- 1Mengchao Hepatobiliary Hospital of Fujian Medical University, Fujian, China
- 2Fuzong Clinical Medical College of Fujian Medical University, Fujian, China
- 3900th Hospital of Joint Logistics Support Forces of the Chinese PLA, Fujian, China
The role of genetic factors in the occurrence and progression of CHB (CHB) is still not fully explored. In recent years, genome-wide association studies on CHB patients have demonstrated that a large number of CHB-associated single nucleotide polymorphisms exist in the gene intron, which may regulate expression at the transcriptional level. Modification of RNA m6A methylation is one of the key mechanisms regulating gene expression. Here we show that METTL16, an m6A regulator involved in mRNA intron splicing, is differentially expressed in CHB the tissue of patients who has definite diagnosis of mild and severe fibrosis. At the same time, there are also significant differences in the expression of CHB-associated genes such as HLA-DPA1 and HLA-DPB1. The expression of HLA-DPB1 is related to METTL16. Furthermore, analyses of RNA binding of METTL16 and HLA-DPB1 show that the silencing of METTL16 in astrocytes downregulates m6A and expression of HLA-DPB1. In conclusion, METTL16 participates in the progression of CHB fibrosis by regulating the m6A level and expression of HLA-DPB1.
Introduction
Chronic hepatitis B (CHB) is a chronic inflammatory disease in patients with hepatitis B virus (HBV) infection. The incidence of CHB ranks first among all kinds of infectious diseases (Lok, 2002). More than 1.3 billion people in global are infected with HBV, about 260 million are with CHB, which causes about 1 million deaths every year (Perz et al., 2006; Schweitzer et al., 2015). CHB has become a very serious health and social problem.
Heredity, the virus, and the environment are important factors in the pathogenesis of CHB, which leads to high heterogeneity in clinic. From the perspective of population susceptibility to CHB and disease progression, genetic variation can lead to differences in clinical manifestations among individuals. Since the publication of the first genome-wide association study (GWAS) of CHB in 2009, genetic studies on patients with CHB have revealed many single nucleotide polymorphisms (SNPs) associated with susceptibility to CHB (Raza et al., 2007). Several studies have confirmed that these SNPs are mainly concentrated in a series of human leukocyte antigen (HLA) loci, including HLA-DP, HLA-DQ, HLA-C, and HLA-DOA (Lau et al., 2011; Yamada et al., 2014; Akcay et al., 2018). Among them, Mbared et al. found that the SNP rs9277535 with the most significant association with CHB in a Japanese population was located in the 3′ untranslated region of HLA-DPB1. The SNP was also identified in Korean, Thai, and Han populations with different significance. Moreover, rs3077, a representative CHB-associated SNP in different populations, is located in the 3′ untranslated region of HLA-DPA1. In addition, SNPs located in EHMT2, TCF19, UBE2l3, CFB, FDX1, and other gene regions are also associated with susceptibility to CHB in different regions. However, CHB progresses to liver cirrhosis and liver cancer. GWAS shows that variation in the SNP of HLA gene closely related to the progression of CHB to liver cirrhosis and participates in the occurrence of liver cancer. Previous studies have shown that the cytotoxicity of HLA class I and class II play an critical role in the spontaneous clearance of HBV. However, the clinical heterogeneity of CHB cannot be fully analyzed from only the level of genetic variation. The associated SNPs vary in different populations, and some findings are difficult to replicate, or even show the opposite results. In the results of GWAS, the genes that play an important role in CHB is not statistically significant. Most SNPs located at HLA loci are located in the untranslated region. The functional mechanism is not clear, which may be related to mRNA expression of the gene. SNP loci associated with hepatitis are distributed in the intron region of the gene. From the perspective of SNP–amino acid protein function, the mechanism of action of these SNPs cannot be deeply analyzed. Although it is believed that these SNPs can affect the pathogenesis of CHB by altering gene expression, their key mechanism of action has not been revealed.
