- 1Department of Medical Oncology, The Affiliated Huaian No.1 People's Hospital of Nanjing Medical University, Huai'an, China
- 2Department of Medical Oncology, Huai'an Hongze District People's Hospital, Huai'an, China
A 78 years old Chinese woman with five different cancer types and a family history of malignancy was the subject of this study. Pancreatic adenocarcinoma and gingival squamous cell carcinoma tissues were obtained from the patient and sequenced using Whole Exome Sequencing. Whole exome sequencing identified 20 mutation sites in six candidate genes. Sanger Sequencing was used for further validation. The results verified six mutations in three genes, OBSCN, TTN, and RPGRIP1L, in at least one cancer type. Immunohistochemistry was used to verify protein expression. mRNA expression analysis using The Cancer Genome Atlas database revealed that RPGRIP1L was highly expressed in several cancer types, especially in pancreatic adenocarcinoma, and correlated with patient survival and sensitivity to paclitaxel, probably through the TGF-β signaling pathway. The newly identified somatic mutations in RPGRIP1L might contribute to pathogenesis in the patients. Protein conformation simulation demonstrated that the alterations had caused the binding pocket at position 708 to change from concave to convex, which could restrict contraction and extension, and interfere with the physiological function of the protein. Further studies are required to determine the implication of RPGRIP1L in this family and in multiple primary tumors.
Introduction
Multiple primary tumors (MPTs) are a phenomenon rarely clinically seen. MPTs are defined as two or more malignancies simultaneously or successively occurring in an individual (Vogt et al., 2017). The incidence of MPTs is higher in the elderly, especially in people aged 50–60 (Patrascu et al., 2010). Despite understanding that multiple factors including unhealthy lifestyle (Hori et al., 2011), chemoradiotherapy (Oeffinger et al., 2013), and genetic mutations (Park et al., 2014), are involved in the development of MPTs, the pathogenesis of MPTs remains unclear. Deleterious germline mutations and defects in DNA repair genes are closely related to MPTs (Tiwari et al., 2016). Recently, Xu et al. identified a homozygous germline insertion mutation in WWOX, a DNA repair-related gene, from a 26 years old female patient with MPTs (Xu et al., 2019). Nevertheless, further investigation is required to clarify the molecular mechanisms underlying MPTs.
Materials and Methods
Clinical Information and Samples in the Study
The patient is a 78 years old Chinese woman, who had successively developed five different cancer types. This patient developed hepatocellular carcinoma in 2000 (for which she received interventional therapy in the liver), colorectal adenocarcinoma in 2002 (for which she received a partial colon resection), invasive breast carcinoma in 2011 (for which she received radical mastectomy of the left breast), gingival squamous cell carcinoma in 2018 (for which she received right side gingival tumor resection), and pancreatic adenocarcinoma in 2018 (for which she received a partial pancreatectomy). Based on the patients' report, none of the cancers were treated with radiotherapy or chemotherapy. The patient was admitted with a right cervical mass in November 2019 and mass biopsy was performed in The Affiliated Huaian No.1 People's Hospital of Nanjing Medical University. The pathology results showed metastatic squamous cell carcinoma that had originated from the gingival tumor and the patient received chemoradiotherapy in our hospital. Moreover, the patient had a very complicated family history of malignancy, which showed that the patient's mother died of esophageal squamous cell carcinoma, and her two brothers (the patient's uncles) also died of malignancies (with no exact details). The patient had two older sisters, two older brothers, and a younger brother. Her oldest brother died of leukemia, and her second oldest brother, who had a history of cardia carcinoma, was alive. Her younger brother died of cardia carcinoma and had a history of gingival squamous cell carcinoma. However, the rest of her family, including her father, two sisters, and her four children, had no cancer history (Supplementary Figure 1). Therefore, it is valuable to discuss the etiology and pathogenesis of this case. This study was approved by the Clinical Research Ethics Committee of The First Affiliated Hospital of University of Science and Technology of China (Protocol number: P-015). The patient and her family members provided written informed consent for study participation. A written, informed consent was obtained from the participant for the publication of this case report. Our study followed the institutional ethical guidelines approved by The Affiliated Huaian No.1 People's Hospital of Nanjing Medical University (Huai'an, China).
DNA Extraction
Sample loss and damage meant that only two cancer samples (gingival and pancreatic) were available for further sequencing. The patient's blocks of gingival squamous cell carcinoma and pancreatic adenocarcinoma after resection, as well as her peripheral blood samples were collected. Genomic DNA of the samples was isolated using the FastPure FFPE DNA Isolation Kit (Vazyme, Nanjing, China), and purified using 1% Sepharose electrophoresis. DNA purity and concentration were measured using a NanoPhotometer® spectrophotometer (IMPLEN, CA, USA) and Qubit® 3.0 Flurometer (Life Technologies, CA, USA).
