- 1Institution of Genomics and Bioinformatics, South China Agricultural University, Guangzhou, China
- 2Plant Germplasm and Genomics Center, Germplasm Bank of Wild Species in Southwest China, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 3University of Chinese Academy of Sciences, Beijing, China
The WRKY gene family is widespread in plants, which plays key roles in plant development and stress response. Although WRKY transcription factors have been widely characterized in many plants, a genome-wide analysis of the WRKY gene family is lacking in Oryza rufipogon. In this study, we identified 101 O. rufipogon WRKY (OrWRKY) transcription factors, which were further classified into eight subgroups. Phylogenetic analysis showed that OrWRKY transcription factors were supported by highly conserved motifs and gene structures. Chromosomal distribution of OrWRKYs indicated that most genes were dispersed on all 12 chromosomes, especially enriched on Chromosome 1. Syntenic analysis revealed that 69 (68.3%) genes were derived from either segmental (49) or tandem duplication events (20), suggesting a pivotal role of segmental duplications. We totally characterized 39 orthologous gene pairs between Oryza sativa ssp. japonica WRKY (OsjWRKY) and OrWRKY genes. We performed quantitative real-time polymerase chain reaction (qRT-PCR) experiments to validate tissue-specific and differential expression of the OrWRKYs and corresponsive expression of the OrWRKYs in response to salt stresses in leaves and roots. This study provides a new insight on the evolution of the WRKY gene family in O. rufipogon and will help further functional characterization of candidate genes toward wild rice germplasm exploration for rice genetic improvement programs.
Introduction
Among the genus Oryza comprising 23 species and nine recognized genome types (Vaughan et al., 2003), common wild rice, Oryza rufipogon Griff., is, besides the assumed annual wild progenitor Oryza nivara, another presumed perennial wild progenitor of Asian cultivated rice, Oryza sativa (Oka, 1988; Khush, 1997; Cheng et al., 2003; Fuller et al., 2010; Huang X. et al., 2012), which feeds half of the human population in the world. As a result of the domestication under intensive human cultivation, O. sativa has undergone considerable phenotypic and physiological changes, leading to an extensive loss of genetic diversity through successive bottlenecks and artificial selection for agronomic traits compared to its wild progenitors (Kovach et al., 2007; Xu et al., 2012). Oryza rufipogon spans a broad geographical range of global pantropical regions (Morishima and Barbier, 1990) and, for example, extensively grows in diverse natural habitats in South China (Gao et al., 1996; Gao, 2004). As a promising donor for genetic improvement of cultivated rice, many of alien genes associated with morphological traits, quality traits, abiotic stress- and disease resistance-related traits have successfully been introduced through introgression lines from O. rufipogon, which has helped expand the rice gene pool important to the generation of environmentally resilient and high-yielding varieties (Brar and Ramos, 2008).
Salt stress is one of the most devastating abiotic stresses in rice, and the salt-affected soils currently account for about 20% of the total paddy rice planting area (Zhou Y. et al., 2016). More seriously, the increased salinization of arable land is expected to have devastating global impacts, resulting in 30% land loss within the next 25 years and up to 50% by the year 2050 (Wang et al., 2003). In recent years, lots of studies have shown valuable insight into the molecular and cellular mechanisms by which rice responds to and tolerate salinity (Schmidt et al., 2013; Zhu et al., 2015). However, the regulatory mechanisms involved in coordinating salt stress tolerance and plant growth are not fully understood. Plants have developed a series of complex, effective systems to keep themselves from numerous adverse environmental fluctuations during growth. And studies have shown that these coping mechanisms mainly function via specific transcriptional activation or inhibition of transcription-related genes. Transcription factors play an essential role in this process by binding to specific DNA regions (Singh et al., 2002).
The WRKY transcription factors family, one of the largest families in higher plants, plays an essential role in plant development and stress processes. Since the first WRKY gene (SPF1) was identified and cloned in potato (Huang S. et al., 2012), a large number of WRKY genes in other plants were increasingly reported through genome-wide analyses (Zhang and Wang, 2005; Bi et al., 2016; Zhou H. et al., 2016). WRKY proteins, which can identify and bind to W-box cis-regulatory elements, are about 60 amino acids in length and contain one or two highly conserved WRKYGQK motifs and a typical zinc-finger structure (Eulgem et al., 2000). Besides the conserved sequences of a WRKYGQK motif, some variants including WRKYGEK, WKKYGQK, and WRKYDHK can also be found in plants. And the zinc-finger motif is either Cx4-5Cx22-23HxH (C2H2) or Cx7Cx23HxC (C2HC) (Tang et al., 2013; Guo et al., 2014). Previous studies suggested that the WRKY gene family could be divided into three subgroups based on the number of WRKY domains and the type of zinc-finger motif. Group I contains two WRKY domains and one zinc-finger structure, which can be classified into Ia and Ib subgroups based on the variants in zinc-finger motifs. Ia harbors two WRKY domains and C2H2 structure, while Ib has two WRKY domains and C2HC motif (Xu et al., 2016). Group II has one WRKY domain and a C2H2 (Cx4-5Cx22-23HxH) type of zinc-finger structure. On the basis of variants in zinc-finger motif, Group II can also be further divided into five subgroups, including II-a, II-b, II-c, II-d, and II-e. Proteins from Group III have only one WRKY domain but with a C2HC motif (Eulgem et al., 2000).
WRKY transcription factors are involved in morphology forming processes, such as seed development, germination, and flower induction. For example, ScWRKY1, isolated from Solanum chacoense, was related to seed development and highly expressed in fertilized ovules at late torpedo-staged embryos (Eulgem et al., 1999). In Arabidopsis, the mutant with AtWRKY39 knockdown had less germination compared with wild-type plants (Li et al., 2010). A total of 18 DlWRKY genes in longan showed a specific expression during three stages of flower induction, indicating that these genes may be involved in the flower induction and the genetic control of the perpetual flowering trait (Jue et al., 2018). Except for plant development, WRKY genes also play important roles in biotic and abiotic stress responses including fungal-induced defense programs and responses to heat/cold, drought, and salt stresses (Raineri et al., 2015; Yang et al., 2018; Jimmy and Babu, 2019). As one of the most important functions, the WRKY gene family seems to regulate the response to salt stress. For example, ZmWRKY33 was induced by salt treatments and overexpressed in Arabidopsis, which was more salt tolerant than the wild type (Li et al., 2013). GarWRKY17 and GarWRKY104 overexpression plants in Arabidopsis showed enhanced salt tolerance during different developmental stages (Fan et al., 2015). Besides, the transgenic Nicotiana tabacum seedlings expressing VvWRKY2 from grapevine exhibited strong resistance to salt treatments (Mzid et al., 2018).
Due to diverse and important functions of WRKY genes in various physiological and developmental processes, many WRKY genes have been functionally studied in rice (Xie et al., 2005; Wu et al., 2009; Xu et al., 2016). However, little is known about WRKY TFs and their responses to salt stresses in O. rufipogon. In this study, we comprehensively characterized the WRKY gene family with an emphasis on genome-wide identification of salt-responsive members of the WRKY family in O. rufipogon. The results of this study enhance our understanding of the OrWRKY gene family, which will facilitate the genetic improvement of salt stress tolerance in rice.
Materials and Methods
Identification of Putative OrWRKY Genes
The genome sequences of O. rufipogon, which was sampled in Yuanjiang, Yunnan Province, China, were generated by our lab (Li et al., 2020), and the HMM file (PF03106) of WRKY domain was obtained from the Pfam database1. To comprehensively identify the OrWRKYs, HMMER 3.0 software2 was used to search against the rice protein sequences with default parameters. The incorrect and redundant predicted sequences were manually removed, and then all putative OrWRKY genes were further verified using Pfam database3. The molecular weight (MW) and isoelectric point (PI) were evaluated using ExPASy-ProtParam online software4. Meanwhile, the protein sequences of O. sativa ssp. japonica cv. Nipponbare and O. sativa ssp. indica were downloaded from Phytozome5 and plantfdb (see text footnote 3), respectively. We employed the same method as described above to identify the candidate WRKY genes. The deduced WRKY genes of O. nivara were named as OnWRKY1 to OnWRKY125 according to Xu et al. (2016). The sequences of Arabidopsis WRKY genes were downloaded from TAIR6. To name the WRKY genes of O. rufipogon and O. sativa ssp. indica (OsiWRKY), we employed the same method (McCouch, 2008; Group, 2012). The proteins of all candidate OrWRKY and OsiWRKY genes were aligned against the OsjWRKY proteins via BLASTP program with E value 1e–13 and then named the OrWRKY and OsiWRKY genes according to their homology to O. sativa ssp. japonica cv. Nipponbare.