Recent studies have found that modification of N6 methyl adenosine (m6A) is an important way of controlling gene expression by eukaryotic mRNA. m6A modification is mainly distributed in introns and the 3′ untranslated region, especially in region near the stop codon and splice site, which is involved in RNA processing and metabolic function (Liu and Zhang, 2018). m6A modification takes part in different stages of development of mRNA (Imam et al., 2018), including RNA folding, stability, splicing, nuclear output, translation regulation, and degradation, to regulate RNA biological function, protein translation, and life activity (Zhao et al., 2021; Tong et al., 2022). m6A modification of precursor mRNA mainly takes place in the untranslated region, and m6A methylase and reader proteins located in the nucleus. Thus, it can be inferred that m6A modification mainly occurs in the nucleus and affects mRNA splicing (Meyer et al., 2012; Zhao et al., 2014; Xu et al., 2017). Knockout of METTL3 results in the downregulation of introns. In addition, m6A demethylase FTO preferentially binds to the RNA intron region, downregulates m6A modification on the one hand, but prevents RNA from binding to splicing protein SRSF2 on the other hand, resulting in abnormal splicing (Dominissini et al., 2012). These studies show that m6A modification of RNA in untranslated regions could affects gene expression by regulating RNA processing and metabolism. This phenomenon provides clues for analyzing the role of SNPs in the untranslated region in the pathogenesis of CHB. We speculate that SNPs in the untranslated region impact the occurrence and development of CHB by affecting m6A modification and regulating gene expression.
In addition, many studies have shown that m6A modification can change expression of important viral genes. Researchers have proven that modification of m6A methylation is widely involved in replication of the HBV virus, inflammatory response, immune regulation, and fibrosis and plays a role in liver injury, tumors, and organ failure (Kostyusheva et al., 2021). Imam h et al. mapped the m6A site in HBV RNA (Qu et al., 2021; Cheng et al., 2022; Kim et al., 2022; Kim and Siddiqui, 2022; Zhao et al., 2022). m6A modification is necessary for efficient reverse transcription of the viral genome and can also regulate the stability of HBV RNA (Kim and Siddiqui, 2021a). Chronic infection with HBV and hepatitis C virus is the main cause of hepatocellular carcinoma (Xiao et al., 2016; Xu et al., 2017). There is increasing evidence that hepatocellular carcinoma oncoproteins induced by both virus are controlled by m6A modification. Recent works found that m6A modification involves the regulation of hepatocellular carcinoma through METTL3 and METTL14. First, Chen et al. (2018) observed the expression of METTL3 increased abnormally in liver cancer and increased cell proliferation in vitro, resulting in promoted tumorigenicity in vivo (Xu et al., 2017). METTL3 is significantly upregulated in hepatocellular carcinoma and promotes tumor progression. It inhibits SOCS2 expression and promotes cancer cell proliferation and metastasis through the m6A-YTHDF2 mechanism. Chen et al. (2018) found interference with METTL3 reduce the expression of SOCS2 mRNA. Second, it was reported that METTL14 is downregulated in liver cancer, and thereby regulates the development of liver cancer (Bartosovic et al., 2017; Ma et al., 2017). Together these evidences suggest that m6A modification has a key role in liver-related diseases through various m6A-related proteins (Wu et al., 2019; Wu et al., 2020; Kim and Siddiqui, 2021b; Wang and Zhou, 2022). Modification of m6A methylation is involved in the pathogenesis of liver injury, organ failure, and fibrosis. However, it is unclear whether it is involved in the development of CHB.
Here, we investigated the expression of m6A regulator in different stages of CHB, examined the relationship between m6A and CHB-associated genes, and checked the change in m6A and expression of gene loci with CHB-associated SNPs.
Materials and methods
Patients
The ethical approval was approved by the ethics committee of Mengchao Hepatobiliary Hospital of Fujian Medical University and all study participants obtained informed consent. Clinical data were collected from patients with CHB diagnosed by liver biopsy in our hospital in 2019 or 2020. The diagnostic criteria were in accordance with the guidelines for the prevention and treatment of CHB (2019 Edition), and study subjects provided informed consent before enrollment. Inclusion criteria were 1) being HBsAg positive for more than 6 months and HBsAb negative and 2) being between 18 and 60 years old. Exclusion criteria were 1) the presence of acute hepatitis B, liver failure, or primary liver cancer, in combination with drug liver, alcoholic liver, or fatty liver, in combination with any other viral infection and other serious disease; 2) use of antiviral drugs up to 3 months before enrollment; 3) receipt of immunosuppressant and immunomodulator treatment up to 6 months before enrollment; 4) autoimmune liver disease and systemic autoimmune disease; and 5) pregnancy.
Specimens
A BARD puncture biopsy gun (with a sampling length of 2.2 cm) and 16 g disposable cutting biopsy needle were used for the liver puncture biopsy. One tissue specimen was stained with he, Masson, and reticular fibers, and a single pathologist read the film uniformly according to the pathological diagnostic criteria. The other specimen was kept in the refrigerator at −80°C.
