- 1 Department of Oral Biology, University of Oslo, Oslo, Norway
- 2 Department of Biomaterials, Institute for Clinical Dentistry, University of Oslo, Oslo, Norway
- 3 The Biotechnology Centre of Oslo, University of Oslo, Oslo, Norway
The aim of this study was to describe the expression of genes, including ameloblastin (Ambn), amelogenin X chromosome (Amelx), and enamelin (Enam) during early (pre-secretory) tooth development. The expression of these genes has predominantly been studied at post-secretory stages. Deoxyoligonucleotide microarrays were used to study gene expression during development of the murine first molar tooth germ at 24 h intervals, starting at the 11th embryonic day (E11.5), and up to the 7th day after birth (P7). The profile search function of Spotfire software was used to select genes with similar expression profile as the enamel genes (Ambn, Amelx, and Enam). Microarray results where validated using real-time reverse transcription-polymerase chain reaction (real-time RT-PCR), and translated proteins identified by Western-blotting. In situ localization of the Ambn, Amelx, and Enam mRNAs were monitored from E12.5 to E17.5 using deoxyoligonucleotide probes. Bioinformatics analysis was used to associate biological functions with differentially expressed (DE; p ≤ 0.05) genes. Microarray results showed a total of 4362 genes including Ambn, Amelx, and Enam to be significant DE throughout the time-course. The expression of the three enamel genes was low at pre-natal stages (E11.5–P0) increasing after birth (P1–P7). Profile search lead to isolation of 87 genes with significantly similar expression to the three enamel proteins. These mRNAs were expressed in dental epithelium and epithelium derived cells. Although expression of Ambn, Amelx, and Enam were lower during early tooth development compared to secretory stages enamel proteins were detectable by Western-blotting. Bioinformatic analysis associated the 87 genes with multiple biological functions. Around 35 genes were associated with 15 transcription factors.
Introduction
Interactions between oral epithelium and neural crest derived mesenchyme are considered essential for tooth development. Cells of the epithelium expand and proliferate, invaginating into the condensing mesenchyme, and subsequently forms the tooth germ (Thesleff, 1995; Chai et al., 2000; Sharpe, 2001), this process been modulated by several growth factors (Thesleff and Mikkola, 2002; Zhang et al., 2005). As the invaginating epithelium expands it is surrounded by condensing mesenchyme transforming into a bud and cap, subsequently developing into the bell stage (Tucker and Sharpe, 1999; Fleischmannova et al., 2008). Mesenchymal cells facing the basement membrane differentiate into dentin producing odontoblasts, while the adjacent layer of epithelial cells differentiates into ameloblasts which secrete the organic enamel matrix (Thesleff and Hurmerinta, 1981).
Ameloblastin (encoded by Ambn) is expressed in the mineralizing matrix of bones and teeth. This matrix protein inhibits ameloblast proliferation and is essential for ameloblast adhesion affecting thickness of the enamel layer (Zhang et al., 2011).
Amelogenins (encoded by Amel) are expressed in epithelium derived cells, bone marrow, and mesenchymal stem cells (MSCs). Amelogenins are the main component in the developing enamel matrix and essential for normal enamel thickness and structure (Feng et al., 2012).
Enamelin (encoded by Enam) is a minor constituent of the extracellular matrix but plays a critical role in normal enamel formation. Enamelin is required for the deposition of tooth enamel, but it is also necessary to maintain the ameloblast phenotype, as is the case for ameloblastin (Hu et al., 2011).
The expression of about 300 genes has been mapped1 and has contributed to our understanding of tooth development. The expression of a substantially higher number of genes is likely to be involved. The use of microarrays facilitates the global mapping of genes at various stages of tooth development.
Recent results obtained from gene expression profiling, between two developmental stages (E15.5 and P2) of murine tooth germs using microarrays indicated that Amelx, Ambn, and Enam exhibited low levels of expression at the studied pre-natal stage (E15.5; Osmundsen et al., 2007). We investigated if these three genes were expressed prior to tooth mineralization and if other genes followed the same expression pattern; low levels of expression at pre-natal stages followed by increase at post-natal stages, during murine tooth development. The study of global gene expression during both pre- and post-natal stages (16 time points) of murine molar tooth development using microarrays should provide data capable of capturing dynamic gene expression profiles during tooth development, combined with bioinformatics might help our understanding of cellular processes underlying development of the murine tooth germ.
Materials and Methods
Experimental Animals
The day of the vaginal plug was set to embryonic day E0.5 in pregnant female CD-1 mice (n = 3–5). The embryo/fetus developmental stage was assessed using the Theiler criteria (Kaufman, 1992).
The animal house had 12 h light/dark cycle and was thermostated at 21°C with relative humidity at 55 ± 5%. Water and fodder was provided ad lib. The animals were housed according to the regulations of the Norwegian Gene Technology Act of 1994.
Female mice pregnant at various stages were sacrificed by cervical dislocation and embryos quickly removed from the amnion sac and decapitated.