Classification of OrWRKY Genes
Based on the number of WRKY domains and the type of zinc-finger motif, the WRKY genes can be classified into three groups (Xie et al., 2005; Ross et al., 2007). Group I contains two WRKY domains and can be divided into two subgroups. Ia has two WRKY domains with C2H2 zinc-finger motifs, while Ib owns C2HC structures. Group II contains one WRKY domain with a C2H2 zinc-finger motif. Based on a specific sequence variation in zinc-finger motifs, Group II can be further divided into five subgroups. Subgroup II-a contains CX5CPVKKK(L/V)Q structure, II-b CX5CPVRKQVQ, II-c CX4C, II-d CX5CPARKHVE, and II-e CX5CPARK(Q/M)V(E/D). Group III has one WRKY domain with a C2HC zin-finger motif. The WRKYs with incomplete domain are assigned to Group IV.
Sequence Alignment and Phylogenetic Analysis
Multiple sequence alignment of the WRKY domains and full-length proteins was performed by CLUSTAL (Larkin et al., 2007). The phylogenetic tree was constructed with MEGA5.0 using a neighbor-joining (NJ) method (Tamura et al., 2011). The bootstrap values were calculated for 1,000 iterations.
Characterization of the Structure and Motif of OrWRKYs
The available information of exons and introns was retrieved from the O. rufipogon genome sequences and then visualized by the GSDS (version 2.0)7 online program with coding sequences and genomic sequences. The motifs of each deduced OrWRKY protein were analyzed by MEME suit software (version 4.12.0)8 (Bailey et al., 2009) with parameters as follows: maximum number of motifs, 10. The upstream 1.5-Kb sequences of WRKY genes were extracted with an in-house Perl script to determine the cis-elements by using the PlantCARE database9.
Determination of Chromosomal Distribution, Gene Duplication, and Synteny
The chromosomal distribution of OrWRKY genes was determined from the annotation file (GFF3) of O. rufipogon. The synteny and segmental and tandem duplications were analyzed using a previously reported method (Tang et al., 2013) and visualized (including gene positions) using the Circos v0.69 (Krzywinski et al., 2009). To date OrWRKY genes derived from duplication events, Ka and Ks values were estimated using the maximum likelihood method implemented in codeml program (Yang, 2007). The divergence time (T) was estimated as T = Ks/2r × 10–6 MYA (million years ago) according to the approximate substitution rate r = 6.5 × 10–9 (Yu et al., 2005). And the branch-site model A in EasyCodeML v1.21 (Gao et al., 2019) was used to further assess positive selection in OrWRKY genes.
Identification of Orthologous WRKY Genes Between O. rufipogon and O. sativa ssp. japonica
In order to identify the putative orthologous WRKY genes between O. rufipogon and O. sativa ssp. japonica, a reciprocal best hit (RBH) method was employed (Xia et al., 2014). Briefly speaking, each sequence from OrWRKY was initially searched against all sequences from OsjWRKYs using Blast, and each sequence of OsjWRKY was in turn searched against all sequences from OrWRKYs. We selected pairs of gene sequences that were the best hits for each other as putative orthologs. Due to the limitation of RBH method, potential paralogs may not be completely excluded from the ortholog dataset. To remove the possible paralogs, we first searched all pairs of gene sequences against all plant protein sequences available in GeneBank and selected only pairs of gene sequences unambiguously mapped to the same protein with an E-value < 1e–13 as orthologous genes. The CDS sequences of each orthologous gene were employed to calculate the synonymous (Ks) and non-synonymous (Ka) substitution rate by codeml program (Yang, 2007) using the maximum likelihood method. To validate whether the orthologous WRKY genes between O. rufipogon and O. sativa ssp. japonica have undergone positive selection, we used the branch-site model A in EasyCodeML v1.21 (Gao et al., 2019).
Quantitative Real-Time PCR Experimental Validation
The O. rufipogon plants were grown in the greenhouse at 28°C to collect expression profiles of WRKY genes under salt treatments. They were transferred into containers filled with 200 mM NaCl solution. Then, the leaves and roots were harvested at 0, 1, 2, 4, and 8 h after treatments and immediately frozen in liquid nitrogen until used. Three seedlings were sampled for each treatment with the three biological replicates. Total RNA was extracted with Trizol reagent (Invitrogen) and treated with RNase-free DNase I (Boehringer Mannheim). And then, the cDNA was synthesized by reverse transcription of 1.0 μg total RNA using MMLV reverse transcriptase (Toyobo) according to manufacturer’s instructions. Gene-specific primers of OrWRKY genes were designed using Primer Premier 5.0 software (Supplementary Table S1). The designed quantitative real-time polymerase chain reaction (qRT-PCR) primer pairs were first verified using BLAST On Line at NCBI, and a dissociation curve was then analyzed after the PCR reaction to confirm their specificity. RT-PCR was performed with a multicolor RT-PCR detection system (Bio-Rad). Each reaction contained 10 μl SYBR, 1.0 μl samples, and 0.8 μl of each primer pair in a final volume of 20 μl. The RT-PCR cycle was as follows: 95°C for 30 s; 40 cycles of 95°C for 5 s, and 60°C for 30 s. The Actin gene was selected as an internal standard for normalization, and three technical replicates were conducted for each sample. The 2–ΔΔCT method (Livak and Schmittgen, 2001) was performed to analyze RT-PCR data, and we used 0 h as an untreated control (expression = 1.0) to calculate the fold change in the expression level of the relevant genes.
Statistic Analysis
All data were expressed as the mean ± SD after normalization. The Fisher’s least significant difference (LSD) analysis at P = 0.05 level was performed by SPSS software (version 18.0).
Results
Identification of OrWRKY Genes
The PF03106 file and HMMER 3.0 software were used to comprehensively identify the OrWRKY genes. We totally detected 101 non-redundant deduced OrWRKY gene members, which were further named as OrWRKY1 to OrWRKY121 according to names of their OsjWRKY orthologs (Supplementary Table S2). Among them, WRKYGQK domain was a highly conserved motif, while other members also contained WRKYGKK, WRKYGEK, WSKYEQK, WRMCGQK, WRMCGQN, WRMCGQS, WRMTGQS, and WIKYGQK domains. In addition, three genes, OrWRKY87.2, OrWRKY96.2, and OrWRKY105, were found to lose the WRKYGQK domain, and the four genes (OrWRKY27, OrWRKY39, OrWRKY49, and OrWRKY116) were found to lose zinc-finger motif. The lengths of the OrWRKY proteins spanned from 97 to 1,209 amino acids, and the detailed information about each OrWRKY gene was shown in Supplementary Table S2.
Classification of the OrWRKY Genes
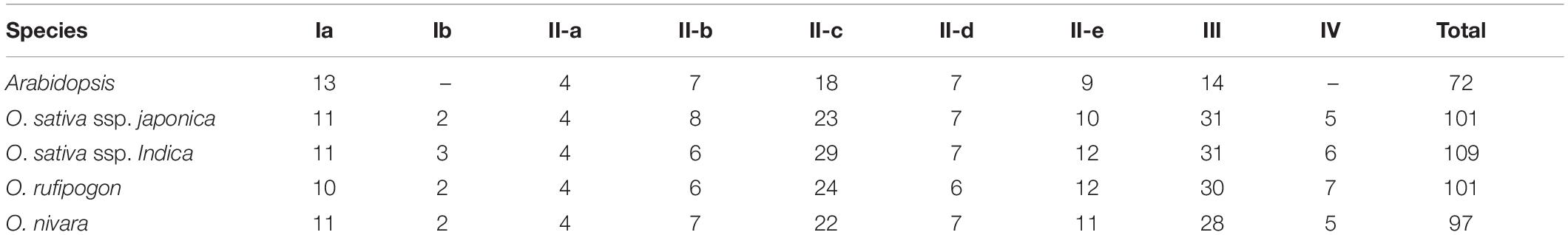
All of the putative WRKY genes could be divided into four groups (i.e., I, II, III, and IV). Table 1 presented a summary of classification for the WRKY gene family from this wild rice genome. Groups I, II, III, and IV had 12, 52, 30, and 7 members, respectively. Group II comprised of five subgroups, and each of subgroup II-a, II-b, II-c, II-d, and II-e contained 4, 6, 24, 6, and 12 members, respectively. In addition, based on the phylogenetic relationships of both WRKY domains and full-proteins (Supplementary Figures S1, S2), all the seven members (OrWRKY27, OrWRKY39, OrWRKY49, OrWRKY87.2, OrWRKY96.2, OrWRKY105, and OrWRKY116) in Group IV could be classified into the other subgroups (Groups II-b, II-e/II-c, II-c, II-c, Ia, II-c, and III, respectively). Detailed information about the classification of OrWRKY genes and the WRKY domains could be found in Supplementary Table S2 and Supplementary Figure S3.