Tandem mass spectrometry (LC/MS)
After total RNA is extracted with Trizol, mRNA can be enriched with Oligo (dT) magnetic beads. RNA was digested from a single strand into a single base with nuclease P1. Alkaline phosphatase and ammonium bicarbonate were added, the sample was allowed to incubate for several hours, and then the sample was injected into a liquid chromatograph. Finally, the overall degree of m6A methylation on mRNA was calculated according to the ratio of m6A to total adenine.
Real-time fluorescence quantitative PCR
Tissues or cells were digested and lysed by Trizol reagent. After Trizol was added to cells or tissues, total RNA was extracted with chloroform isopropanol extraction. cDNA was synthesized by reverse transcription with a one-step PrimeScript cDNA synthesis kit. Quantitative PCR was performed with a one-step SYBR PrimeScript RT-PCR kit. GAPDH was used as the internal reference gene, and the quantitative results were 2−ΔΔCT indicates. The primer information was in (Supplemental Table S1).
meIP-PCR
The combination of immunoprecipitation (ChIP) and PCR technology can be utilized to efficiently determine the interaction in vivo. RNA was isolated and broken into small fragments by ultrasounication. An specific antibody was added, and the antibody formed an immune binding complex with the target protein. De crosslinking, RNA purification and qPCR were further processed.
Statistical analysis
SPSS 20.0 was used for statistical the analysis. The measurement data conforming to normal distribution adopts mean ± standard deviation (±s). t tests were used for pairwise comparisons of normally distributed data. Single-factor analysis of variance was used for multigroup comparisons. Spearman correlation analysis was used to analyze correlations between various factors and the occurrence and degree of liver fibrosis in patients with CHB.
Results
SNPs associated with susceptibility to CHB are located in different genes
GWAS has identified 102 SNP sites related to susceptibility to or progression of CHB (Figure 1A and Supplemental Table S2). We discovered that only three SNPs were distributed in the exon region of the gene, nearly 26 were distributed in the intron region of the gene, and the rest were distributed in the 3′ and 5′ untranslated regions (Figure 1B).
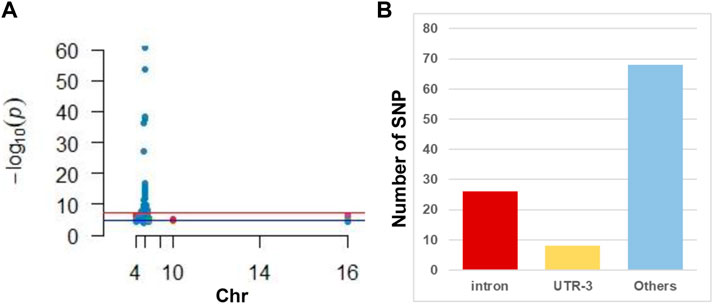
FIGURE 1. Illustration of GWAS studies in CHB. (A) Manhattan plot of CHB associated SNP reviewed from published literature. Note that most SNPs located in the chromosome 6. (B) Distribution of SNP in gene different regions. Note that intron is hot regions where CHB associated SNP frequently located.
Patients show different levels of liver fibrosis
A BARD puncture biopsy gun (with a sampling length of 2.2 cm) and 16 g disposable cutting biopsy needle were used for the liver puncture biopsy. Two complete liver tissues with a length of about 1.5–2.0 cm were taken. One tissue sample was sectioned consecutively into five pieces; and stained with conventional HE staining, Masson staining, and reticular fibers. A single pathologist read the film uniformly according to the pathological diagnostic criteria and divided the films into a mild fibrosis group (s1–s2) and a severe fibrosis group (s4–s5) according to Ishak scoring criteria (Figure 2).
