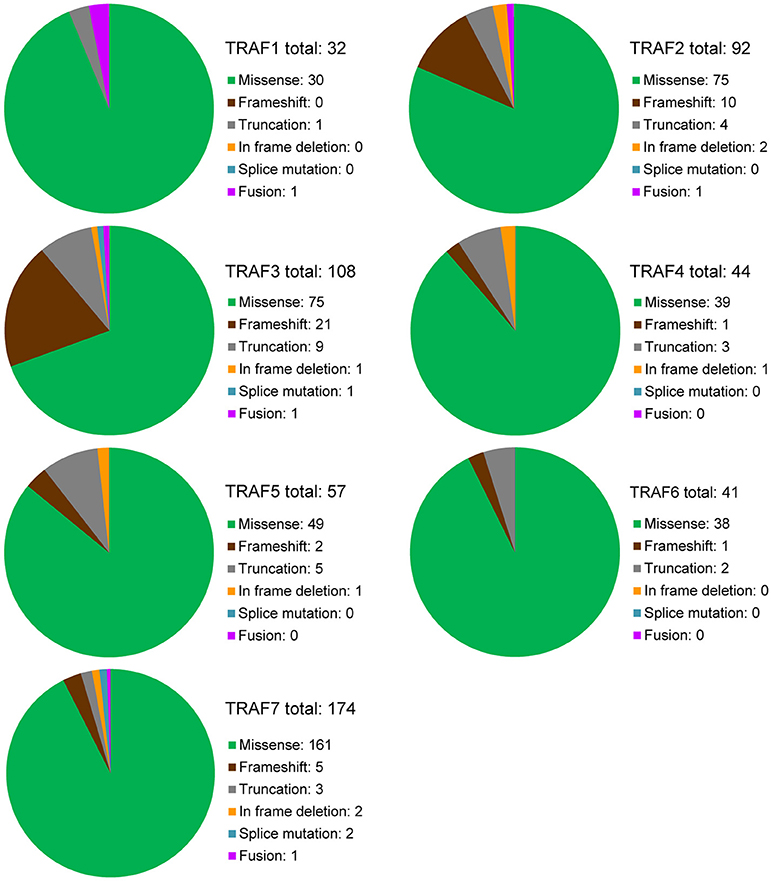
Figure 2. Overview of recurrent mutations of the TRAF family in human cancers. Recurrent mutations of the TRAF family that are identified in at least 2 cancer patients are summarized in this figure. The composition of recurrent mutation types are shown in a pie graph for each TRAF gene. The total count of recurrent mutations and the actual count of each category of recurrent mutation for each TRAF gene are indicated in each pie graph.
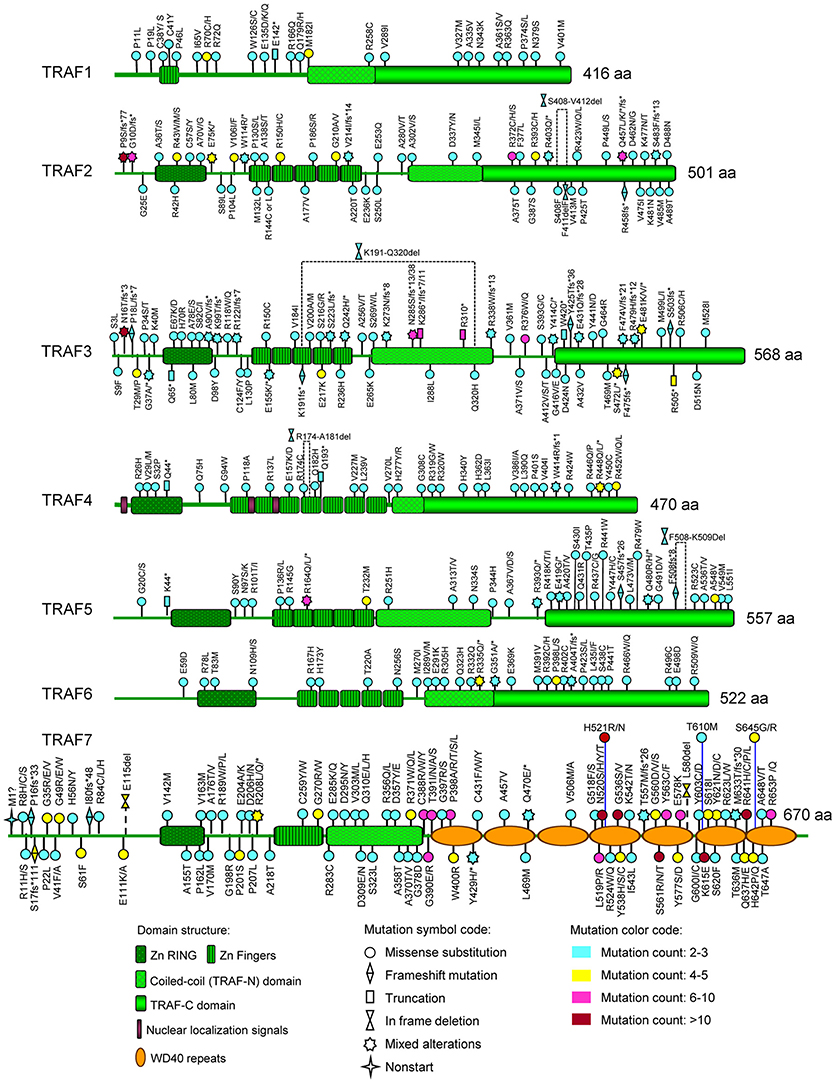
Figure 3. Map of recurrent TRAF mutations of human cancers on the TRAF proteins. The distribution of recurrent mutations on the domain structure of each TRAF protein is depicted in this figure. The domain structures of TRAF proteins shown include the zinc RING (Zn RING), zinc fingers (Zn Fingers), coiled-coil (TRAF-N) domain, TRAF-C domain, nuclear localization signals, and WD40 repeats. For each recurrent mutation, the nature of the mutation is indicated by a mutation symbol code and the patient count is indicated by a color code as shown at the bottom legend of the figure. The actual amino acid changes are also given for each recurrent mutation: letter change, missense mutation; *, Nonsense mutation (truncation); fs*, frameshift insertion or deletion; del, in frame deletion.
Fusion
There is only one fusion of the TRAF1 gene detected in human cancers, the TRAF1-ALK fusion that has been detected in five patients with anaplastic large cell lymphoma (ALCL) (16–19). All five cases contain the identical in frame fusion of TRAF1 and ALK that generates a chimeric protein linking the N-terminal 1–294 aa of TRAF1 to the entire intracellular domain of ALK (1,058–1,620 aa), including its kinase domain (16–19). Interestingly, expression of the TRAF1-ALK fusion protein leads to constitutive activation of the ALK and NF-κB pathways as demonstrated by the elevated levels of phosphorylated ALK (pALK) and STAT3 (pSTAT3) as well as nuclear p50 NF-κB1 and p52 NF-κB2 in ALCL cells (18). Similar to wild type (WT) TRAF1, the TRAF1-ALK fusion protein also binds to TRAF2 in co-immunoprecipitation experiments (18), suggesting the involvement of TRAF2 in the activation of NF-κB pathways. Furthermore, treatment of patient ALCL cells expressing the TRAF1-ALK fusion protein with proteasome inhibitors that decrease NF-κB1/2 or a selective ALK inhibitor (CEP28122) results in significant inhibition on lymphoma growth but could not eradicate lymphoma cells (18). Thus, constitutive activation of NF-κB1/2 pathways contributes to the neoplastic phenotype of TRAF1-ALK-expressing ALCL.
In vivo Causal Oncogenic Roles
Gene amplification is the most common TRAF1 genetic alteration in human cancers. TRAF1 expression is ubiquitously elevated in skin squamous cell carcinoma (SSCC), non-small cell lung cancer (NSCLC), Hodgkin lymphomas (HLs) and non-Hodgkin lymphomas (NHLs) (20–25). Notably, TRAF1 protein is consistently elevated in B cell leukemias and lymphomas without evidence of gene amplification (1, 23). In this case, TRAF1 upregulation might be the result of epigenetic alterations and/or aberrant activation of NF-κB1/2, as TRAF1 is a direct target gene of NF-κB (23, 26, 27). Interestingly, TRAF1 expression levels are increased in chronic lymphocytic leukemia (CLL) cells from patients with refractory disease, suggesting a role for TRAF1 in the progression of this disease and in the development of chemoresistance (23). Furthermore, genetic association studies identify TRAF1 as a susceptibility gene for risk of CLL (28). Thus, human evidence implicates TRAF1 as a candidate oncogene. Indeed, in vivo evidence obtained from mouse models demonstrates the causal oncogenic roles of TRAF1 in the skin, lung, T cells, and B cells (Table 2). TRAF1−/− mice exhibit increased skin sensitivity to TNFα-induced necrosis and reduced skin tumor formation induced by DMBA/chronic solar UV radiation (UVR) (20, 29). Mechanistically, TRAF1 enhances the ubiquitination of ERK5 and is required for UVR-induced ERK5 phosphorylation and the expression of AP-1 family members (c-Fos/c-Jun) in keratinocytes and epithelial cells (20). TRAF1−/− mice also show reduced lung tumorigenesis induced by i.p. administration of urethane (30). In this lung cancer model, TRAF1 affects TRAF2-mediated K48-linked ubiquitination and degradation of BRAF, and thereby promotes the survival and proliferation of lung cancer cells (30). Consistent with studies of the TRAF1-ALK fusion protein in ALCL, transgenic mice overexpressing TRAF1 in T cells exhibit decreased antigen-induced apoptosis of CD8 T cells (35), while TRAF1−/− mice display impaired survival and altered proliferation of T cells in response to the 4-1BB-NF-κB2 and T cell receptor (TCR)-NF-κB1 signaling pathways, respectively (29, 31–34). In line with the evidence of TRAF1 overexpression in HLs and NHLs, TRAF1 deficiency inhibits the spontaneous development of small B cell lymphoma in a transgenic mouse model that expresses the human lymphoma-associated NF-κB2 mutant p80HT specifically in lymphocytes (p80HT tg mice) (Table 2) (27). Taken together, these findings identify TRAF1 as a therapeutic target in skin cancer, lung cancer, and T cell and B cell lymphomas.
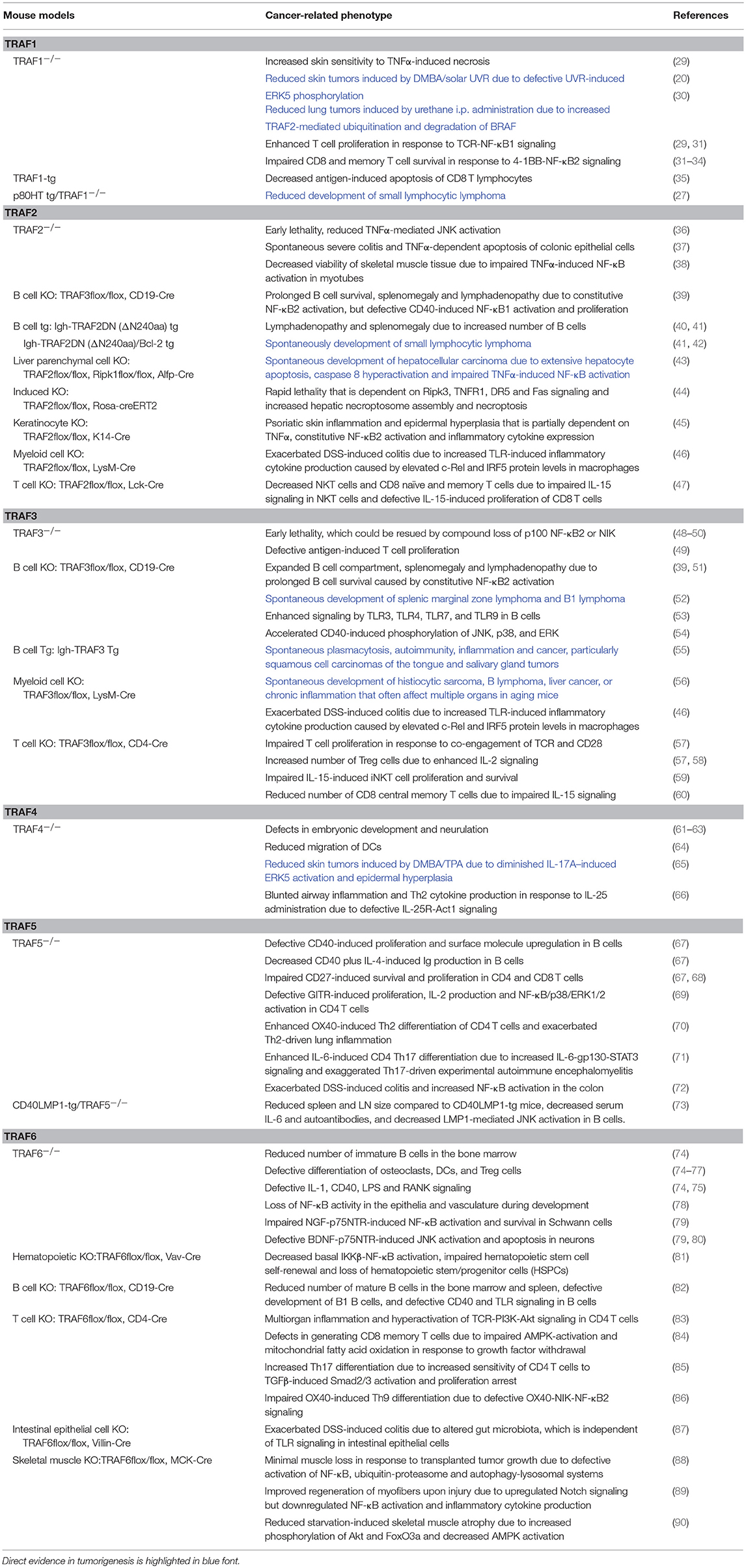
Table 2. In vivo evidence of the causal roles of genetic alterations of the TRAF family in cancer pathogenesis.
Key Oncogenic Pathways
In addition to the above TRAF1-dependent oncogenic pathways (ERK5-AP1, BRAF-ERK, NF-κB1, and NF-κB2) that have been verified in both human cancers and in vivo mouse models, several oncogenic pathways involving TRAF1 have been suggested by studies using patient samples, cultured human cancer cells or xenograft models. These include: (1) CD30-TRAF1 in HL and ALCL tumors (22, 91); (2) TNF-R1/2-TRAF1/TRAF2-JNK/NF-κB in cervical and colon cancer cells (92); (3) Wnt/β-catenin-NF-κB-TRAF1/iNOS in colon, breast and liver cancer cells (93, 94); and (4) TWEAK-Fn14-TRAF1 in solid tumors (95–97). Further investigation of these signaling pathways using TRAF1−/− or TRAF1-transgenic animal models would provide new insights on the roles and mechanisms of TRAF1 in cancer pathogenesis.
TRAF2
Landscape of Genetic Alterations
The frequency of genetic alterations of TRAF2 is generally <6% in human cancers (Figure 1A) based on the TCGA and COSMIC datasets of sample size n > 180. The eight human cancers with relatively higher genetic alterations of TRAF2 are prostate cancer (5.5%) (98), ovarian cancer (5.1%) (TCGA, Provisional), uterine cancer (4.4%) (TCGA, PanCancer Atlas), esophageal cancer (3.9%) (TCGA, PanCancer Atlas), skin cutaneous melanoma (3.4%) (TCGA, PanCancer Atlas), head and neck squamous cell carcinoma (HNSCC, 3.2%) (TCGA, Provisional), bladder cancer (3.2%) (TCGA, PanCancer Atlas), and stomach cancer (3.1%) (8). Notably, although not cataloged in TCGA, mutations of TRAF2 are recognized as one of the most frequent somatic mutations in mantle cell lymphoma (MCL, 6.1%, 10/165) (99–101) and diffuse large B-cell lymphoma (DLBCL, 6%, 6/101) (Figure 1B) (102). In addition, TRAF2 has been identified as an oncogene that is recurrently amplified and rearranged in 15% of human epithelial cancers (Figure 1B) (103). Thus, the most common genetic alterations of TRAF2 are deep deletion, gene amplification and mutation (Figure 1). Truncation and fusion of TRAF2 are relatively rare but also detected in human cancers (Figure 1).
Overview and Map of Recurrent Mutations
There are 237 different mutations of TRAF2 detected in human cancers, comprising 86% (205/237) mutations that change the protein sequence of TRAF2 and 14% (32/237) coding silent mutations (Table 1). Notably, 45% (92/205) of the coding-altering mutations of TRAF2 are recurrently detected in at least two cancer patients. Recurrent mutations of TRAF2 are more complex than those of TRAF1, including not only missense mutations (82%, 75/92), but also frameshifts (11%, 10/92), truncations (4%, 4/92), in frame deletions (2%, 2/92) and fusion (1%, 1/92) (Table 1 and Figure 2). TRAF2 recurrent mutations are identified at 52 different amino acids that are almost evenly distributed in all the structural motifs and domains of the TRAF2 protein (Figure 3). Interestingly, four mutation hotspots of TRAF2 are detected in more than 5 cancer patients, specifically P9, G10, R372, and Q457 (Figure 3). In particular, the frameshift deletion occurred at P9 (P9fs*77) is found in 16 patients with colon cancer, colorectal cancer (CRC), uterine cancer, stomach cancer, and sarcoma, and an additional missense mutation at P9 (P9S) is also detected in a CRC patient (TCGA) (12, 104–108). The amino acid right next to P9, G10, also exhibits similar frameshift deletion (G10fs*76) or insertion (G10fs*70) or missense mutation (G10D) in five patients with colon cancer, CRC, gallbladder cancer, and glioblastoma (TCGA) (105, 106, 109). Missense mutations at R372 (R372C, H or S) of the TRAF-C domain of TRAF2 are detected in eight patients with HNSCC, melanoma, and prostate, uterine, cervical, stomach, and liver cancers (TCGA; COSMIC) (110–113). Another amino acid of the TRAF-C domain, Q457, shows complex mutations, including a truncation (Q457*), a frameshift insertion (Q457fs*277), and missense mutations (Q457K or L) in six patients of HNSCC, oral squamous cell carcinoma (OSCC), stomach cancer, melanoma, and breast cancer (TCGA; COSMIC) (8, 114). Frameshift mutations occurring at P9 and G10 are functionally equivalent to deletion of TRAF2. Missense mutations at R372 and the complex mutations at Q457 of the TRAF-C domain of TRAF2 are predicted to result in inactivation of the TRAF2 protein (99–102).
Fusion
There are five different fusions of the TRAF2 gene detected in human cancers, including TRAF2-CCDC183 in breast and bladder cancers, TRAF2-CACNA1B in bladder cancer, TMEM141-TRAF2 in breast cancer (TCGA), TRAF2-NOTCH1 in ovarian cancer (106) and NTRK2-TRAF2 in melanoma (115). Among these, only the TRAF2-CCDC183 fusion is recurrently detected in two patients with breast cancer and bladder cancer (TCGA). Functional contribution of these TRAF2 fusions to cancer pathogenesis is currently unclear.
In vivo Tumor Suppressive Roles
Inactivating mutations of TRAF2 are frequently detected in human MCL and DLBCL, resulting in elevated activation of NF-κB1 and NF-κB2 in malignant B cells (99–102). TRAF2 is also involved in human MALT lymphomagenesis induced by the oncogenic cIAP2-MALT1 fusion protein through the interaction between TRAF2 and the BIR1 domain of cIAP2 portion of the fusion protein, leading to activation of the TRAF2-RIP1-NF-κB pathway (116). Consistent with the human evidence, B cell-specific TRAF2−/− (B-TRAF2−/−) mice exhibit expanded B cell compartment in lymphoid organs due to constitutive NF-κB2 activation and survival advantage independent of the B cell survival factor BAFF (Table 2) (117). Similarly in TRAF2DN-tg mice that express a dominant negative form of TRAF2 specifically in lymphocytes (Igh-TRAF2DN), inhibition of TRAF2 also leads to splenomegaly and lymphadenopathy due to constitutive NF-κB2 activation and increased numbers of B cells (40, 42). Remarkably, TRAF2DN/Bcl-2 double-transgenic mice spontaneously develop small B cell lymphoma progressing to leukemia with many similarities to human CLL (Table 2) (41, 42). Thus, TRAF2 acts as a tumor suppressor in B lymphocytes primarily by inhibiting the NF-κB2 pathway through the well-established cIAP1/2-TRAF2-TRAF3-NIK axis (48, 54, 118).
Genetic alterations of TRAF2 are detected in 1–2% of human liver cancers, including deletion, mutation and amplification (TCGA, PanCancer Atlas) (119). In human hepatocellular carcinoma (HCC), low expression of TRAF2 and its interacting partner RIP1 is associated with an unfavorable prognosis (43). In line with human evidence, deletion of both TRAF2 and RIP1 in liver parenchymal cells (LPC) leads to spontaneous development of hepatocellular carcinoma, which results from extensive hepatocyte apoptosis due to hyperactivation of caspase-8 but impaired NF-κB activation induced by TNFα (Table 2) (43). Interestingly, TRAF2 also suppresses TNFα-induced necroptosis in hepatocytes by constitutively interacting with MLKL, thereby disrupting the TNFα-induced RIPK3-MLKL association and necroptosome formation. Induced TRAF2 deletion in adult mice results in rapid lethality, in conjunction with increased hepatic necroptosome assembly (Table 2) (44). Therefore, TRAF2 protects hepatocytes from death and tumorigenesis by inhibiting both the TNFα-TNFR1-TRADD-FADD-caspase 8 apoptosis and TNFα-TNFR1-RIPK1-RIPK3-MLKL necroptosis pathways.