Whole Exome Sequencing and Data Analysis
To prepare the Illumina sequencing libraries, the SureSelect Human All Exon kit V6 (Agilent Technologies, Santa Clara, CA, USA) was used. Genomic DNA samples (3 μg) were randomly fragmented into 150–200 bp fragments using a Covaris S2 sonicator (Covaris, Woburn, MA, USA) and subjected to library preparation according to the Sure Select XT Target Enrichment System (Agilent Technologies, Santa Clara, CA, USA). In brief, DNA fragments had deoxyadenosine bases added to their 3′ ends, were ligated with paired-end adaptors, and amplified by PCR. The enriched libraries were sequenced with the Illumina NovaSeq 6000 platform and 150-bp paired-end reads were generated. The reads were aligned to the GRCh37/hg19 reference genome using BWA (http://bio-bwa.sourceforge.net/bwa.shtml). Picard tools (picard.sourceforge.net, version 1.46, MarkDuplicates) were applied to remove duplicates. The base quality of the reads was recalibrated using the GATK Haplotype Caller, which was also used to identify germline and somatic mutations and variations. Called SNV and InDel variants were annotated using the ANNOVAR package. FastQCv0.11.7 software was used to assess the sequencing data quality. Candidate somatic mutations were filtered based on international filtering criteria. Then, the filtered SNVs and InDels were screened with an alternative allele frequency <1% in public databases including the 1000 Genome Project and ESP 6500. Synonymous mutations were removed and deleterious mutations were retained with standard SIFT Score ≤ 0.05, PolyPhen-2 Sore ≥ 0.909, and Mutation Taster Score ≥ 0.85.
Sanger Sequencing Validation
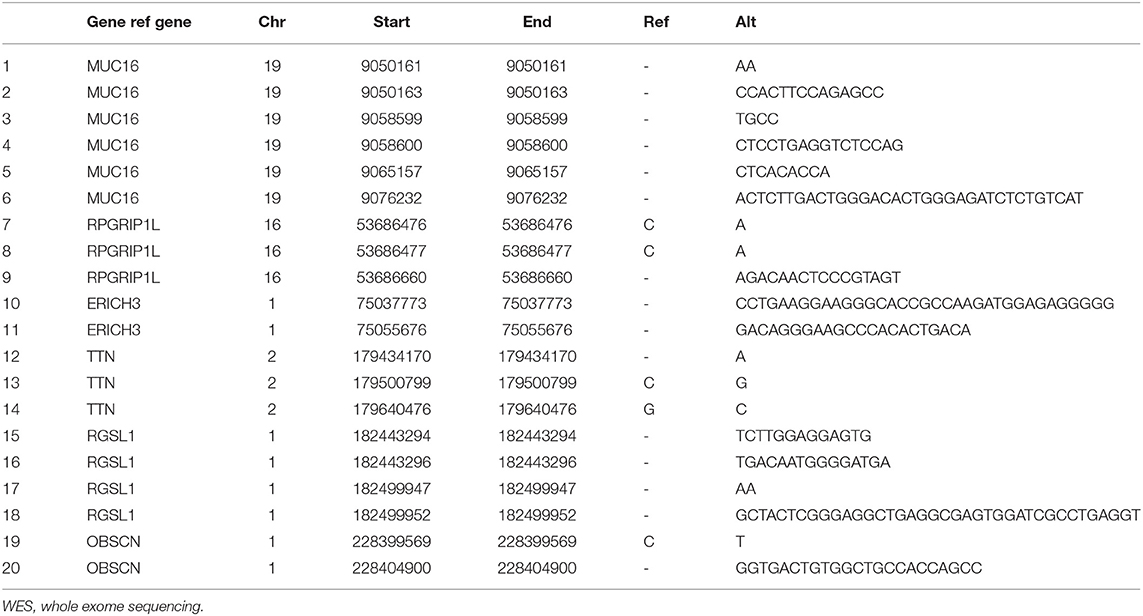
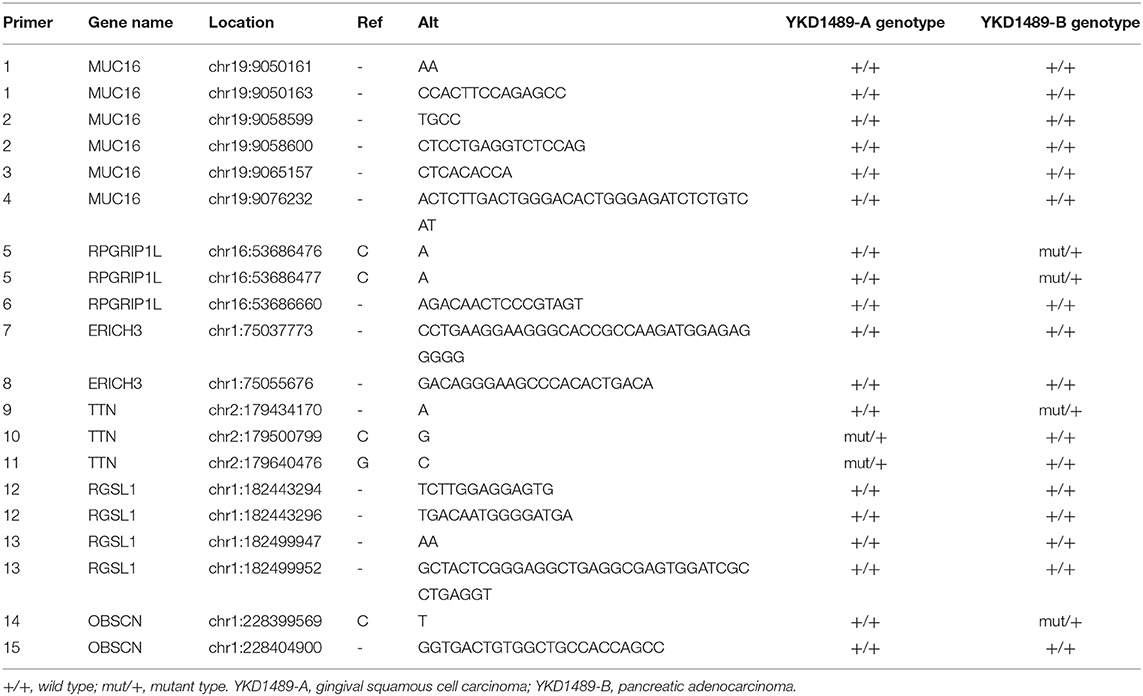
Shared non-synonymous SNVs and InDels identified in the two cancers were amplified using PCR Master Mix (Illumina, San Diego, CA, USA) and subjected to Sanger sequencing. Among the 20 mutation sites, five of them were close to another five target sites which could be amplified by the same primers. Therefore, we designed 15 primer pairs. Sequencing primers were designed using Primer3 (v.0.4.0) software and were generated by Sangon Biotech (Shanghai, China; Table 1).