Exon-Intron Organization of OrWRKY Genes
To investigate the evolution of OrWRKY genes, the exon/intron boundaries were analyzed. As illustrated in Figure 1, exon numbers highly varied among OrWRKY genes. The number of exons spanned from 1 to 12, and 52 OrWRKY genes (51.4%) contained three exons, accounting for the largest proportion. Fifteen OrWRKYs had four exons, and 15 OrWRKYs retained two exons, while only one OrWRKY gene contained 12 exons. Note that OrWRKY genes in the same group seemed to have a similar number of exons. For example, most genes in Groups II-b contained five introns, while most II-d and II-e genes had three introns. And Group III contained 30 members, and 23 genes had three introns. These results showed a strong correlation between exon–intron structure and phylogenetic relationship, additionally supporting the classification of the OrWRKY gene family. Based on the position of introns inserted into the conserved WRKY structure, they could be divided into two groups (PR and VQR type). The N-terminal domain of Ia OrWRKYs all contained a VQR type of intron, while the C-terminal domain had a PR-type intron. On the contrary, the two Ib proteins had a PR-type intron in the N-terminal domain. The PR-type intron was widely distributed among the six other subgroups, the same cases were also found in Arabidopsis, grape, and strawberry (Eulgem et al., 2000; Guo et al., 2014; Zhou H. et al., 2016).
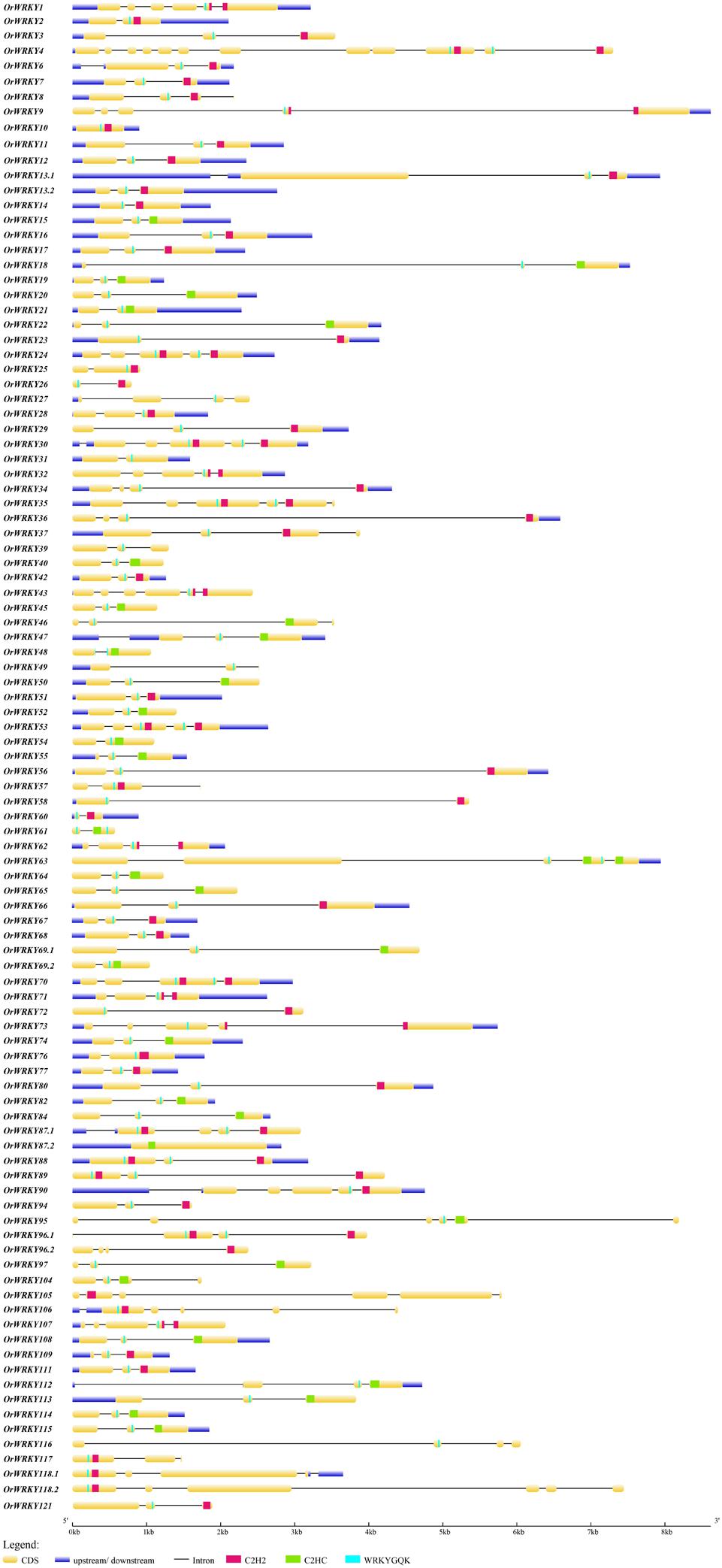
Figure 1. The exon–intron structures of OrWRKY genes. The exons and introns are indicated by yellow rectangles and black lines, respectively. The untranslated regions (UTRs) are indicated by blue rectangles. The WRKY motifs are represented by light blue vertical bars. The C2H2 zinc fingers are represented by red bars. The C2HC zing fingers are represented by green bars.
Phylogenetic Analysis of WRKY Genes Among Arabidopsis, O. sativa, O. nivara, and O. rufipogon
In order to investigate the phylogenetic relationships and evolutionary history of OrWRKYs, WRKY domains from the five plant genomes (Arabidopsis, O. sativa ssp. japonica, O. sativa ssp. indica, O. rufipogon, and O. nivara) were combined into the data analysis. The genome data of cultivated rice were retrieved from Phytozome and plantfdb. WRKY genes of Arabidopsis were downloaded from TAIR, and WRKY sequences of O. nivara were obtained from a previous study (Xu et al., 2016). Sequences with variations in WRKY domains were excluded from the analysis. Finally, an unrooted phylogenetic tree was constructed using the NJ method with MEGA from the 531 conserved WRKY domains from these five genomes. As shown in Supplementary Figure S4, all WRKY domains could be divided into eight subgroups. The N-terminal and C-terminal domains of Group Ia were separately clustered in the phylogenetic tree, suggesting that the N-terminal motifs might not be duplicated by the C-terminal motifs. However, both C-terminal and N-terminal motifs of major Ib were clustered with Group III, showing a relatively closed relationship to it. In addition, similar to what was found in other plants, our results further support that II-a and II-b were clustered into a single subgroup and subgroups II-d and II-e were clustered into the other clade.
When the number of WRKY genes was compared with those of Arabidopsis, O. sativa ssp. japonica, O. sativa ssp. indica, and O. nivara (Table 1), we observed that the number of Group III in all four rice species genomes greatly increased and they had approximately the same number of WRKY genes. The results indicated that a large-scale expansion of Group III members in rice seemingly occurred before the divergence of the three rice species, evidenced by a number of the rice WRKY domains exclusively clustered together in Group III.
Chromosomal Location, Gene Duplication, and Genomic Synteny of OrWRKY Genes
To examine genomic distribution of OrWRKY genes, we analyzed their approximate locations on chromosomes. Almost all of the 101 OrWRKY genes were mapped on the chromosomes except for OrWRKY96.2 because of missing gaps in the genome assembly. Among the 12 chromosomes, as shown in Figure 2, Chr1 contained the most number of OrWRKY (22, 21.7%) genes, followed by Chr 5 (17, 16.8%), Chr 8 (9, 8.9%), and Chr 11 (9, 8.9%). However, Chr 10 only had two OrWRKYs (OrWRKY2 and OrWRKY18).