Tissue Preparation
The first mandibular molar tooth germs was dissected for microarray and real-time RT-PCR analysis from CD-1 embryos at various developmental stages starting at the 11th embryonic day (E11.5) and ending at the 7th day after birth (P7).
For in situ hybridization (ISH) embryos heads were dissected and fixed in 4% (v/v) cold, neutral buffered formalin (NBF; AppliChem, Darmstadt, Germany). Heads dissected at P2 up to P7 were decalcified in 12.5% (w/v) ethylenediaminetetraacetic acid (EDTA), 2.5% (v/v) formalin. Paraffin embedding was carried out essentially as described by Hougaard et al. (1997). Serial sections 8 μm thick were used.
Batches of three to nine molar tooth germs were dissected for Western blot analysis at each of the various developmental time points used in this investigation. The tooth germs were lysed using CelLytic MT (Sigma, St Louis, MO, USA) and Halt Protease Inhibitor Cocktail (Pierce Biotechnology, Rockford, IL, USA). Protein concentrations were assayed using Bio-Rad Protein Assay kit (Bio-Rad Laboratories, Hercules, CA, USA).
Microarray Analysis of mRNAs Isolated from Tooth Germs
Total RNA was extracted from tooth germs as described previously by Osmundsen et al. Murine deoxyoligonucleotide (30 k)-microarrays printed with Operon murine v.3 oligo-set (Qiagen GmbH, Hilden, Germany) were purchased from the NTNU Microarray Core Facility (Norwegian University of Science and Technology, Trondheim, Norway). Spikes from A. thaliana (purchased from Stratagene, La Jolla, CA, USA) were used to normalize the fluorescence within each microarray and to monitor the quality of the hybridization. Complementary DNA synthesis, labeling, and hybridization were carried out as described previously (Osmundsen et al., 2007).
After hybridization and scanning the resulting expression data (48 arrays) was assembled into a single data file. Cy3 and Cy5 channels of each slide were treated as single channel data as if derived from single color arrays to facilitate statistical and bioinformatic analysis. The genes exhibiting a net fluorescence fewer than 200 were excluded. LOESS normalized fluorescence intensities (median values, with background subtracted) from each of the two channels were converted to log2-scale, and the log2-values were subjected to z-score normalization (Cheadle et al., 2003).
Statistical analysis of microarray data was carried out using Spotfire v. 9™ Decision Site for Microarray Analysis (Spotfire, MA, USA) from sets of three arrays at each time point.
The ANOVA facility of the Spotfire program was used to select genes which exhibited statistically significant differences in levels of expression (p < 0.05) between various developmental stages. False discovery rate (FDR; 0.05; Benjamini and Hochberg, 1995; Reiner et al., 2003) was used to correct selection of genes for false positives.
Experimental design and resulting microarray files have been deposited in the MIAME database with reference E-MEXP-3581.
Isolation of Genes Using Profile Search
Profile search function of Spotfire software v.9 Decision Site for Microarray analysis software (TIBCO Spotfire, Somerville, MA, USA) was used to select differentially expressed (DE) genes with a similar expression pattern to that of pre-selected genes Ambn, Amelx, and Enam. The mean time-course for Ambn, Amelx, and Enam genes (normalized data) throughout the studied time-course was used as search criteria (master profile). The resulting expression profile was subjected to hierarchical clustering and the result presented as heat map. Unknown genes without an Entrez ID were omitted from this analysis.
Real-Time RT-PCR
Triplicates of tooth germs for each time point were used in cDNA synthesis (Fermentas, St. Leon Route, Germany). The subsequent real-time PCR was carried out in a Stratagene MX3005P (Stratagene, La Jolla, CA, USA), using SYBR Green-based assay Ampliqon III (Ampliqon, Rødovre, Denmark). Real-time RT-PCR data was analyzed using the 2−ΔΔCt method 2[−Delta Delta C(T); Pfaffl, 2001]. Where ΔΔCt = (Cttarget − CtRpl27)Time point x − (Cttarget − CtRpl27)E11.5. Where time x = E11.5 up to P7. All time points where compared to E11.5 and normalized against ribosomal protein L27 (Rpl27; Pfaffl, 2001). The primer sequences are listed in Table 1.
Bioinformatic Analysis of Differentially Expressed Genes
Bioinformatic analysis using Ingenuity Pathway Analysis (IPA; Ingenuity Systems Inc., Redwood City, CA, USA) was carried out to identify significant associations (Fisher’s Exact Test, p ≤ 0.05) with canonical pathways, signaling pathways, transcription factors, molecular, and cellular functions for the genes DE, isolated using the profile search function of the Spotfire software. Transcription factors with p-value of overlap <0.01 where considered to be significantly associated with the DE expressed genes.