Genetic alterations of TRAF2 are detected in 3–4% of human HNSCC and melanoma (Figure 1A). In cultured HNSCC cell lines, TRAF2 is required for cellular proliferation by acting in the TNFα-TNFR1-TRADD-TRAF2-RIPK1-TAK1-IKK-NF-κB pathway (120). In primary human keratinocytes, exposure to UV light triggers association of TRAF2 with TNF-R1 to induce NF-κB activation and inflammation (121). Keratinocyte-specific TRAF2−/− mice exhibit psoriatic skin inflammation associated with apoptotic death and epidermal hyperplasia, which is dependent on TNFα, constitutive NF-κB2 activation and inflammatory cytokine production (45). Further in support of a role for TRAF2 in skin tumorigenesis, mutations of the TRAF2-deubiquitinating enzyme CYLD are identified in patients with familial cylindromatosis, a condition that results in benign tumors of skin appendages, and CYLD−/− mice are highly susceptible to chemically induced skin tumors (122). Similarly, genetic alterations of TRAF2 are also identified in 2.7% (12/439) of human colon cancers (TCGA, PanCancer Atlas). In cultured primary human colon cancer cells, TRAF2 mediates the apoptosis by acting in the AMPK-ASK1-TRAF2-JNK-p53 axis in response to chemotherapies (123). Consistent with a potential role of TRAF2 in colon tumorigenesis, germline TRAF2−/− mice spontaneously develop severe colitis, which results from TNFα-TNFR1-mediated apoptosis of TRAF2−/− colonic epithelial cells and altered colonic microbiota (37). Interestingly, myeloid cell-specific ablation of TRAF2 markedly exacerbates DSS-induced colitis in mice due to enhanced TLR-induced proinflammatory cytokine expression in macrophages (46). This is caused by constitutively elevated levels of the transcription factors c-Rel and IRF5 that are targeted for proteasome-dependent degradation by the cIAP1/2-TRAF2-TRAF3 E3 ubiquitin ligase complex (46). Together, the above evidence consistently supports a suppressive role for TRAF2 in skin and colon tumorigenesis.
It is also noteworthy that genetic alterations of TRAF2 are detected in 2.6% (7/265) of human sarcomas (TCGA) and TRAF2−/− mice display decreased viability of skeletal muscle tissue because of defective TNFα-induced NF-κB activation in myotubes (Table 2) (38). Additionally, specific deletion of TRAF2 in T cells results in decreased numbers of CD8 naïve and memory T cells as well as NKT cells, due to impaired IL-15-induced signaling in these cells (Table 2). However, evidence of TRAF2 genetic alteration in T cell neoplasms is still lacking. Potential causal roles of TRAF2 dysregulation in muscle or T cell tumorigenesis remain to be elucidated.
Key Signaling Pathways in Cancer Pathogenesis
In addition to the above TRAF2-dependent tumor suppressive pathways verified in both human cancers and In Vivo mouse models, several important tumor suppressive pathways involving TRAF2 have been suggested by evidence obtained from cultured human cancer cells or xenograft models. These are: (1) IRE1α-TRAF2-ASK1-JNK in the apoptosis of melanoma, lung cancer and OSCC cells induced by chemotherapies or ER stress (124–126); (2) the TRAF2-caspase-2 complex in mediating DNA damage- or chemotherapy-induced apoptosis of breast, cervical and lung cancer cells, in which TRAF2-mediated ubiquitination of caspase-2 stabilizes the caspase-2 dimer complex and enhances its activity to fully commit the cell to apoptosis (127, 128); (3) TRAF2-mediated inhibition of constitutive NF-κB2 activation, cell proliferation, and anchorage-independent growth in pancreatic cancer, and a similar TRAF2-mediatied inhibition of the Eva1-induced NF-κB2-Sox2/CD15/CD49f pathway in the stemness of glioblastoma (129, 130); and (4) TRAF2-mediated K63-linked ubiquitination of MLST8 that disrupts the MLST8-SIN1-mTORC2-Akt pathway in the Kras-driven lung tumorigenesis (131). Together, these data suggest that TRAF2 is a tumor suppressor in many human cancers.
Interestingly however, increasing evidence indicates that TRAF2 also plays oncogenic roles in epithelial cancers and some other neoplasms. Consistent with the frequent amplification of TRAF2 detected in human epithelial cancers (Figure 1B) (103), TRAF2 expression is higher in prostate cancer (133), pancreatic cancer (132), lung cancer (134), stomach cancer (135), colon cancer (136), glioblastoma (137) than in normal tissues. Increased TRAF2 expression is recognized as a prognostic factor in pancreatic cancer (132), stomach cancer (135), and glioblastoma (137). Importantly, suppression of TRAF2 in cancer cells harboring a TRAF2 copy number gain inhibits proliferation, NF-κB activation, anchorage-independent growth, and tumorigenesis (103). Knockdown of TRAF2 also enhances TRAIL-induced apoptosis in prostate cancer (133) and inhibits the growth but induces radiosensitization of lung cancer and glioblastoma cells (134). Thus, TRAF2 is required for the maintenance of the malignant state in certain cancer cells containing TRAF2 amplification or overexpression, and TRAF2 protein levels also regulate the sensitivity of cancer cells to chemotherapy and radiotherapy.
A variety of TRAF2-dependent oncogenic pathways have been reported based on studies of patient samples, cultured human cancer cells or xenograft models. Examples include: (1) TRAF2-NEMO-p65-NF-κB1-Bcl2/XIAP/Survivin/TNFα/IL-1/IL-8/HIF-1α in the migration, invasion, metastasis, or drug resistance of breast, stomach and pancreatic cancer cells as well as DLBCL (135, 138–140); (2) EGF-EGFR-TRAF2-RSK2-AP1 in the growth of colon cancer cells (136) and EGFR-TRAF2-RIP1-IKK-NF-κB1 in the resistance to chemotherapy (EGFR inhibitors) in lung cancer cells (141); (3) cIAP1-cIAP2-TRAF2-IKKε-TBK1-IRF3/7/NF-κB1/STAT3 in the tumorigenesis of breast cancer, in which IKKε is amplified in 30% of patients (142, 143); (4) Although TRAF2 is generally considered as a K63-specific E3 ubiquitin ligase (144), a few studies reported TRAF2-mediated K48-linked ubiquitination and degradation of Caspase 8 in the switch of the DR5-Caspase 8 apoptotic pathway to the DR5-Cbl-TRAF2-JNK-AP1-MMP1 invasion/metastasis pathway or the cytoprotective TRAF2-RIPK1-JNK autophagic survival pathway following TRAIL treatment in HNSCC, prostate, lung, stomach, colorectal, and bladder cancer cells (145–148); (5) S100A9-CD147-TRAF2-cdc42 in the metastasis of melanoma (149); (6) TNFα-TRAF2-NF-κB1/AP1-COX2/IL-6/IL-8-PGE2-NOS2 and NOS2-NO-IRE1α-TRAF2-NF-κB1/AP1-COX2/IL-6/IL-8-PGE2 in the growth of breast cancer (150); (7) CD95-TRAF2-NF-κB1/AP1-IL-8/uPA in the invasion of pancreatic cancer (132); and (8) TWEAK-Fn14-TRAF2-SGEF-RhoG-Rac1 in the migration and invasion of glioma (151). Taken together, it is perplexing that both tumor suppressive and oncogenic roles of TRAF2 have been reported in the same type of human cancers. The exact roles of TRAF2 may be dependent on the genetic alteration context and malignant stage of the cancer cells as well as the nature of the environmental cue and treatment regimen.
TRAF3
Landscape of Genetic Alterations
The frequency of genetic alterations of TRAF3 is generally <6% in human cancers (Figure 1A) according to the TCGA and COSMIC datasets of sample size n > 250. The eight human cancers with relatively higher genetic alterations of TRAF3 are HNSCC (5.4%) (113), lung cancer (5.3%) (TCGA, PanCancer Atlas), cervical cancer (4.7%) (TCGA, PanCancer Atlas), uterine cancer (4.5%) (TCGA, PanCancer Atlas), stomach cancer (4.1%) (TCGA, PanCancer Atlas), bladder cancer (3.6%) (152), ovarian cancer (3.4%) (TCGA, PanCancer Atlas), and skin cutaneous melanoma (3.4%) (TCGA, PanCancer Atlas). Interestingly however, a subgroup among the 279 cases of HNSCC cataloged in TCGA, the human papilloma virus-positive (HPV+) HNSCC tumors, has much higher frequency (22%, 8/36) of deep deletions and truncations of TRAF3 than the HPV- HNSCC tumors (Figure 1B) (113). Notably, although not cataloged in TCGA, deletions and mutations of TRAF3 are recognized as one of the most frequent genetic alterations in a variety of B cell malignancies (153), including gastric marginal zone lymphoma (MZL, 21%) (154), multiple myeloma (MM, 17%) (155, 156), HL (15%) (157), DLBCL (14.3%) (158), splenic MZL (10%) (159), and Waldenstrom's macroglobulinemia (WM, 5.3%) (160) (Figure 1B). Furthermore, somatic mutations of TRAF3 are also frequently detected in human nasopharyngeal cancer (NPC, 8.6%) (161) (Figure 1B). Together, the most common genetic alteration of TRAF3 is deep deletion, followed by mutation and then amplification. Truncation and fusion of TRAF3 are less common but also detected in several different types of human cancers (Figure 1).
Overview and Map of Recurrent Mutations
There are 280 different mutations of TRAF3 detected in human cancers, comprising 90% (253/280) mutations that change the protein sequence of TRAF3 and 10% (27/280) coding silent mutations (Table 1). Approximately 43% (108/253) of the coding-altering mutations of TRAF3 are recurrently detected in at least two cancer patients. Among all the TRAF genes, TRAF3 recurrent mutations exhibit the most complex pattern and include the highest frequencies of frameshift mutations (19%, 21/108) and truncations (8%, 9/108). TRAF3 recurrent mutations also include 69% (75/108) missense mutations, 1% (1/108) in frame deletion, 1% splice mutation, and 1% fusion (Table 1 and Figure 2). These recurrent mutations occurred at 67 amino acid positions that are distributed in almost the entire length of the TRAF3 protein (Figure 3).
Five mutation hotspots of TRAF3 are identified in more than 5 cancer patients, specifically N16, N285, K286, R310, and R376 (Figure 3). TRAF3 mutations at N16 have the highest patient count, including the missense mutation (N16T) identified in 10 patients with HNSCC (COSMIC) and the frameshift deletion (N16fs*3) detected in a patient with splenic MZL (162, 163). Mutations at the two consecutive amino acids N285 and K286 of the coiled-coil domain of TRAF3 exhibit the most complex pattern. N285 contains frameshift deletion (N285fs*38), frameshift insertion (N285fs*13) and missense mutation (N285S) identified in 8 patients with HNSCC, MZL, NPC, CRC, stomach cancer and uterine cancer (TCGA; COSMIC) (12, 107, 161, 164, 165). Similarly, K286 exhibits frameshift deletion (K286fs*7 or fs*11) and truncation (K286*) detected in six patients with B cell malignancies, including MM, CLL and WM (155, 160, 166, 167). A third amino acid of the coiled-coil domain, R310, is consistently targeted by truncation (R310*) as detected in 8 patients with DLBCL, MM, HNSCC, cervical cancer and uterine cancer (TCGA) (113, 155, 158, 166, 168). Missense mutations at R376 (R376W or Q) located in the linker between the coiled-coil and TRAF-C domains of TRAF3 are detected in six patients with lung cancer, CRC, SSCC, and melanoma (TCGA; COSMIC) (14, 108, 169). Many of these truncations, frameshifts and missense mutations have been shown to result in inactivation of TRAF3 by disrupting its interaction with NIK, thereby inducing constitutive NF-κB2 activation (155, 156, 158, 159, 170). Thus, most of the recurrent genetic alterations of TRAF3 identified in human cancers cause complete loss or inactivation of the TRAF3 protein.
Fusion
There are 14 different fusions of TRAF3 detected in human cancers, including TRAF3-WDR20 in stomach and uterine cancers, four fusions of TRAF3-MYO16, TRAF3-RCOR1, TRAF3-KLC1, and EVL-TRAF3 in breast cancer, TRAF3-SFXN1 in cervical cancer, UBR5-TRAF3 in HNSCC, two fusions of TRAF3-ZNF839 and TRAF3-MARK3 in kidney cancer, two fusions of TRAF3-BMP3 and SLC22A23-TRAF3 in lung cancer, TRAF3-IFNL1 in ovarian cancer, TRAF3-ITPK1 in pheochromocytoma and TRAF3-SIVA1 in stomach cancer (TCGA). Among the 14 fusions, only the TRAF3-WDR20 fusion is recurrently detected in two patients with stomach cancer and uterine cancer (TCGA, PanCancer Atlas). However, the functional significance of these TRAF3 fusions is currently unknown.
In vivo Causal Roles in Cancer Pathogenesis
Similar to TRAF2 and also consistent with the frequent deletions and inactivating mutations of TRAF3 identified in human B cell malignancies (Figure 1B), a tumor suppressive role for TRAF3 in B lymphocytes has been demonstrated by in vivo evidence obtained from mouse models. As shown for B-TRAF2−/− mice, B cell-specific TRAF3−/− (B-TRAF3−/−) mice also exhibit severe peripheral B cell hyperplasia due to prolonged survival of mature B cells independent of BAFF, which results from constitutive NF-κB2 activation (39, 51). These B-TRAF3−/− mice spontaneously develop splenic MZL or B1 lymphoma at high incidence (52). Interestingly, B-TRAF3−/− mice also have doubled number of plasma cells due to enhanced responsiveness to IL-6 (171). Mechanistically, TRAF3 inhibits the IL-6-IL-6R-JAK1-STAT3 survival and differentiation pathway in plasma cells by facilitating the association of PTPN22 with JAK1 (171). Furthermore, the EBV-encoded oncoprotein LMP1 sequesters TRAF3 to produce functional TRAF3 deficiency in human and mouse B lymphoma cells (172, 173). Intriguingly, lymphocyte-specific TRAF3 transgenic mice also develop plasmacytosis, autoimmunity, inflammation, and cancers, which are likely caused by hyper-responsiveness of B cells to antigens and TLR agonists (55). Thus, TRAF3 acts as a tumor suppressor in naïve B cells, but an appropriate and balanced level, neither too high nor too low, of TRAF3 is required to maintain the homeostasis of plasma cells and protect them from tumorigenesis.
Interestingly, specific deletion of TRAF3 from myeloid cells (granulocytes, monocytes, and macrophages) leads to spontaneous development of histiocytic sarcomas derived from TRAF3−/− tissue-resident macrophages in aging mice (56, 174). The pathogenic mechanisms are likely related to the enhanced TLR-induced inflammatory responses observed in TRAF3−/− macrophages through constitutive activation of NF-κB2, c-Rel, and IRF5, as described for TRAF2−/− macrophages (56, 174). Two other mouse models with functional relevance to TRAF3, Dok1−/−Dok2−/−Dok3−/− mice and humanized TLR7/TLR8 transgenic mice, also spontaneously develop histiocytic sarcomas (175, 176). DOK3, a negative regulator of protein tyrosine kinase (PTK)-mediated signaling, has recently been identified as a TRAF3-interacting protein (177). Similar to TRAF3−/− macrophages, DOK3−/− macrophages are defective in the TLR3-IRF3-IFNβ antiviral pathway (177). TRAF3 is also a transducer of TLR7 and TLR8 signaling through direct interaction with MyD88 (1). Transgenic expression of human TLR7/TLR8 in mice deficient for endogenous TLR7/TLR8 drives inflammation and proliferative histiocytosis, which can be reversed by compound deletion of MyD88 (176). Collectively, the above in vivo evidence indicates that TRAF3 is a tumor suppressor in macrophages and that dysregulation of the TLR-MyD88-TRAF3-Dok3 axis in macrophages plays causal roles in the pathogenesis of histiocytic sarcoma. However, because histiocytic sarcoma in humans is a rare malignancy with sparse pathologic and cytogenetic data (178, 179), potential TRAF3 genetic alterations in human histiocytic sarcomas require future investigation.
In addition to the phenotype of histiocytic sarcoma, aging myeloid cell-specific TRAF3−/− (M-TRAF3−/−) mice spontaneously develop chronic inflammation and other cancers that often affect multiple organs including the gastrointestinal tract (56). Similar to M-TRAF2−/− mice, young adult M-TRAF3−/− mice exhibit exacerbated DSS-induced colitis with increased levels of inflammatory cytokines produced by TRAF3−/− macrophages in response to TLR signaling (46). Notably, another mouse model with functional relevance to TRAF3, NLRP12−/− mice, is highly susceptible to colitis and colitis-associated colon cancer (180). NLRP12 interacts with both TRAF3 and NIK, and NLRP12−/− cells have constitutively activated NF-κB2 associated with a decreased protein level of TRAF3 (180). Interestingly, both NLRP12−/− hematopoietic and non-hematopoietic cells contribute to inflammation, but the latter dominantly contributes to colon tumorigenesis (180). In line with the in vivo data, mutations and deletions of TRAF3 are detected in 2.3% (10/439) of human colon cancers (TCGA, PanCancer Atlas). Furthermore, miR-32-TRAF3-mediated inhibition of the NIK-NF-κB2 pathway protects human colorectal epithelium against colorectal cancer in response to a diet of non-digestible carbohydrates (181). Thus, TRAF3 appears to act in both epithelial cells and myeloid cells to suppress colon tumorigenesis by inhibiting the NF-κB2 and TLR-induced inflammatory pathways.
Although most evidence identifies TRAF3 as a tumor suppressor, studies of the T cell-specific TRAF3−/− (T-TRAF3−/−) mouse model suggest an oncogenic role for TRAF3 in T cells. Despite their constitutive NF-κB2 activation, TRAF3−/− T cells exhibit impaired proliferation and activation in response to TCR and CD28 co-stimulation (39, 57). T-TRAF3−/− mice show defects in T cell-mediated immunity and IL-15-induced proliferation and survival of iNKT cells, and also have reduced number of CD8 central memory T cells (Table 2) (57, 59, 60). Consistent with these in vivo data, TRAF3 is required for the proliferation of human neoplastic ALCL cells in culture (182). Silencing of TRAF3 in ALCL cells not only results in aberrant activation of the NIK-NF-κB2 pathway, but also affects the continued PI3K-AKT and JAK-STAT signaling (182). Therefore, distinct tumor suppressive and oncogenic roles of TRAF3 in different cellular contexts have been revealed from studies of both human and mouse models.
Key Tumor Suppressive Pathways
In addition to the above TRAF3-dependent tumor suppressive pathways verified in both human cancers and in vivo mouse models, several additional tumor suppressive pathways involving TRAF3 have been suggested by studies of cultured human cancer cells or xenograft models. These include: (1) TRAF3-mediated inhibition of the oncogenic RelB-SMAD4 association that represses TGFβ-SMAD target gene expression to promote the tumorigenesis of lung cancer, in which TRAF3 is targeted by RAS-NDP52-mediated autophagic degradation via the NDP52-TRAF3 interaction (183); (2) LIGHT-LTβR-TRAF3/TRAF5-ROS-ASK1-Caspase3 in the apoptosis of human colon cancer and hepatoma cells (184); (3) membrane-bound CD40L-CD40-TRAF3-p40phox-ROS-ASK1-MKK4-JNK-AP1-caspase 9/3/Bax/Bak in the apoptosis of human bladder and CRC cells but not normal epithelial cells (185–187); and (4) TRAF3-mediated inhibition of the oncogenic RIP2-NF-κB1/NF-κB2/p38-Bcl-xL pathway in the survival and proliferation of glioblastoma cells (188). Taken together, available evidence supports that TRAF3 acts as a tumor suppressor in a variety of cell types, but we cannot rule out that TRAF3 upregulation might also alter normal cell homeostasis in the same or other cell types and therefore contribute to transformation, as it has been observed in B cells and T cells.
TRAF4
Landscape of Genetic Alterations
The frequency of genetic alterations of TRAF4 is generally <11% in human cancers (Figure 1A) based on the TCGA and COSMIC datasets of sample size n > 100. The eight human cancers with relatively higher genetic alterations of TRAF4 are pancreatic cancer (10.1%) (7), bladder cancer (7.3%) (152), breast cancer (5.5%) (189), uterine cancer (5.1%) (TCGA, PanCancer Atlas), esophageal cancer (3.2%) (TCGA, Provisional), lung cancer (2.6%) (190), melanoma (2.5%) (191), and ovarian cancer (2.4%) (TCGA, PanCancer Atlas). The most common genetic alteration of TRAF4 is amplification, followed by mutation (Figure 1). Deep deletion, truncation and fusion of TRAF4 are relatively rare in human cancers.
Overview and Map of Recurrent Mutations
There are 123 different mutations of TRAF4 detected in human cancers, comprising 85% (105/123) mutations that cause changes in the amino acid sequence of TRAF4 and 15% (18/123) coding silent mutations (Table 1). About 42% (44/105) of the coding-altering mutations of the TRAF4 gene are recurrent and detected in at least two cancer patients, including mostly missense mutations (89%, 39/44), 3 truncations, 1 frameshift deletion, and 1 in frame deletion (Table 1 and Figure 2). TRAF4 recurrent mutations occurred at 32 different amino acids that are distributed in the entire length of the TRAF4 protein but are relatively enriched in the TRAF-C domain (Figure 3). Only two specific amino acids, R448 and R452 located at the C-terminal TRAF-C domain, are mutated in more than 3 patients (Figure 3). For R448, mixed missense mutations (R448Q or L) and a truncation (R448*) are identified in 4 patients with prostate cancer, uterine cancer, HNSCC, and OSCC (192–195). For R452, missense mutations (R452W or Q or L) are detected in four patients with uterine, colorectal and lung cancers (10, 108, 193). Further studies are needed to determine whether such missense mutations in the TRAF-C domain result in loss- or gain-of-function of TRAF4.
Fusion
There is only one fusion of TRAF4 detected in human cancers, the TRAF4-FASN fusion identified in a glioma patient (TCGA), with currently unknown functional significance.