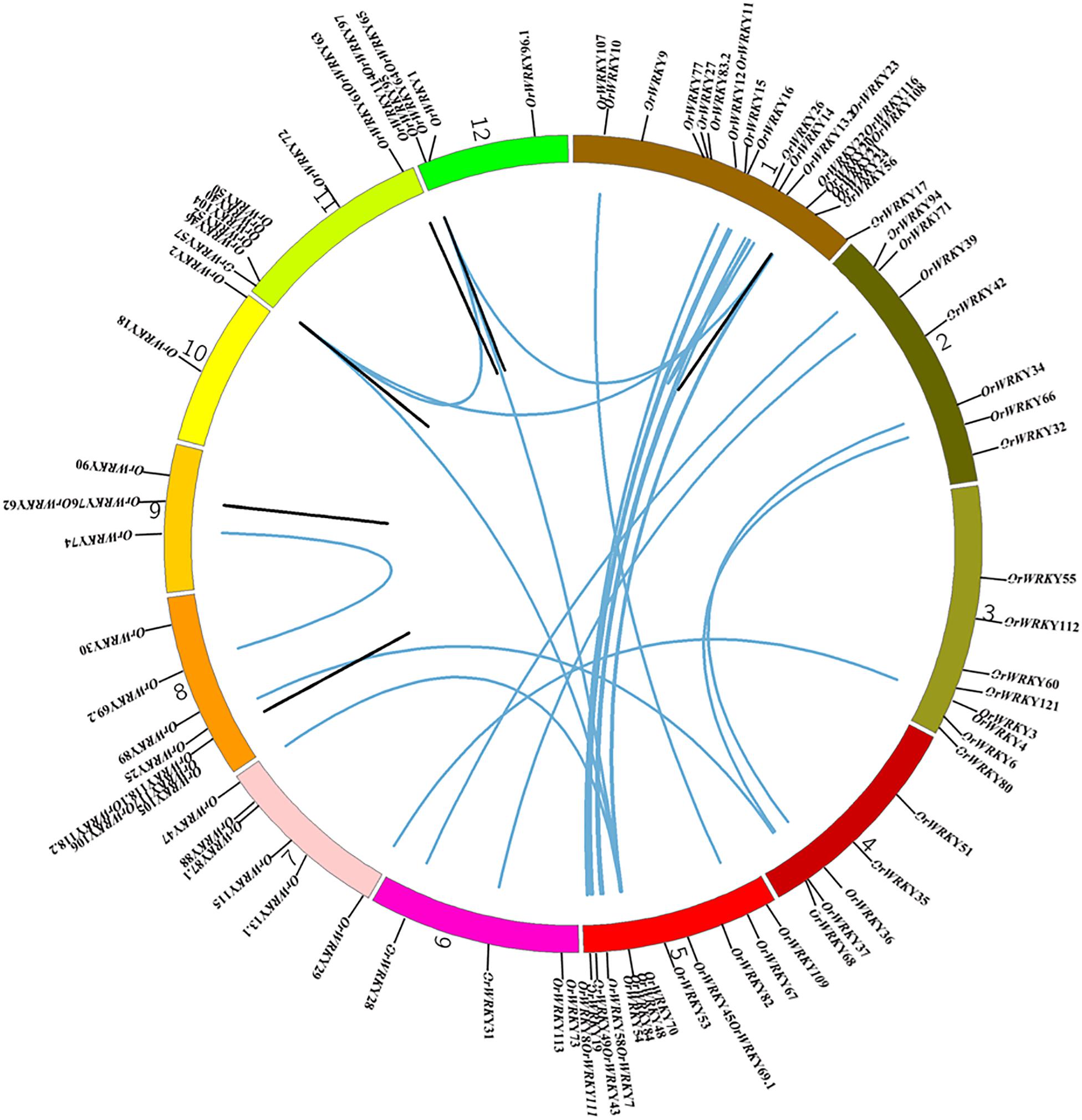
Figure 2. Chromosome distribution and synteny analysis of OrWRKY genes. Chromosomes 1–12 are shown with different colors, and the approximate location of each OrWRKY gene is marked. Blue lines indicate duplicated OrWRKY genes, and black lines in the circle indicate tandemly duplicated genes.
Previous studies suggest that gene duplication events and subsequent positive selection may have played vital roles in gene family expansion and the creation of novel biological functions (Vision et al., 2000; Davidson et al., 2013). In order to investigate the role of gene duplication in the OrWRKY gene family, segmental and tandem duplications were detected throughout the O. rufipogon genome assembly. Among the 101 OrWRKY genes, we found 49 segmentally duplicated genes that formed 29 pairs (Figure 2 and Supplementary Table S3), indicating that segmental duplication events acted as a major force to drive the evolution of the OrWRKY gene family. As shown in Figure 2, chromosomes 1 and 5 had most of WGD-derived OrWRKY duplicated genes, while Chromosome 10 had none of these OrWRKY genes. In addition, pairs of OrWRKY duplicated genes belonged to the same subgroup, most of which (48.3%; 14/29) were clustered into Group III. Moreover, we identified 11 gene pairs generated from tandem duplication events (Figure 2 and Supplementary Table S4), and more than half of them were clustered into Group III.
To further examine adaptive evolution of OrWRKY genes, the Ka/Ks (non-synonymous/synonymous) ratios of the duplicated gene pairs were calculated. The Ka/Ks ratio is often used to estimate selective pressure acting on protein-coding genes. Ka/Ks < 1 shows a sign of purifying (negative) selection, Ka/Ks = 1 means neutral evolution, and Ka/Ks > 1 suggests adaptive (positive selection) (Hurst, 2002). In this study, we found that most of the segmentally duplicated gene pairs had a Ka/Ks value < 1 with an average Ka/Ks value of 0.9, indicating that the majority of the segmentally duplicated genes underwent negative selection. However, we observed that six pairs of duplicated genes were under positive selection with Ka/Ks > 1, and only one pair had Ka/Ks > 2 (OrWRKY16 and OrWRKY49). To further validate whether the OrWRKY genes have undergone strong selection pressures, we also used the branch-site model [Model A (Model 2, NSites = 2, ncatG = ignored) versus Model null] in EasyCodeML software to detect positive selection sites. We failed to detect any genes under positive selection (Supplementary Table S5). Moreover, the divergence of the duplicated OrWRKY genes was also dated using an evolutionary rate of 6.5 × 10–9 substitutions per site per year. Our results showed that the divergence of segmental duplicated genes spanned from 7.11 to 153.22 million years, and most of the duplicated WRKY genes occurred ∼40–70 million years ago.
Identification of Orthologous WRKY Genes Between O. rufipogon and O. sativa and Sequence Divergence
To identify orthologous genes between O. rufipogon and O. sativa, an RBH method was used with further relatively strict filters. We totally found 39 putative orthologous WRKY genes with an average length of 442 bp and an average sequence similarity of 98.02%. To examine the molecular evolution of orthologous WRKY genes, Ks and Ka of each orthologous gene pair were also evaluated (Supplementary Table S6). Of the 39 orthologous gene pairs, the majority of the orthologous gene pairs (84.6%; 33/39) had ratios of Ka/Ks < 1, indicating that they underwent purifying selection. However, we also found six orthologous WRKY genes with ratios of Ka/Ks > 1, indicating that they may be under positive evolution. Moreover, we used branch-site model A in EasyCodeML to further confirm whether the WRKY genes were under positive selection. Our results showed that these genes had no sign of positive selection (Supplementary Table S7).
Expression Profiles of OrWRKY Genes
The OrWRKY genes could be classified into seven major subgroups (I, II-a–II-d, and III), and thus, a total of 10 WRKY genes were randomly selected to represent all these subgroups to exhibit their expression profiling in O. rufipogon. The expression patterns of these 10 OrWRKY genes were examined in two tissues (leaves and roots) through qRT-PCR. As shown in Figure 3, these OrWRKY genes exhibited distinct expression patterns, many of which were differentially expressed between the two tissues. OrWRKY4, OrWRKY23, OrWRKY24, OrWRKY37, OrWRKY45, OrWRKY62, and OrWRKY96.2 were expressed in leaves and roots, which were relatively highly expressed in leaves but lowly expressed in roots. In contrast, OrWRKY32, OrWRKY71, and OrWRKY114 were relatively highly expressed in roots but lowly expressed in leaves. We found that OrWRKY genes belonging to different subgroups showed a distinct expression pattern, such as OrWRKY62 (II-a) and OrWRKY71 (II-a) as well as OrWRKY45 (III) and OrWRKY114 (III).