In situ Hybridization
In situ hybridization was used to visualize microarray and real-Time RT-PCR results for the genes of the expression profile and was carried out using a protocol essentially as described by Farquharson et al. (1990). ISH was performed in a Discovery XT hybridization instrument (Ventana Medical Systems, AZ, USA) according to the Ventana recommendations.2 The Ambn anti-sense probe, 5′-ggtttcagtcctggctggtgaggctgcaaggatggctgctgggtttca-3′, hybridizes to nucleotides 378–425 located within the coding sequence of NM_009664.
The Amelx probe, 5′-ggacaggggctgcatggagaacagtggaggcagaggtggctgtggtgc-3′, hybridizes to nucleotides 544–591 within the coding sequence of NM_009666. The Enam probe, 5′-tgtggctgtggctcttgtggattggtctggttgggtttggcggtctcc-3′ hybridizes to nucleotides 672–719 located within the coding sequence of NM_017468. The probe concentration was 0. 45 μg μl −1. The hybridization temperature was set to 57°C. Antibodies (diluted 1:2000) purchased from Jacksons Immuno (Jackson Immuno Research Europe, Suffolk, UK) and BlueMap kit (NBT/BCIP; Ventana Medical Systems) was used for detection and visualization. No counter-staining was used following ISH.
The quality of mRNA in the tissue was assessed using digoxigenin (DIG)-labeled poly dT deoxyoligonucleotide probes. The nature of the hybridization signals was ascertained by treating separate tissue sections prior to pre-hybridization with either RNAse (RNAseA, Qiagen, Hilden, Germany) or DNAseI (DNA-free™ Ambion, CA, USA). RNAse was used at a final concentration of 20 μg RNAseA ml−1, the tissue sections being incubated for 30–120 min at 37°C. The DNAse treatment was performed for 15–30 min at 90°C using a final concentration of 5 μg DNAse I μl−1. ISH micrographs were obtained using a Nicon Elipse E400 instrument (Nikon Corporation, Tokyo, Japan).
Western-Blotting
Aliquots containing 100 μg of total protein were separated by sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using ReadyGel (4–20%) with Tris-HCl buffer (Bio-Rad Laboratories). Proteins were blotted onto 0.45 μm Trans-blot nitrocellulose membranes (Bio-Rad Laboratories) and incubated with antibodies (1:500) against Amelogenin X (SC-32892), Ameloblastin (N-18; SC-33100), or Enamelin (C-18; SC-33107; Santa Cruz Biotechnology, CA, USA), using goat anti-rabbit as secondary antibody (1:5000; Vector Laboratories, CA, USA). Enzymatic activity was detected by Enhanced Chemifluorescence (ECF) substrate (GE Healthcare, Bucks, UK) using the Storm 860 imaging scanner (GE Healthcare). The specificity of the antibodies was tested incubating with only the secondary antibody.
Results
Microarray Analysis of mRNAs Isolated from Tooth Germs
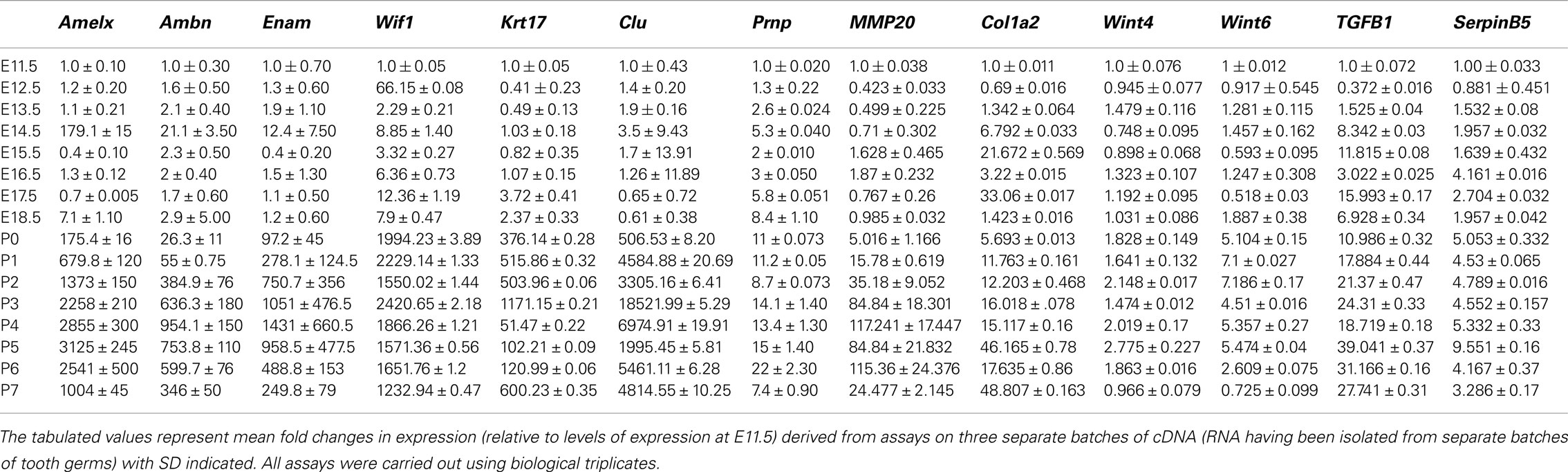
Ambn, Amelx, and Enam had similar expression pattern throughout the developmental stages (Figure 1), showing low pre-natal levels of expression (net fluorescence intensities of about 800). At P1–P5 about 33-fold increase in fluorescence intensity was observed, further increasing to 100-fold at P6–P7, compared to the pre-natal stages (Figure 1).