Immunohistochemistry
Samples of the patients' pancreatic adenocarcinoma and gingival squamous cell carcinoma were further analyzed by immunohistochemistry. Paraffin-embedded sections were deparaffinized and rehydrated, and sections were covered with Tris-EDTA (TE) buffer and heated for 10 min for antigen retrieval. Sections were incubated overnight at 4°C with an anti-RPGRIP1L antibody (1:200; 55160-1-AP, Proteintech, Wuhan, China), rinsed with phosphate buffered saline, and incubated with a secondary antibody (anti-rabbit) at 37°C for 30 min. The sections were finally incubated with 3,3′-diaminobenzidine and stained with hematoxylin. RPGRIP1L expression was identified by pathologists using microscopy.
Bioinformatics Analysis
The Cancer Genome Atlas (TCGA) database was used to verify the expression of candidate genes in different cancers. The prognostic value of the hub genes was analyzed by TCGA, KM-plotter and an online tool called OncoLnc (http://www.oncolnc.org/). The pancreatic adenocarcinoma patients from the online database were divided into two groups at the optimal node based on the P value distribution curve. Co-expression analysis was performed with the condition of absolute correlation coefficient > 0.3 and P < 0.001. Gene-set enrichment analysis (GESA) and gene set variation analysis (GSVA) were conducted to identify possible signaling pathways, biological processes, and drug resistances related to the gene function. Statistical analysis was performed using R (version 3.6) and a P-value < 0.05 was considered significant.
Protein Conformation Simulation
The RPGRIP1L structure information was obtained from the UniProt database (https://ebi10.uniprot.org/uniprot/Q68CZ1/), which showed existing crystal structure data obtained using the NMR method (PDB Entry: 2YRB, Chain: A, Positions: 595–737). The NMR structure of RPGRIP1L was acquired from the PDB database. The PRGRIP1L amino acid alterations were p.G708C and p.G708V. Protein conformation simulations were performed using the SWISS-MODEL online tool (https://swissmodel.expasy.org/). PyMOL software was used for structure visualization.
Results
Five Distinct Cancers Successively Developed in an Individual Patient
This 78-year-old Chinese woman was first admitted to our hospital on November 2nd, 2019 with a right cervical mass. This mass was confirmed to be the result of metastasis of her previously diagnosed gingival squamous cell carcinoma. While taking her history, we discovered that she had a history of five different cancer types and a complicated family history. The five different cancer types were hepatocellular carcinoma in 2000, colorectal adenocarcinoma in 2002, invasive breast carcinoma in 2011, gingival squamous cell carcinoma in 2018, and pancreatic adenocarcinoma in 2018. Unfortunately, all of her cancers were treated in different hospitals and the first three cancer sample blocks were lost or damaged. Diagnosis of her first three cancers was based on her previous medical records and imaging examinations. The latter two cancer types, gingival squamous cell carcinoma and pancreatic adenocarcinoma, were confirmed by immunohistochemical methods (Figure 1).
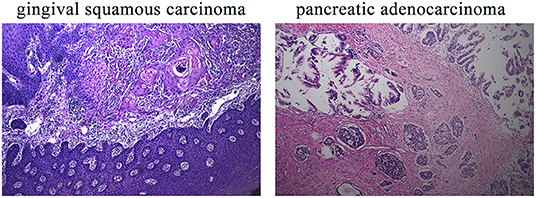
Figure 1. IHC results of the patient's gingival squamous carcinoma and pancreatic adenocarcinoma. IHC, immunohistochemistry. Magnification for all images: 20×.
Candidate Variants Detected From WES
DNA extracted from the patient's samples was sequenced using WES. The results of her peripheral blood were applied to deduction of background variants. This identified 221 SNVs and InDels in her gingival squamous cell carcinoma and 281 SNVs and InDels in her pancreatic adenocarcinoma. Her peripheral blood results were used as a control. Comparison and analysis of the results revealed six common candidate genes with 20 mutation sites for further validation (Table 2).
Validation of Mutation Sites by Sanger Sequencing
WES analysis identified a total of 20 variants in six genes, ERICH3, OBSCN, RGSL1, TTN, RPGRIP1L, and MUC16. These variants were subjected to confirmation using Sanger sequencing. Sanger Sequencing results confirmed two mutation sites in RPGRIP1L, one of TTN, and one of OBSCN in the patient's pancreatic adenocarcinoma, and two mutation sites in TTN in her gingival squamous cell carcinoma (Table 3).