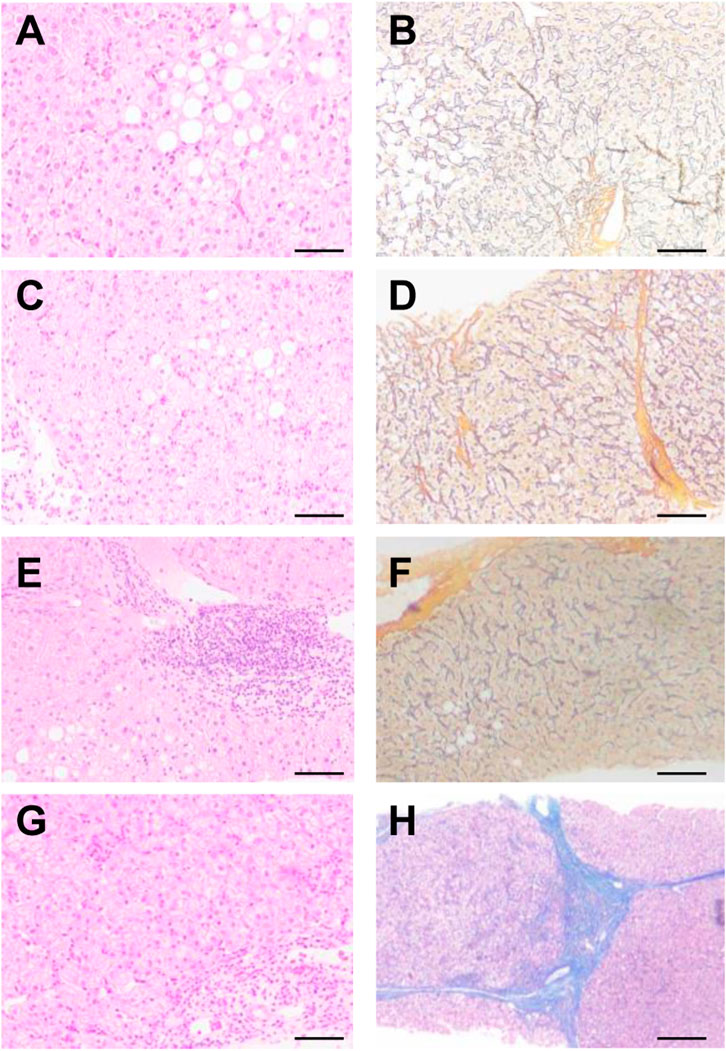
FIGURE 2. Pathological analysis of patients with different levels of liver fibrosis. According to Ishak scoring (A,B) s1, (C,D) s2, (E,F) s4, (G,H) s5. (A,C,E,G) HE, (B,D,F,H) Masson.
METTL16 is differentially expressed in the mild and severe fibrosis groups
Quantitative PCR was carried out to detect the expression level of a series of m6A methyltransferase regulator genes. METTL16 expression was significantly higher in the severe group than in the mild group (Figure 3A). The expression of other m6A demethyl regulators were also checked, and there was no statistically significant differences. Then we detected the m6A modification level of total RNA in the two groups by LC/MS and found that it was significantly (more than 2 times) higher in the severe group than in the mild group (Figure 3B).
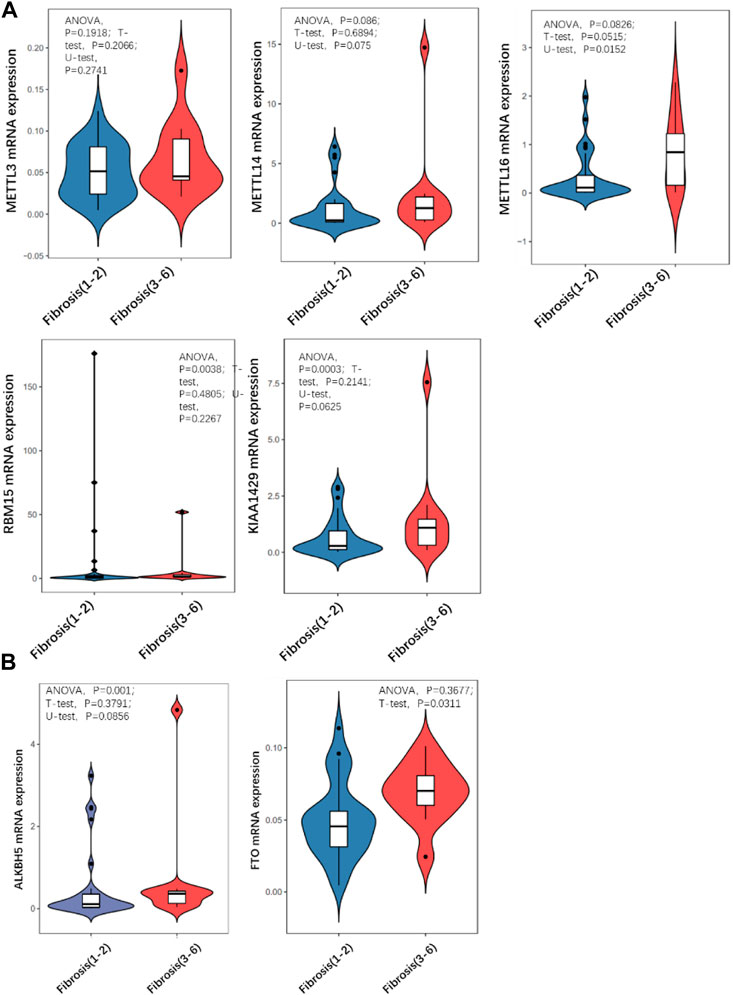
FIGURE 3. Comparison of expression level of m6A regulator in mild and severe fibrosis groups. (A) writers, (B) erasers. Note that METTL16 is significantly up-regulated in severe fibrosis group.