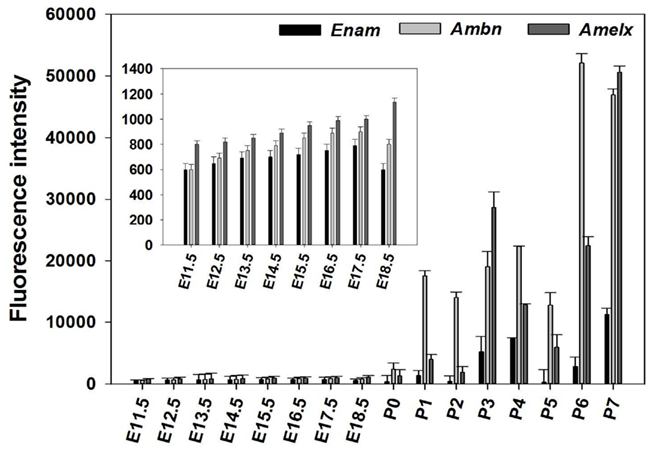
Figure 1. Expression pattern of Amelx, Ambn, and Enam during development of the molar tooth germ. Expression of Ambn, Amelx, and Enam mRNA monitored using deoxyoligonucleotide microarrays at the various stages of murine tooth development. The plotted normalized net fluorescence intensities (raw fluorescence intensity minus background) are means derived from biological triplicates at each time point with SD indicated. Expression pattern showed low fluorescence intensities at the pre-natal stages (E11.5–P0), with increasing signal throughout the post-natal stages (P1–P7).
The microarray results showed a total of 4362 genes to be differentially (p ≤ 0.05) expressed at every time point studied (E11.5–P7), 1169 of which being without an Entrez ID and were consequently not used during further analysis. The remaining 2441 genes were used in a profile search using the mean time-course of expression for Ambn, Amelx, and Enam (Figure 2A). From the 2441 genes screened with the profile search function in Spotfire, 87 genes (Figure 2B) exhibited at time-course of expression similar that of Amelx, Ambn, and Enam.
Figure 2. Profile search results. The mean net normalized fluorescence intensities for Ambn, Amelx, and Enam genes (A) at each time point was used as search criteria (master profile) for profile search. The resulting 87 genes were hierarchically clustered using the Euclidean distance and the result is presented as heat map (B). Columns represent different developmental stages and rows represent each gene. Each cell in the matrix represents the expression level of a gene at a certain time point/developmental stage. Green and red in cells reflect low and high expression levels, respectively.
Real-Time RT-PCR
The microarray results of mRNA expression for the enamel genes Ambn, Amelx, Enam, and some random genes from the profile search clusterin (Clu), Wnt inhibitory factor 1 (Wif1), keratin 17 (Krt17), prion protein (Prnp), matrix metallopeptidase 20 (Mmp20), Collagen type I, alpha 2 (Col1a2), transforming growth factor, beta 1(Tgfb1), wingless-related MMTV integration site 4 (Wint4), wingless-related MMTV integration site 6 (Wint6), and serine (or cysteine) peptidase inhibitor, clade B, member 5(Serpinb5), were verified using real-time RT-PCR. The results show good agreement with the results obtained from microarray (Figure 3 and Table 2), confirming that pre-natal levels of mRNAs of Amelx, Ambn, Enam, Wif1, Krt17, Clu, Prnp, Mmp20, Col1a1, Tgfb1, Wint4, Wint6, and Serpinb5 are markedly lower compared to post-natal levels (Table 2). The microarray and real-time RT-PCR results showed good agreement.