In vivo Causal Oncogenic Roles
Available human evidence indicates that gene amplification is the most common TRAF4 genetic alteration in cancers and that TRAF4 expression is ubiquitously elevated in many human cancers (196–204). This suggests that TRAF4 overexpression may play causal roles in cancer initiation, progression and metastasis. Similar to classical oncogenes (such as c-Myc and K-ras), TRAF4 is also required for ontogenic processes and TRAF4−/− mice show defects in embryonic development and neurulation (61, 62, 205). Interestingly, TRAF4−/− dendritic cells (DCs) derived from the null mice exhibit reduced in vivo and in vitro migration (64). Furthermore, recent in vivo evidence obtained from mouse models demonstrates the causal oncogenic roles of TRAF4 in skin tumorigenesis (Table 2) (65). TRAF4 deficiency substantially diminishes IL-17A-induced ERK5 activation and epidermal hyperplasia in mice. In the DMBA/TPA-induced skin cancer model, TRAF4−/− mice exhibit remarkably reduced tumor incidence and tumor numbers. Mechanistically, TRAF4 bridges the interaction between Act1 and MEKK3 in response to IL-17A signaling, and therefore is required for the activation of the downstream MEK5-ERK5-Steap4/p63 pathway. The transcription factor p63 directly induces TRAF4 expression in keratinocytes, promoting positive feedback on TRAF4 in the epidermis and thus sustaining the activation of the TRAF4-ERK5 axis to induce keratinocyte proliferation and skin tumorigenesis (65). These in vivo findings are reinforced by the examination of human SSCC samples, which also demonstrates the presence of the IL-17A-Act1-TRAF4-MEKK3-MEK5-ERK5-Steap4/p63 pathway (65). Together, both human and in vivo mouse evidence supports an oncogenic role for TRAF4.
Key Oncogenic Pathways
In addition to the established IL-17A-TRAF4-ERK5 axis, a variety of potential TRAF4-dependent oncogenic pathways have been suggested by studies of patient samples, cultured human cancer cells or their xenografts in immunodeficient mice. These include: (1) TRAF4-Akt/NF-κB-Glut1/HK2/RSK4/Slug in the proliferation and metastasis of lung and breast cancer cells as well as the migration and epithelial-mesenchymal transition (EMT) of hepatocellular carcinoma cells (HCC) (199, 203, 206); (2) TGFβ-TβRI-TRAF4-Smurf1/Smurf2/USP15-SMAD2/TAK1-N-cadherin/Fibronectin/Vimentin/SMA in the migration, EMT, and metastasis of breast cancer cells (200, 207); (3) SRC3-TRAF4-mediated inhibition of the USP7-p53 interaction, leading to the loss of p53 deubiquitination/stabilization and thus the resistance to cytotoxic drugs and stress in breast cancer (208); (4) NGF-TrkA-TRAF4-Akt/p38-IL-6/Integrins/COX2 in the metastasis of prostate cancer cells (204); (5) TNFα-TRAF4/TRAF2-NF-κB1 in the survival and proliferation of breast cancer cells (209); (6) TRAF4-mediated up-regulation and nuclear translocation of β-catenin in the Wnt/β-catenin-cyclin D1/c-myc/Bcl-2/MMPs pathway that promote the growth and migration of OSCC and breast cancer cells (210, 211); (7) TRAF4-mTOR-p70S6K-S6 in the proliferation of breast cancer cells (212); and (8) the TRAF4-phosphoinositide (PIP) interaction at tight junctions that favors breast cancer cell migration (213). It would be interesting to verify these TRAF4-dependent oncogenic pathways using in vivo models.
TRAF5
Landscape of Genetic Alterations
The landscape of TRAF5 genetic alterations is similar to that of TRAF4. The frequency of genetic alterations of TRAF5 is generally <13% in human cancers (Figure 1A) according to the TCGA and COSMIC datasets of sample size n > 140. The eight human cancers with relatively higher genetic alterations of TRAF5 are breast cancer (12.2%) (189), liver cancer (8.4%) (TCGA, Provisional), uterine cancer (6.4%) (TCGA, PanCancer Atlas), lung cancer (5.3%) (TCGA, Provisional), ovarian cancer (5.1%) (TCGA, Provisional), melanoma (4.0%) (TCGA, Provisional), esophageal cancer (3.8%) (TCGA, Provisional), and prostate cancer (3.3%) (214). As described for TRAF4, the most common genetic alteration of TRAF5 is also amplification, followed by mutation (Figure 1A). Deep deletion, truncation and fusion of TRAF5 are rare events in human cancers.
Overview and Map of Recurrent Mutations
There are 188 different mutations of TRAF5 detected in human cancers, comprising 85% (160/188) mutations that alter the amino acid sequence of TRAF5 and 15% (28/188) coding silent mutations (Table 1). Approximately 36% (57/160) of the coding-altering mutations of TRAF5 are recurrent in human cancers. Similar to TRAF4, TRAF5 recurrent mutations also include mostly missense mutations (85%, 49/57), but also some truncations (9%, 5/57), frameshift deletions (4%, 2/57), and an in frame deletion (2%, 1/57) (Table 1 and Figure 2). These recurrent mutations occurred at 36 different amino acids that are mainly enriched in the TRAF-C domain but also scattered in other regions of the TRAF5 protein (Figure 3). Mutations of three specific amino acids, R164, T232, and A548, are detected in more than three patients (Figure 3). Complex alterations of R164 of the zinc finger motif, including truncation (R164*) and missense mutations (R164Q or L), are detected in six patients with uterine, colon and bile duct cancers and DLBCL (TCGA) (12, 193, 215). Another missense mutation of the zinc finger motif, T232M, is detected in four patients with colon, breast, and prostate cancers (TCGA; COSMIC) (98). Missense mutation A548V of the TRAF-C domain is identified in four patients with uterine, cervical, stomach, and breast cancers (TCGA) (107). The functional consequences of these recurrent TRAF5 mutations await further investigation.
In vivo Causal Oncogenic Roles
Although not cataloged in TCGA, TRAF5 mutations are detected in 5% (5/101) of human DLBCL (102). TRAF5 expression is upregulated in human splenic MZL (216). In addition, apoptosis-resistant B cell-acute lymphoblastic leukemia (B-ALL) cells have aberrantly higher protein level of TRAF5 and TRAF6 in response to irradiation than apoptosis-proficient B-ALL cells (217). The above evidence suggests that TRAF5 may be oncogenic in B cells. Consistent with human evidence, B cells of TRAF5−/− mice show defects in CD40-induced proliferation and up-regulation of surface molecules and activation markers as well as CD40 plus IL-4-induced Ig production (Table 2) (67). Using a chimeric CD40-LMP1 transgenic (CD40LMP1-tg) mouse model that mimics the B cell hyperactivation induced by the EBV-encoded oncoprotein LMP1 (218), Kraus et al. demonstrated that TRAF5 is a critical mediator of the in vivo functions of LMP1 (73). TRAF5 deficiency reverses the CD40-LMP1-induced enlargement of the spleen and lymph nodes, decreases the serum levels of IL-6 and autoantibodies that are elevated by CD40-LMP1-tg expression, and also inhibits LMP1-mediated JNK activation in B lymphocytes (Table 2) (73). Together, both human and mouse evidence supports an oncogenic role for TRAF5 in B cells that appears to be required for LMP1-mediated B lymphomagenesis and is likely also involved in spontaneous B lymphomagenesis initiated by genetic alterations.
Additionally, available in vivo evidence indicates the importance of TRAF5 in the survival, proliferation and differentiation of different T cell subsets as detailed in Table 2, suggesting that TRAF5 malfunction may contribute to T cell malignancies. However, the evidence of TRAF5 alterations in human T cell lymphomas/leukemias is still lacking.
TRAF5-Dependent Signaling Pathways in Human Cancer Cells
In addition to the signaling pathways of B cells and T cells revealed by the in vivo studies of TRAF5−/− mice, a number of TRAF5-dependent signaling pathways have been proposed based on the studies of patient samples, cultured human cancer cells or their xenografts in immunodeficient mice. These include: (1) CD30-TRAF2/TRAF5-NIK-IKKα-NF-κB-IL-13 in the survival of Hodgkin-Reed-Sternberg (H-RS) cells of HL (219, 220) and a similar CD30v-TRAF2/TRAF5-NIK-NF-κB pathway in acute myeloid leukemia (AML) and ALL (221); (2) LIGHT-LTβR-TRAF3/TRAF5-ROS-ASK1-Caspase 3 in the apoptosis of human colon cancer and hepatoma cells (184); (3) upregulated TRAF5-NF-κB in the migration and invasion of glioma cells, in which TRAF5 is directly targeted for degradation by the tumor suppressor Numbl (222); (4) TRAF5/TRAF6-NF-κB-Vimentin/TWIST1/SNAIL2/MMP13/IL-11 in the EMT and metastasis of prostate cancer cells, in which TRAF5 is directly targeted for downregulation by the tumor suppressive miR-141-3p (223); and (5) TRAF5-MEK1/2-ERK1/2-Bcl2 in the survival and proliferation of melanoma cells, in which TRAF5 is directly targeted for downregulation by tumor suppressive MiR-26b (224). The above evidence supports the hypothesis that TRAF5 plays oncogenic roles in various human cancer cells primarily by inducing NF-κB activation but also by activating the ERK1/2 pathway.
TRAF6
Landscape of Genetic Alterations
The frequency of genetic alterations of TRAF6 is generally <7% in human cancers (Figure 1A) based on the TCGA and COSMIC datasets of sample size n > 120. The eight human cancers with relatively higher genetic alterations of TRAF6 are breast cancer (6.9%) (225), uterine cancer (4.5%) (TCGA, PanCancer Atlas), stomach cancer (4.1%) (8), HNSCC (3.6%) (113), lung cancer (3.4%) (10), bladder cancer (3.1%) (226), sarcoma (3%) (TCGA, Provisional), and ovarian cancer (2.8%) (TCGA, Provisional). Although not listed in TCGA, TRAF6 amplification is recognized as one of the most frequent genomic alterations in human lung cancer (9.2%, 24/261) (227) and OSCC (10%, 2/20) (228). Consistent with the frequent amplification of TRAF6 in human cancers, TRAF6 is overexpressed in many human cancers such as breast cancer, HCC, colon cancer, esophageal cancer, and melanoma (229–233). TRAF6 overexpression is also identified as a prognostic factor for breast and esophageal cancers (229, 232). Together, the most common genetic alterations of TRAF6 are mutation and amplification (Figure 1A). Deep deletion of TRAF6 is less common but also detected in several different types of human cancers. Truncation and fusion of TRAF6 are rare in human cancers.
Overview and Map of Recurrent Mutations
There are 178 different mutations of TRAF6 detected in human cancers, comprising 85% (152/178) mutations that alter the protein sequence of TRAF6 and 15% (26/178) coding silent mutations (Table 1). Only 27% (41/152) of the coding-altering mutations of TRAF6 are recurrently detected in at least two cancer patients. Similar to TRAF1, TRAF6 recurrent mutations also have the simplest composition and are almost exclusively missense mutations (93%, 38/41) except 2 truncations and 1 frameshift insertion (Table 1 and Figure 2). These recurrent mutations occurred at 30 different amino acids that are distributed in all the major domains but are enriched in the coiled-coil and TRAF-C domains of the TRAF6 protein (Figure 3). Mutations of only two specific amino acids, R335 and P398, are detected in more than three patients (Figure 3). A truncation (R335*) and missense mutation (R335Q) at R335 within the coiled-coil domain of TRAF6 are detected in five patients with colon and uterine cancers (TCGA) (12, 234). Missense mutations at P398 (P398L or S) of the TRAF-C domain are identified in 4 patients with uterine, lung, and stomach cancers (TCGA) (8, 193, 235). Functional significance of these TRAF6 recurrent mutations in cancer pathogenesis remains to be elucidated.
In vivo Evidence of Potential Roles for TRAF6 in Tumorigenesis
Causal roles of TRAF6 in tumorigenesis have not been directly demonstrated with TRAF6 knockout or transgenic mice yet. However, available in vivo evidence supports potential contributions of TRAF6 dysregulation in tumorigenesis. Consistent with the genetic alterations (mainly amplification and mutation) and frequent overexpression of TRAF6 detected in human epithelial cancers such as breast cancer and uterine cancer (Figure 1A) (229–233), deletion of TRAF6 in mice results in loss of NF-κB activity in epithelia and vasculature during mouse development (Table 2) (78). Corroborating initial evidence, intestinal epithelial cell-specific TRAF6−/− mice exhibit exacerbated DSS-induced colitis (87). In line with the in vivo data, knockdown of TRAF6 or inhibition of TRAF6 E3 ligase activity suppresses the survival, proliferation, migration, and metastasis of many human epithelial cancers, including breast, lung, liver, and colon cancers as well as HNSCC (230–232, 236–240). In the majority of these cases, the TRAF6-NF-κB axis is identified as the main oncogenic pathway, which is constitutively activated by TRAF6 overexpression or hyperactivated by upstream receptors such as TNFα, RANK, and TLR4/3 (236–240).
Similar findings have been obtained in the hematopoietic/lymphoid system. Hematopoietic-specific deletion of TRAF6 in mice leads to decreased tonic IKKβ-NF-κB activation, impaired hematopoietic stem cell (HSC) self-renewal and loss of hematopoietic stem/progenitor cells (HSPCs) in the bone marrow (BM) (Table 2) (81). Knockdown of TRAF6 in human AML cell lines or patient samples results in rapid apoptosis and impaired malignant HSPC function as well as increased sensitivity to bortezomib (241). In the lymphoid lineage, TRAF6 mutations have been detected in 2.1% human DLBCL (TCGA) and 2.4% human cutaneous T cell lymphoma (CTCL) (242). TRAF6−/− mice have reduced numbers of immature B cells in the BM and B cells from the null mice show defects in CD40 and LPS-induced proliferation and NF-κB activation (Table 2) (74, 75). B cell-specific TRAF6−/− mice (B-TRAF6−/−) mice also exhibit reduced numbers of mature B cells in the BM and spleen as well as defective B1 B cell development (Table 2) (82). Knockdown of TRAF6 or inhibition of the TRAF6-NF-κB axis induces apoptosis and cell cycle arrest in DLBCL, and also inhibits the proliferation and bone resorption of MM (243, 244). Interestingly, T cell-specific TRAF6−/− mice (T-TRAF6−/−) mice show multiorgan inflammation due to hyperactivation of the PI3K-Akt pathway in T cells (Table 2) (83). T-TRAF6−/− mice also exhibit increased Th17 differentiation due to enhanced sensitivity of CD4 T cells to TGFβ signaling (85), but have defects in generating CD8 memory T cells caused by defective AMPK activation in activated CD8 T cells (84). In addition, T-TRAF6−/− mice exhibit impaired OX40-induced CD4 Th9 differentiation, which requires TRAF6-mediated activation of the NIK-NF-κB2 pathway in CD4 T cells (Table 2) (86). In support of a role for TRAF6 in T cell tumorigenesis, inhibition of the TRAF6-NF-κB-c-Myc axis through miR-146a/b-mediated downregulation of TRAF6 delays PTEN−/− T cell lymphomagenesis in mice (245). Furthermore, inhibition of the IRAK1/4-TRAF6 axis sensitizes human T cell ALL (T-ALL) to chemotherapies (246). Collectively, the above evidence is consistent with the hypothesis that TRAF6 may serve as an oncoprotein in epithelial cancers and hematopoietic/lymphoid neoplasms mainly through inducing aberrant NF-κB activation.
Interestingly, in vivo evidence also indicates the functional importance of TRAF6 in the brain and muscle (Table 2). TRAF6−/− mice show defective neural tube closure and exencephaly (80). Mechanistically, TRAF6 interacts with the p75 neurotrophin receptor (p75NTR), and thus is required for NGF-induced NF-κB activation and survival in Schwann cells as well as BDNF-induced JNK activation and apoptosis in superior cervical ganglia neurons (79). In skeletal muscle, TRAF6 deficiency prevents muscle loss and cancer cachexia in response to transplanted tumor growth, improves regeneration of myofibers upon injury and reduces skeletal muscle atrophy upon starvation through regulating NF-κB activation/ubiquitin-proteasome/autophagy-lysosomal systems, Akt/FoxO3a/AMPK activation and Notch signaling, respectively (88–90). In line with the mouse data, genetic alterations of TRAF6, including amplification, mutation and deletion, are detected in 1% of human glioblastoma (247, 248) and 3% of human sarcoma (TCGA) (9). It would be interesting to test potential causal roles of TRAF6 in brain and muscle tumorigenesis in future studies.
Key Signaling Pathways in Human Cancers
In addition to the TRAF6-NF-κB axis that has been verified in both human cancers and in vivo mouse models, numerous TRAF6-dependent oncogenic pathways have been reported with studies of patient samples, cultured human cancer cells or their xenografts in mice. Examples are: (1) the TRAF6-p53 crosstalk, in which TRAF6 promotes K63-linked ubiquitination of p53 and limits the tumor suppressive function of p53 in cancer development and resistance to chemotherapy and radiotherapy (249); (2) the Ras-TRAF6-NF-κB axis in the tumorigenesis of lung and pancreatic cancers (227, 250, 251); (3) the TRAF6-Akt axis in the tumorigenesis of glioblastoma, HNSCC, prostate cancer, oral cancer, and CRC (252–255); (4) the EGFR-TRAF6 axis in the growth, migration and metastasis of lung cancer and cutaneous SCC (256, 257); (5) the TRAF6-HIF1α axis in the tumorigenesis, angiogenesis, and metastasis of breast and colon cancers (258, 259); (6) the TGFβ-TβRI/II-TRAF6 axis in the proliferation, migration, and invasion of prostate cancer (260–263); (7) TRAF6-AEP-HSP90α in the invasion and metastasis of breast cancer (229); (8) nutrients-MEKK3-MEK3/6-p38δ-p62-TRAF6-mTORC1 in the growth of prostate cancer (264); (9) pVHL inactivation-CARD9-BCL10-TRAF6-TAK1-MKK4-JNK-AP1-Twist in the EMT of renal cell carcinoma (265); (10) ADAM10-p75NTR ICD-TRAF6 in the metastasis and chemoresistance of HNSCC and breast cancer (266); (11) TRAF6-DNMT1-DNA methylation in chemoresistance of breast cancer (267); and (12) IL-1β-TRAF6-TNFα and TRAF6-Cdc42-F-actin in the invasion of SCC (268). Interestingly, the importance of TRAF6-dependent oncogenic pathways in human cancers is also underscored by the findings that TRAF6 mRNA is the direct target of tumor suppressive mi-RNAs, including miR-146a (269–274), and miR-141-3p (223). Thus, most evidence indicates that TRAF6 is an oncogene in human cancers.
Intriguingly, several TRAF6-dependent tumor suppressive pathways have also been described for human cancers in the literature. Examples are: (1) TRAF6-p62-mediated inhibition of the HK2 glycolytic activity and the growth of liver cancer, in which TRAF6 catalyzes the K63-linked ubiquitination of HK2 and targets HK2 for p62-mediated autophagic degradation (275); (2) TRAF6-mediated suppression of the EZH2-H3K27me3 pathway and the progression of prostate cancer, in which TRAF6 mediates K63-linked ubiquitination and degradation of EZH2 (276); and (3) TRAF6-mediated decrease of the H3K4me3 level and thus the tumorigenesis of prostate cancer, in which TRAF6 mediates K63-linked ubiquitination of JARID1B to increase the demethylase activity of JARID1B on H3K4me3 (277). Taken together, both tumor suppressive and oncogenic roles of TRAF6 have been reported in human liver cancer and prostate cancer. As discussed for TRAF2, this phenomenon may be related to the mutational profile and malignant stage of the cancer cells as well as the nature of the environmental cue or treatment regimen.
TRAF7
Landscape of Genetic Alterations
TRAF7 lacks the TRAF homology domain and does not directly interact with any member of the TNFR superfamily, two defining features of the TRAF family (278, 279), and is therefore still controversial to be considered as a genuine member of the TRAF family. The frequency of genetic alterations of TRAF7 is generally <7% in human cancers (Figure 1A) according to the TCGA and COSMIC datasets of sample size n > 150. The eight human cancers with relatively higher genetic alterations of TRAF7 are breast cancer (6%) (189), prostate cancer (5.1%) (280), stomach cancer (4.8%) (8), sarcoma (3.8%) (TCGA, Provisional), esophageal cancer (3.3%) (TCGA, PanCancer Atlas), uterine cancer (3.2%) (TCGA, PanCancer Atlas), melanoma (3.1%) (TCGA, PanCancer Atlas), and liver cancer (2.4%) (TCGA, PanCancer Atlas). However, it should be noted that although not yet cataloged in TCGA, the rate of TRAF7 mutation is overwhelmingly high in patients with adenomatoid tumors of the male and female genital tracts (100%, 31/31) (281), secretory meningiomas (97%, 29/30) (282), intraneural perineuriomas (62.5%, 10/16) (283), and meningiomas 23% (182/775) (284) (Figure 1B). In particular, high frequencies (15–26%) of TRAF7 mutations has been reproducibly detected in multiple studies (284–288), and knowledge of TRAF7 mutations has contributed significantly to improving the diagnosis, classification, prognosis, and treatment of patients with meningiomas (282, 286, 289–291). Additionally, deletion of TRAF7 is detected in 67% (18/27) of malignant mesothelioma patients' malignant cells in pleural fluids (292). The most common genetic alteration of TRAF7 is mutation, followed by amplification and then deep deletion (Figure 1). Truncation and fusion of TRAF7 are rarely detected in human cancers.