HLA-DPB1 is differentially expressed in fibrosis groups
As mentioned earlier, SNPs related to CHB are located in different genes in the genome according to GWAS. The expression of 15 genes was detected in each sample by quantitative RT-PCR. A total of eight genes were significantly differentially expressed in the two groups of samples. That is, HLA-DPA1, HLA-DPB1, HLA-DPB2, HLA-DQB2, ITPR3, and NUP205 were upregulated in the severe group. In contrast, HSD17B8, RING1, and SKIV2L were downregulated in the severe group (Figure 4A).
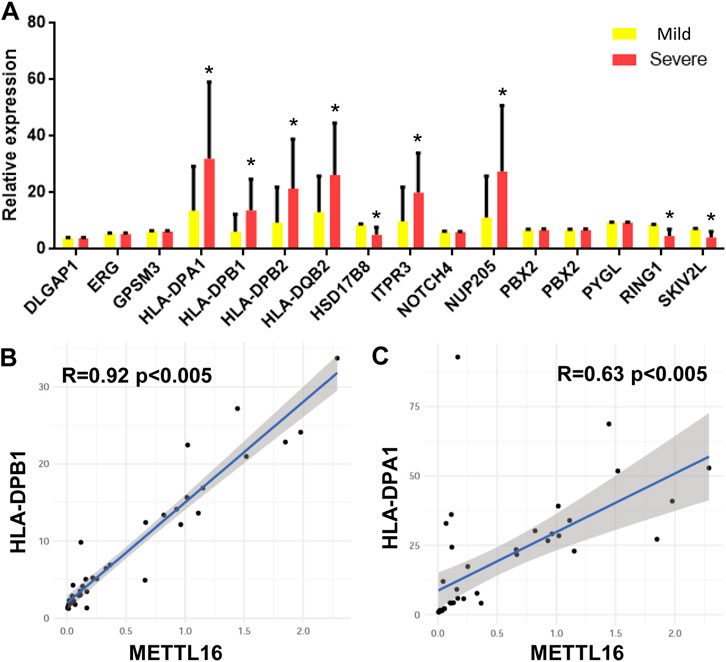
FIGURE 4. CHB GWAS genes differentially expressed between mild and severe fibrosis groups. (A) expression level of CHB GWAS genes. Note that HLA-DPB1 is up-regulated in severe fibrosis group. (B,C) METTL16 is co-expressed with HLA-DPB1 and HLA-DPA1.
The relationships between these differentially expressed genes and the expression of m6A regulators were analyzed by Pearson correlation analysis. mettl16 was significantly positively correlated with HLA-DPB1 and HLA-DPA1 (Figures 4B,C).
There are different levels of m6A on HLA-DPB1 in the mild and severe fibrosis groups
It was suggesting that the expression of HLA-DPB1 is related to the level of RNA m6A. The m6A level of HLA-DPB1 in each sample was detected by MeIP qPCR. The level of m6A on HLA-DPB1 mRNA was significantly increased in the severe group than in the mild group (Figure 5A).
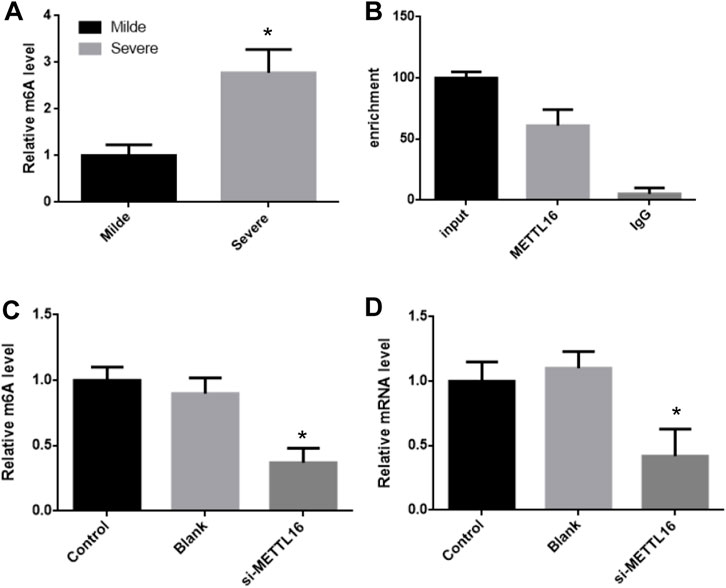
FIGURE 5. METTL16 control HLA-DPB1 expression through regulating m6A level. (A) m6A level of HLA-DPB1 mRNA between mild and severe fibrosis groups. (B) METTL16 interact with HLA-DPB1 mRNA. (C,D) knock-down METTL16 reduced m6A and mRNA of HLA-DPB1 in hepatic stellate cells.