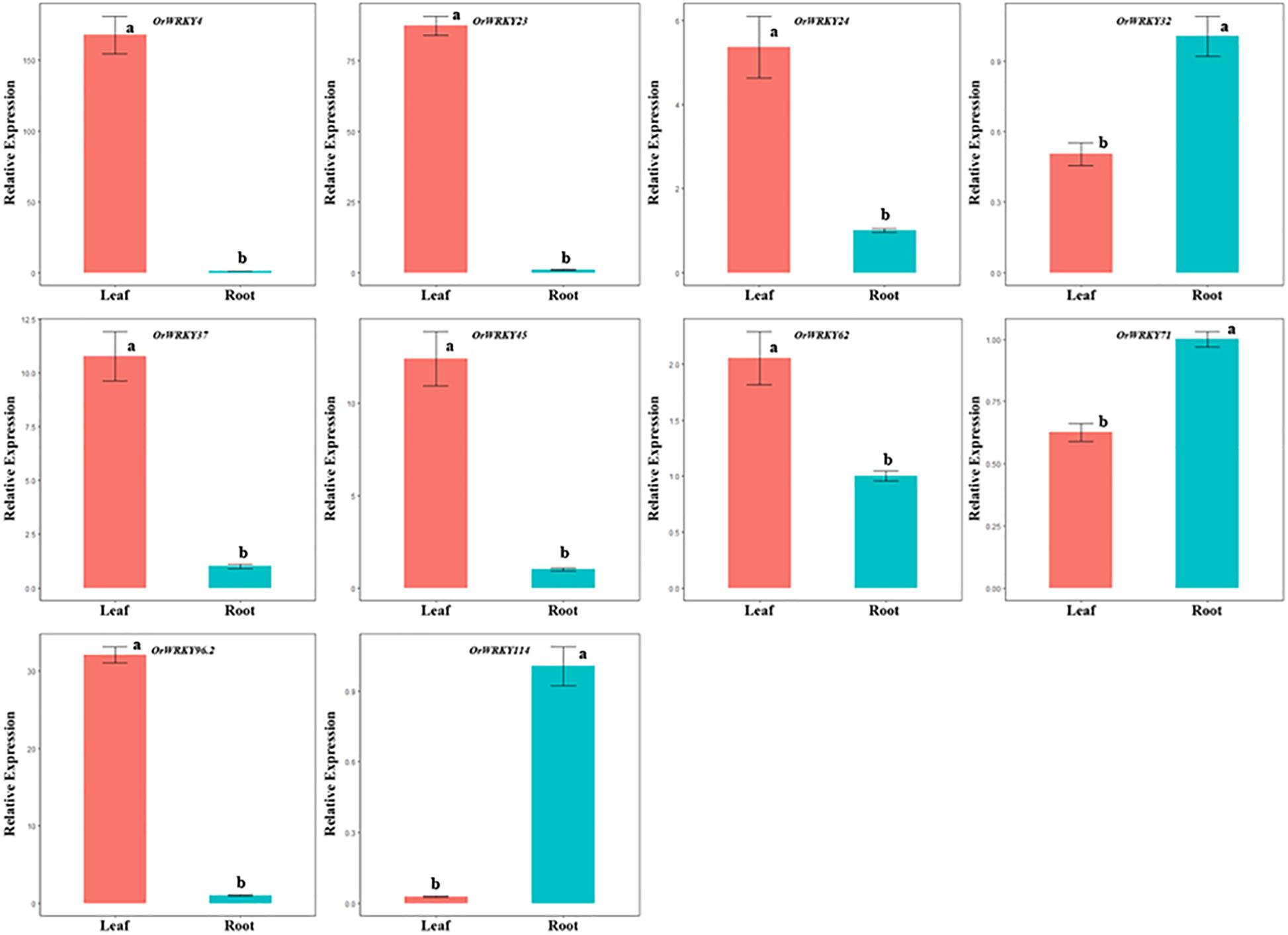
Figure 3. Expression patterns of 10 OrWRKY genes in leaves (red) and roots (green) tissues. The y-axis represents the relative expression levels of OrWRKY genes using 2–ΔΔCT method. The x-axis shows different tissues. Error bars indicate standard deviations (mean ± SD) of three independent replicates. The histogram bars labeled with different letters (a and b) above histogram bars show significantly different [least significant difference (LSD) test, P < 0.05].
Previous studies reported that WRKY genes were involved in salt stress (Liu et al., 2014; Fan et al., 2015). To investigate expression patterns of OrWRKY genes under salt stress, the O. rufipogon seedlings were grown in 200 mM NaCl solution. Expression patterns of the 10 selected OrWRKY genes from all subgroups were examined under salt stresses. As shown in Figure 4, these selected OrWRKY genes that were sensitive to salt stresses, each of which exhibited different expression profiling in the two tissues after treatments. Some OrWRKY genes in roots seemed more sensitive to salt stresses. For example, expression levels of OrWRKY23 in roots initially increased, reached the maximum level after 1 h and then decreased. However, OrWRKY23 was not sensitive to salt treatment in leaves. Some genes in leaves seemed to be more sensitive to salt stress. The expression level of OrWRKY114 in leaves was evidently upregulated by about 50-fold after 8 h, whereas the expression level of OrWRKY114 in roots increased only by almost seven times at 2 h. These genes showed a different expression profiling under salt stress, suggesting that the OrWRKY genes might respond to salt stress through different genetic networks between leaves and roots. Gene expression divergence was also examined by comparing expression levels of OrWRKY duplicated genes. Our results showed that the OrWRKY genes functionally exhibited similar responses to salt stress but presented differential expression. For example, OrWRKY62 (II-a) in roots showed relatively sensitive response to salt stress and increased by almost 105 times after 2 h, whereas OrWRKY62 increased by almost 17 times in leaves after 1 h. However, OrWRKY71 (II-a) in leaves seemed to be more sensitive to salt stress and raised 39 times after 8 h.
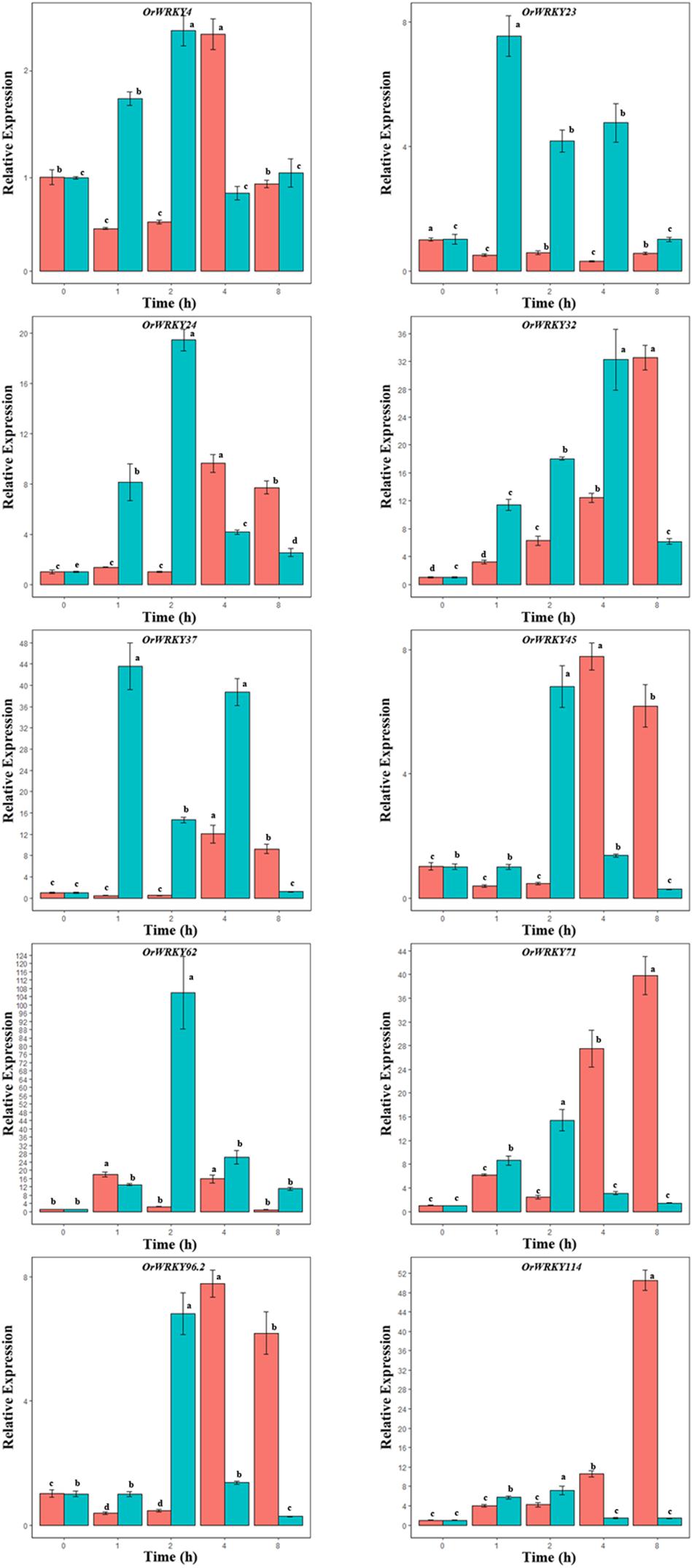
Figure 4. The relative expression levels of the 10 selected genes based on quantitative real-time polymerase chain reaction (qRT-PCR) between leaves (red) and roots (green) tissues under salt stress treatment. Samples were collected at 0, 1, 2, 4, and 8 h after the treatment with salt solution. Note that the time point of 0 h was for the seedlings which were not treated with salt solution. Error bars indicate standard deviations (mean ± SD) of three independent replicates. The histogram bars labeled with different letters (a, b, c, d, and e) above histogram bars are significantly different [least significant difference (LSD) test, P < 0.05].