Overview and Map of Recurrent Mutations
In the TRAF family, TRAF7 has the highest counts of total and recurrent mutations. There are 376 different mutations of TRAF7 detected in human cancers, including 87% (326/376) coding-altering mutations and 13% (50/376) coding silent mutations (Table 1). Over half (53%, 174/326) of the TRAF7 coding-altering mutations are recurrently detected in at least two cancer patients. TRAF7 recurrent mutations are mostly missense mutations (92%, 161/174). Small percentages of other recurrent mutations include 5 frameshifts, 3 truncations, 2 in frame deletions, 2 splice mutations, and 1 fusion (Table 1 and Figure 2). These recurrent mutations occurred at 89 different amino acids covering different regions of the entire length but highly enriched in the last 4 WD40 repeats of the TRAF7 protein (Figure 3). Of particular interest, missense mutations of six specific amino acids located within the C-terminal WD40 repeats, N520, H521, G536, S561, K615, and R641, are identified as mutation hotspots of TRAF7 detected in more than 15 cancer patients (Figure 3). N520 mutations (N520S, H, Y, or T) are found in 31 patients with meningioma, mesothelioma, sarcoma and colon cancer (12, 106, 282, 284, 285, 293, 294). Mutations of the next amino acid H521 (H521R or N) are identified in 15 patients with adenomatoid tumor, perineurioma, and meningioma (281, 283, 284). G536 mutations (G536S or V) are detected in 16 patients with meningioma, pancreatic cancer, mesothelioma and stomach cancer (106, 282, 284, 285, 293–295). S561 mutations (S561R, N or T) are identified in 19 patients with adenomatoid tumor, perineurioma and meningioma (281, 283, 284). K615E mutations are detected in 15 patients with meningioma and OSCC (284, 296). R641 mutations (R641H, C, P, or L) are detected in 24 patients with uterine, bile duct, colon, stomach and lung cancers and meningioma (TCGA, PanCancer Atlas) (8, 106, 282, 284, 285, 294, 297–299). Although the functional significance of most TRAF7 mutations is currently unclear, the exceptionally high recurrence and clustering of missense mutations implicate TRAF7 malfunction as a critical pathogenic event in relevant human cancers.
Fusion
There are six different fusions of TRAF7 and other genes detected in human cancers, including TRAF7-LRRC1 in lung cancer, GFER-TRAF7 in mesothelioma, CORO7-TRAF7 in glioma, TRAF7-MAPK8IP3 in bladder cancer, and TRAF7-RAB26 and E4F1-TRAF7 in ovarian cancer (TCGA) (106). Among these, only the TRAF7-LRRC1 fusion is recurrently detected in two patients with lung cancer (TCGA). However, all the TRAF7 gene fusions have not been verified at the protein level and their functional consequence is unknown.
In vivo Evidence of Potential Roles in Cancer Pathogenesis
No TRAF7−/− or TRAF7-tg mouse model has been published yet. Importantly however, Tokita et al. recently reported that de novo missense mutations in TRAF7 cause developmental abnormalities and other clinical symptoms in seven unrelated patients, including developmental delay (5/5), congenital heart defects (6/7), limb and digital anomalies (7/7), and dysmorphic facial features (7/7) (300). TRAF7 mutations identified in this study include a recurrent R655Q mutation found in four patients, and another 3 single mutations each identified in one patient, including K346E, R371G, and T601A (300). Interestingly, R371 recurrent mutations are also detected in human cancers (Figure 3). K346 is a ubiquitination site of TRAF7 (301). Both K346 and R371 are located in the coiled-coil domain of TRAF7 that is important for TRAF7 homodimerization (302, 303). The recurrent R655Q mutation has also been previously identified as a de novo event in an autism patient (304). Both T601 and R665 are located in the C-terminal WD40 repeats of TRAF7, which contain most mutation hotspots of TRAF7 detected in human cancers (Figure 3) and are known to mediate the interaction of TRAF7 with MEKK3 or c-Myb (302, 305). Tokita et al. further revealed that transfection of the R665Q, T601A, or R371G mutants of TRAF7 into HEK293T cells results in significantly reduced levels of ERK1/2 phosphorylation, both basal and in response to TNFα signaling (300). Consistent with this biochemical evidence, conditional ERK2−/− mice show a phenotype mirroring that observed in the seven patients with TRAF7 mutations, including craniofacial abnormalities, cardiovascular malformations and limb defects (306). These highly interesting findings warrant further investigation of the in vivo functions of TRAF7 mutations in cancer pathogenesis using animal models.
TRAF7-Mediated Signaling Pathways
Compared to other TRAF proteins, the signaling mechanisms of TRAF7 are understudied and remain poorly defined (289, 307). In addition to the above TRAF7-ERK1/2 pathway revealed by studying TRAF7 mutants of patients with developmental defects, the following TRAF7 signaling pathways have been proposed based on in vitro studies. (1) Transfection of tumor-derived TRAF7 mutants (H521R, Y538S, or S561R) but not WT TRAF7 in 293T cells causes increased phosphorylation of RelA and expression of the NF-κB target gene L1CAM, which is also elevated in adenomatoid tumors (281). (2) Overexpression of TRAF7 or TNFα induces caspase-dependent apoptosis in HEK293 and HeLa cells via the TRAF7-MEKK3-NF-κB/p38/JNK-AP1/CHOP pathway, in which TRAF7 interacts with MEKK3 and potentiates the kinase activity of MEKK3 (302, 303). (3) TRAF7 mediates TNFα-induced apoptosis in Jurkat and HeLa cells via promoting the K29-, K33-, and K63-linked ubiquitination and lysosomal degradation of c-FLIP, an inhibitor of caspase activation (308). (4) TRAF7 represses TNFα-induced NF-κB activation to enhance apoptosis in HEK293 cells by promoting K29-linked ubiquitination and lysosomal degradation of NEMO and RelA (309). Paradoxically, TRAF7 is identified as an activator of the NEMO-RelA-NF-κB-cyclin D1 pathway in mouse myoblasts and thus a suppressor of myoblast differentiation (310). (5) TRAF7 participates in TLR2-induced production of inflammatory cytokines (TNFα, IL-1β, and IL-8) in A549 and HeLa cells by acting in the TLR2-TRAF6/TRAF7-IKK1/2/NEMO-NF-κB and TLR2-TRAF6/TRAF7-MKK3/6-p38 pathways (311). (6) TRAF7 inhibits the transcriptional activity of the oncoprotein c-Myb in M1 mouse leukemia cells and DND39 human Burkitt's B lymphoma cells by directly interacting with c-Myb and stimulating the sumoylation of c-Myb to sequester c-Myb in the cytoplasm (305). (7) TRAF7 mediates K48-linked ubiquitination of p53 as demonstrated by an in vitro ubiquitination assay, which likely induces p53 degradation. Correspondingly, TRAF7 protein is downregulated and p53 protein is upregulated in a panel of breast cancer specimens, and TRAF7 downregulation correlates with poor prognosis in breast cancer (312). In summary, WT TRAF7 appears to be a tumor suppressor that promotes cell apoptosis. TRAF7 mutations or downregulated protein levels may lead to aberrant NF-κB activation or altered signaling of ERK1/2, p38, JNK, c-FLIP, c-Myb, or p53 to drive tumorigenesis. Further studies are required to clarify the roles and mechanisms of TRAF7 alterations in cancer pathogenesis.
Combined Genetic Alterations of All TRAFs in the Same Human Cancer
After analyzing the genetic alterations of each TRAF gene in human cancers individually, we next examined the combined genetic alterations of all TRAFs in the same type of human cancer using the TCGA tool. Although the frequency of the genetic alterations of each TRAF is generally low (usually <5%), their combined rate is substantially increased to 10–35% in many types of human cancers (Figure 4) (TCGA). For example, the combined frequency of gene amplification of all seven TRAFs is 35% (709/2015) in breast cancer (313, 314). The combined frequency of genetic alterations of all seven TRAFs is 23% (71/311) in ovarian cancer (TCGA, Provisional), 19% (77/408) in bladder cancer (152), 19% (45/240) in uterine cancer (193), 17% (81/469) in lung cancer (TCGA, PanCancer Atlas), 15% (41/265) in oesophageal cancer (315), 14% (48/353) in liver cancer (TCGA, PanCancer Atlas), 13% (35/279) in HNSCC (113), 13% (36/278) in cervical cancer (TCGA, PanCancer Atlas), 13% (58/438) in melanoma (TCGA, PanCancer Atlas), 12% (46/389) in colon cancer (TCGA, PanCancer Atlas), and 10% (106/1013) in prostate cancer (98). It is interesting that genetic alterations of different TRAFs are often mutually exclusive in the same cancer patient and simultaneous genetic alterations of two or three different TRAFs in the same cancer patient are generally rare events (Figure 4) except for ovarian cancer (TCGA, Provisional), uterine cancer and melanoma (TCGA, PanCancer Atlas).
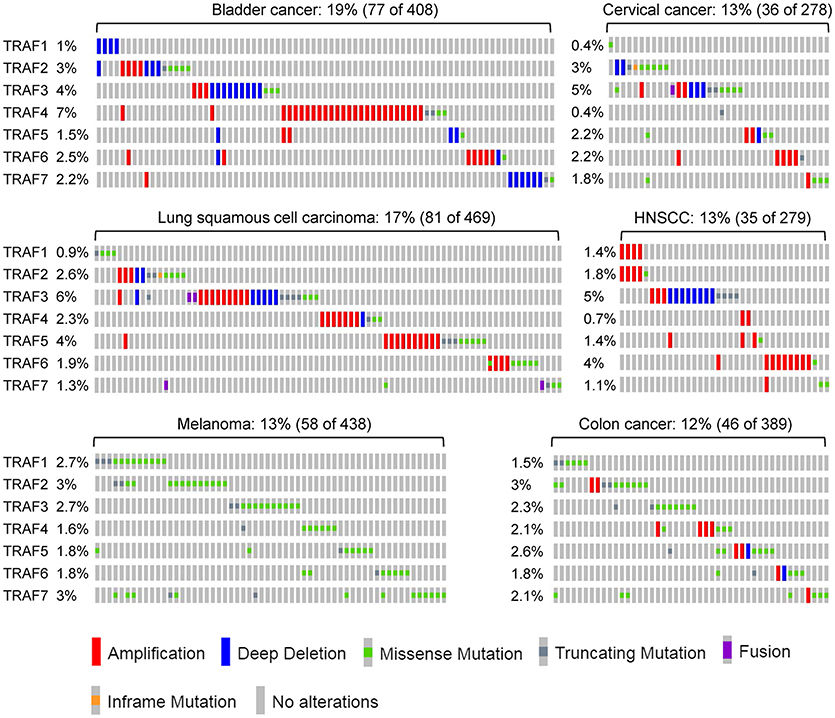
Figure 4. Combined genetic alterations of the TRAF family in human cancers. Representative results of the combined genetic alterations of the TRAF family in individual human cancers are retrieved from TCGA, specifically bladder cancer, lung cancer, melanoma, cervical cancer, HNSCC, and colon cancer. The sample size and the number of patients containing genetic alterations of TRAFs as well as the frequency of each TRAF alteration are indicated for each type of cancer in the figure. The nature of TRAF genetic alteration identified in each patient is indicated by a mutation symbol as shown at the bottom legend of the figure.
We summarize key oncogenic pathways that involve multiple TRAF proteins in skin carcinogenesis as depicted in Figure 5 as well as in B cell malignancies as depicted in Figure 6, both of which have been verified by studies of human cancers and in vivo mouse models. However, we believe current understanding only represents “the tip of the iceberg” of oncogenic mechanisms involving TRAF proteins. Given the often mutually exclusive genetic alterations of different TRAFs in the same cancer, it is very likely that all seven TRAFs may have non-overlapping and distinct contributions to different aspects or at different stages of the initiation, progression and metastasis of the same cancer. These unanswered questions represent fascinating areas for future exploration.
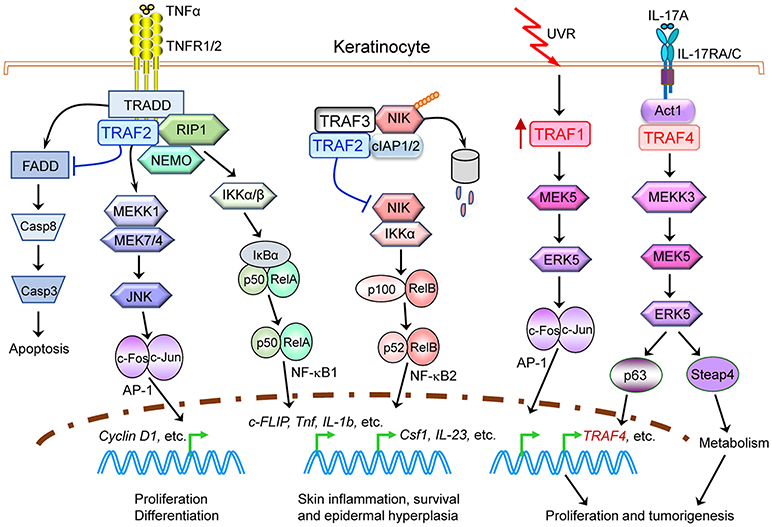
Figure 5. Causal roles and signaling mechanisms of TRAF proteins in skin carcinogenesis. Evidence of both genetic alterations of TRAFs in human patients as well as in vivo TRAF−/− mouse models indicates that alterations of multiple TRAF proteins, specifically TRAF1, TRAF2, and TRAF4, play causal roles in skin carcinogenesis. Oncogenic TRAF proteins (TRAF1 and TRAF4) are depicted in red, while tumor suppressive TRAF2 proteins are depicted in blue. This figure depicts a simplified model of keratinocyte-intrinsic, TRAF-dependent signaling mechanisms in skin carcinogenesis. Only key TRAF-dependent receptors, TRAF-interacting proteins and downstream kinases and transcription factors that have been verified in both human cancers and in vivo mouse models are shown. Keratinocyte-extrinsic, indirect mechanisms of TRAF proteins in skin carcinogenesis are not depicted in the figure, including the known roles of TRAFs in tumor immunity, inflammation and bone resorption and thus their indirect contributions in tumorigenesis and metastasis.
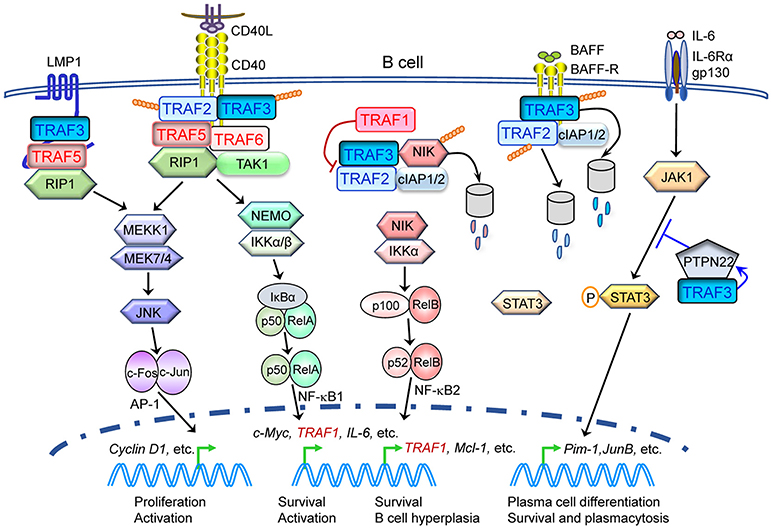
Figure 6. Complex protective and pathogenic roles as well as signaling mechanisms of TRAF proteins in B cell malignancies. Evidence of both genetic alterations of TRAFs in human patients as well as in vivo TRAF knockout and transgenic mouse models indicates that alterations of multiple TRAF proteins, specifically TRAF1, TRAF2, TRAF3, TRAF5, and TRAF6, play causal roles in the pathogenesis of B cell malignancies, such as B lymphomas and multiple myeloma. Tumor suppressive TRAF proteins (TRAF2 and TRAF3) are depicted in blue, while oncogenic TRAF proteins (TRAF1, TRAF5, and TRAF6) are depicted in red. This figure depicts a simplified model of B cell-intrinsic, TRAF-dependent signaling mechanisms in B cell malignancies. Only key TRAF-dependent receptors, TRAF-interacting proteins and downstream kinases and transcription factors that have been verified in both human cancers and in vivo mouse models are shown. Potential contribution of TRAF1, TRAF2, and TRAF6 in LMP1 signaling, TRAF6 in B cell receptor (BCR) signaling and TRAFs in CD40- and LMP1-induced activation of PI-3K-Akt to B cell tumorigenesis are not included in the figure.
TRAF Proteins in Pathogen-Induced Carcinogenesis
The importance of TRAF proteins in cancer pathogenesis is strengthened by mounting evidence that demonstrates their involvement in pathogen-mediated carcinogenesis in certain human cancers. For example, chronic infection with the bacteria Helicobacter pylori (H. pylori) is a major cause of gastric cancer. H. pylori infection induces TRAF1 overexpression and the expression of the transcription factor Cdx2 in both human gastric epithelial cells and mice, which are mainly driven by NF-κB activation. TRAF1 overexpression plays an antiapoptotic role in H. pylori-infected gastric cells (316). Induction of Cdx2 contributes to intestinal metaplasia, a precursor event to gastric cancer. Interestingly, TRAF3 inhibits H. pylori infection-induced NF-κB activation and Cdx2 expression, and is required to resist the infection by acting in the NOD1-RIP2-TRAF3 pathway in gastric epithelial cells (317, 318). Furthermore, the oncoprotein cag PAI of H. pylori activates NF-κB and induces IL-8 secretion through the TRAF2/TRAF6-NIK-IKK pathway in gastric cancer cells (319). Another carcinogenic factor, Tip-α, of H. pylori activates NF-κB by inhibiting the expression of miR-3178, which directly targets TRAF3 mRNA for downregulation, in gastric epithelial cells (320). Therefore, H. pylori chronic infection-induced gastric tumorigenesis involves activation of TRAF1, TRAF2, and TRAF6 as well as inhibition of TRAF3 (Table 3).
A variety of viral infections have also been linked to cancer development. DNA viruses, such as Epstein-Barr virus (EBV), Kaposi's sarcoma-associated herpesvirus (KSHV), human cytomegalovirus (HCMV) and human papilloma virus (HPV), cause NPC, B lymphomas, breast cancer, glioma, cervical cancer, and HNSCC in the host (335, 336). RNA viruses, such as hepatitis C virus (HCV) and human T-cell leukemia virus type 1 (HTLV-1), may lead to HCC and T cell leukemia, respectively, in an infected individual (335, 336). Notably, oncogenic proteins of these viruses exploit or target one or multiple TRAF proteins for their signal transduction. These include the EBV-encoded oncoprotein LMP1, v-FLIP of KSHV, pUL48 of HCMV, E2 and E6 of HPV, Core protein of HCV and Tax protein of HTLV-1. In particular, consistent with the high frequency of TRAF3 deletions and mutations in HPV+ HNSCC, overexpression of TRAF3 inhibits the growth, migration and chemoresistance of HPV+ HNSCC by decreasing HPV E6 oncoprotein and increasing p53 and RB tumor suppressors (328). We briefly summarize the TRAF-dependent signaling mechanisms of pathogen-encoded proteins that contribute to human carcinogenesis as detailed in Table 3.
Indirect Mechanisms of TRAFs in Human Cancers
Although beyond the scope of this review, we would like to point out that as critical regulators of adaptive immunity, innate immunity, and inflammation (1–4), TRAF proteins may indirectly contribute to the development, progression, and metastasis of various cancers by affecting tumor surveillance, tumor immunity, chronic inflammation and the tumor microenvironment. For example, disorders of innate antibacterial response are of fundamental importance in the development of gastrointestinal cancers, including pancreatic cancer, and increased expression of TRAF6, TLR4, and NOD1 are detected in peripheral blood leukocytes of pancreatic cancer patients (337). Specific deletion of TRAF3 from myeloid cells leads to development of B lymphomas and liver cancer in mice (56, 174). Similarly, lymphocyte-specific TRAF3 transgenic mice develop autoimmunity, inflammation and cancers (such as squamous cell carcinomas of the tongue, salivary gland tumors, and hepatoma) (55). TRAF2 regulates inflammatory cytokine production in tumor-associated macrophages, which facilitates tumor growth (46). TRAF4 promotes lung cancer aggressiveness by modulating the tumor microenvironment in normal fibroblasts via the TRAF4-NOX2/NOX4/p47-phox-ROS pathway (338). Importantly, TRAFs are also recognized as potential targets or modulators of cancer immunotherapy. For example, the immune adjuvants dsRNA such as Sendai Virus, poly-I:C, and rintatolimod all activate the TLR3-TRAF3-IRF3 axis to promote CD8 cytotoxic T lymphocytes chemotaxis to the tumor microenvironment in cancer immunotherapy (339). Anti-GITR immunotherapy-induced tumor-specific Th9 cells, which are highly effective in eradicating advanced tumors in vivo, display a unique hyperproliferative feature driven by the Pu.1-TRAF6-NF-κB axis (340, 341). Furthermore, TRAF3 and TRAF6 are crucial for osteoclast differentiation, and therefore can regulate bone metastasis of various cancers (4, 342). We shall witness rapid advancement in these exciting areas in the coming years.
Conclusions
In this article, we have analyzed the current evidence of genetic alterations of the TRAF family in human cancers. The results revealed that genetic alterations of all seven TRAF genes are present in various human cancers and that recurrent mutations of each TRAF gene have been detected in cancer patients. In particular, loss-of-function genetic alterations of TRAF2 and TRAF3 are frequently detected in B cell malignancies, and the rates of missense mutations of TRAF7 are overwhelmingly high in adenomatoid tumors, secretory meningiomas and perineuriomas. Gain-of-function alterations (gene amplification and overexpression) are common for TRAF1, TRAF4, TRAF5, and TRAF6 in human cancers, and are also identified for TRAF2 in epithelial cancers. Corroborating human evidence, direct causal roles of TRAF genetic alterations (except TRAF7) in tumorigenesis have been demonstrated in vivo with genetically engineered mouse models that have each TRAF gene deleted or overexpressed in specific cell types. Importantly, however, the functional significance of most TRAF point mutations identified in human cancers remains to be assessed in future studies. A number of interesting TRAF-dependent oncogenic and tumor suppressive pathways have been elucidated from both in vivo and in vitro studies, although current understanding is still far from complete and further investigation is required. The significance of TRAFs in cancer pathogenesis is reinforced by the evidence that TRAF proteins also participate in pathogen-induced carcinogenesis, including bacteria and viruses. Furthermore, emerging evidence indicates that TRAF proteins can indirectly contribute to tumorigenesis and metastasis by affecting tumor immunity, chronic inflammation, bone resorption, and the tumor microenvironment. In conclusion, the information presented in this article provides a rationale for the development of novel immunotherapies and other strategies to manipulate TRAF proteins or TRAF-dependent signaling pathways in human cancers by precision medicine, which represents the next primary challenge in the field.