METTL16 interacts with HLA-DPB1 mRNA
The m6A level of HLA-DPB1 mRNA was consistent with its expression in each group and was also related to the expression of mettl16. This implies that mettl16 may be one of the causes of the difference in m6A level and expression of HLA-DPB1. First RNAip experiments showed that mettl16 could bind to HLA-DPB1 mRNA (Figure 5B).
Then we silenced the expression of METTL16 in hepatic stellate cells and detected the expression of HLA-DPB1 and the degree of m6A modification. In the METTL16 silencing group, the m6A level of HLA-DPB1 mRNA was significantly downregulated by more than 2 times (Figures 5C,D).
Discussion
Molecular genetics research on CHB has revealed a large amount of genetic information that is of great value for obtaining a complete understanding of the pathogenesis of CHB and the development of innovative treatments. Especially in the past 2 decades of population genetics research, a large number of SNPs related to susceptibility to and progression of CHB have been found through GWAS. Most of these studies have been conducted in Asian populations, and their conclusions are well targeted. The high prevalence of CHB in Asia can be further understood from these research results. The SNPs found in these GWASs are mainly concentrated in HLA loci, including HLA-DPA1, -DPB1, -DQB2, and -DPB2. As an important gene group that regulates the body’s immune response, the HLA complex participates in the anti-HBV immune response, affects the chronicity of HBV infection and the strength of the immune response, and participates in the progression of CHB to cirrhosis and liver cancer. Therefore, the expression of these genes is likely closely related to CHB. In our study, we found that HLA-DPA1 and HLA-DPB1 differed significantly in groups with different degrees of liver fibrosis. This result suggests that the expression of these two genes may be involved in mediating the progression of CHB. In addition, we found that other CHB-related loci, such as HSD17b8, ITPR3, NUP205, RING1, and SKIV2l, were upregulated or downregulated in different ways in the groups with different degrees of liver fibrosis. This shows that controlling the expression of CHB-related genes at the transcriptional level is of great significance for regulating the progression of CHB. However, we found a large number of CHB-associated SNPs found in GWAS were located in the noncoding region of the locus, which suggests that these genes may be involved in regulating CHB at the transcriptional level rather than the function of the encoded protein. In conclusion, our data show that genes with CHB-associated SNPs can participate in the mechanism of CHB through transcriptional regulation.
m6A modification plays an vital role in transcriptional regulation in eukaryotes. The stability, transportation, splicing, and translation efficiency of mRNA are closely related to the degree of m6A modification. This modification is regulated by the complex. METTL3, METTL14, WTAP, and KIAA1429 form the “writer,” whereas alkbh4 and FTO form the “eraser.” These usually regulate the modification of mRNA in the coding region and the 3′ or 5′ end. Recent studies have found that RNA has m6A modifications in the intron region, which affects the splicing of mRNA. Mettl16 is a key methyltransferase whose precursor mrnam6a modification affects intron cleavage. In our study, key regulatory factors of m6A, especially mettl16, were differentially expressed in tissues with different degrees of liver fibrosis, although other m6A regulators did not differ significantly. This shows that m6A participates in the regulation of CHB mainly through mett16. However, the GWASs summarized above found that SNPs associated with CHB are mainly located in the noncoding region of the gene. This is consistent with the function of mettl16. We further found that mettl16 could bind to HLA-DPB1 mRNA and change its m6A modification level and expression. In clinical samples, the expression of METTL16 was also correlated with HLA-DPB1. All these findings suggest that mettl16 may affect CHB by regulating the expression of these CHB-associated loci, a new mechanism in the process of CHB that needs to be analyzed further.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by the ethics committee of Mengchao Hepatobiliary Hospital of Fujian Medical University. The patients/participants provided their written informed consent to participate in this study.