Discussion
In this study, we identified a total of 101 WRKY genes from the O. rufipogon genome, which could be divided into three groups. In this study, almost all of the OrWRKY genes shared the highly conserved WRKYGQK domain. However, we also found some variants, such as WRKYGEK, WRKYGKK, WRMCGQK, and WRMCGQS motifs. Such slight variations in this region had also been reported in other plants. For example, the five unusual VvWRKY domains changed in grape (Guo et al., 2014), and 15 OnWRKY domains varied in O. nivara (Xu et al., 2016). Previous studies demonstrated that WRKYGQK domain can bind to w-box cis-elements with a C/TTGACC/T core sequence to activate downstream genes (Eulgem et al., 2000). The variation of the WRKYGQK domain could affect the specificity of DNA binding, and the WRKY genes lost the motif may bind to other specific sequences (Ciolkowski et al., 2008). In Arabidopsis, AtWRKY59 protein, containing a WRKYGKK motif, could not recognize and bound to TTGAC (W-box) (Dong et al., 2003). Moreover, NtWRKY12 with a WRKYGKK domain in tobacco could not recognize the w-box motif, but it may bind to wk-box element (van Verk et al., 2008). Further efforts are required to experimentally validate the function and binding specificities of the 22 proteins with motif variance. In addition, compared with O. rufipogon (101 OrWRKYs; genome size 439 Mb), a comparable number of WRKYs was identified in O. sativa (101; genome size 389 Mb) and O. nivara (97; genome size 448 Mb), although the number of WRKYs was fewer in carrot (67; genome size 473 Mb) and tea tree (56; genome size between 2.9 and 3.1 Gb) and more in Brassica oleracea (148; genome size 630 Mb) (International Rice Genome Sequencing Project, 2005; Zuccolo et al., 2007; Yao et al., 2015; Iorizzo et al., 2016; Xia et al., 2017; Nan and Gao, 2019; Wang et al., 2019). These results indicate that the number of WRKY genes may not be associated with the genome size and phylogenetically closed plants may harbor comparable numbers of WRKY genes.
Phylogenetic analysis suggested that OrWRKYs could be classified into three major groups in accordance with the classification of WRKYs in Arabidopsis, grape, carrot, tea tree, and cabbage (Wu et al., 2005; Guo et al., 2014; Yao et al., 2015; Nan and Gao, 2019; Wang et al., 2019). Several hypotheses have been proposed to explain how the WRKY genes evolved in plants. In wild rice O. nivara, Xu et al. (2016) suggested that ancient II-c WRKY genes were the ancestors of all WRKY genes. Our study further supports that the ancient Group II-c was the progenitor of WRKY genes in O. rufipogon. Moreover, we found that OrWRKY genes in the same group seemed to have similar conserved motifs and exon–intron structures. This phenomenon was also observed in apple (Lui et al., 2017), kiwifruit (Jing and Liu, 2018), and Salix suchowensis (Bi et al., 2016). All these results indicated that the WRKY genes, to a certain extent, were functionally conserved among higher plants.
Gene duplication events, regarded as the major evolutionary force in higher plants, play a crucial role in gene family expansion. The origin of new genes in plants during evolution is mainly caused by gene duplication (Ohno, 1970). Almost 80% of WRKY genes in cultivated rice (O. sativa ssp. japonica) were located in duplicated regions, and it was gene duplication events that lead to generating new genes (Ramamoorthy et al., 2008). In this study, we found that OrWRKY genes were derived from either segmental or tandem duplications, indicating that high segmental but low tandem duplications resulted in OrWRKY genes, consistent with wheat (Ning et al., 2017), peanut (Song et al., 2016b), grapevine (Wang et al., 2014), and soybean (Song et al., 2016a). Thus, we can infer that compared to tandem duplication events, segmental duplication events have mainly contributed to the expansion of OrWRKY genes.
Soil salinity is one of major abiotic stresses to limit plant growth and development. Enhancement of grain yield stability under salt stress would be of great importance. WRKY genes participate in the control of a wide variety of biological processes, some of which are associated with salt stress. For example, the overexpression of one WRKY gene, GmWRKY54, could improve salt stress tolerance in soybean (Zhou et al., 2008). In Arabidopsis, the expression of AtWRKY25 and AtWRKY33 was dramatically induced by sodium chloride (NaCl) treatment (Li et al., 2011). Our results showed that the expression of 10 selected OrWRKY genes was upregulated or downregulated by NaCl in either of these two tissues, suggesting that they might be related to salt responses. Previous study showed that there were two kinds of OsWRKY45 genes. Under salt treatment, the expression of OsWRKY45-1 was first induced and then suppressed, whereas the expression of OsWRKY45-2 was first suppressed, then induced, and later suppressed, indicating their different functions under salt stress (Tao et al., 2011). In this study, the expression pattern OrWRKY45 was first suppressed, then induced, and later suppressed, suggesting that it might have the same functions with OsWRKY45-2. It was reported that the G-box, ABRE, and CRE are the major cis-acting elements for salt-responsive gene expression (Narusaka et al., 2003; Ahmad et al., 2015). Our study showed that nine of the 10 sampled OrWRKY genes had G-boxes, and half of them had ABREs. Additionally, we found that the same gene exhibited a distinct expression trend between leaves and roots under salt stresses, suggesting that it might respond to abiotic stresses through different genetic networks.
Conclusion
In this study, 101 OrWRYK genes were comprehensively identified in the O. rufipogon genome assembly. Phylogenetic analysis suggested that OrWRKY transcription factors were conserved, supported by the identification of highly conserved motifs and gene structures. Genomic synteny analysis suggested that segmental duplications may have played a major role in the OrWRKY gene family evolution, and most duplicated WRKY genes underwent purifying selection. We showed tissue-specific expression of the OrWRKYs and their differential expression in response to salt stresses, implying the existence of a complicated molecular regulatory network response to salt stresses in O. rufipogon. We obtain novel insights into the evolutionary and functional divergence of the WRKY gene family, which will greatly help functional genomic studies of candidate WRKY genes in O. rufipogon for the purpose of rice genetic improvement.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
HN, WL, and YL performed the data analysis and experiments. HN drafted the manuscript. LG served as the principal investigator, facilitated the project, and revised the manuscript.
Funding
This study was supported by the Project of Innovation Team of Yunnan Province to LG.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate the reviewers for their comments on this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00359/full#supplementary-material
FIGURE S1 | Phylogenetic analysis of the WRKY domains of OrWRKY genes. The phylogenetic tree was constructed using Neighbor-Joining method based on WRKY domains. Reliability of the predicted trees was tested using bootstrapping with 1,000 replicates.
FIGURE S2 | Phylogenetic analysis of WRKY proteins in the O. rufipogon. The phylogenetic tree was constructed using Neighbor-Joining method based on WRKY full-length proteins. Reliability of the predicted trees was tested using bootstrappingwith 1,000 replicates.
FIGURE S3 | Multiple sequence alignment of domains among the OrWRKY proteins. The sequences were aligned using Clustal W. Dashes indicate gaps, and ‘N’ and ‘C’ mean the N-terminal and C-terminal WRKY domains of a specific WRKY protein.