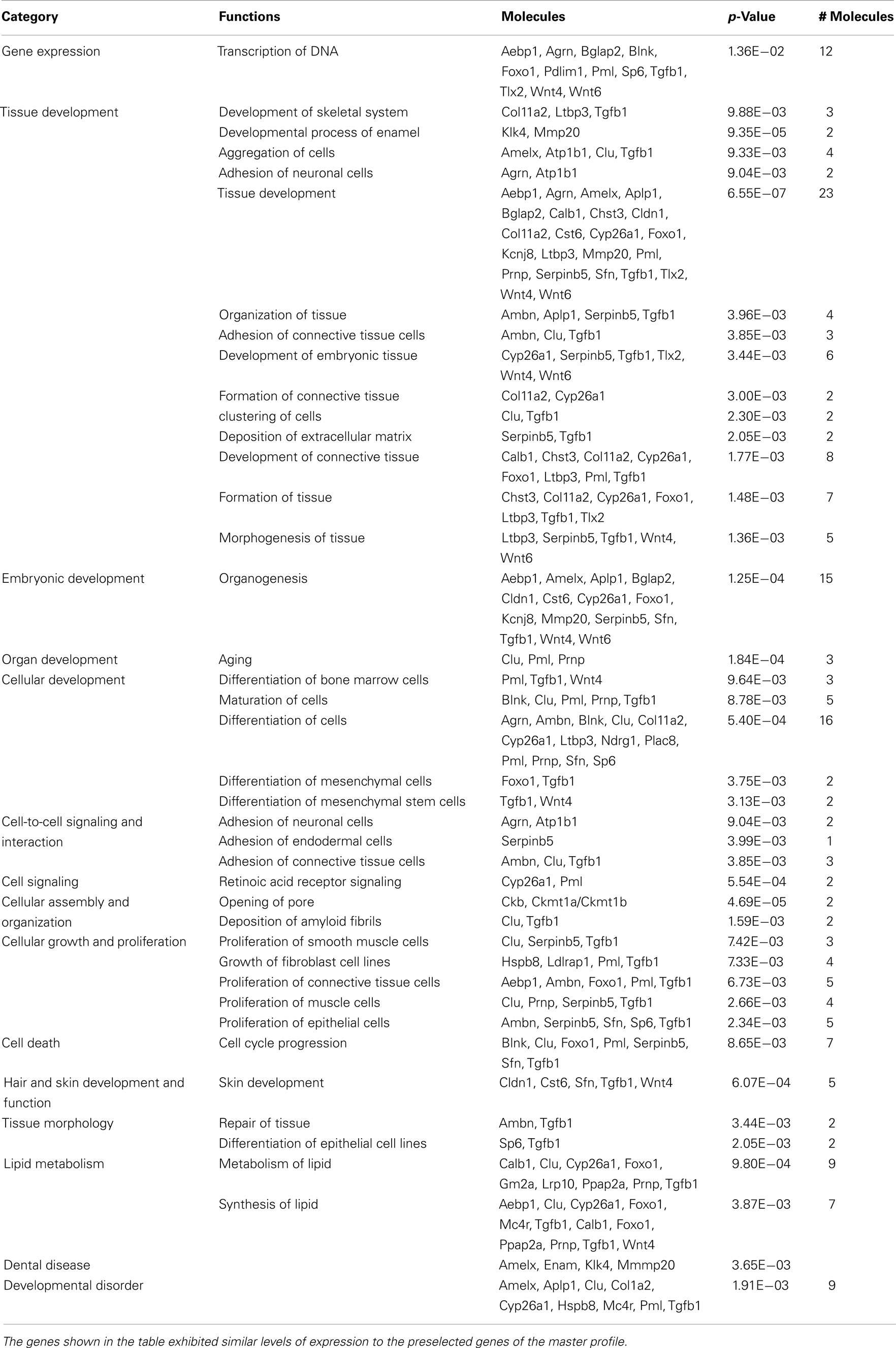
Table 3. The 87 genes isolated using profile search were investigated using IPA to determine significant associations (p = 0.01) with “Category” or with “Functions” as judged by Fisher’s exact test.
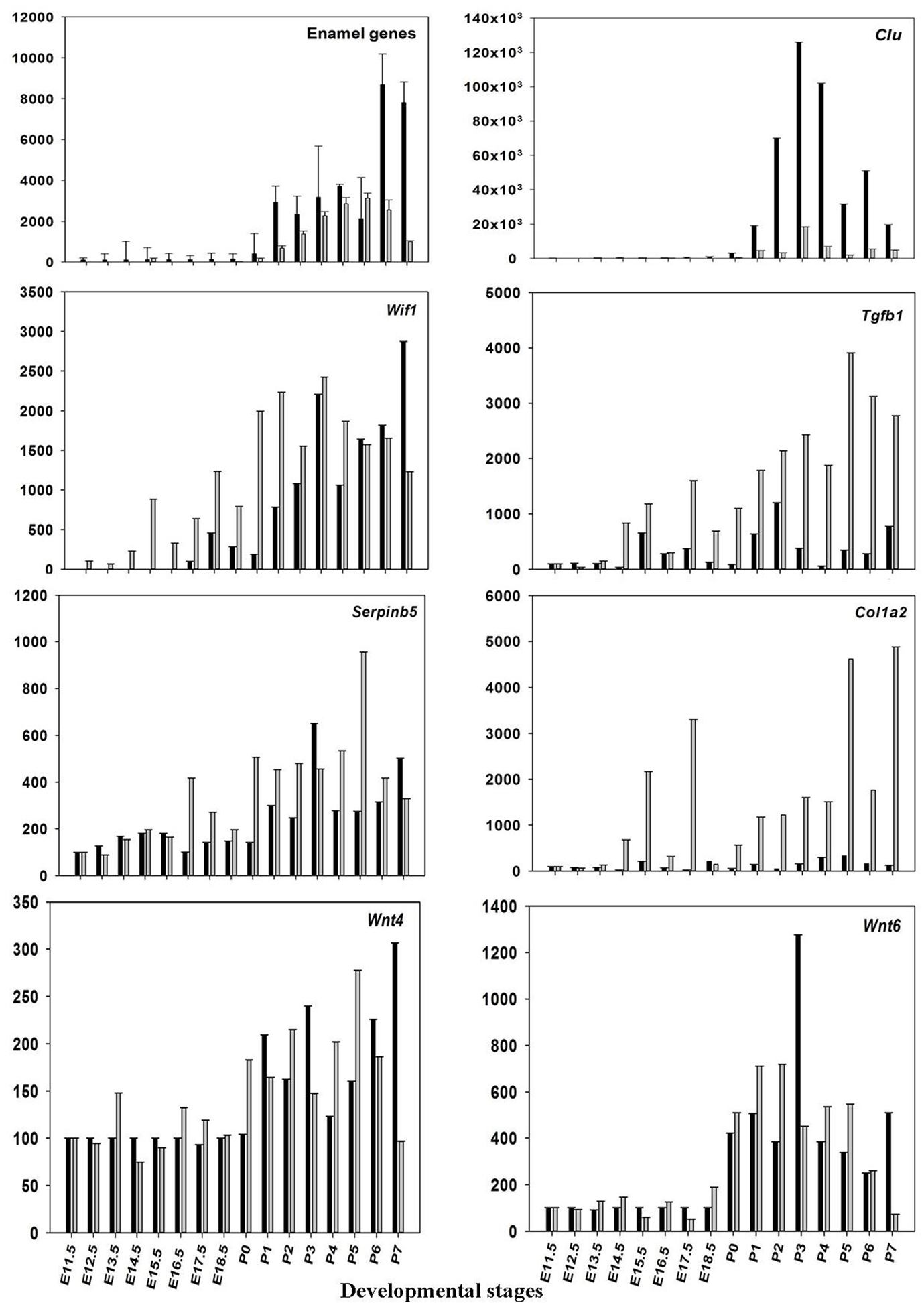
Figure 3. Validation of microarray data. The mean ratios (expression at E11.5 up to P7/expression at E11.5) for some of the differentially expressed (DE) genes (Amelx, Ambn, Enam, Wif1, Clu, Prnp, Mmp20, Col1a1, Tgfb1, Wint6, and Serpinb5) from the microarray data (black) and real-time RT-PCR (gray). Plotted microarray and real-time RT-PCR data are means derived from at least three biological triplicates at each time point and presented with SD.
Bioinformatic Analysis of Differentially Expressed Genes
These 87 genes (Figure 2B) submitted to bioinformatic analysis using IPA were found to be significantly associated with the Ingenuity categories called “gene expression,” “tissue development” (Table 2), “embryonic, organ, and cellular development,” “cell-to-cell signaling and interaction” (Tgfb1, Wnt4, Agrn, Atp1b1, Serpinb5, Ambn, Clu, Tgfb1), “cellular growth and proliferation,” “hair and skin development and function” (Cldn1, Cst6, Sfn, Tgfb1, Wnt4), “dental disease” (Amelx, Enam, Klk4, Mmp20; Table 3). Transcription factor analysis suggested that 15 transcription regulators.
(Egr1, Fos, Folsl2, Hif1a, Nfel2, Nfya Runx2, Smad1, Ap1 Sp1,Sp3, Stat4, Tp53, Tp63, VitaminD3-VDR-RXR) exhibited a p-value of overlap <0.01, proposing significant regulatory associations to several of the 87 cluster genes (Figure 4).