Author contributions
HG, ML, and DL designed the project and wrote the manuscript. HG, XW, and HM did almost molecular experiments. SL, DZ, WW, ZL, MC, and QL helped data analysis and revised manuscript.
Funding
Fujian Medical Innovation Project (2020CXB038); Fujian Natural Science Foundation (2021J011289); Fuzhou Science and technology plan project (2021-S-099; 2021-S-114; 2020-WS-130).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.996245/full#supplementary-material
References
Akcay, I. M., Katrinli, S., Ozdil, K., Doganay, G. D., and Doganay, L. (2018). Host genetic factors affecting Hepatitis B infection outcomes: Insights from genome-wide association studies. World J. Gastroenterol. 24 (30), 3347–3360. doi:10.3748/wjg.v24.i30.3347
Bartosovic, M., Molares, H. C., Gregorova, P., Hrossova, D., Kudla, G., and Vanacova, S. (2017). N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res. 45 (19), 11356–11370. doi:10.1093/nar/gkx778
Chen, M., Wei, L., Law, C. T., Tsang, F. H., Shen, J., Cheng, C. L., et al. (2018). RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatol. Baltim. Md) 67 (6), 2254–2270. doi:10.1002/hep.29683
Cheng, D., Wu, C., Li, Y., Liu, Y., Mo, J., Fu, L., et al. (2022). METTL3 inhibition ameliorates liver damage in mouse with Hepatitis B virus-associated acute-on-chronic liver failure by regulating miR-146a-5p maturation. Biochim. Biophys. Acta. Gene Regul. Mech. 1865 (3), 194782. doi:10.1016/j.bbagrm.2021.194782
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Imam, H., Khan, M., Gokhale, N. S., McIntyre, A. B. R., Kim, G. W., Jang, J. Y., et al. (2018). N6-methyladenosine modification of Hepatitis B virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci. U. S. A. 115 (35), 8829–8834. doi:10.1073/pnas.1808319115
Kim, G. W., Moon, J. S., and Siddiqui, A. (2022). N6-methyladenosine modification of the 5' epsilon structure of the HBV pregenome RNA regulates its encapsidation by the viral core protein. Proc. Natl. Acad. Sci. U. S. A. 119 (7), e2120485119. doi:10.1073/pnas.2120485119
Kim, G. W., and Siddiqui, A. (2022). Hepatitis B virus X protein expression is tightly regulated by N6-methyladenosine modification of its mRNA. J. Virol. 96 (4), e0165521. doi:10.1128/JVI.01655-21
Kim, G. W., and Siddiqui, A. (2021). Hepatitis B virus X protein recruits methyltransferases to affect cotranscriptional N6-methyladenosine modification of viral/host RNAs. Proc. Natl. Acad. Sci. U. S. A. 118 (3), e2019455118. doi:10.1073/pnas.2019455118
Kim, G. W., and Siddiqui, A. (2021). The role of N6-methyladenosine modification in the life cycle and disease pathogenesis of Hepatitis B and C viruses. Exp. Mol. Med. 53 (3), 339–345. doi:10.1038/s12276-021-00581-3
Kostyusheva, A., Brezgin, S., Glebe, D., Kostyushev, D., and Chulanov, V. (2021). Host-cell interactions in HBV infection and pathogenesis: The emerging role of m6A modification. Emerg. Microbes Infect. 10 (1), 2264–2275. doi:10.1080/22221751.2021.2006580
Lau, K. C., Lam, C. W., Law, C. Y., Lai, S. T., Tsang, T. Y., Siu, C. W., et al. (2011). Non-invasive screening of HLA-DPA1 and HLA-DPB1 alleles for persistent Hepatitis B virus infection: Susceptibility for vertical transmission and toward a personalized approach for vaccination and treatment. Clin. Chim. Acta. 412 (11-12), 952–957. doi:10.1016/j.cca.2011.01.030
Liu, Z., and Zhang, J. (2018). Human C-to-U coding RNA editing is largely nonadaptive. Mol. Biol. Evol. 35 (4), 963–969. doi:10.1093/molbev/msy011
Lok, A. S. (2002). Chronic Hepatitis B. N. Engl. J. Med. 346 (22), 1682–1683. doi:10.1056/NEJM200205303462202
Ma, J. Z., Yang, F., Zhou, C. C., Liu, F., Yuan, J. H., Wang, F., et al. (2017). METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatol. Baltim. Md) 65 (2), 529–543. doi:10.1002/hep.28885