FIGURE S4 | Phylogenetic analysis of the WRKY genes in Arabidopsis, O. sativa ssp. japonica, O. sativa ssp. indica, O. rufipogon, and O. nivara. The phylogenetic tree was constructed using Neighbor-Joining method based on 531 WRKY domain sequences. Reliability of the predicted tree was tested using bootstrapping with 1,000 replicates.
TABLE S1 | Primer pairs of the 10 OrWRKY genes identified in this study.
TABLE S2 | OrWRKY genes identified in Oryza rufipogon and their orthologs in Oryza sativa ssp. japonica.
TABLE S3 | Ka/Ks calculation and divergence times of the duplicated OrWRKY gene pairs in syntenic blocks.
TABLE S4 | Tandem duplicated gene pairs identified in O. rufipogon.
TABLE S5 | The detection of positive selection by the likelihood method with branch-site model for OrWRKY genes.
TABLE S6 | Orthologous WRKY genes between O. rufipogon and O. sativa ssp. japonica.
TABLE S7 | Detection of positive selection by the likelihood method with branch-site model for OrWRKY genes between O. rufipogon and O. sativa.
Footnotes
- ^ http://pfam.xfam.org/
- ^ http://hmmer.janelia.org/
- ^ http://planttfdb.cbi.pku.edu.cn/
- ^ http://web.expasy.org/protparam/
- ^ https://phytozome.jgi.doe.gov/pz/portal.html#
- ^ https://www.arabidopsis.org/
- ^ http://gsds.cbi.pku.edu.cn
- ^ http://meme-suite.org/tools/meme
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Ahmad, A., Niwa, Y., Goto, S., Ogawa, T., Shimizu, M., Suzuki, A., et al. (2015). bHLH106 integrates functions of multiple genes through their G-box to confer salt tolerance on Arabidopsis. PLoS One 10:e0126872. doi: 10.1371/journal.pone.0126872
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37(Suppl. 2), W202–W208. doi: 10.1093/nar/gkp335
Bi, C., Xu, Y., Ye, Q., Yin, T., and Ye, N. (2016). Genome-wide identification and characterization of WRKY gene family in Salix suchowensis. Peerj 4:e2437. doi: 10.7717/peerj.2437
Brar, D., and Ramos, J. (2008). “Wild species of Oryza: a rich reservoir of genetic variability for rice improvement,” in Charting New Pathways To C4 Rice, Ed. B. Hardy (Singapore: World Scientific), 351–359.
Cheng, C., Motohashi, R., Tsuchimoto, S., Fukuta, Y., Ohtsubo, H., and Ohtsubo, E. (2003). Polyphyletic origin of cultivated rice: based on the interspersion pattern of SINEs. Mol. Biol. Evol. 20, 67–75. doi: 10.1093/molbev/msg004
Ciolkowski, I., Wanke, D., Birkenbihl, R.P., and Somssich, I. E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68, 81–92. doi: 10.1007/s11103-008-9353-1
Davidson, C. J., Guthrie, E. E., and Lipsick, J. S. (2013). Duplication and maintenance of the Myb genes of vertebrate animals. Biol. Open 2, 101–110. doi: 10.1242/bio.20123152
Dong, J. X., Chen, C. H., and Chen, Z. X. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant. Mol. Biol. 51, 21–37. doi: 10.1023/A:1020780022549
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Eulgem, T., Rushton, P. J., Schmelzer, E., Hahlbrock, K., and Somssich, I. E. (1999). Early nuclear events in plant defense: rapid gene activation by WRKY transcription factors. EMBO J. 18, 4689–4699.
Fan, X., Guo, Q., Xu, P., Gong, Y., Shu, H., Yang, Y., et al. (2015). Transcriptome-wide identification of salt-responsive members of the WRKY gene family in Gossypium aridum. PLoS One 10:e0126148–e0126148. doi: 10.1371/journal.pone.0126148
Fuller, D. Q., Sato, Y. I., Castillo, C., Qin, L., Weisskopf, A. R., Kingwell-Banham, E. J., et al. (2010). Consilience of genetics and archaeobotany in the entangled history of rice. Archaeol. Anthrop. Sci. 2, 115–131. doi: 10.1007/s12520-010-0035-y
Gao, F., Chen, C., Arab, D. A., Du, Z., He, Y., and Ho, S. Y. W. (2019). EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol. Evol. 9, 3891–3898. doi: 10.1002/ece3.5015
Gao, L. (2004). Population structure and conservation genetics of wild rice Oryza rufipogon (Poaceae): a region-wide perspective from microsatellite variation. Mol. Ecol. 13, 1009–1024.
Gao, L., Zhang, S., Zhou, Y., Ge, S., and Hong, D. (1996). A survey of the current status of wild rice in China. Biol. Sci. 4, 160–166.
Group, R. W. W. (2012). Nomenclature report on rice WRKY’s - Conflict regarding gene names and its solution. Rice 5, 1–3. doi: 10.1186/1939-8433-5-3
Guo, C., Guo, R., Xu, X., Gao, M., Li, X., Song, J., et al. (2014). Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 65, 1513–1528. doi: 10.1093/jxb/eru007
Huang, S., Gao, Y., Liu, J., Peng, X., Niu, X., Fei, Z., et al. (2012). Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genomics 287, 495–513. doi: 10.1007/s00438-012-0696-6
Huang, X., Kurata, N., Wei, X., Wang, Z. X., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Hurst, L. D. (2002). The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18, 486–487.
International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436, 793–800. doi: 10.1038/nature03895
Iorizzo, M., Ellison, S., Senalik, D., Zeng, P., Satapoomin, P., Huang, J., et al. (2016). A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 48, 657–666. doi: 10.1038/ng.3565
Jimmy, J. L., and Babu, S. (2019). Variations in the structure and evolution of rice WRKY genes in indica and japonica genotypes and their co-expression network in mediating disease resistance. Evol Bioinform Online 15, 1176934319857720. doi: 10.1177/1176934319857720
Jing, Z., and Liu, Z. (2018). Genome-wide identification of WRKY transcription factors in kiwifruit (Actinidia spp.) and analysis of WRKY expression in responses to biotic and abiotic stresses. Genes Genom. 40, 429–446. doi: 10.1007/s13258-017-0645-1
Jue, D., Sang, X., Liu, L., Shu, B., Wang, Y., Liu, C., et al. (2018). Identification of WRKY gene family from Dimocarpus longan and its expression analysis during flower induction and abiotic stress responses. Int. J. Mol. Sci. 19:2169. doi: 10.3390/ijms19082169
Khush, G. S. (1997). Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35, 25–34.
Kovach, M. J., Sweeney, M. T., and McCouch, S. R. (2007). New insights into the history of rice domestication. Trends Genet. 23, 578–587. doi: 10.1016/j.tig.2007.08.012
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Larkin, M. A., Blackshields, G., Brown, N., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, H., Gao, Y., Xu, H., Dai, Y., Deng, D., and Chen, J. (2013). ZmWRKY33, a WRKY maize transcription factor conferring enhanced salt stress tolerances in Arabidopsis. Plant Growth Regul. 70, 207–216. doi: 10.1007/s10725-013-9792-9
Li, S., Fu, Q., Chen, L., Huang, W., and Yu, D. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252. doi: 10.1007/s00425-011-1375-2
Li, S., Zhou, X., Chen, L., Huang, W., and Yu, D. (2010). Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells 29, 475–483. doi: 10.1007/s10059-010-0059-2
Li, W., Li, K., Huang, Y., Shi, C., Hu, W. S., Zhang, Y., et al. (2020). SMRT sequencing of the Oryza rufipogon genome reveals the genomic basis of rice adaptation. bioRxiv [preprint] doi: 10.1101/2020.01.14.905281
Liu, Q. L., Xu, K. D., Pan, Y. Z., Jiang, B. B., Liu, G. L., Jia, Y., et al. (2014). Functional analysis of a novel chrysanthemum WRKY transcription factor gene involved in salt tolerance. Plant Mol. Biol. Rep. 32, 282–289. doi: 10.1007/s11105-013-0639-3
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lui, S., Luo, C., Zhu, L., Sha, R., Qu, S., Cai, B., et al. (2017). Identification and expression analysis of WRKY transcription factor genes in response to fungal pathogen and hormone treatments in apple (Malus domestica). J. Plant Biol. 60, 215–230. doi: 10.1007/s12374-016-0577-3
McCouch, S. R. (2008). Gene nomenclature system for rice. Rice 1, 72–84. doi: 10.1007/s12284-008-9004-9
Morishima, H., and Barbier, P. (1990). Mating system and genetic structure of natural populations in wild rice Oryza rufipogon. Plant Spec. Biol. 5, 31–39.