Author Contributions
PX and SZ have taken the leading roles in designing and writing this manuscript, and all co-authors (JJ, SG, AL, HS, and JF) have also made significant contributions to writing this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Institutes of Health grants R01 CA158402 and R21 AI128264 (PX), the Department of Defense grant W81XWH-13-1-0242 (PX), a Busch Biomedical Grant (PX), and a Pilot Award from the Cancer Institute of New Jersey through Grant Number P30CA072720 from the National Cancer Institute (PX).
References
1. Xie P. TRAF molecules in cell signaling and in human diseases. J Mol Signal. (2013) 8:7. doi: 10.1186/1750-2187-8-7
2. So T, Nagashima H, Ishii N. TNF Receptor-Associated Factor (TRAF) signaling network in CD4(+) T-lymphocytes. Tohoku J Exp Med. (2015) 236:139–54. doi: 10.1620/tjem.236.139
3. Yang XD, Sun SC. Targeting signaling factors for degradation, an emerging mechanism for TRAF functions. Immunol Rev. (2015) 266:56–71. doi: 10.1111/imr.12311
4. Lalani AI, Zhu S, Gokhale S, Jin J, Xie P. TRAF molecules in inflammation and inflammatory diseases. Curr Pharmacol Rep. (2018) 4:64–90. doi: 10.1007/s40495-017-0117-y
5. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. (2013) 45:1113–20. doi: 10.1038/ng.2764
6. Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr Protoc Hum Genet. (2008) Chapter 10:Unit 10.11. doi: 10.1002/0471142905.hg1011s57
7. Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. (2015) 6:6744. doi: 10.1038/ncomms7744
8. Network TCGAR. Comprehensive molecular characterization of gastric adenocarcinoma. Nature (2014) 513:202–9. doi: 10.1038/nature13480
9. Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. (2010) 42:715–21. doi: 10.1038/ng.619
10. Network TCGAR. Comprehensive genomic characterization of squamous cell lung cancers. Nature (2012) 489:519–25. doi: 10.1038/nature11404
11. Kim TM, Jung SH, Kim MS, Baek IP, Park SW, Lee SH, et al. The mutational burdens and evolutionary ages of early gastric cancers are comparable to those of advanced gastric cancers. J Pathol. (2014) 234:365–74. doi: 10.1002/path.4401
12. Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature (2012) 487:330–7. doi: 10.1038/nature11252
13. Sanz-Pamplona R, Lopez-Doriga A, Pare-Brunet L, Lazaro K, Bellido F, Alonso MH, et al. Exome sequencing reveals AMER1 as a frequently mutated gene in colorectal cancer. Clin Cancer Res. (2015) 21:4709–18. doi: 10.1158/1078-0432.CCR-15-0159
14. Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. (2014) 4:94–109. doi: 10.1158/2159-8290.CD-13-0617
15. Ljungstrom V, Cortese D, Young E, Pandzic T, Mansouri L, Plevova K, et al. Whole-exome sequencing in relapsing chronic lymphocytic leukemia: clinical impact of recurrent RPS15 mutations. Blood (2016) 127:1007–16. doi: 10.1182/blood-2015-10-674572
16. Feldman AL, Vasmatzis G, Asmann YW, Davila J, Middha S, Eckloff BW, et al. Novel TRAF1-ALK fusion identified by deep RNA sequencing of anaplastic large cell lymphoma. Genes Chromosomes Cancer (2013) 52:1097–102. doi: 10.1002/gcc.22104
17. Takeoka K, Okumura A, Honjo G, Ohno H. Variant translocation partners of the anaplastic lymphoma kinase (ALK) gene in two cases of anaplastic large cell lymphoma, identified by inverse cDNA polymerase chain reaction. J Clin Exp Hematop. (2014) 54:225–35. doi: 10.3960/jslrt.54.225
18. Abate F, Todaro M, van der Krogt JA, Boi M, Landra I, Machiorlatti R, et al. A novel patient-derived tumorgraft model with TRAF1-ALK anaplastic large-cell lymphoma translocation. Leukemia (2015) 29:1390–401. doi: 10.1038/leu.2014.347
19. Lawrence K, Berry B, Handshoe J, Hout D, Mazzola R, Morris SW, et al. Detection of a TRAF1-ALK fusion in an anaplastic large cell lymphoma patient with chemotherapy and ALK inhibitor-resistant disease. BMC Res Notes (2015) 8:308. doi: 10.1186/s13104-015-1277-7
20. Yamamoto H, Ryu J, Min E, Oi N, Bai R, Zykova TA, et al. TRAF1 is critical for DMBA/Solar UVR-induced skin carcinogenesis. J Invest Dermatol. (2017) 137:1322–32. doi: 10.1016/j.jid.2016.12.026
21. Wen X WB, Feng T, Yuan W, Zhou J, Fang T. TNF receptor-associated factor 1 as a biomarker for assessment of non-small cell lung cancer metastasis and overall survival. Clin Respir J. (2018) 12:2197–203. doi: 10.1111/crj.12789
22. Durkop H, Hirsch B, Hahn C, Foss HD, Stein H. Differential expression and function of A20 and TRAF1 in Hodgkin lymphoma and anaplastic large cell lymphoma and their induction by CD30 stimulation. J Pathol. (2003) 200:229–39. doi: 10.1002/path.1351
23. Zapata JM, Krajewska M, Krajewski S, Kitada S, Welsh K, Monks A, et al. TNFR-associated factor family protein expression in normal tissues and lymphoid malignancies. J Immunol. (2000) 165:5084-96. doi: 10.4049/jimmunol.165.9.5084
24. Munzert G, Kirchner D, Stobbe H, Bergmann L, Schmid RM, Dohner H, et al. Tumor necrosis factor receptor-associated factor 1 gene overexpression in B-cell chronic lymphocytic leukemia: analysis of NF-kappa B/Rel-regulated inhibitors of apoptosis. Blood (2002) 100:3749–56. doi: 10.1182/blood.V100.10.3749
25. Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De Leval L, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood (2003) 102:3871–9. doi: 10.1182/blood-2003-06-1841
26. Zapata JM, Reed JC. TRAF1: lord without a RING. Sci STKE (2002) 2002:pe27. doi: 10.1126/stke.2002.133.pe27
27. Zhang B, Wang Z, Li T, Tsitsikov EN, Ding HF. NF-kappaB2 mutation targets TRAF1 to induce lymphomagenesis. Blood (2007) 110:743–51. doi: 10.1182/blood-2006-11-058446
28. Slager SL, Kay NE, Fredericksen ZS, Wang AH, Liebow M, Cunningham JM, et al. Susceptibility genes and B-chronic lymphocytic leukaemia. Br J Haematol. (2007) 139:762–71. doi: 10.1111/j.1365-2141.2007.06872.x
29. Tsitsikov EN, Laouini D, Dunn IF, Sannikova TY, Davidson L, Alt FW, et al. TRAF1 is a negative regulator of TNF signaling. enhanced TNF signaling in TRAF1-deficient mice. Immunity (2001) 15:647–57. doi: 10.1016/S1074-7613(01)00207-2
30. Wang Q, Gao G, Zhang T, Yao K, Chen H, Park MH, et al. TRAF1 is critical for regulating the BRAF/MEK/ERK pathway in non-small cell lung carcinogenesis. Cancer Res. (2018) 78:3982–94. doi: 10.1158/0008-5472.CAN-18-0429
31. McPherson AJ, Snell LM, Mak TW, Watts TH. Opposing roles for TRAF1 in the alternative versus classical NF-κB pathway in T cells. J Biol Chem. (2012) 287:23010–9. doi: 10.1074/jbc.M112.350538
32. Sabbagh L, Srokowski CC, Pulle G, Snell LM, Sedgmen BJ, Liu Y, et al. A critical role for TNF receptor-associated factor 1 and Bim down-regulation in CD8 memory T cell survival. Proc Natl Acad Sci USA. (2006) 103:18703–8. doi: 10.1073/pnas.0602919103
33. Sabbagh L, Pulle G, Liu Y, Tsitsikov EN, Watts TH. ERK-dependent Bim modulation downstream of the 4-1BB-TRAF1 signaling axis is a critical mediator of CD8 T cell survival in vivo. J Immunol. (2008) 180:8093–101. doi: 10.4049/jimmunol.180.12.8093
34. Wang C, McPherson AJ, Jones RB, Kawamura KS, Lin GH, Lang PA, et al. Loss of the signaling adaptor TRAF1 causes CD8+ T cell dysregulation during human and murine chronic infection. J Exp Med. (2012) 209:77–91. doi: 10.1084/jem.20110675
35. Speiser DE, Lee SY, Wong B, Arron J, Santana A, Kong YY, et al. A regulatory role for TRAF1 in antigen-induced apoptosis of T cells. J Exp Med. (1997) 185:1777–83.
36. Yeh WC, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, et al. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity (1997) 7:715–25.
37. Piao JH, Hasegawa M, Heissig B, Hattori K, Takeda K, Iwakura Y, et al. Tumor necrosis factor receptor-associated factor (TRAF) 2 controls homeostasis of the colon to prevent spontaneous development of murine inflammatory bowel disease. J Biol Chem. (2011) 286:17879–88. doi: 10.1074/jbc.M111.221853
38. MacLachlan TK, Giordano A. TRAF2 expression in differentiated muscle. J Cell Biochem. (1998) 71:461–6.
39. Gardam S, Sierro F, Basten A, Mackay F, Brink R. TRAF2 and TRAF3 signal adapters act cooperatively to control the maturation and survival signals delivered to B cells by the BAFF receptor. Immunity (2008) 28:391–401. doi: 10.1016/j.immuni.2008.01.009
40. Lee SY, Reichlin A, Santana A, Sokol KA, Nussenzweig MC, Choi Y. TRAF2 is essential for JNK but not NF-kappaB activation and regulates lymphocyte proliferation and survival. Immunity (1997) 7:703–13.
41. Zapata JM, Krajewska M, Morse HC III, Choi Y, Reed JC. TNF receptor-associated factor (TRAF) domain and Bcl-2 cooperate to induce small B cell lymphoma/chronic lymphocytic leukemia in transgenic mice. Proc Natl Acad Sci USA. (2004) 101:16600–5. doi: 10.1073/pnas.0407541101
42. Perez-Chacon G, Llobet D, Pardo C, Pindado J, Choi Y, Reed JC, et al. TNFR-associated factor 2 deficiency in B lymphocytes predisposes to chronic lymphocytic leukemia/small lymphocytic lymphoma in mice. J Immunol. (2012) 189:1053–61. doi: 10.4049/jimmunol.1200814
43. Schneider AT, Gautheron J, Feoktistova M, Roderburg C, Loosen SH, Roy S, et al. RIPK1 suppresses a TRAF2-dependent pathway to liver cancer. Cancer Cell (2017) 31:94–109. doi: 10.1016/j.ccell.2016.11.009
44. Petersen SL, Chen TT, Lawrence DA, Marsters SA, Gonzalvez F, Ashkenazi A. TRAF2 is a biologically important necroptosis suppressor. Cell Death Differ. (2015) 22:1846–57. doi: 10.1038/cdd.2015.35
45. Etemadi N, Chopin M, Anderton H, Tanzer MC, Rickard JA, Abeysekera W, et al. TRAF2 regulates TNF and NF-κB signalling to suppress apoptosis and skin inflammation independently of Sphingosine kinase 1. Elife (2015) 4:e10592. doi: 10.7554/eLife.10592
46. Jin J, Xiao Y, Hu H, Zou Q, Li Y, Gao Y, et al. Proinflammatory TLR signalling is regulated by a TRAF2-dependent proteolysis mechanism in macrophages. Nat Commun. (2015) 6:5930. doi: 10.1038/ncomms6930
47. Villanueva JE, Malle EK, Gardam S, Silveira PA, Zammit NW, Walters SN, et al. TRAF2 regulates peripheral CD8(+) T-cell and NKT-cell homeostasis by modulating sensitivity to IL-15. Eur J Immunol. (2015) 45:1820–31. doi: 10.1002/eji.201445416
48. Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. (2008) 9:1371–8. doi: 10.1038/ni.1676
49. Xu Y, Cheng G, Baltimore D. Targeted disruption of TRAF3 leads to postnatal lethality and defective T-dependent immune responses. Immunity (1996) 5:407–15.
50. He JQ, Zarnegar B, Oganesyan G, Saha SK, Yamazaki S, Doyle SE, et al. Rescue of TRAF3-null mice by p100 NF-kappa B deficiency. J Exp Med. (2006) 203:2413–8. doi: 10.1084/jem.20061166
51. Xie P, Stunz LL, Larison KD, Yang B, Bishop GA. Tumor necrosis factor receptor-associated factor 3 is a critical regulator of B cell homeostasis in secondary lymphoid organs. Immunity (2007) 27:253–67. doi: 10.1016/j.immuni.2007.07.012
52. Moore CR, Liu Y, Shao CS, Covey LR, Morse HC 3rd, Xie P. Specific deletion of TRAF3 in B lymphocytes leads to B lymphoma development in mice. Leukemia (2012) 26:1122–7. doi: 10.1038/leu.2011.309
53. Xie P, Poovassery J, Stunz LL, Smith SM, Schultz ML, Carlin LE, et al. Enhanced Toll-like receptor (TLR) responses of TNFR-associated factor 3 (TRAF3)-deficient B lymphocytes. J Leukoc Biol. (2011) 90:1149–57. doi: 10.1189/jlb.0111044
54. Gardam S, Turner VM, Anderton H, Limaye S, Basten A, Koentgen F, et al. Deletion of cIAP1 and cIAP2 in murine B lymphocytes constitutively activates cell survival pathways and inactivates the germinal center response. Blood (2011) 117:4041–51. doi: 10.1182/blood-2010-10-312793
55. Zapata JM, Llobet D, Krajewska M, Lefebvre S, Kress CL, Reed JC. Lymphocyte-specific TRAF3-transgenic mice have enhanced humoral responses and develop plasmacytosis, autoimmunity, inflammation, and cancer. Blood (2009) 113:4595–603. doi: 10.1182/blood-2008-07-165456
56. Lalani AI, Moore CR, Luo C, Kreider BZ, Liu Y, Morse HC 3rd, et al. Myeloid cell TRAF3 regulates immune responses and inhibits inflammation and tumor development in mice. J Immunol. (2015) 194:334–48. doi: 10.4049/jimmunol.1401548
57. Xie P, Kraus ZJ, Stunz LL, Liu Y, Bishop GA. TNF receptor-associated factor 3 is required for T cell-mediated immunity and TCR/CD28 signaling. J Immunol. (2011) 186:143–55. doi: 10.4049/jimmunol.100029
58. Yi Z, Lin WW, Stunz LL, Bishop GA. The adaptor TRAF3 restrains the lineage determination of thymic regulatory T cells by modulating signaling via the receptor for IL-2. Nat Immunol. (2014) 15:866–74. doi: 10.1038/ni.2944
59. Yi Z, Stunz LL, Bishop GA. TNF receptor associated factor 3 plays a key role in development and function of invariant natural killer T cells. J Exp Med. (2013) 210:1079–86. doi: 10.1084/jem.20122135
60. Yi Z, Stunz LL, Lin WW, Bishop GA. TRAF3 regulates homeostasis of CD8+ central memory T cells. PLoS ONE (2014) 9:e102120. doi: 10.1371/journal.pone.0102120
61. Shiels H, Li X, Schumacker PT, Maltepe E, Padrid PA, Sperling A, et al. TRAF4 deficiency leads to tracheal malformation with resulting alterations in air flow to the lungs. Am J Pathol. (2000) 157:679–88. doi: 10.1016/S0002-9440(10)64578-6
62. Regnier CH, Masson R, Kedinger V, Textoris J, Stoll I, Chenard MP, et al. Impaired neural tube closure, axial skeleton malformations, and tracheal ring disruption in TRAF4-deficient mice. Proc Natl Acad Sci USA. (2002) 99:5585–90. doi: 10.1073/pnas.052124799
63. Blaise S, Kneib M, Rousseau A, Gambino F, Chenard MP, Messadeq N, et al. in vivo evidence that TRAF4 is required for central nervous system myelin homeostasis. PLoS ONE (2012) 7:e30917. doi: 10.1371/journal.pone.0030917
64. Cherfils-Vicini J, Vingert B, Varin A, Tartour E, Fridman WH, Sautes-Fridman C, et al. Characterization of immune functions in TRAF4-deficient mice. Immunology (2008) 124:562–74. doi: 10.1111/j.1365-2567.2008.02810.x
65. Wu L, Chen X, Zhao J, Martin B, Zepp JA, Ko JS, et al. A novel IL-17 signaling pathway controlling keratinocyte proliferation and tumorigenesis via the TRAF4-ERK5 axis. J Exp Med. (2015) 212:1571–87. doi: 10.1084/jem.20150204
66. Zepp JA, Wu L, Qian W, Ouyang W, Aronica M, Erzurum S, et al. TRAF4-SMURF2-mediated DAZAP2 degradation is critical for IL-25 signaling and allergic airway inflammation. J Immunol. (2015) 194:2826–37. doi: 10.4049/jimmunol.1402647
67. Nakano H, Sakon S, Koseki H, Takemori T, Tada K, Matsumoto M, et al. Targeted disruption of Traf5 gene causes defects in CD40- and CD27-mediated lymphocyte activation. Proc Natl Acad Sci USA. (1999) 96:9803–8.
68. Kraus ZJ, Haring JS, Bishop GA. TNF receptor-associated factor 5 is required for optimal T cell expansion and survival in response to infection. J Immunol. (2008) 181:7800–9. doi: 10.4049/jimmunol.181.11.7800
69. Esparza EM, Lindsten T, Stockhausen JM, Arch RH. Tumor necrosis factor receptor (TNFR)-associated factor 5 is a critical intermediate of costimulatory signaling pathways triggered by glucocorticoid-induced TNFR in T cells. J Biol Chem. (2006) 281:8559–64. doi: 10.1074/jbc.M512915200
70. So T, Salek-Ardakani S, Nakano H, Ware CF, Croft M. TNF receptor-associated factor 5 limits the induction of Th2 immune responses. J Immunol. (2004) 172:4292–7. doi: 10.4049/jimmunol.172.7.4292
71. Nagashima H, Okuyama Y, Asao A, Kawabe T, Yamaki S, Nakano H, et al. The adaptor TRAF5 limits the differentiation of inflammatory CD4(+) T cells by antagonizing signaling via the receptor for IL-6. Nat Immunol. (2014) 15:449–56. doi: 10.1038/ni.2863
72. Shang J, Li L, Wang X, Pan H, Liu S, He R, et al. Disruption of tumor necrosis factor receptor-associated factor 5 exacerbates murine experimental colitis via regulating T helper cell-mediated inflammation. Mediators Inflamm. (2016) 2016:9453745. doi: 10.1155/2016/9453745
73. Kraus ZJ, Nakano H, Bishop GA. TRAF5 is a critical mediator of in vitro signals and in vivo functions of LMP1, the viral oncogenic mimic of CD40. Proc Natl Acad Sci USA. (2009) 106:17140–5. doi: 10.1073/pnas.0903786106
74. Naito A, Azuma S, Tanaka S, Miyazaki T, Takaki S, Takatsu K, et al. Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells (1999) 4:353–62.
75. Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, et al. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. (1999) 13:1015–24.
76. Kobayashi T, Walsh PT, Walsh MC, Speirs KM, Chiffoleau E, King CG, et al. TRAF6 is a critical factor for dendritic cell maturation and development. Immunity (2003) 19:353–63. doi: 10.1016/S1074-7613(03)00230-9
77. Shimo Y, Yanai H, Ohshima D, Qin J, Motegi H, Maruyama Y, et al. TRAF6 directs commitment to regulatory T cells in thymocytes. Genes Cells (2011) 16:437–47. doi: 10.1111/j.1365-2443.2011.01500.x
78. Dickson KM, Bhakar AL, Barker PA. TRAF6-dependent NF-kB transcriptional activity during mouse development. Dev Dyn. (2004) 231:122–7. doi: 10.1002/dvdy.20110
79. Yeiser EC, Rutkoski NJ, Naito A, Inoue J, Carter BD. Neurotrophin signaling through the p75 receptor is deficient in traf6−/− mice. J Neurosci (2004) 24:10521–9. doi: 10.1523/JNEUROSCI.1390-04.2004
80. Lomaga MA, Henderson JT, Elia AJ, Robertson J, Noyce RS, Yeh WC, et al. Tumor necrosis factor receptor-associated factor 6 (TRAF6) deficiency results in exencephaly and is required for apoptosis within the developing CNS. J Neurosci. (2000) 20:7384–93. doi: 10.1523/JNEUROSCI.20-19-07384.2000
81. Fang J, Muto T, Kleppe M, Bolanos LC, Hueneman KM, Walker CS, et al. TRAF6 mediates basal activation of NF-κB necessary for hematopoietic stem cell homeostasis. Cell Rep. (2018) 22:1250–62. doi: 10.1016/j.celrep.2018.01.013
82. Kobayashi T, Kim TS, Jacob A, Walsh MC, Kadono Y, Fuentes-Panana E, et al. TRAF6 is required for generation of the B-1a B cell compartment as well as T cell-dependent and -independent humoral immune responses. PLoS ONE (2009) 4:e4736.