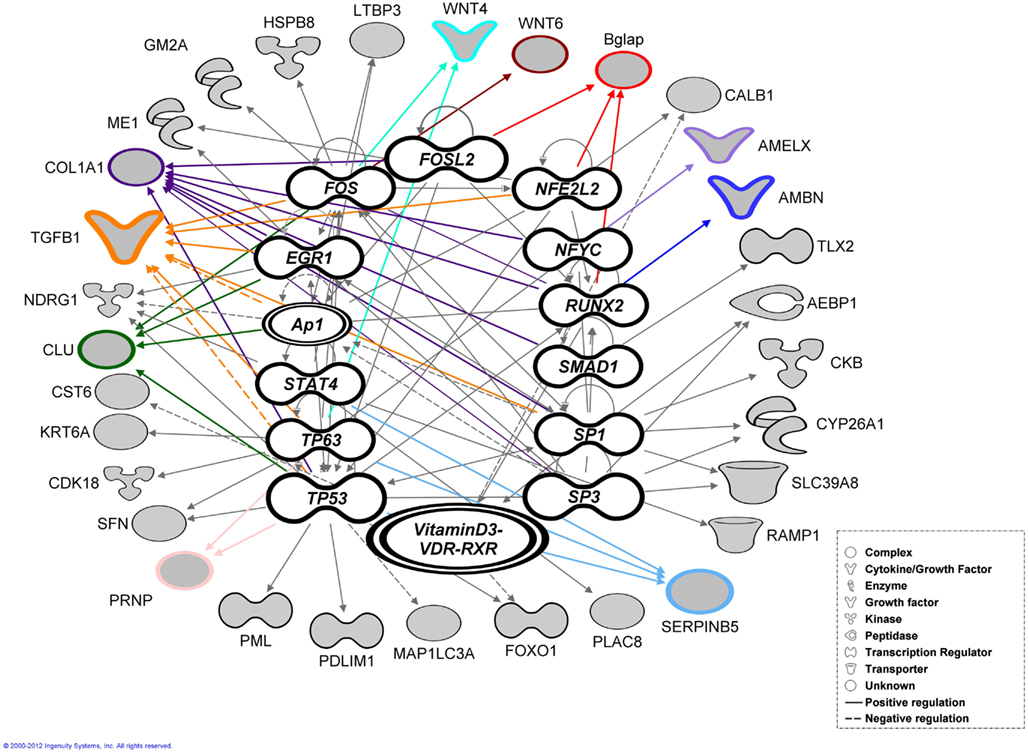
Figure 4. Bioinformatic analysis. Transcription factor analysis performed using ingenuity (IPA) for the 87 genes exhibited a p-value of overlap <0.01 associating significantly 35 genes with 15 transcription factors. The genes verified by real-time RT-PCR highlighted with colored outlines and lines together with most of the associated genes (shaded gray).
In situ Hybridization
During early odontogenesis Amelx, Ambn, and Enam mRNAs showed similar patterns of localization and were mainly observed in cells derived from dental epithelium (Figure 5). These three mRNAs were, however, also detected in the mesenchyme and mesenchyme-derived tissues. Expression of Enam and Ambn mRNA were also observed in cells known to form facial bone structures (Figure 5 arrows). In general the signal from the Amelx probe appeared stronger than signals from the Ambn or Enam probes. During the cap stage the area of the enamel knot exhibited a hybridization signal with Amelx only (Figure 5, E14.5). Treatment with DNase did not alter the hybridization signal. Hybridization with the sense probes consistently showed absence of positive signals (results not shown). Sections treated with RNAse showed no hybridization signals, suggesting the signal to originate from hybridization with RNA in the tissue (results not shown).
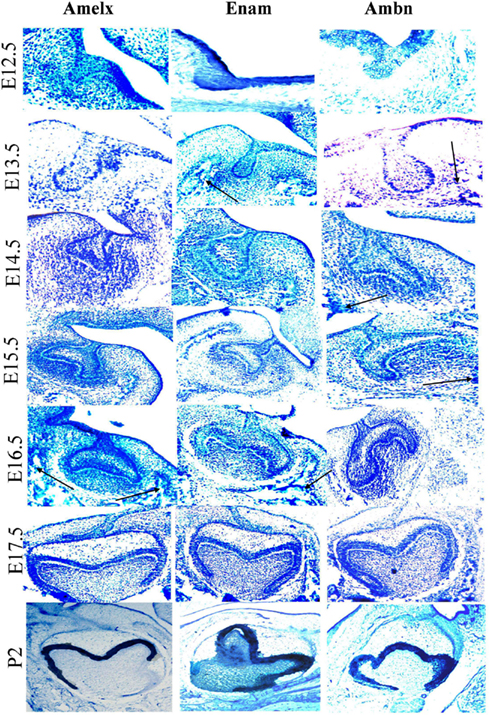
Figure 5. Localization of expression of Ambn, Amelx, and Enam mRNAs during early odontogenesis as detected by in situ hybridization. Detection of Amelx, Enam, and Ambn mRNA was carried out in dissected sections of tooth germs at various stages of development (E12.5–P2). The black arrows indicate mRNA expressed at future facial bone features. The magnifications were ×200 for E12.5–E13.5, ×100 for E14.5 and E15.5–P2.
Western-Blotting
Amelogenin was identified as a band of approximately 50 kDa detected at all developmental stages investigated, whereas an additional band of amelogenin of molecular weight of about 25 kDa was detected at post-natal stages, being the predominant protein band at P2–4 (Figures 6A,B).