Expression of the RPGRIP1L Protein in Cancers
Sanger sequencing confirmed that RPGRIP1L, TTN, and OBSCN were mutated in this patient. We used the Human Protein Atlas (https://www.proteinatlas.org/) to analyze the level of protein expression from the three genes in multiple cancers. This analysis showed that RPGRIP1L is highly expressed in liver cancer, breast cancer, and pancreatic cancer, while expression of TTN and OBSCN was not markedly observed (Figure 2). The image sources are listed in Table 4. We also analyzed RPGRIP1L, TTN, and OBSCN mRNA expression using Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia2.cancer-pku.cn/) tools. Results of this analysis were consistent with the observed protein expression results and showed that RPGRIP1L mRNA was more highly expressed than both TTN, and OBSCN mRNA in cancers (Figure 4A). Identification of high levels of RPGRIP1L expression in pancreatic cancer prompted us to extend this finding by analyzing RPGRIP1L protein expression in the patients' cancer samples. Immunostaining for RPGRIP1L in her pancreatic cancer tissues revealed predominantly cytoplasmic staining in most of the cancer cells, which demonstrated that RPGRIP1L protein expression was higher in her pancreatic adenocarcinoma tissues than adjacent pancreatic tissues, but was slightly lower in her gingival squamous cell carcinoma tissues than in normal gingival tissues (Figure 3).
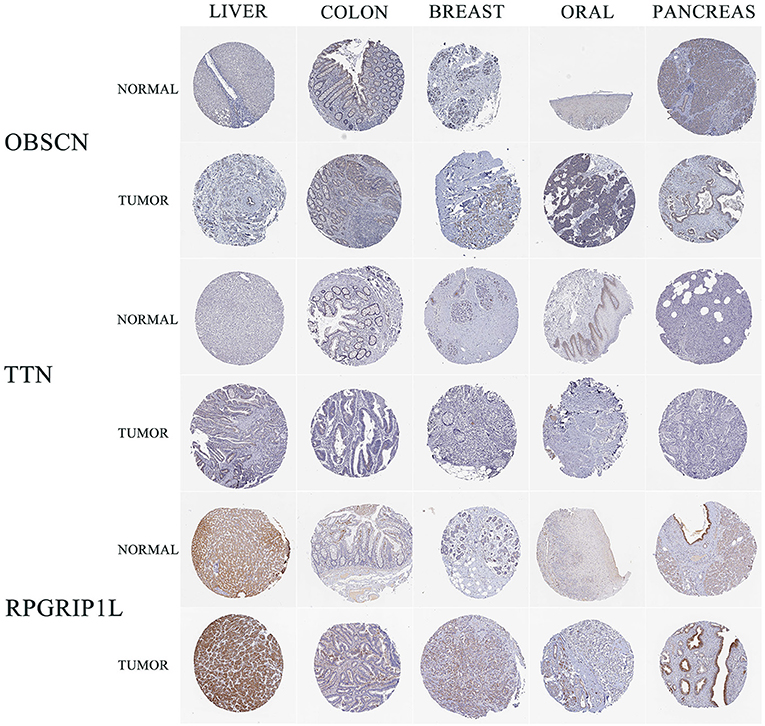
Figure 2. IHC results of three genes in five cancer types from HPA. IHC, immunohistochemistry; HPA, Human Protein Atlas; LIHC, liver hepatocellular carcinoma; COAD, colon adenocarcinoma; BRCA, breast invasive carcinoma; HNSC, head and neck squamous cell carcinoma; PAAD, pancreatic adenocarcinoma.
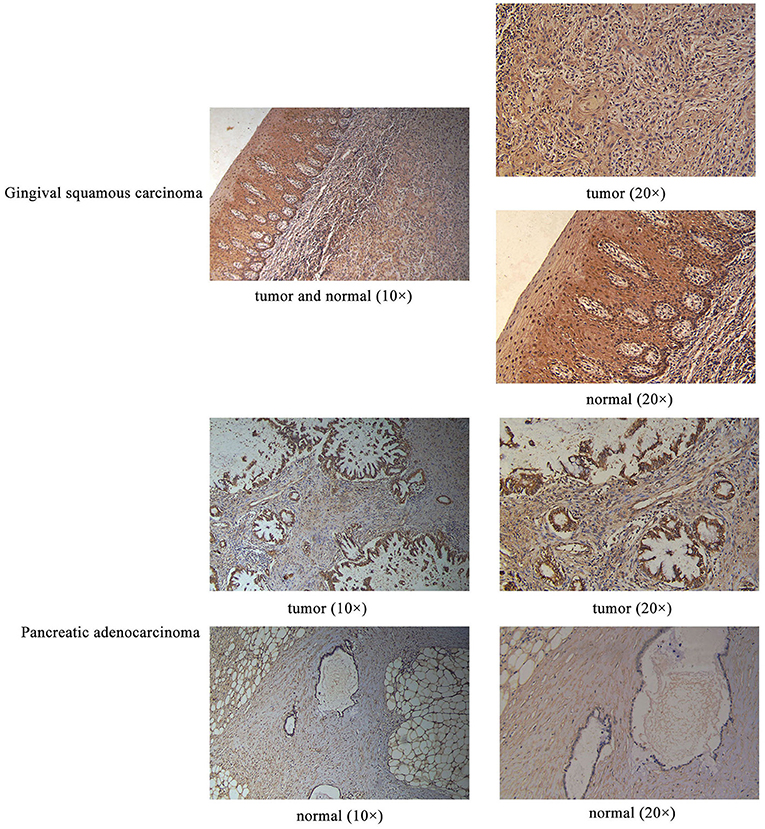
Figure 3. IHC results of RPGRIP1L protein expression in the patient's gingival squamous carcinoma and pancreatic adenocarcinoma. Magnification for all images: 20×.