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Perz, J. F., Armstrong, G. L., Farrington, L. A., Hutin, Y. J., and Bell, B. P. (2006). The contributions of Hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J. Hepatol. 45 (4), 529–538. doi:10.1016/j.jhep.2006.05.013
Qu, S., Jin, L., Huang, H., Lin, J., Gao, W., and Zeng, Z. (2021). A positive-feedback loop between HBx and ALKBH5 promotes hepatocellular carcinogenesis. BMC Cancer 21 (1), 686. doi:10.1186/s12885-021-08449-5
Raza, S. A., Clifford, G. M., and Franceschi, S. (2007). Worldwide variation in the relative importance of Hepatitis B and hepatitis C viruses in hepatocellular carcinoma: A systematic review. Br. J. Cancer 96 (7), 1127–1134. doi:10.1038/sj.bjc.6603649
Schweitzer, A., Horn, J., Mikolajczyk, R. T., Krause, G., and Ott, J. J. (2015). Estimations of worldwide prevalence of chronic Hepatitis B virus infection: A systematic review of data published between 1965 and 2013. Lancet (London, Engl. 386 (10003), 1546–1555. doi:10.1016/S0140-6736(15)61412-X
Tong, J., Zhang, W., Chen, Y., Yuan, Q., Qin, N. N., and Qu, G. (2022). The emerging role of RNA modifications in the regulation of antiviral innate immunity. Front. Microbiol. 13, 845625. doi:10.3389/fmicb.2022.845625
Wang, Y., and Zhou, X. (2022). N(6)-methyladenosine and its implications in viruses. Genomics Proteomics Bioinforma. S1672-0229(22), 00083–3. doi:10.1016/j.gpb.2022.04.009
Wu, F., Cheng, W., Zhao, F., Tang, M., Diao, Y., and Xu, R. (2019). Association of N6-methyladenosine with viruses and related diseases. Virol. J. 16 (1), 133. doi:10.1186/s12985-019-1236-3
Wu, F., Cheng, W., Zhao, F., Tang, M., Diao, Y., and Xu, R. (2020). Association of N6-methyladenosine with viruses and virally induced diseases. Front. Biosci. 25 (6), 1184–1201. doi:10.2741/4852
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61 (4), 507–519. doi:10.1016/j.molcel.2016.01.012
Xu, K., Yang, Y., Feng, G. H., Sun, B. F., Chen, J. Q., Li, Y. F., et al. (2017). Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 27 (9), 1100–1114. doi:10.1038/cr.2017.100
Yamada, N., Shigefuku, R., Sugiyama, R., Kobayashi, M., Ikeda, H., Takahashi, H., et al. (2014). Acute Hepatitis B of genotype H resulting in persistent infection. World J. Gastroenterol. 20 (11), 3044–3049. doi:10.3748/wjg.v20.i11.3044
Zhao, B., Wang, W., Zhao, Y., Qiao, H., Gao, Z., and Chuai, X. (2021). Regulation of antiviral immune response by N (6)-methyladenosine of mRNA. Front. Microbiol. 12, 789605. doi:10.3389/fmicb.2021.789605
Zhao, T., Qi, J., Liu, T., Wu, H., and Zhu, Q. (2022). N6-Methyladenosine modification participates in the progression of hepatitis B virus-related liver fibrosis by regulating immune cell infiltration. Front. Med. 9, 821710. doi:10.3389/fmed.2022.821710
Keywords: M6A, METTL16, CHB (chronic hepatitis B), HBV-hepatitis B virus, GWAS
Citation: Gao H, Wang X, Ma H, Lin S, Zhang D, Wu W, Liao Z, Chen M, Li Q, Lin M and Li D (2022) METTL16 regulates m6A methylation on chronic hepatitis B associated gene HLA-DPB1 involved in liver fibrosis. Front. Genet. 13:996245. doi: 10.3389/fgene.2022.996245
Received: 17 July 2022; Accepted: 19 October 2022;
Published: 04 November 2022.
Edited by:
Xiao Han, Fuzhou University, ChinaReviewed by:
Lili Xu, Beijing Children’s Hospital, Capital Medical University, ChinaXianlin Han, Peking Union Medical College Hospital (CAMS), China
Copyright © 2022 Gao, Wang, Ma, Lin, Zhang, Wu, Liao, Chen, Li, Lin and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongliang Li, dongliangli93@163.com; Minghua Lin, drlmh543@126.com
†These authors have contributed equally to this work
 Haibing Gao
Haibing Gao Xiangmei Wang1†
Xiangmei Wang1† Shenglong Lin
Shenglong Lin