Mzid, R., Zorrig, W., Ben Ayed, R., Ben Hamed, K., Ayadi, M., Damak, Y., et al. (2018). The grapevine VvWRKY2 gene enhances salt and osmotic stress tolerance in transgenic Nicotiana tabacum. 3 Biotech 8:277. doi: 10.1007/s13205-018-1301-4
Nan, H., and Gao, L. Z. (2019). Genome-wide analysis of WRKY genes and their response to hormone and mechanic stresses in carrot. Front. Genet. 10:363. doi: 10.3389/fgene.2019.00363
Narusaka, Y., Nakashima, K., Shinwari, Z. K., Sakuma, Y., Furihata, T., Abe, H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34, 137–148.
Ning, P., Liu, C., Kang, J., and Lv, J. (2017). Genome-wide analysis of WRKY transcription factors in wheat (Triticum aestivum L.) and differential expression under water deficit condition. PeerJ 5:e3232. doi: 10.7717/peerj.3232
Ohno, S. (1970). “The creation of a new gene from a redundant duplicate of an old gene,” in Evolution by Gene Duplication, (Berlin: Springer Berlin Heidelberg), 71–82.
Raineri, J., Wang, S., Peleg, Z., Blumwald, E., and Chan, R. L. (2015). The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress. Plant Mol. Biol. 88, 401–413. doi: 10.1007/s11103-015-0329-7
Ramamoorthy, R., Jiang, S. Y., Kumar, N., Venkatesh, P. N., and Ramachandran, S. (2008). A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 49, 865–879. doi: 10.1093/pcp/pcn061
Ross, C. A., Liu, Y., and Shen, Q. J. (2007). The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 49, 827–842. doi: 10.1111/j.1672-9072.2007.00504.x
Schmidt, R., Mieulet, D., Hubberten, H. M., Obata, T., Hoefgen, R., Fernie, A. R., et al. (2013). Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25, 2115–2131. doi: 10.1105/tpc.113.113068
Singh, K., Foley, R. C., and Onate-Sanchez, L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5, 430–436.
Song, H., Wang, P., Hou, L., Zhao, S., Zhao, C., Xia, H., et al. (2016a). Global analysis of WRKY genes and their response to dehydration and salt stress in soybean. Front. Plant Sci. 7:9. doi: 10.3389/fpls.2016.00009
Song, H., Wang, P., Lin, J. Y., Zhao, C., Bi, Y., and Wang, X. (2016b). Genome-wide identification and characterization of WRKY gene family in peanut. Front. Plant Sci. 7:534. doi: 10.3389/fpls.2016.00534
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tang, J., Wang, F., Hou, X. L., Wang, Z., and Huang, Z. N. (2013). Genome-wide fractionation and identification of WRKY transcription factors in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals collinearity and their expression patterns under abiotic and biotic stresses. Plant Mol. Biol. Rep. 32, 781–795. doi: 10.1007/s11105-013-0672-2
Tao, Z., Kou, Y., Liu, H., Li, X., Xiao, J., and Wang, S. (2011). OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J. Exp. Bot. 62, 4863–4874. doi: 10.1093/jxb/err144
van Verk, M. C., Pappaioannou, D., Neeleman, L., Bol, J. F., and Linthorst, H. J. M. (2008). A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 146, 1983–1995. doi: 10.1104/pp.107.112789
Vaughan, D. A., Morishima, H., and Kadowaki, K. (2003). Diversity in the Oryza genus. Curr. Opin. Plant Biol. 6, 139–146.
Vision, T. J., Brown, D. G., and Tanksley, S. D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117.
Wang, M., Vannozzi, A., Wang, G., Liang, Y. H., Tornielli, G. B., Zenoni, S., et al. (2014). Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic. Res. 1:14016. doi: 10.1038/hortres.2014.16
Wang, P., Yue, C., Chen, D., Zheng, Y., Zhang, Q., Yang, J., et al. (2019). Genome-wide identification of WRKY family genes and their response to abiotic stresses in tea plant (Camellia sinensis). Genes Genom. 41, 17–33. doi: 10.1007/s13258-018-0734-9
Wang, W., Vinocur, B., and Altman, A. (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14. doi: 10.1007/s00425-003-1105-5
Wu, K. L., Guo, Z. J., Wang, H. H., and Li, J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12, 9–26. doi: 10.1093/dnares/12.1.9
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xia, E. H., Jiang, J. J., Huang, H., Zhang, L. P., Zhang, H. B., and Gao, L. Z. (2014). Transcriptome analysis of the oil-rich tea plant, Camellia oleifera, reveals candidate genes related to lipid metabolism. PLoS One 9:e104150. doi: 10.1371/journal.pone.0104150
Xia, E. H., Zhang, H. B., Sheng, J., Li, K., Zhang, Q. J., Kim, C., et al. (2017). The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant 10, 866–877. doi: 10.1016/j.molp.2017.04.002
Xie, Z., Zhang, Z. L., Zou, X., Huang, J., Ruas, P., Thompson, D., et al. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. doi: 10.1104/pp.104.054312
Xu, H., Watanabe, K. A., Zhang, L., and Shen, Q. J. (2016). WRKY transcription factor genes in wild rice Oryza nivara. DNA Res. 23, 311–323. doi: 10.1093/dnares/dsw025
Xu, X., Liu, X., Ge, S., Jensen, J. D., Hu, F., Li, X., et al. (2012). Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30:105. doi: 10.1038/nbt.2050
Yang, X., Li, H., Yang, Y., Wang, Y., Mo, Y., Zhang, R., et al. (2018). Identification and expression analyses of WRKY genes reveal their involvement in growth and abiotic stress response in watermelon (Citrullus lanatus). PLoS One 13:e0191308. doi: 10.1371/journal.pone.0191308
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yao, Q. Y., Xia, E. H., Liu, F. H., and Gao, L. Z. (2015). Genome-wide identification and comparative expression analysis reveal a rapid expansion and functional divergence of duplicated genes in the WRKY gene family of cabbage, Brassica oleracea var. capitata. Gene 557, 35–42. doi: 10.1016/j.gene.2014.12.005
Yu, J., Wang, J., Lin, W., Li, S., Li, H., Zhou, J., et al. (2005). The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3:e38. doi: 10.1371/journal.pbio.0030038
Zhang, Y., and Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5:1. doi: 10.1186/1471-2148-5-1
Zhou, H., Li, Y., Zhang, Q., Ren, S., Shen, Y., Qin, L., et al. (2016). Genome-wide analysis of the expression of WRKY family genes in different developmental stages of wild strawberry (Fragaria vesca) fruit. PLoS One 11:e0154312. doi: 10.1371/journal.pone.0154312
Zhou, Y., Yang, P., Cui, F., Zhang, F., Luo, X., and Xie, J. (2016). Transcriptome analysis of salt stress responsiveness in the seedlings of dongxiang wild rice (Oryza rufipogon Griff.). PLoS One 11:e0146242. doi: 10.1371/journal.pone.0146242
Zhou, Q. Y., Tian, A. G., Zou, H. F., Xie, Z. M., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Zhu, N., Cheng, S., Liu, X., Du, H., Dai, M., Zhou, D. X., et al. (2015). The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 236, 146–156. doi: 10.1016/j.plantsci.2015.03.023
Keywords: WRKY transcription factor, wild rice, Oryza rufipogon, expression divergence, whole genome duplication, salt stress
Citation: Nan H, Li W, Lin Y-l and Gao L-z (2020) Genome-Wide Analysis of WRKY Genes and Their Response to Salt Stress in the Wild Progenitor of Asian Cultivated Rice, Oryza rufipogon. Front. Genet. 11:359. doi: 10.3389/fgene.2020.00359
Received: 13 June 2019; Accepted: 24 March 2020;
Published: 08 May 2020.
Edited by:
Longjiang Fan, Zhejiang University, ChinaReviewed by:
Serena Aceto, University of Naples Federico II, ItalyLongbiao Guo, China National Rice Research Institute (CAAS), China
Hui Song, Qingdao Agricultural University, China
Fantao Zhang, Jiangxi Normal University, China
Copyright © 2020 Nan, Li, Lin and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-zhi Gao, Lgaogenomics@163.com
†These authors have contributed equally to this work
 Hong Nan
Hong Nan Wei Li1†
Wei Li1† Li-zhi Gao
Li-zhi Gao