Bioinformatic Analyses Identify RPGRIP1L as a Potential Susceptibility Gene
Bioinformatics analyses were applied to explore the role of RPGRIP1L in pancreatic adenocarcinoma. In combination with the TCGA dataset, expression analysis revealed that RPGRIP1L mRNA was significantly upregulated in 179 pancreatic adenocarcinoma patients, (Figure 4A). The patients were then divided into two groups based on the minimum P-value of the curve. Using the Kaplan-Meier Plotter (KM, http://www.kmplot.com/) tool, survival analysis showed that RPGRIP1L expression was closely related to survival in patients with pancreatic adenocarcinoma (Figure 4B). Next, we performed GESA (Subramanian et al., 2005) and GSVA (Hanzelmann et al., 2013). The GESA results showed that RPGRIP1L expression corelated with three pathways: circadian rhythm, ECM-receptor interaction, and TGF-β signaling pathways (Figures 4D,F). GSVA showed that RPGRIP1L expression was associated with several biological processes including apoptosis, angiogenesis, and in part of classic signaling pathways involved in cancer development including the TGF-β, KRAS, PI3K-Akt-mTOR, p53, and Wnt/β-catenin signaling pathways (Figure 4E). As chemotherapy is commonly used to treat pancreatic adenocarcinoma, we estimated the chemotherapeutic response of patients with pancreatic adenocarcinoma using the R package “pRRophetic” (Geeleher et al., 2014). This analysis was based on the largest pharmacogenomics database GDSC (https://www.cancerrxgene.org/). This estimation was based on the half maximal inhibitory concentration (IC50), which revealed that RPGRIP1L expression was markedly associated with paclitaxel, one of the most widely used clinical anti-tumor drugs in cancer chemotherapy (Figure 4C). Protein conformation simulation was conducted using information from UniProt and PDB databases, and revealed that, in this patient, the glycine (Gly) at RPGRIP1L protein position 708 had changed to cysteine (Cys) or valine (Val) due to the somatic mutations in RPGRIP1L. The simulation demonstrated that these alterations changed the binding pocket structure from concave to convex. This could restrict contraction and extension, and interfere with the physiological function of the protein (Figure 5).
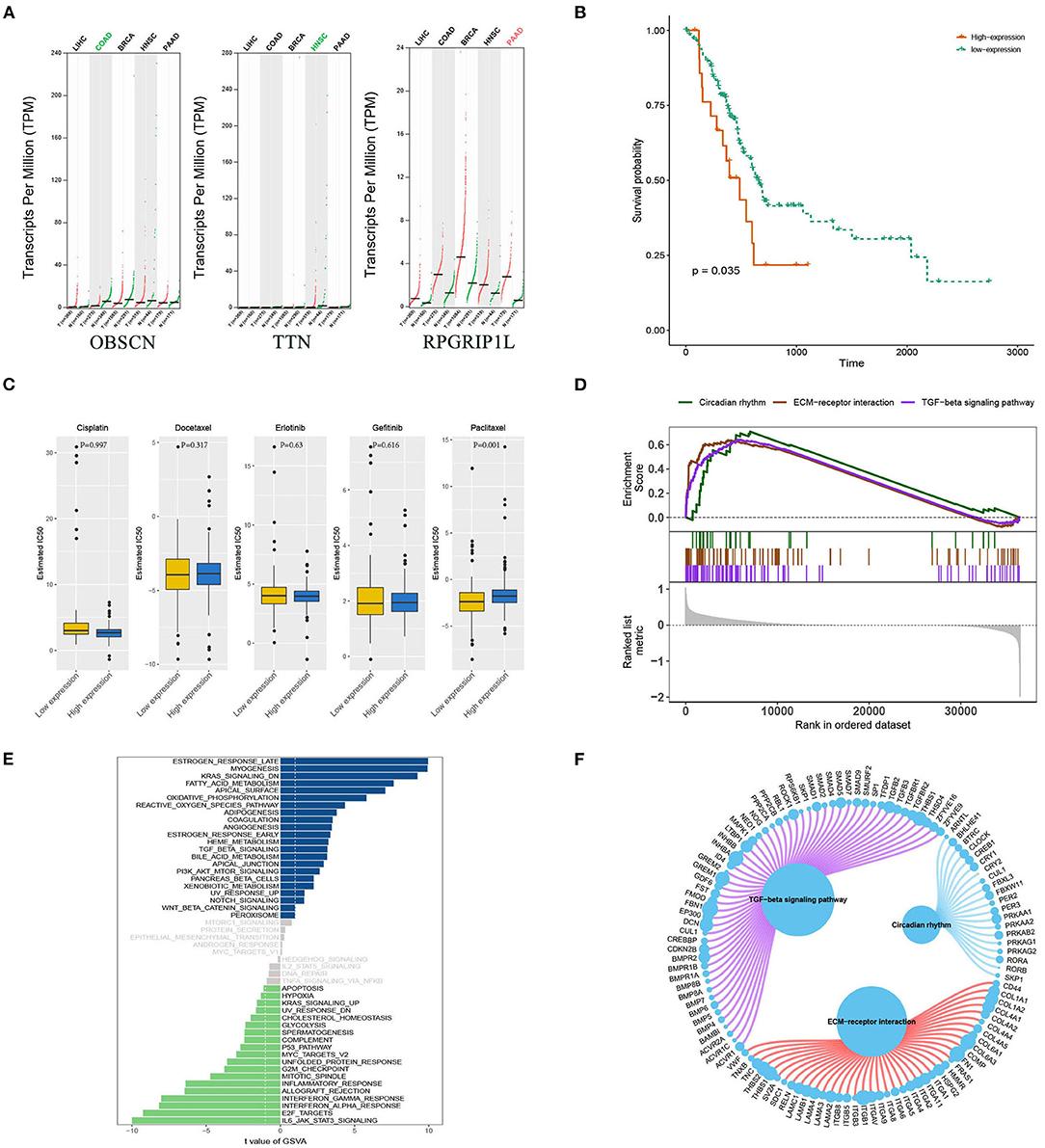
Figure 4. Bioinformatic analyses of RPGRIP1L gene expression and function. (A) Analysis of OBSCN, TTN, and RPGRIP1L mRNA expressions in five cancer types from TCGA database. The red and green colors of cancer types mean significantly difference (red, upregulated in tumor; green, downregulated in tumor, all compared with normal tissues). (B) The RPGRIP1L expression is related with PAAD patients' survival. (C) The RPGRIP1L expression is related with patients' sensitivity to paclitaxel. (D) GESA results showed that the RPGRIP1L expression was corelated with three pathways including circadian rhythm, ECM-receptor interaction, and TGF-β signaling pathways. (E) GSVA showed that RPGRIP1L expression was associated with several biological processes. Blue part means low expression, green part means high expression, gray part means no difference of RPGRIP1L expression. (F) GESA results of predicted RPGRIP1L binding targets in circadian rhythm, ECM-receptor interaction, and TGF-β signaling pathways. TCGA, The Cancer Genome Atlas; PAAD, pancreatic adenocarcinoma; GESA, gene-set enrichment analysis; GSVA, gene set variation analysis.