83. King CG, Kobayashi T, Cejas PJ, Kim T, Yoon K, Kim GK, et al. TRAF6 is a T cell-intrinsic negative regulator required for the maintenance of immune homeostasis. Nat Med. (2006) 12:1088–92. doi: 10.1038/nm1449
84. Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature (2009) 460:103–7. doi: 10.1038/nature08097
85. Cejas PJ, Walsh MC, Pearce EL, Han D, Harms GM, Artis D, et al. TRAF6 inhibits Th17 differentiation and TGF-beta-mediated suppression of IL-2. Blood (2010) 115:4750–7. doi: 10.1182/blood-2009-09-242768
86. Xiao X, Balasubramanian S, Liu W, Chu X, Wang H, Taparowsky EJ, et al. OX40 signaling favors the induction of T(H)9 cells and airway inflammation. Nat Immunol. (2012) 13:981–90. doi: 10.1038/ni.2390
87. Vlantis K, Polykratis A, Welz PS, van Loo G, Pasparakis M, Wullaert A. TLR-independent anti-inflammatory function of intestinal epithelial TRAF6 signalling prevents DSS-induced colitis in mice. Gut (2016) 65:935–43. doi: 10.1136/gutjnl-2014-308323
88. Paul PK, Gupta SK, Bhatnagar S, Panguluri SK, Darnay BG, Choi Y, et al. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J Cell Biol. (2010) 191:1395–411. doi: 10.1083/jcb.201006098
89. Hindi SM, Paul PK, Dahiya S, Mishra V, Bhatnagar S, Kuang S, et al. Reciprocal interaction between TRAF6 and notch signaling regulates adult myofiber regeneration upon injury. Mol Cell Biol. (2012) 32:4833–45. doi: 10.1128/MCB.00717-12
90. Paul PK, Bhatnagar S, Mishra V, Srivastava S, Darnay BG, Choi Y, et al. The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Mol Cell Biol. (2012) 32:1248–59. doi: 10.1128/MCB.06351-11
91. Guo F, Sun A, Wang W, He J, Hou J, Zhou P, et al. TRAF1 is involved in the classical NF-kappaB activation and CD30-induced alternative activity in Hodgkin's lymphoma cells. Mol Immunol. (2009) 46:2441–8. doi: 10.1016/j.molimm.2009.05.178
92. Wicovsky A, Henkler F, Salzmann S, Scheurich P, Kneitz C, Wajant H. Tumor necrosis factor receptor-associated factor-1 enhances proinflammatory TNF receptor-2 signaling and modifies TNFR1-TNFR2 cooperation. Oncogene (2009) 28:1769–81. doi: 10.1038/onc.2009.29
93. Deng J, Miller SA, Wang HY, Xia W, Wen Y, Zhou BP, et al. β-catenin interacts with and inhibits NF-κ B in human colon and breast cancer. Cancer Cell (2002) 2:323-34. doi: 10.1016/S1535-6108(02)00154-X
94. Du Q, Zhang X, Cardinal J, Cao Z, Guo Z, Shao L, et al. Wnt/beta-catenin signaling regulates cytokine-induced human inducible nitric oxide synthase expression by inhibiting nuclear factor-kappaB activation in cancer cells. Cancer Res. (2009) 69:3764–71. doi: 10.1158/0008-5472.CAN-09-0014
95. Carmona Arana JA, Seher A, Neumann M, Lang I, Siegmund D, Wajant H. TNF receptor-associated factor 1 is a major target of soluble TWEAK. Front Immunol. (2014) 5:63. doi: 10.3389/fimmu.2014.00063
96. Lassen UN, Meulendijks D, Siu LL, Karanikas V, Mau-Sorensen M, Schellens JH, et al. A phase I monotherapy study of RG7212, a first-in-class monoclonal antibody targeting TWEAK signaling in patients with advanced cancers. Clin Cancer Res. (2015) 21:258–66. doi: 10.1158/1078-0432.CCR-14-1334
97. Meulendijks D, Lassen UN, Siu LL, Huitema AD, Karanikas V, Mau-Sorensen M, et al. Exposure and tumor Fn14 expression as determinants of pharmacodynamics of the anti-TWEAK monoclonal antibody RG7212 in patients with Fn14-positive solid tumors. Clin Cancer Res. (2016) 22:858–67. doi: 10.1158/1078-0432.CCR-15-1506
98. Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet. (2018) 50:645–51. doi: 10.1038/s41588-018-0078-z
99. Rahal R, Frick M, Romero R, Korn JM, Kridel R, Chan FC, et al. Pharmacological and genomic profiling identifies NF-κB-targeted treatment strategies for mantle cell lymphoma. Nat Med. (2014) 20:87–92. doi: 10.1038/nm.3435
100. Meissner B, Kridel R, Lim RS, Rogic S, Tse K, Scott DW, et al. The E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell lymphoma. Blood (2013) 121:3161–4. doi: 10.1182/blood-2013-01-478834
101. Saba N, Wiestner A. Do mantle cell lymphomas have an 'Achilles heel'? Curr Opin Hematol. (2014) 21:350–7. doi: 10.1097/MOH.0000000000000057
102. Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature (2009) 459:717–21. doi: 10.1038/nature07968
103. Shen RR, Zhou AY, Kim E, O'Connell JT, Hagerstrand D, Beroukhim R, et al. TRAF2 is an NF-κB-activating oncogene in epithelial cancers. Oncogene (2015) 34:209–16. doi: 10.1038/onc.2013.543
104. Giannakis M, Hodis E, Jasmine Mu X, Yamauchi M, Rosenbluh J, Cibulskis K, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. (2014) 46:1264–6. doi: 10.1038/ng.3127
105. Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell (2018) 33:125–36.e3. doi: 10.1016/j.ccell.2017.12.004
106. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. (2017) 23:703–13. doi: 10.1038/nm.4333
107. Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. (2014) 46:573–82. doi: 10.1038/ng.2983
108. Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. (2016) 15:857–65. doi: 10.1016/j.celrep.2016.03.075
109. Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell (2016) 164:550–63. doi: 10.1016/j.cell.2015.12.028
110. Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. (2016) 22:298–305. doi: 10.1038/nm.4045
111. Jones S, Stransky N, McCord CL, Cerami E, Lagowski J, Kelly D, et al. Genomic analyses of gynaecologic carcinosarcomas reveal frequent mutations in chromatin remodelling genes. Nat Commun. (2014) 5:5006. doi: 10.1038/ncomms6006
112. Chen K, Yang D, Li X, Sun B, Song F, Cao W, et al. Mutational landscape of gastric adenocarcinoma in Chinese: implications for prognosis and therapy. Proc Natl Acad Sci USA. (2015) 112:1107–12. doi: 10.1073/pnas.1422640112
113. Network TCGA. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature (2015) 517:576–82. doi: 10.1038/nature14129
114. Galante PA, Parmigiani RB, Zhao Q, Caballero OL, de Souza JE, Navarro FC, et al. Distinct patterns of somatic alterations in a lymphoblastoid and a tumor genome derived from the same individual. Nucleic Acids Res. (2011) 39:6056–68. doi: 10.1093/nar/gkr221
115. Lezcano C, Shoushtari AN, Ariyan C, Hollmann TJ, Busam KJ. Primary and metastatic melanoma with NTRK fusions. Am J Surg Pathol. (2018) 42:1052–8. doi: 10.1097/PAS.0000000000001070
116. Garrison JB, Samuel T, Reed JC. TRAF2-binding BIR1 domain of c-IAP2/MALT1 fusion protein is essential for activation of NF-kappaB. Oncogene (2009) 28:1584–93. doi: 10.1038/onc.2009.17
117. Grech AP, Amesbury M, Chan T, Gardam S, Basten A, Brink R. TRAF2 differentially regulates the canonical and noncanonical pathways of NF-kappaB activation in mature B cells. Immunity (2004) 21:629–42. doi: 10.1016/j.immuni.2004.09.011
118. Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H, et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol. (2008) 9:1364–70. doi: 10.1038/ni.1678
119. Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM, Sung CO, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology (2014) 60:1972–82. doi: 10.1002/hep.27198
120. Jackson-Bernitsas DG, Ichikawa H, Takada Y, Myers JN, Lin XL, Darnay BG, et al. Evidence that TNF-TNFR1-TRADD-TRAF2-RIP-TAK1-IKK pathway mediates constitutive NF-kappaB activation and proliferation in human head and neck squamous cell carcinoma. Oncogene (2007) 26:1385–97. doi: 10.1038/sj.onc.1209945
121. Tobin D, van Hogerlinden M, Toftgard R. UVB-induced association of tumor necrosis factor (TNF) receptor 1/TNF receptor-associated factor-2 mediates activation of Rel proteins. Proc Natl Acad Sci USA. (1998) 95:565–9.
122. Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell (2006) 125:665–77. doi: 10.1016/j.cell.2006.03.041
123. Chen MB, Zhang Y, Wei MX, Shen W, Wu XY, Yao C, et al. Activation of AMP-activated protein kinase (AMPK) mediates plumbagin-induced apoptosis and growth inhibition in cultured human colon cancer cells. Cell Signal. (2013) 25:1993–2002. doi: 10.1016/j.cellsig.2013.05.026
124. Mhaidat NM, Thorne R, Zhang XD, Hersey P. Involvement of endoplasmic reticulum stress in Docetaxel-induced JNK-dependent apoptosis of human melanoma. Apoptosis (2008) 13:1505–12. doi: 10.1007/s10495-008-0276-8
125. Zhang J, Liang Y, Lin Y, Liu Y, YouYou, Yin W. IRE1α-TRAF2-ASK1 pathway is involved in CSTMP-induced apoptosis and ER stress in human non-small cell lung cancer A549 cells. Biomed Pharmacother. (2016) 82:281–9. doi: 10.1016/j.biopha.2016.04.050
126. Chen H, Yang H, Pan L, Wang W, Liu X, Ren X, et al. The molecular mechanisms of XBP-1 gene silencing on IRE1α-TRAF2-ASK1-JNK pathways in oral squamous cell carcinoma under endoplasmic reticulum stress. Biomed Pharmacother. (2016) 77:108–13. doi: 10.1016/j.biopha.2015.12.010
127. Lamkanfi M, D'Hondt K, Vande Walle L, van Gurp M, Denecker G, Demeulemeester J, et al. A novel caspase-2 complex containing TRAF2 and RIP1. J Biol Chem. (2005) 280:6923–32. doi: 10.1074/jbc.M411180200
128. Robeson AC, Lindblom KR, Wojton J, Kornbluth S, Matsuura K. Dimer-specific immunoprecipitation of active caspase-2 identifies TRAF proteins as novel activators. EMBO J. (2018) 37:e97072. doi: 10.15252/embj.201797072
129. Doppler H, Liou GY, Storz P. Downregulation of TRAF2 mediates NIK-induced pancreatic cancer cell proliferation and tumorigenicity. PLoS ONE (2013) 8:e53676. doi: 10.1371/journal.pone.0053676
130. Ohtsu N, Nakatani Y, Yamashita D, Ohue S, Ohnishi T, Kondo T. Eva1 maintains the stem-like character of glioblastoma-initiating cells by activating the noncanonical NF-κB signaling pathway. Cancer Res. (2016) 76:171–81. doi: 10.1158/0008-5472.CAN-15-0884
131. Wang B, Jie Z, Joo D, Ordureau A, Liu P, Gan W, et al. TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature (2017) 545:365–9. doi: 10.1038/nature22344
132. Trauzold A, Roder C, Sipos B, Karsten K, Arlt A, Jiang P, et al. CD95 and TRAF2 promote invasiveness of pancreatic cancer cells. FASEB J. (2005) 19:620–2. doi: 10.1096/fj.04-2984fje
133. Wei B, Ruan J, Mi Y, Hu J, Zhang J, Wang Z, et al. Knockdown of TNF receptor-associated factor 2 (TRAF2) modulates in vitro growth of TRAIL-treated prostate cancer cells. Biomed Pharmacother. (2017) 93:462–9. doi: 10.1016/j.biopha.2017.05.145
134. Zheng M, Morgan-Lappe SE, Yang J, Bockbrader KM, Pamarthy D, Thomas D, et al. Growth inhibition and radiosensitization of glioblastoma and lung cancer cells by small interfering RNA silencing of tumor necrosis factor receptor-associated factor 2. Cancer Res. (2008) 68:7570–8. doi: 10.1158/0008-5472.CAN-08-0632
135. Zhao J, Li H, Min L, Han X, Shu P, Yang Y, et al. High expression of tumor necrosis factor receptor-associated factor 2 promotes tumor metastasis and is associated with unfavorable prognosis in gastric cancer. J Gastroenterol Hepatol. (2018) 33:431–42. doi: 10.1111/jgh.13818
136. Peng C, Zhu F, Wen W, Yao K, Li S, Zykova T, et al. Tumor necrosis factor receptor-associated factor family protein 2 is a key mediator of the epidermal growth factor-induced ribosomal S6 kinase 2/cAMP-responsive element-binding protein/Fos protein signaling pathway. J Biol Chem. (2012) 287:25881–92. doi: 10.1074/jbc.M112.359521
137. Zhang W, Sun Y, Liu L, Li Z. Prognostic significance of TNFR-associated factor 1 and 2 (TRAF1 and TRAF2) in glioblastoma. Med Sci Monit. (2017) 23:4506–12. doi: 10.12659/MSM.903397
138. Peramuhendige P, Marino S, Bishop RT, de Ridder D, Khogeer A, Baldini I, et al. TRAF2 in osteotropic breast cancer cells enhances skeletal tumour growth and promotes osteolysis. Sci Rep. (2018) 8:39. doi: 10.1038/s41598-017-18327-5
139. Yu C, Chen S, Guo Y, Sun C. Oncogenic TRIM31 confers gemcitabine resistance in pancreatic cancer via activating the NF-κB signaling pathway. Theranostics (2018) 8:3224–36. doi: 10.7150/thno.23259
140. Chiche J, Pommier S, Beneteau M, Mondragon L, Meynet O, Zunino B, et al. GAPDH enhances the aggressiveness and the vascularization of non-Hodgkin's B lymphomas via NF-κB-dependent induction of HIF-1α. Leukemia (2015) 29:1163–76. doi: 10.1038/leu.2014.324
141. Blakely CM, Pazarentzos E, Olivas V, Asthana S, Yan JJ, Tan I, et al. NF-κB-activating complex engaged in response to EGFR oncogene inhibition drives tumor cell survival and residual disease in lung cancer. Cell Rep. (2015) 11:98–110. doi: 10.1016/j.celrep.2015.03.012
142. Zhou AY, Shen RR, Kim E, Lock YJ, Xu M, Chen ZJ, et al. IKKε-mediated tumorigenesis requires K63-linked polyubiquitination by a cIAP1/cIAP2/TRAF2 E3 ubiquitin ligase complex. Cell Rep. (2013) 3:724–33. doi: 10.1016/j.celrep.2013.01.031
143. Shen RR, Zhou AY, Kim E, Lim E, Habelhah H, Hahn WC. IκB kinase ε phosphorylates TRAF2 to promote mammary epithelial cell transformation. Mol Cell Biol. (2012) 32:4756–68. doi: 10.1128/MCB.00468-12
144. Martinez-Forero I, Rouzaut A, Palazon A, Dubrot J, Melero I. Lysine 63 polyubiquitination in immunotherapy and in cancer-promoting inflammation. Clin Cancer Res. (2009) 15:6751–7. doi: 10.1158/1078-0432.CCR-09-1225
145. Gonzalvez F, Lawrence D, Yang B, Yee S, Pitti R, Marsters S, et al. TRAF2 Sets a threshold for extrinsic apoptosis by tagging caspase-8 with a ubiquitin shutoff timer. Mol Cell (2012) 48:888–99. doi: 10.1016/j.molcel.2012.09.031
146. Oh YT, Yue P, Wang D, Tong JS, Chen ZG, Khuri FR, et al. Suppression of death receptor 5 enhances cancer cell invasion and metastasis through activation of caspase-8/TRAF2-mediated signaling. Oncotarget (2015) 6:41324–38. doi: 10.18632/oncotarget.5847
147. He W, Wang Q, Xu J, Xu X, Padilla MT, Ren G, et al. Attenuation of TNFSF10/TRAIL-induced apoptosis by an autophagic survival pathway involving TRAF2- and RIPK1/RIP1-mediated MAPK8/JNK activation. Autophagy (2012) 8:1811–21. doi: 10.4161/auto.22145
148. Xu L, Zhang Y, Qu X, Che X, Guo T, Li C, et al. DR5-Cbl-b/c-Cbl-TRAF2 complex inhibits TRAIL-induced apoptosis by promoting TRAF2-mediated polyubiquitination of caspase-8 in gastric cancer cells. Mol Oncol. (2017) 11:1733–51. doi: 10.1002/1878-0261.12140
149. Hibino T, Sakaguchi M, Miyamoto S, Yamamoto M, Motoyama A, Hosoi J, et al. S100A9 is a novel ligand of EMMPRIN that promotes melanoma metastasis. Cancer Res. (2013) 73:172–83. doi: 10.1158/0008-5472.CAN-11-3843
150. Basudhar D, Glynn SA, Greer M, Somasundaram V, No JH, Scheiblin DA, et al. Coexpression of NOS2 and COX2 accelerates tumor growth and reduces survival in estrogen receptor-negative breast cancer. Proc Natl Acad Sci USA. (2017) 114:13030–5. doi: 10.1073/pnas.1709119114
151. Fortin Ensign SP, Mathews IT, Eschbacher JM, Loftus JC, Symons MH, Tran NL. The Src homology 3 domain-containing guanine nucleotide exchange factor is overexpressed in high-grade gliomas and promotes tumor necrosis factor-like weak inducer of apoptosis-fibroblast growth factor-inducible 14-induced cell migration and invasion via tumor necrosis factor receptor-associated factor 2. J Biol Chem. (2013) 288:21887–97. doi: 10.1074/jbc.M113.468686
152. Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell (2017) 171:540–56.e25. doi: 10.1016/j.cell.2017.09.007
153. Moore CR, Edwards SK, Xie P. Targeting TRAF3 downstream signaling pathways in b cell neoplasms. J Cancer Sci Ther. (2015) 7:67–74. doi: 10.4172/1948-5956.1000327
154. Hyeon J, Lee B, Shin SH, Yoo HY, Kim SJ, Kim WS, et al. Targeted deep sequencing of gastric marginal zone lymphoma identified alterations of TRAF3 and TNFAIP3 that were mutually exclusive for MALT1 rearrangement. Mod Pathol. (2018) 31:1418–28. doi: 10.1038/s41379-018-0064-0
155. Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell (2007) 12:131–44. doi: 10.1016/j.ccr.2007.07.003
156. Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. (2015) 33:3911–20. doi: 10.1200/JCO.2014.59.1503
157. Otto C, Giefing M, Massow A, Vater I, Gesk S, Schlesner M, et al. Genetic lesions of the TRAF3 and MAP3K14 genes in classical Hodgkin lymphoma. Br J Haematol. (2012) 157:702–8. doi: 10.1111/j.1365-2141.2012.09113.x
158. Zhang B, Calado DP, Wang Z, Frohler S, Kochert K, Qian Y, et al. An oncogenic role for alternative NF-κB signaling in DLBCL revealed upon deregulated BCL6 expression. Cell Rep. (2015) 11:715–26. doi: 10.1016/j.celrep.2015.03.059
159. Rossi D, Deaglio S, Dominguez-Sola D, Rasi S, Vaisitti T, Agostinelli C, et al. Alteration of BIRC3 and multiple other NF-κB pathway genes in splenic marginal zone lymphoma. Blood (2011) 118:4930–4. doi: 10.1182/blood-2011-06-359166
160. Braggio E, Keats JJ, Leleu X, Van Wier S, Jimenez-Zepeda VH, Valdez R, et al. Identification of copy number abnormalities and inactivating mutations in two negative regulators of nuclear factor-kappaB signaling pathways in Waldenstrom's macroglobulinemia. Cancer Res. (2009) 69:3579–88. doi: 10.1158/0008-5472.CAN-08-3701
161. Li YY, Chung GT, Lui VW, To KF, Ma BB, Chow C, et al. Exome and genome sequencing of nasopharynx cancer identifies NF-κB pathway activating mutations. Nat Commun. (2017) 8:14121. doi: 10.1038/ncomms14121
162. Martin D, Abba MC, Molinolo AA, Vitale-Cross L, Wang Z, Zaida M, et al. The head and neck cancer cell oncogenome: a platform for the development of precision molecular therapies. Oncotarget (2014) 5:8906–23. doi: 10.18632/oncotarget.2417
163. Rossi D, Trifonov V, Fangazio M, Bruscaggin A, Rasi S, Spina V, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. (2012) 209:1537–51. doi: 10.1084/jem.20120904
164. Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. (2015) 21:632–41. doi: 10.1158/1078-0432.CCR-13-3310
165. Clipson A, Wang M, de Leval L, Ashton-Key M, Wotherspoon A, Vassiliou G, et al. KLF2 mutation is the most frequent somatic change in splenic marginal zone lymphoma and identifies a subset with distinct genotype. Leukemia (2014) 29:1177–85. doi: 10.1038/leu.2014.330
166. Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell (2007) 12:115–30. doi: 10.1016/j.ccr.2007.07.004
167. Puente XS, Bea S, Valdes-Mas R, Villamor N, Gutierrez-Abril J, Martin-Subero JI, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature (2015) 526:519–24. doi: 10.1038/nature14666
168. Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. (2011) 43:830–7. doi: 10.1038/ng.892
169. South AP, Purdie KJ, Watt SA, Haldenby S, den Breems N, Dimon M, et al. NOTCH1 mutations occur early during cutaneous squamous cell carcinogenesis. J Invest Dermatol. (2014) 134:2630–8. doi: 10.1038/jid.2014.154
170. Demchenko YN, Glebov OK, Zingone A, Keats JJ, Bergsagel PL, Kuehl WM. Classical and/or alternative NF-kappaB pathway activation in multiple myeloma. Blood (2010) 115:3541–52. doi: 10.1182/blood-2009-09-243535
171. Lin WW, Yi Z, Stunz LL, Maine CJ, Sherman LA, Bishop GA. The adaptor protein TRAF3 inhibits interleukin-6 receptor signaling in B cells to limit plasma cell development. Sci Signal. (2015) 8:ra88. doi: 10.1126/scisignal.aaa5157
172. Xie P, Hostager BS, Bishop GA. Requirement for TRAF3 in signaling by LMP1 But not CD40 in B lymphocytes. J Exp Med. (2004) 199:661–71. doi: 10.1084/jem.20031255
173. Bangalore-Prakash P, Stunz LL, Mambetsariev N, Whillock AL, Hostager BS, Bishop GA. The oncogenic membrane protein LMP1 sequesters TRAF3 in B-cell lymphoma cells to produce functional TRAF3 deficiency. Blood Adv. (2017) 1:2712–23. doi: 10.1182/bloodadvances.2017009670
174. Lalani AI, Luo C, Han Y, Xie P. TRAF3: a novel tumor suppressor gene in macrophages. Macrophage (2015) 2:e1009. doi: 10.14800/macrophage.1009
175. Mashima R, Honda K, Yang Y, Morita Y, Inoue A, Arimura S, et al. Mice lacking Dok-1, Dok-2, and Dok-3 succumb to aggressive histiocytic sarcoma. Lab Invest. (2010) 90:1357–64. doi: 10.1038/labinvest.2010.121
176. Snyder JM, Treuting PM, Nagy L, Yam C, Yi J, Brasfield A, et al. Humanized TLR7/8 expression drives proliferative multisystemic histiocytosis in C57BL/6 mice. PLoS ONE (2014) 9:e107257. doi: 10.1371/journal.pone.0107257
177. Kim SS, Lee KG, Chin CS, Ng SK, Pereira NA, Xu S, et al. DOK3 is required for IFN-β production by enabling TRAF3/TBK1 complex formation and IRF3 activation. J Immunol. (2014) 193:840–8. doi: 10.4049/jimmunol.1301601
178. Carrasco DR, Fenton T, Sukhdeo K, Protopopova M, Enos M, You MJ, et al. The PTEN and INK4A/ARF tumor suppressors maintain myelolymphoid homeostasis and cooperate to constrain histiocytic sarcoma development in humans. Cancer Cell (2006) 9:379–90. doi: 10.1016/j.ccr.2006.03.028
179. Durham BH, Diamond EL, Abdel-Wahab O. Histiocytic neoplasms in the era of personalized genomic medicine. Curr Opin Hematol. (2016) 23:416–25. doi: 10.1097/MOH.0000000000000256
180. Allen IC, Wilson JE, Schneider M, Lich JD, Roberts RA, Arthur JC, et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-κB signaling. Immunity (2012) 36:742–54. doi: 10.1016/j.immuni.2012.03.012
181. Malcomson FC, Willis ND, McCallum I, Xie L, Lagerwaard B, Kelly S, et al. Non-digestible carbohydrates supplementation increases miR-32 expression in the healthy human colorectal epithelium: a randomized controlled trial. Mol Carcinog. (2017) 56:2104–11. doi: 10.1002/mc.22666
182. Muro I, Fang G, Gardella KA, Mahajan IM, Wright CW. The TRAF3 adaptor protein drives proliferation of anaplastic large cell lymphoma cells by regulating multiple signaling pathways. Cell Cycle (2014) 13:1918–27. doi: 10.4161/cc.28895
183. Newman AC, Kemp AJ, Drabsch Y, Behrends C, Wilkinson S. Autophagy acts through TRAF3 and RELB to regulate gene expression via antagonism of SMAD proteins. Nat Commun. (2017) 8:1537. doi: 10.1038/s41467-017-00859-z
184. Chen MC, Hwang MJ, Chou YC, Chen WH, Cheng G, Nakano H, et al. The role of apoptosis signal-regulating kinase 1 in lymphotoxin-beta receptor-mediated cell death. J Biol Chem. (2003) 278:16073–81. doi: 10.1074/jbc.M208661200
185. Georgopoulos NT, Steele LP, Thomson MJ, Selby PJ, Southgate J, Trejdosiewicz LK. A novel mechanism of CD40-induced apoptosis of carcinoma cells involving TRAF3 and JNK/AP-1 activation. Cell Death Differ. (2006) 13:1789–801. doi: 10.1038/sj.cdd.4401859
186. Elmetwali T, Young LS, Palmer DH. CD40 ligand-induced carcinoma cell death: a balance between activation of TNFR-associated factor (TRAF) 3-dependent death signals and suppression of TRAF6-dependent survival signals. J Immunol. (2010) 184:1111–20. doi: 10.4049/jimmunol.0900528
187. Dunnill CJ, Ibraheem K, Mohamed A, Southgate J, Georgopoulos NT. A redox state-dictated signalling pathway deciphers the malignant cell specificity of CD40-mediated apoptosis. Oncogene (2017) 36:2515–28. doi: 10.1038/onc.2016.401
188. Cai X, Yang Y, Xia W, Kong H, Wang M, Fu W, et al. RIP2 promotes glioma cell growth by regulating TRAF3 and activating the NFkappaB and p38 signaling pathways. Oncol Rep. (2018) 39:2915–23. doi: 10.3892/or.2018.6397
189. Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell (2015) 163:506–19. doi: 10.1016/j.cell.2015.09.033
190. Network TCGAR. Comprehensive molecular profiling of lung adenocarcinoma. Nature (2014) 511:543–50. doi: 10.1038/nature13385
191. Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell (2012) 150:251–63. doi: 10.1016/j.cell.2012.06.024
192. Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature (2012) 487:239–43. doi: 10.1038/nature11125
193. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, et al. Integrated genomic characterization of endometrial carcinoma. Nature (2013) 497:67–73. doi: 10.1038/nature12113
194. Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science (2011) 333:1154–7. doi: 10.1126/science.1206923
195. Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. (2013) 3:770–81. doi: 10.1158/2159-8290.CD-12-0537
196. Bieche I, Tomasetto C, Regnier CH, Moog-Lutz C, Rio MC, Lidereau R. Two distinct amplified regions at 17q11-q21 involved in human primary breast cancer. Cancer Res. (1996) 56:3886–90.