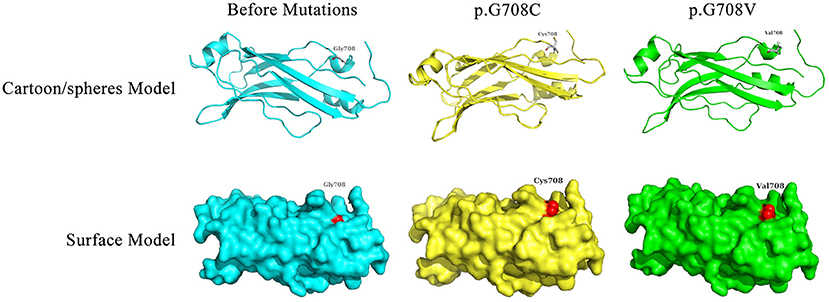
Figure 5. Display of the predicted protein structures before/after mutations of RPGRIP1L at position 708. The red color indicated the amino acid mutation site. Gly, glycine; Cys, cysteine; Val, valine.
Discussion
This study investigated the molecular alterations in a case of MPTs. RPGRIP1L was identified as a candidate gene with nonsynonymous SNVs (chr16:53686476, C>A; chr16:53686477, C>A) as a somatic mutation candidate. Moreover, bioinformatic analyses revealed that RPGRIP1L expression was related to patient' survival and chemosensitivity to paclitaxel in pancreatic adenocarcinoma, presumably occurring through the TGF-β signaling pathway.
Recently, great achievements have been made in the diagnosis and treatment of cancers. These advances have contributed to more satisfactory survival times and higher incidence of MPTs. It is generally accepted that inherited genetic defects are most likely to cause MPTs (Chan et al., 2018). A recent paper reported a rare variant within a PARP4 pseudogene (PARP4P2) in a patient with MPTs and familial cancer history. They suggested that the PARP4P2 pseudogene variant could induce PARP4 down-regulation, which might confer susceptibility to the development of multiple metachronous cancers (Cirello et al., 2019). In this study, our results represent newly discovered somatic mutations in the RPGRIP1L gene of a patient with MPTs and family history.
The retinitis pigmentosa GTPase regulator interacting protein 1-like (RPGRIP1L), also known as Ftm, localizes to the basal body-centrosome complex or to primary cilia and centrosomes in ciliated cells, is highly conserved (Wiegering et al., 2018). RPGRIP1L negative embryos show a variety of defects caused by cilia dysfunction (Vierkotten et al., 2007; Gerhardt et al., 2015). Defects in this gene can affect the development of several organs (Chen et al., 2015; Andreu-Cervera et al., 2019; Wang et al., 2019) and result in multiple diseases, including Joubert syndrome (JBTS) (Arts et al., 2007) and Meckel syndrome (MKS) (Delous et al., 2007). Previous studies revealed that RPGRIP1L plays an important role in the assembly of the transition zone, a region of the cilia (Jensen et al., 2015). RPGRIP1L was previously shown to govern the function of the Proteasome 26S Subunit, Non-ATPase 2 (PSMD2) by interacting with it and controlling the ciliary signaling through affecting ciliary proteasome activity (Gerhardt et al., 2015). Furthermore, it was recently demonstrated that RPGRIP1L deficiency impairs Hedgehog (Hh)/Gli signaling. RPGRIP1L can region-specifically influence mouse forebrain development through Hh signaling (Andreu-Cervera et al., 2019). RPGRIP1L was also reported to affect autophagy activity. Struchtrup et al. reported that absence of RPGRIP1L inhibits the initiation and later steps in the autophagy process, e.g., the generation of autophagosomes, which occurs via cilia-mediated mTOR signaling activation (Struchtrup et al., 2018). Recently, mutations in RPGRIP1L were reported in obesity and brain function. One study showed that rs13334070, in RPGRIP1L intron 4, has a significant association with obesity (Javanrouh et al., 2019). Reble et al. recently identified a SNV, rs7203525, that influences an alternative splicing event in RPGRIP1L, increasing exon 20 inclusion and potentially impacting brain function (Reble et al., 2020).
Using WES, our study discovered two novel RPGRIP1L SNVs in a case with MPTs, which were further validated by Sanger sequencing. Since the two SNVs were found only in the pancreatic adenocarcinoma tissues, and that the mutations occurred in exons, they are likely to influence translation efficiency, alternative splicing, and DNA copy number (El Marabti and Younis, 2018), and may be related to RPGRIP1L protein expression levels. However, the relationship between the SNVs and the observed expression differences needed to be validated further.
Previous studies had reported downregulation of RPGRIP1L in human hepatocellular carcinoma and suggested it was a tumor suppressor gene. Downregulation of RPGRIP1L increased Mitotic arrest deficient 2 (Mad2) protein levels, resulting in tumor cell transformation (Lin et al., 2009). Moreover, interaction between RPGRIP1L and Myosin Va, which was reported to be increased in several cancers, was detected at the ciliary base, indicating that RPGRIP1L might regulate the amount of Myosin Va and suppress tumorigenesis (Assis et al., 2017; Wiegering et al., 2018). Conversely, based on the above analysis and description, we contended that RPGRIP1L might act as a tumor promoter gene in pancreatic adenocarcinoma, since high RPGRIP1L expression was observed, and was closely related to patient's survival, TGF-β signaling pathway, and sensitivity to the chemotherapeutic drug paclitaxel. Additionally, protein conformation simulation analysis revealed that the identified mutations might affect the binding ability of the protein and impact downstream targets. In combination with the previous studies, our results suggest that RPGRIP1L might play a promoter or suppresser role in a cancer type-specific manner.
Finally, our study has some limitations. Samples of the patient's other three cancer tissues were not available, and the novel RPGRIP1L mutations were not found in gingival squamous cell carcinoma. One conceivable explanation is that her gingival squamous cell carcinoma might be caused by dietary habit and lack of oral care. Indeed, people from this region have a higher incidence of esophageal squamous cell carcinoma because of their preference for pickled and hot foods. The patient also suffered from diabetes, which appeared to be linked to oral cancer (Mekala et al., 2020). Moreover, samples from her family members were not available as well. Due to her complicated family history of malignancy, we could not exclude the possibility that there were germline mutations inherited by the patient. Further investigations are required to clarify the potential role of RPGRIP1L mutations in tumorigenesis and its value as a therapeutic target.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Clinical Research Ethics Committee of The First Affiliated Hospital of University of Science and Technology of China. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
MH and XS designed the research. JG and YY performed the experiments. ZJ provided the clinical samples. ZJ, MY, and XX analyzed and interpreted the data. JG wrote the manuscript. MH critically commented and edited the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the Six-One project in Jiangsu Province (No. LGY2017051).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank professor Juncheng Dai from Department of Epidemiology, School of Public Health, Nanjing Medical University, who offered technical support and guidance for this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.620472/full#supplementary-material
Supplementary Figure 1. The pedigree tree of the family.
References
Andreu-Cervera, A., Anselme, I., Karam, A., Laclef, C., Catala, M., and Schneider-Maunoury, S. (2019). The ciliopathy gene Ftm/Rpgrip1l controls mouse forebrain patterning via region-specific modulation of hedgehog/gli signaling. J. Neurosci. 39, 2398–2415. doi: 10.1523/JNEUROSCI.2199-18.2019
Arts, H. H., Doherty, D., van Beersum, S. E., Parisi, M. A., Letteboer, S. J., Gorden, N. T., et al. (2007). Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat. Genet. 39, 882–888. doi: 10.1038/ng2069
Assis, L. H., Silva-Junior, R. M., Dolce, L. G., Alborghetti, M. R., Honorato, R. V., Nascimento, A. F., et al. (2017). The molecular motor Myosin Va interacts with the cilia-centrosomal protein RPGRIP1L. Sci. Rep. 7:43692. doi: 10.1038/srep43692
Chan, G. H. J., Ong, P. Y., Low, J. J. H., Kong, H. L., Ow, S. G. W., Tan, D. S. P., et al. (2018). Clinical genetic testing outcome with multi-gene panel in Asian patients with multiple primary cancers. Oncotarget 9, 30649–30660. doi: 10.18632/oncotarget.25769
Chen, J., Laclef, C., Moncayo, A., Snedecor, E. R., Yang, N., Li, L., et al. (2015). The ciliopathy gene Rpgrip1l is essential for hair follicle development. J. Invest. Dermatol. 135, 701–709. doi: 10.1038/jid.2014.483
Cirello, V., Colombo, C., Pogliaghi, G., Proverbio, M. C., Rossi, S., Mussani, E., et al. (2019). Genetic variants of PARP4 gene and PARP4P2 pseudogene in patients with multiple primary tumors including thyroid cancer. Mutat. Res. 816–818:111672. doi: 10.1016/j.mrfmmm.2019.111672
Delous, M., Baala, L., Salomon, R., Laclef, C., Vierkotten, J., Tory, K., et al. (2007). The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat. Genet. 39, 875–881. doi: 10.1038/ng2039
El Marabti, E., and Younis, I. (2018). The cancer spliceome: reprograming of alternative splicing in cancer. Front. Mol. Biosci. 5:80. doi: 10.3389/fmolb.2018.00080
Geeleher, P., Cox, N., and Huang, R. S. (2014). pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE 9:e107468. doi: 10.1371/journal.pone.0107468