197. Krajewska M, Krajewski S, Zapata JM, Van Arsdale T, Gascoyne RD, Berern K, et al. TRAF-4 expression in epithelial progenitor cells. Analysis in normal adult, fetal, and tumor tissues. Am J Pathol. (1998) 152:1549–61.
198. Camilleri-Broet S, Cremer I, Marmey B, Comperat E, Viguie F, Audouin J, et al. TRAF4 overexpression is a common characteristic of human carcinomas. Oncogene (2007) 26:142–7. doi: 10.1038/sj.onc.1209762
199. Li W, Peng C, Lee MH, Lim D, Zhu F, Fu Y, et al. TRAF4 is a critical molecule for Akt activation in lung cancer. Cancer Res. (2013) 73:6938–50. doi: 10.1158/0008-5472.CAN-13-0913
200. Zhang L, Zhou F, Garcia de Vinuesa A, de Kruijf EM, Mesker WE, Hui L, et al. TRAF4 promotes TGF-β receptor signaling and drives breast cancer metastasis. Mol Cell (2013) 51:559–72. doi: 10.1016/j.molcel.2013.07.014
201. Yang K, Wang F, Han JJ. TRAF4 promotes the growth and invasion of colon cancer through the Wnt/β-catenin pathway. Int J Clin Exp Pathol. (2015) 8:1419–26.
202. Yao W, Wang X, Cai Q, Gao S, Wang J, Zhang P. TRAF4 enhances osteosarcoma cell proliferation and invasion by Akt signaling pathway. Oncol Res. (2014) 22:21–8. doi: 10.3727/096504014X14077751730351
203. Liu K, Wu X, Zang X, Huang Z, Lin Z, Tan W, et al. TRAF4 regulates migration, invasion, and epithelial-mesenchymal transition via PI3K/AKT signaling in hepatocellular carcinoma. Oncol Res. (2017) 25:1329–40. doi: 10.3727/096504017X14876227286564
204. Singh R, Karri D, Shen H, Shao J, Dasgupta S, Huang S, et al. TRAF4-mediated ubiquitination of NGF receptor TrkA regulates prostate cancer metastasis. J Clin Invest. (2018) 128:3129–43. doi: 10.1172/JCI96060
205. Kedinger V, Rio MC. TRAF4, the unique family member. Adv Exp Med Biol. (2007) 597:60–71. doi: 10.1007/978-0-387-70630-6_5
206. Zhu L, Zhang S, Huan X, Mei Y, Yang H. Down-regulation of TRAF4 targeting RSK4 inhibits proliferation, invasion and metastasis in breast cancer xenografts. Biochem Biophys Res Commun. (2018) 500:810–6. doi: 10.1016/j.bbrc.2018.04.164
207. Wang X, Jin C, Tang Y, Tang LY, Zhang YE. Ubiquitination of tumor necrosis factor receptor-associated factor 4 (TRAF4) by Smad ubiquitination regulatory factor 1 (Smurf1) regulates motility of breast epithelial and cancer cells. J Biol Chem. (2013) 288:21784–92. doi: 10.1074/jbc.M113.472704
208. Yi P, Xia W, Wu RC, Lonard DM, Hung MC, O'Malley BW. SRC-3 coactivator regulates cell resistance to cytotoxic stress via TRAF4-mediated p53 destabilization. Genes Dev. (2013) 27:274–87. doi: 10.1101/gad.203760.112
209. Zhang X, Wen Z, Sun L, Wang J, Song M, Wang E, et al. TRAF2 regulates the cytoplasmic/nuclear distribution of TRAF4 and its biological function in breast cancer cells. Biochem Biophys Res Commun. (2013) 436:344–8. doi: 10.1016/j.bbrc.2013.05.107
210. Yang J, Wei D, Wang W, Shen B, Xu S, Cao Y. TRAF4 enhances oral squamous cell carcinoma cell growth, invasion and migration by Wnt-β-catenin signaling pathway. Int J Clin Exp Pathol. (2015) 8:11837–46.
211. Wang A, Wang J, Ren H, Yang F, Sun L, Diao K, et al. TRAF4 participates in Wnt/β-catenin signaling in breast cancer by upregulating β-catenin and mediating its translocation to the nucleus. Mol Cell Biochem. (2014) 395:211–9. doi: 10.1007/s11010-014-2127-y
212. Ren HY, Wang J, Yang F, Zhang XL, Wang AL, Sun LL, et al. Cytoplasmic TRAF4 contributes to the activation of p70s6k signaling pathway in breast cancer. Oncotarget (2015) 6:4080–96. doi: 10.18632/oncotarget.2977
213. Rousseau A, McEwen AG, Poussin-Courmontagne P, Rognan D, Nomine Y, Rio MC, et al. TRAF4 is a novel phosphoinositide-binding protein modulating tight junctions and favoring cell migration. PLoS Biol. (2013) 11:e1001726. doi: 10.1371/journal.pbio.1001726
214. Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell (2015) 161:1215–28. doi: 10.1016/j.cell.2015.05.001
215. Morin RD, Assouline S, Alcaide M, Mohajeri A, Johnston RL, Chong L, et al. Genetic landscapes of relapsed and refractory diffuse large B-cell lymphomas. Clin Cancer Res. (2016) 22:2290–300. doi: 10.1158/1078-0432.CCR-15-2123
216. Ruiz-Ballesteros E, Mollejo M, Rodriguez A, Camacho FI, Algara P, Martinez N, et al. Splenic marginal zone lymphoma: proposal of new diagnostic and prognostic markers identified after tissue and cDNA microarray analysis. Blood (2005) 106:1831–8. doi: 10.1182/blood-2004-10-3898
217. Weston VJ, Austen B, Wei W, Marston E, Alvi A, Lawson S, et al. Apoptotic resistance to ionizing radiation in pediatric B-precursor acute lymphoblastic leukemia frequently involves increased NF-κB survival pathway signaling. Blood (2004) 104:1465–73. doi: 10.1182/blood-2003-11-4039
218. Stunz LL, Busch LK, Munroe ME, Sigmund CD, Tygrett LT, Waldschmidt TJ, et al. Expression of the cytoplasmic tail of LMP1 in mice induces hyperactivation of B lymphocytes and disordered lymphoid architecture. Immunity (2004) 21:255–66. doi: 10.1016/j.immuni.2004.07.008
219. Horie R, Watanabe T, Morishita Y, Ito K, Ishida T, Kanegae Y, et al. Ligand-independent signaling by overexpressed CD30 drives NF-kappaB activation in Hodgkin-Reed-Sternberg cells. Oncogene (2002) 21:2493–503. doi: 10.1038/sj.onc.1205337
220. Horie R, Watanabe T, Ito K, Morisita Y, Watanabe M, Ishida T, et al. Cytoplasmic aggregation of TRAF2 and TRAF5 proteins in the Hodgkin-Reed-Sternberg cells. Am J Pathol. (2002) 160:1647–54. doi: 10.1016/S0002-9440(10)61112-1
221. Horie R, Gattei V, Ito K, Imajo-Ohmi S, Tange T, Miyauchi J, et al. Frequent expression of the variant CD30 in human malignant myeloid and lymphoid neoplasms. Am J Pathol. (1999) 155:2029–41.
222. Tao T, Cheng C, Ji Y, Xu G, Zhang J, Zhang L, et al. Numbl inhibits glioma cell migration and invasion by suppressing TRAF5-mediated NF-κB activation. Mol Biol Cell (2012) 23:2635–44. doi: 10.1091/mbc.E11-09-0805
223. Huang S, Wa Q, Pan J, Peng X, Ren D, Huang Y, et al. Downregulation of miR-141-3p promotes bone metastasis via activating NF-κB signaling in prostate cancer. J Exp Clin Cancer Res. (2017) 36:173. doi: 10.1186/s13046-017-0645-7
224. Li M, Long C, Yang G, Luo Y, Du H. MiR-26b inhibits melanoma cell proliferation and enhances apoptosis by suppressing TRAF5-mediated MAPK activation. Biochem Biophys Res Commun. (2016) 471:361–7. doi: 10.1016/j.bbrc.2016.02.021
225. Lefebvre C, Bachelot T, Filleron T, Pedrero M, Campone M, Soria JC, et al. Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med. (2016) 13:e1002201. doi: 10.1371/journal.pmed.1002201
226. Network TCGAR. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature (2014) 507:315–22. doi: 10.1038/nature12965
227. Starczynowski DT, Lockwood WW, Delehouzee S, Chari R, Wegrzyn J, Fuller M, et al. TRAF6 is an amplified oncogene bridging the RAS and NF-κB pathways in human lung cancer. J Clin Invest. (2011) 121:4095–105. doi: 10.1172/JCI58818
228. Al-Hebshi NN, Li S, Nasher AT, El-Setouhy M, Alsanosi R, Blancato J, et al. Exome sequencing of oral squamous cell carcinoma in users of Arabian snuff reveals novel candidates for driver genes. Int J Cancer (2016) 139:363–72. doi: 10.1002/ijc.30068
229. Lin Y, Qiu Y, Xu C, Liu Q, Peng B, Kaufmann GF, et al. Functional role of asparaginyl endopeptidase ubiquitination by TRAF6 in tumor invasion and metastasis. J Natl Cancer Inst. (2014) 106:dju012. doi: 10.1093/jnci/dju012
230. Li JJ, Luo J, Lu JN, Liang XN, Luo YH, Liu YR, et al. Relationship between TRAF6 and deterioration of HCC: an immunohistochemical and in vitro study. Cancer Cell Int. (2016) 16:76. doi: 10.1186/s12935-016-0352-z
231. Sun H, Li X, Fan L, Wu G, Li M, Fang J. TRAF6 is upregulated in colon cancer and promotes proliferation of colon cancer cells. Int J Biochem Cell Biol. (2014) 53:195–201. doi: 10.1016/j.biocel.2014.04.010
232. Liu X, Wang Z, Zhang G, Zhu Q, Zeng H, Wang T, et al. High TRAF6 expression is associated with esophageal carcinoma recurrence and prompts cancer cell invasion. Oncol Res. (2017) 25:485–93. doi: 10.3727/096504016X14749340314441
233. Luo Z, Zhang X, Zeng W, Su J, Yang K, Lu L, et al. TRAF6 regulates melanoma invasion and metastasis through ubiquitination of Basigin. Oncotarget (2016) 7:7179–92. doi: 10.18632/oncotarget.6886
234. Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, et al. Recurrent R-spondin fusions in colon cancer. Nature (2012) 488:660–4. doi: 10.1038/nature11282
235. Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell (2012) 150:1107–20. doi: 10.1016/j.cell.2012.08.029
236. Min Y, Kim MJ, Lee S, Chun E, Lee KY. Inhibition of TRAF6 ubiquitin-ligase activity by PRDX1 leads to inhibition of NFKB activation and autophagy activation. Autophagy (2018) 14:1347–58. doi: 10.1080/15548627.2018.1474995
237. Wang Y, Xu H, Jiao H, Wang S, Xiao Z, Zhao Y, et al. STX2 promotes colorectal cancer metastasis through a positive feedback loop that activates the NF-κB pathway. Cell Death Dis. (2018) 9:664. doi: 10.1038/s41419-018-0675-x
238. Chen L, Li YC, Wu L, Yu GT, Zhang WF, Huang CF, et al. TRAF6 regulates tumour metastasis through EMT and CSC phenotypes in head and neck squamous cell carcinoma. J Cell Mol Med. (2018) 22:1337–49. doi: 10.1111/jcmm.13439
239. Zhan Z, Xie X, Cao H, Zhou X, Zhang XD, Fan H, et al. Autophagy facilitates TLR4- and TLR3-triggered migration and invasion of lung cancer cells through the promotion of TRAF6 ubiquitination. Autophagy (2014) 10:257–68. doi: 10.4161/auto.27162
240. Sambandam Y, Ethiraj P, Hathaway-Schrader JD, Novince CM, Panneerselvam E, Sundaram K, et al. Autoregulation of RANK ligand in oral squamous cell carcinoma tumor cells. J Cell Physiol. (2018) 233:6125–34. doi: 10.1002/jcp.26456
241. Fang J, Rhyasen G, Bolanos L, Rasch C, Varney M, Wunderlich M, et al. Cytotoxic effects of bortezomib in myelodysplastic syndrome/acute myeloid leukemia depend on autophagy-mediated lysosomal degradation of TRAF6 and repression of PSMA1. Blood (2012) 120:858–67. doi: 10.1182/blood-2012-02-407999
242. da Silva Almeida AC, Abate F, Khiabanian H, Martinez-Escala E, Guitart J, Tensen CP, et al. The mutational landscape of cutaneous T cell lymphoma and Sezary syndrome. Nat Genet. (2015) 47:1465–70. doi: 10.1038/ng.3442
243. Dai YH, Hung LY, Chen RY, Lai CH, Chang KC. ON 01910.Na inhibits growth of diffuse large B-cell lymphoma by cytoplasmic sequestration of sumoylated C-MYB/TRAF6 complex. Transl Res. (2016) 175:129–43.e13. doi: 10.1016/j.trsl.2016.04.001
244. Chen H, Li M, Sanchez E, Wang CS, Lee T, Soof CM, et al. Combined TRAF6 targeting and proteasome blockade has anti-myeloma and anti-bone resorptive effects. Mol Cancer Res. (2017) 15:598–609. doi: 10.1158/1541-7786.MCR-16-0293
245. Burger ML, Xue L, Sun Y, Kang C, Winoto A. Premalignant PTEN-deficient thymocytes activate microRNAs miR-146a and miR-146b as a cellular defense against malignant transformation. Blood (2014) 123:4089–100. doi: 10.1182/blood-2013-11-539411
246. Li Z, Younger K, Gartenhaus R, Joseph AM, Hu F, Baer MR, et al. Inhibition of IRAK1/4 sensitizes T cell acute lymphoblastic leukemia to chemotherapies. J Clin Invest. (2015) 125:1081–97. doi: 10.1172/JCI75821
247. Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature (2008) 455:1061–8. doi: 10.1038/nature07385
248. Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell (2013) 155:462–77. doi: 10.1016/j.cell.2013.09.034
249. Zhang X, Li CF, Zhang L, Wu CY, Han L, Jin G, et al. TRAF6 restricts p53 mitochondrial translocation, apoptosis, and tumor suppression. Mol Cell (2016) 64:803–14. doi: 10.1016/j.molcel.2016.10.002
250. Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell (2008) 13:343–54. doi: 10.1016/j.ccr.2008.02.001
251. Chiu HW, Lin SW, Lin LC, Hsu YH, Lin YF, Ho SY, et al. Synergistic antitumor effects of radiation and proteasome inhibitor treatment in pancreatic cancer through the induction of autophagy and the downregulation of TRAF6. Cancer Lett. (2015) 365:229–39. doi: 10.1016/j.canlet.2015.05.025
252. Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science (2009) 325:1134–8. doi: 10.1126/science.1175065
253. Feng H, Lopez GY, Kim CK, Alvarez A, Duncan CG, Nishikawa R, et al. EGFR phosphorylation of DCBLD2 recruits TRAF6 and stimulates AKT-promoted tumorigenesis. J Clin Invest. (2014) 124:3741–56. doi: 10.1172/JCI73093
254. Shi YF, Yu DJ, Jiang CY, Wang XJ, Zhu YP, Zhao RZ, et al. TRAF6 regulates proliferation of stromal cells in the transition and peripheral zones of benign prostatic hyperplasia via Akt/mTOR signaling. Prostate (2018) 78:193–201. doi: 10.1002/pros.23456
255. Wu YH, Wu WS, Lin LC, Liu CS, Ho SY, Wang BJ, et al. Bortezomib enhances radiosensitivity in oral cancer through inducing autophagy-mediated TRAF6 oncoprotein degradation. J Exp Clin Cancer Res. (2018) 37:91. doi: 10.1186/s13046-018-0760-0
256. Pan D, Jiang C, Ma Z, Blonska M, You MJ, Lin X. MALT1 is required for EGFR-induced NF-κB activation and contributes to EGFR-driven lung cancer progression. Oncogene (2016) 35:919–28. doi: 10.1038/onc.2015.146
257. Zhang X, Wu L, Xiao T, Tang L, Jia X, Guo Y, et al. TRAF6 regulates EGF-induced cell transformation and cSCC malignant phenotype through CD147/EGFR. Oncogenesis (2018) 7:17. doi: 10.1038/s41389-018-0030-1
258. Rezaeian AH, Li CF, Wu CY, Zhang X, Delacerda J, You MJ, et al. A hypoxia-responsive TRAF6-ATM-H2AX signalling axis promotes HIF1α activation, tumorigenesis and metastasis. Nat Cell Biol. (2017) 19:38–51. doi: 10.1038/ncb3445
259. Sun H, Li XB, Meng Y, Fan L, Li M, Fang J. TRAF6 upregulates expression of HIF-1α and promotes tumor angiogenesis. Cancer Res. (2013) 73:4950–9. doi: 10.1158/0008-5472.CAN-13-0370
260. Mu Y, Sundar R, Thakur N, Ekman M, Gudey SK, Yakymovych M, et al. TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer. Nat Commun. (2011) 2:330. doi: 10.1038/ncomms1332
261. Sundar R, Gudey SK, Heldin CH, Landstrom M. TRAF6 promotes TGFβ-induced invasion and cell-cycle regulation via Lys63-linked polyubiquitination of Lys178 in TGFβ type I receptor. Cell Cycle (2015) 14:554–65. doi: 10.4161/15384101.2014.990302
262. Gudey SK, Sundar R, Mu Y, Wallenius A, Zang G, Bergh A, et al. TRAF6 stimulates the tumor-promoting effects of TGFβ type I receptor through polyubiquitination and activation of presenilin 1. Sci Signal. (2014) 7:ra2. doi: 10.1126/scisignal.2004207
263. Hamidi A, Song J, Thakur N, Itoh S, Marcusson A, Bergh A, et al. TGF-β promotes PI3K-AKT signaling and prostate cancer cell migration through the TRAF6-mediated ubiquitylation of p85α. Sci Signal. (2017) 10:eaal4186. doi: 10.1126/scisignal.aal4186
264. Linares JF, Duran A, Reina-Campos M, Aza-Blanc P, Campos A, Moscat J, et al. Amino acid activation of mTORC1 by a PB1-domain-driven kinase complex cascade. Cell Rep. (2015) 12:1339–52. doi: 10.1016/j.celrep.2015.07.045
265. An J, Liu H, Magyar CE, Guo Y, Veena MS, Srivatsan ES, et al. Hyperactivated JNK is a therapeutic target in pVHL-deficient renal cell carcinoma. Cancer Res. (2013) 73:1374–85. doi: 10.1158/0008-5472.CAN-12-2362
266. Bao X, Shi J, Xie F, Liu Z, Yu J, Chen W, et al. Proteolytic release of the p75(NTR) intracellular domain by ADAM10 promotes metastasis and resistance to anoikis. Cancer Res. (2018) 78:2262–76. doi: 10.1158/0008-5472.CAN-17-2789
267. Yu J, Qin B, Moyer AM, Nowsheen S, Liu T, Qin S, et al. DNA methyltransferase expression in triple-negative breast cancer predicts sensitivity to decitabine. J Clin Invest. (2018) 128:2376–88. doi: 10.1172/JCI97924
268. Chaudhry SI, Hooper S, Nye E, Williamson P, Harrington K, Sahai E. Autocrine IL-1β-TRAF6 signalling promotes squamous cell carcinoma invasion through paracrine TNFα signalling to carcinoma-associated fibroblasts. Oncogene (2013) 32:747–58. doi: 10.1038/onc.2012.91
269. Fang J, Barker B, Bolanos L, Liu X, Jerez A, Makishima H, et al. Myeloid malignancies with chromosome 5q deletions acquire a dependency on an intrachromosomal NF-κB gene network. Cell Rep. (2014) 8:1328–38. doi: 10.1016/j.celrep.2014.07.062
270. Liu R, Liu C, Chen D, Yang WH, Liu X, Liu CG, et al. FOXP3 Controls an miR-146/NF-κB negative feedback loop that inhibits apoptosis in breast cancer cells. Cancer Res. (2015) 75:1703–13. doi: 10.1158/0008-5472.CAN-14-2108
271. Liu R, Yi B, Wei S, Yang WH, Hart KM, Chauhan P, et al. FOXP3-miR-146-NF-κB axis and therapy for precancerous lesions in prostate. Cancer Res. (2015) 75:1714–24. doi: 10.1158/0008-5472.CAN-14-2109
272. Zu Y, Yang Y, Zhu J, Bo X, Hou S, Zhang B, et al. MiR-146a suppresses hepatocellular carcinoma by downregulating TRAF6. Am J Cancer Res. (2016) 6:2502–13.
273. Liu J, Xu J, Li H, Sun C, Yu L, Li Y, et al. miR-146b-5p functions as a tumor suppressor by targeting TRAF6 and predicts the prognosis of human gliomas. Oncotarget (2015) 6:29129–42. doi: 10.18632/oncotarget.4895
274. Li C, Miao R, Liu S, Wan Y, Zhang S, Deng Y, et al. Down-regulation of miR-146b-5p by long noncoding RNA MALAT1 in hepatocellular carcinoma promotes cancer growth and metastasis. Oncotarget (2017) 8:28683–95. doi: 10.18632/oncotarget.15640
275. Jiao L, Zhang HL, Li DD, Yang KL, Tang J, Li X, et al. Regulation of glycolytic metabolism by autophagy in liver cancer involves selective autophagic degradation of HK2 (hexokinase 2). Autophagy (2018) 14:671–84. doi: 10.1080/15548627.2017.1381804
276. Lu W, Liu S, Li B, Xie Y, Izban MG, Ballard BR, et al. SKP2 loss destabilizes EZH2 by promoting TRAF6-mediated ubiquitination to suppress prostate cancer. Oncogene (2017) 36:1364–73. doi: 10.1038/onc.2016.300
277. Lu W, Liu S, Li B, Xie Y, Adhiambo C, Yang Q, et al. SKP2 inactivation suppresses prostate tumorigenesis by mediating JARID1B ubiquitination. Oncotarget (2015) 6:771–88. doi: 10.18632/oncotarget.2718
278. Zapata JM, Pawlowski K, Haas E, Ware CF, Godzik A, Reed JC. A diverse family of proteins containing tumor necrosis factor receptor-associated factor domains. J Biol Chem. (2001) 276:24242–52. doi: 10.1074/jbc.M100354200
279. Zapata JM, Martinez-Garcia V, Lefebvre S. Phylogeny of the TRAF/MATH domain. Adv Exp Med Biol. (2007) 597:1–24. doi: 10.1007/978-0-387-70630-6_1
280. Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. (2016) 22:369–78. doi: 10.1038/nm.4053
281. Goode B, Joseph NM, Stevers M, Van Ziffle J, Onodera C, Talevich E, et al. Adenomatoid tumors of the male and female genital tract are defined by TRAF7 mutations that drive aberrant NF-kB pathway activation. Mod Pathol. (2018) 31:660–73. doi: 10.1038/modpathol.2017.153
282. Reuss DE, Piro RM, Jones DT, Simon M, Ketter R, Kool M, et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol. (2013) 125:351–8. doi: 10.1007/s00401-013-1093-x
283. Klein CJ, Wu Y, Jentoft ME, Mer G, Spinner RJ, Dyck PJ, et al. Genomic analysis reveals frequent TRAF7 mutations in intraneural perineuriomas. Ann Neurol. (2017) 81:316–21. doi: 10.1002/ana.24854
284. Clark VE, Harmanci AS, Bai H, Youngblood MW, Lee TI, Baranoski JF, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. (2016) 48:1253–9. doi: 10.1038/ng.3651
285. Clark VE, Erson-Omay EZ, Serin A, Yin J, Cotney J, Ozduman K, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science (2013) 339:1077–80. doi: 10.1126/science.1233009
286. Yuzawa S, Nishihara H, Yamaguchi S, Mohri H, Wang L, Kimura T, et al. Clinical impact of targeted amplicon sequencing for meningioma as a practical clinical-sequencing system. Mod Pathol. (2016) 29:708–16. doi: 10.1038/modpathol.2016.81
287. Abedalthagafi M, Bi WL, Aizer AA, Merrill PH, Brewster R, Agarwalla PK, et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol. (2016) 18:649–55. doi: 10.1093/neuonc/nov316
288. Peyre M, Gaillard S, de Marcellus C, Giry M, Bielle F, Villa C, et al. Progestin-associated shift of meningioma mutational landscape. Ann Oncol. (2018) 29:681–6. doi: 10.1093/annonc/mdx763
289. Zotti T, Scudiero I, Vito P, Stilo R. The emerging role of TRAF7 in tumor development. J Cell Physiol. (2017) 232:1233–8. doi: 10.1002/jcp.25676
290. Karsy M, Azab MA, Abou-Al-Shaar H, Guan J, Eli I, Jensen RL, et al. Clinical potential of meningioma genomic insights: a practical review for neurosurgeons. Neurosurg Focus (2018) 44:E10. doi: 10.3171/2018.2.FOCUS1849
291. Apra C, Peyre M, Kalamarides M. Current treatment options for meningioma. Expert Rev Neurother. (2018) 18:241–9. doi: 10.1080/14737175.2018.1429920
292. Sneddon S, Dick I, Lee YCG, Musk AWB, Patch AM, Pearson JV, et al. Malignant cells from pleural fluids in malignant mesothelioma patients reveal novel mutations. Lung Cancer (2018) 119:64–70. doi: 10.1016/j.lungcan.2018.03.009
293. Bueno R, Stawiski EW, Goldstein LD, Durinck S, De Rienzo A, Modrusan Z, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. (2016) 48:407–16. doi: 10.1038/ng.3520
294. Yuzawa S, Nishihara H, Tanaka S. Genetic landscape of meningioma. Brain Tumor Pathol. (2016) 33:237–47. doi: 10.1007/s10014-016-0271-7
295. Peyre M, Feuvret L, Sanson M, Navarro S, Boch AL, Loiseau H, et al. Diffuse midline skull base meningiomas: identification of a rare and aggressive subgroup of meningiomas. J Neurooncol. (2017) 133:633–9. doi: 10.1007/s11060-017-2480-2
296. Fadlullah MZ, Chiang IK, Dionne KR, Yee PS, Gan CP, Sam KK, et al. Genetically-defined novel oral squamous cell carcinoma cell lines for the development of molecular therapies. Oncotarget (2016) 7:27802–18. doi: 10.18632/oncotarget.8533
297. Zhang X, Jia H, Lu Y, Dong C, Hou J, Wang Z, et al. Exome sequencing on malignant meningiomas identified mutations in neurofibromatosis type 2 (NF2) and meningioma 1 (MN1) genes. Discov Med. (2014) 18:301–11.
298. Kuhn E, Wu RC, Guan B, Wu G, Zhang J, Wang Y, et al. Identification of molecular pathway aberrations in uterine serous carcinoma by genome-wide analyses. J Natl Cancer Inst. (2012) 104:1503–13. doi: 10.1093/jnci/djs345
299. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (2015) 348:124–8. doi: 10.1126/science.aaa1348
300. Tokita MJ, Chen CA, Chitayat D, Macnamara E, Rosenfeld JA, Hanchard N, et al. De novo missense variants in TRAF7 cause developmental delay, congenital anomalies, and dysmorphic features. Am J Hum Genet. (2018) 103:154–62. doi: 10.1016/j.ajhg.2018.06.005
301. Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell (2011) 44:325–40. doi: 10.1016/j.molcel.2011.08.025
302. Xu LG, Li LY, Shu HB. TRAF7 potentiates MEKK3-induced AP1 and CHOP activation and induces apoptosis. J Biol Chem. (2004) 279:17278–82. doi: 10.1074/jbc.C400063200
303. Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. (2004) 6:97–105. doi: 10.1038/ncb1086
304. Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. (2015) 47:582–8. doi: 10.1038/ng.3303
305. Morita Y, Kanei-Ishii C, Nomura T, Ishii S. TRAF7 sequesters c-Myb to the cytoplasm by stimulating its sumoylation. Mol Biol Cell (2005) 16:5433–44. doi: 10.1091/mbc.e05-08-0731
306. Fremin C, Saba-El-Leil MK, Levesque K, Ang SL, Meloche S. Functional redundancy of ERK1 and ERK2 MAP kinases during development. Cell Rep. (2015) 12:913–21. doi: 10.1016/j.celrep.2015.07.011
307. Zotti T, Vito P, Stilo R. The seventh ring: exploring TRAF7 functions. J Cell Physiol. (2012) 227:1280–4. doi: 10.1002/jcp.24011
308. Scudiero I, Zotti T, Ferravante A, Vessichelli M, Reale C, Masone MC, et al. Tumor necrosis factor (TNF) receptor-associated factor 7 is required for TNFα-induced Jun NH2-terminal kinase activation and promotes cell death by regulating polyubiquitination and lysosomal degradation of c-FLIP protein. J Biol Chem. (2012) 287:6053–61. doi: 10.1074/jbc.M111.300137
309. Zotti T, Uva A, Ferravante A, Vessichelli M, Scudiero I, Ceccarelli M, et al. TRAF7 protein promotes Lys-29-linked polyubiquitination of IkappaB kinase (IKKgamma)/NF-kappaB essential modulator (NEMO) and p65/RelA protein and represses NF-kappaB activation. J Biol Chem. (2011) 286:22924–33. doi: 10.1074/jbc.M110.215426
310. Tsikitis M, Acosta-Alvear D, Blais A, Campos EI, Lane WS, Sanchez I, et al. Traf7, a MyoD1 transcriptional target, regulates nuclear factor-κB activity during myogenesis. EMBO Rep. (2010) 11:969–76. doi: 10.1038/embor.2010.154
311. Yoshida H, Jono H, Kai H, Li JD. The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative cross-talk with TRAF6 AND TRAF7. J Biol Chem. (2005) 280:41111–21. doi: 10.1074/jbc.M509526200
312. Wang L, Zhang S, Qu G, Zhang D, Li S, Liu S. Downregulation of ubiquitin E3 ligase TNF receptor-associated factor 7 leads to stabilization of p53 in breast cancer. Oncol Rep. (2013) 29:283–7. doi: 10.3892/or.2012.2121
313. Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature (2012) 486:346–52. doi: 10.1038/nature10983
314. Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. (2016) 7:11479. doi: 10.1038/ncomms11479
315. Network TCGAR. Integrated genomic characterization of oesophageal carcinoma. Nature (2017) 541:169–75. doi: 10.1038/nature20805
316. Wan XK, Yuan SL, Tao HX, Diao LP, Wang YC, Cao C, et al. The upregulation of TRAF1 induced by helicobacter pylori plays an antiapoptotic effect on the infected cells. Helicobacter (2016) 21:554–64. doi: 10.1111/hel.12311
317. Asano N, Imatani A, Watanabe T, Fushiya J, Kondo Y, Jin X, et al. Cdx2 expression and intestinal metaplasia induced by H. pylori infection of gastric cells is regulated by NOD1-mediated innate immune responses. Cancer Res. (2016) 76:1135–45. doi: 10.1158/0008-5472.CAN-15-2272
318. Watanabe T, Asano N, Fichtner-Feigl S, Gorelick PL, Tsuji Y, Matsumoto Y, et al. NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway. J Clin Invest. (2010) 120:1645–62. doi: 10.1172/JCI39481
319. Maeda S, Yoshida H, Ogura K, Mitsuno Y, Hirata Y, Yamaji Y, et al. H. pylori activates NF-κB through a signaling pathway involving IκB kinases, NF-κB-inducing kinase, TRAF2, and TRAF6 in gastric cancer cells. Gastroenterology (2000) 119:97–108. doi: 10.1053/gast.2000.8540
320. Zou M, Wang F, Jiang A, Xia A, Kong S, Gong C, et al. MicroRNA-3178 ameliorates inflammation and gastric carcinogenesis promoted by Helicobacter pylori new toxin, Tip-α, by targeting TRAF3. Helicobacter (2017) 22:e12348. doi: 10.1111/hel.12348
321. Greenfeld H, Takasaki K, Walsh MJ, Ersing I, Bernhardt K, Ma Y, et al. TRAF1 Coordinates polyubiquitin signaling to enhance epstein-barr virus LMP1-mediated growth and survival pathway activation. PLoS Pathog. (2015) 11:e1004890. doi: 10.1371/journal.ppat.1004890
322. Siegler G, Meyer B, Dawson C, Brachtel E, Lennerz J, Koch C, et al. Expression of tumor necrosis factor receptor-associated factor 1 in nasopharyngeal carcinoma: possible upregulation by Epstein-Barr virus latent membrane protein 1. Int J Cancer (2004) 112:265–72. doi: 10.1002/ijc.20367
323. Kung CP, Raab-Traub N. Epstein-Barr virus latent membrane protein 1 modulates distinctive NF-κB pathways through C-terminus-activating region 1 to regulate epidermal growth factor receptor expression. J Virol. (2010) 84:6605–14. doi: 10.1128/JVI.00344-10
324. Schnetzke U, Fischer M, Spies-Weisshart B, Zirm E, Hochhaus A, Muller JP, et al. The E3 ubiquitin ligase TRAF2 can contribute to TNF-α resistance in FLT3-ITD-positive AML cells. Leuk Res. (2013) 37:1557–64. doi: 10.1016/j.leukres.2013.08.004
325. Shirakata M, Imadome KI, Okazaki K, Hirai K. Activation of TRAF5 and TRAF6 signal cascades negatively regulates the latent replication origin of Epstein-Barr virus through p38 mitogen-activated protein kinase. J Virol. (2001) 75:5059–68. doi: 10.1128/JVI.75.11.5059-5068.2001
326. Guasparri I, Wu H, Cesarman E. The KSHV oncoprotein vFLIP contains a TRAF-interacting motif and requires TRAF2 and TRAF3 for signalling. EMBO Rep. (2006) 7:114–9. doi: 10.1038/sj.embor.7400580
327. Kumari P, Saha I, Narayanan A, Narayanan S, Takaoka A, Kumar NS, et al. Essential role of HCMV deubiquitinase in promoting oncogenesis by targeting anti-viral innate immune signaling pathways. Cell Death Dis. (2017) 8:e3078. doi: 10.1038/cddis.2017.461
328. Zhang J, Chen T, Yang X, Cheng H, Spath SS, Clavijo PE, et al. Attenuated TRAF3 fosters alternative activation of NF-κB and reduced expression of anti-viral interferon, TP53, and RB to promote HPV-positive head and neck cancers. Cancer Res. (2018) 78:4613–26. doi: 10.1158/0008-5472.CAN-17-0642
329. Yoshida H, Kato N, Shiratori Y, Otsuka M, Maeda S, Kato J, et al. Hepatitis C virus core protein activates nuclear factor kappa B-dependent signaling through tumor necrosis factor receptor-associated factor. J Biol Chem. (2001) 276:16399–405. doi: 10.1074/jbc.M006671200
330. Khan KA, Abbas W, Varin A, Kumar A, Di Martino V, Dichamp I, et al. HIV-1 Nef interacts with HCV Core, recruits TRAF2, TRAF5 and TRAF6, and stimulates HIV-1 replication in macrophages. J Innate Immun. (2013) 5:639–56. doi: 10.1159/000350517
331. Diani E, Avesani F, Bergamo E, Cremonese G, Bertazzoni U, Romanelli MG. HTLV-1 Tax protein recruitment into IKKepsilon and TBK1 kinase complexes enhances IFN-I expression. Virology (2015) 476:92–9. doi: 10.1016/j.virol.2014.12.005
332. Choi YB, Harhaj EW. HTLV-1 tax stabilizes MCL-1 via TRAF6-dependent K63-linked polyubiquitination to promote cell survival and transformation. PLoS Pathog. (2014) 10:e1004458. doi: 10.1371/journal.ppat.1004458
333. Boulabiar M, Boubaker S, Favre M, Demeret C. Keratinocyte sensitization to tumour necrosis factor-induced nuclear factor kappa B activation by the E2 regulatory protein of human papillomaviruses. J Gen Virol. (2011) 92(Pt 10):2422–7. doi: 10.1099/vir.0.032466-0
334. Bellanger S, Tan CL, Xue YZ, Teissier S, Thierry F. Tumor suppressor or oncogene? A critical role of the human papillomavirus (HPV) E2 protein in cervical cancer progression. Am J Cancer Res. (2011) 1:373–89.
335. McLaughlin-Drubin ME, Munger K. Viruses associated with human cancer. Biochim Biophys Acta (2008) 1782:127–50. doi: 10.1016/j.bbadis.2007.12.005
336. Morales-Sanchez A, Fuentes-Panana EM. Human viruses and cancer. Viruses (2014) 6:4047–79. doi: 10.3390/v6104047
337. Slotwinski R, Slotwinska SM. Dysregulation of signaling pathways associated with innate antibacterial immunity in patients with pancreatic cancer. Cent Eur J Immunol (2016) 41:404–18. doi: 10.5114/ceji.2016.65140
338. Kim E, Kim W, Lee S, Chun J, Kang J, Park G, et al. TRAF4 promotes lung cancer aggressiveness by modulating tumor microenvironment in normal fibroblasts. Sci Rep. (2017) 7:8923. doi: 10.1038/s41598-017-09447-z
339. Theodoraki MN, Yerneni S, Sarkar SN, Orr B, Muthuswamy R, Voyten J, et al. Helicase-driven activation of NFκB-COX2 mediates the immunosuppressive component of dsRNA-driven inflammation in the human tumor microenvironment. Cancer Res. (2018) 78:4292–302. doi: 10.1158/0008-5472.CAN-17-3985
340. Snell LM, Lin GH, McPherson AJ, Moraes TJ, Watts TH. T-cell intrinsic effects of GITR and 4-1BB during viral infection and cancer immunotherapy. Immunol Rev. (2011) 244:197–217. doi: 10.1111/j.1600-065X.2011.01063.x
341. Lu Y, Wang Q, Xue G, Bi E, Ma X, Wang A, et al. Th9 cells represent a unique subset of CD4(+) T cells endowed with the ability to eradicate advanced tumors. Cancer Cell (2018) 33:1048–60.e7. doi: 10.1016/j.ccell.2018.05.004
Keywords: TRAFs, cancer, oncogenes, tumor suppressor genes, NF-κB, MAPK
Citation: Zhu S, Jin J, Gokhale S, Lu AM, Shan H, Feng J and Xie P (2018) Genetic Alterations of TRAF Proteins in Human Cancers. Front. Immunol. 9:2111. doi: 10.3389/fimmu.2018.02111
Received: 29 July 2018; Accepted: 28 August 2018;
Published: 20 September 2018.
Edited by:
Tania H. Watts, University of Toronto, CanadaReviewed by:
Juan M. Zapata, Instituto de Investigaciones Biomédicas Alberto Sols (IIBM), SpainShao-Cong Sun, University of Texas MD Anderson Cancer Center, United States
Copyright © 2018 Zhu, Jin, Gokhale, Lu, Shan, Feng and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Xie, xie@dls.rutgers.edu
 Sining Zhu1,2
Sining Zhu1,2 Ping Xie
Ping Xie