Gerhardt, C., Lier, J. M., Burmuhl, S., Struchtrup, A., Deutschmann, K., Vetter, M., et al. (2015). The transition zone protein Rpgrip1l regulates proteasomal activity at the primary cilium. J. Cell Biol. 210, 115–133. doi: 10.1083/jcb.201408060
Hanzelmann, S., Castelo, R., and Guinney, J. (2013). GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14:7. doi: 10.1186/1471-2105-14-7
Hori, K., Okada, H., Kawahara, Y., Takenaka, R., Shimizu, S., Ohno, Y., et al. (2011). Lugol-voiding lesions are an important risk factor for a second primary squamous cell carcinoma in patients with esosphageal cancer or head and neck cancer. Am. J. Gastroenterol. 106, 858–866. doi: 10.1038/ajg.2010.489
Javanrouh, N., Soltanian, A. R., Tapak, L., Azizi, F., Ott, J., and Daneshpour, M. S. (2019). A novel association of rs13334070 in the RPGRIP1L gene with adiposity factors discovered by joint linkage and linkage disequilibrium analysis in Iranian pedigrees: Tehran Cardiometabolic Genetic Study (TCGS). Genet. Epidemiol. 43, 342–351. doi: 10.1002/gepi.22179
Jensen, V. L., Li, C., Bowie, R. V., Clarke, L., Mohan, S., Blacque, O. E., et al. (2015). Formation of the transition zone by Mks5/Rpgrip1L establishes a ciliary zone of exclusion (CIZE) that compartmentalises ciliary signalling proteins and controls PIP2 ciliary abundance. EMBO J. 34, 2537–2556. doi: 10.15252/embj.201488044
Lin, Y. W., Yan, M. D., Shih, Y. L., and Hsieh, C. B. (2009). The basal body gene, RPGRIP1L, is a candidate tumour suppressor gene in human hepatocellular carcinoma. Eur. J. Cancer 45, 2041–2049. doi: 10.1016/j.ejca.2009.04.012
Mekala, M. R., Bangi, B. B., Lebaka, J. N. R. R., Nadendla, L. K., and Ginjupally, U. (2020). Association of diabetes with oral cancer- an enigmatic correlation. Asian Pac. J. Cancer Prev. 21, 809–814. doi: 10.31557/APJCP.2020.21.3.809
Oeffinger, K. C., Baxi, S. S., Novetsky Friedman, D., and Moskowitz, C. S. (2013). Solid tumor second primary neoplasms: who is at risk, what can we do? Semin. Oncol. 40, 676–689. doi: 10.1053/j.seminoncol.2013.09.012
Park, S. L., Caberto, C. P., Lin, Y., Goodloe, R. J., Dumitrescu, L., Love, S. A., et al. (2014). Association of cancer susceptibility variants with risk of multiple primary cancers: the population architecture using genomics and epidemiology study. Cancer Epidemiol. Biomarkers Prev. 23, 2568–2578. doi: 10.1158/1055-9965.EPI-14-0129
Patrascu, T., Doran, H., Catrina, E., Mihalache, O., Degeratu, D., and Predescu, G. (2010). Synchronous tumors of the gastrointestinal tract. Chirurgia 105, 93–96. doi: 10.1186/s12957-016-0882-9
Reble, E., Feng, Y., Wigg, K. G., and Barr, C. L. (2020). DNA variant in the RPGRIP1L gene influences alternative splicing. Mol. Neuropsychiatry 5(Suppl.1), 97–106. doi: 10.1159/000502199
Struchtrup, A., Wiegering, A., Stork, B., Ruther, U., and Gerhardt, C. (2018). The ciliary protein RPGRIP1L governs autophagy independently of its proteasome-regulating function at the ciliary base in mouse embryonic fibroblasts. Autophagy 14, 567–583. doi: 10.1080/15548627.2018.1429874
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. doi: 10.1073/pnas.0506580102
Tiwari, A. K., Roy, H. K., and Lynch, H. T. (2016). Lynch syndrome in the 21st century: clinical perspectives. QJM 109, 151–158. doi: 10.1093/qjmed/hcv137
Vierkotten, J., Dildrop, R., Peters, T., Wang, B., and Ruther, U. (2007). Ftm is a novel basal body protein of cilia involved in Shh signalling. Development 134, 2569–2577. doi: 10.1242/dev.003715
Vogt, A., Schmid, S., Heinimann, K., Frick, H., Herrmann, C., Cerny, T., et al. (2017). Multiple primary tumours: challenges and approaches, a review. ESMO Open 2:e000172. doi: 10.1136/esmoopen-2017-000172
Wang, L., De Solis, A. J., Goffer, Y., Birkenbach, K. E., Engle, S. E., Tanis, R., et al. (2019). Ciliary gene RPGRIP1L is required for hypothalamic arcuate neuron development. JCI Insight 4:123337. doi: 10.1172/jci.insight.123337
Wiegering, A., Ruther, U., and Gerhardt, C. (2018). The ciliary protein Rpgrip1l in development and disease. Dev. Biol. 442, 60–68. doi: 10.1016/j.ydbio.2018.07.024
Keywords: RPGRIP1L, multiple primary tumors, somatic mutation, pancreatic adenocarcinoma, whole-exome sequencing
Citation: Guo J, Yang Y, Ji Z, Yao M, Xia X, Sha X and Huang M (2021) Case Report: Novel RPGRIP1L Gene Mutations Identified by Whole Exome Sequencing in a Patient With Multiple Primary Tumors. Front. Genet. 12:620472. doi: 10.3389/fgene.2021.620472
Received: 23 October 2020; Accepted: 07 January 2021;
Published: 01 February 2021.
Edited by:
Loredana Bury, University of Perugia, ItalyReviewed by:
Roberta La Starza, University of Perugia, ItalyRick Kamps, Maastricht University, Netherlands
Copyright © 2021 Guo, Yang, Ji, Yao, Xia, Sha and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingde Huang, mdhdoctor@126.com; Xiaofeng Sha, 1045736787@qq.com
†These authors share first authorship
 Jiani Guo
Jiani Guo Yu Yang1†
Yu Yang1†