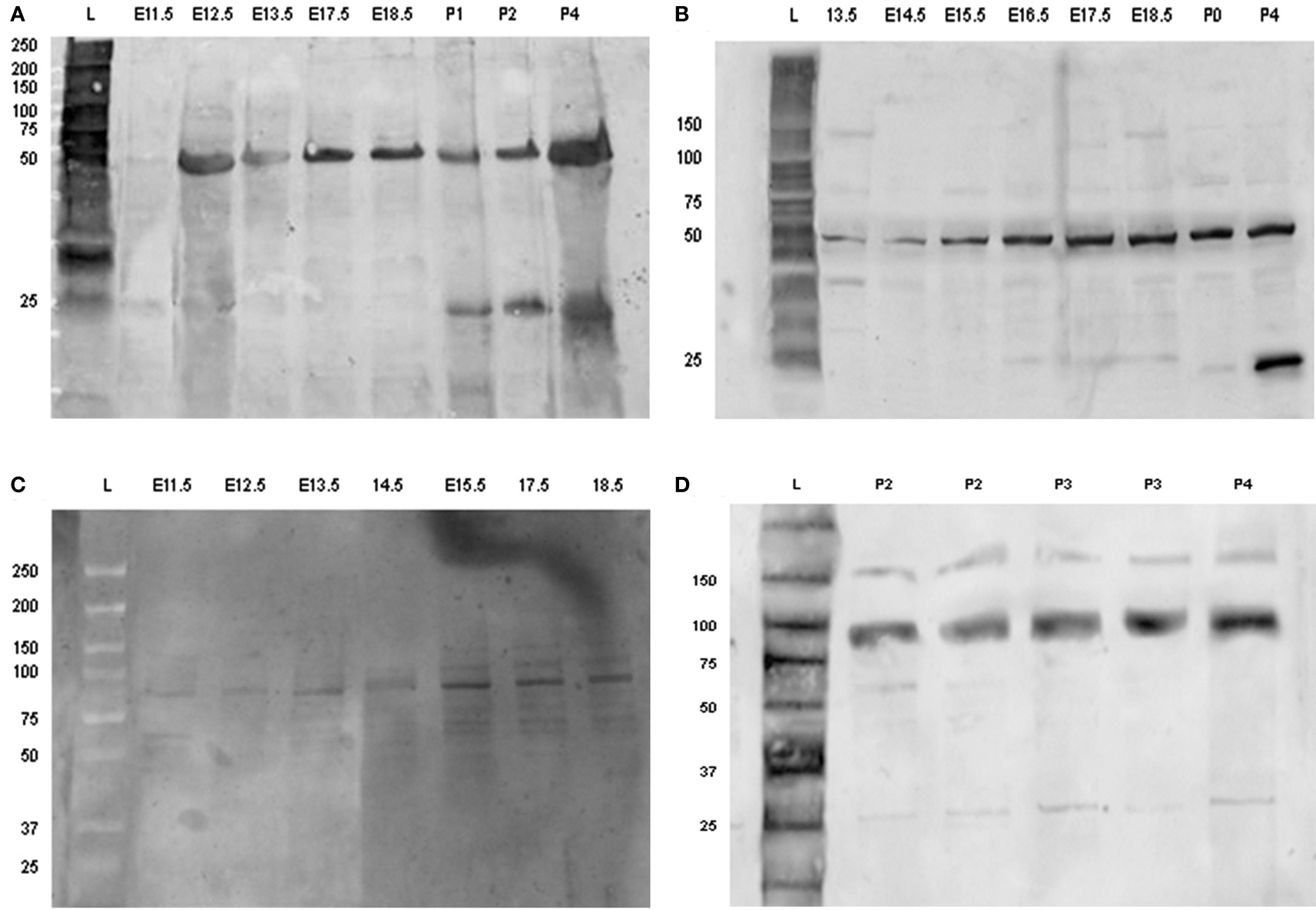
Figure 6. Detection of amelogenin and enamel in using Western-blotting. Western blot of amelogenin (A,B) and enamelin (C,D) from tooth germs dissected from E11.5 up to P5. Each sample loaded on to the gels was derived from separate tooth germs. Abbreviations: L, molecular weight standard ladder; E11.5–E18.5, embryonic days 11.5–18.5; P0, birth; P1–P5, post-natal day 1–5.
Enamelin was detected at pre-secretory stages as a band with molecular mass of about 89 kDa (Figure 6C). The intensity of this band increased at the later stages of tooth development, together with an additional band of molecular mass of about 60–64 kDa (Figure 6D).
The ameloblastin antibody identified a single band of molecular weight of about 48 kDa at pre-secretory stages. This band was relatively weak at all time points (results not shown).
Discussion
To the authors knowledge there are no systematic analysis of gene expression throughout the entire time-course of murine tooth development. Several studies focus on gene expression profiles of selected genes, and or between two time points during murine tooth development (Osmundsen et al., 2007; Kim et al., 2012). Here we present global gene expression across a range of time points (E11.5–P7) during tooth development. The expression of several genes, including Amelx, Ambn, Enam, Wif1, Clu, Prnp, Mmp20, Col1a2, Tgfb1, Wint6, and Serpinb5 at pre-secretory stages has not been described earlier. The resulting data indicate that some of the genes mentioned above are expressed in detectable levels at stages prior to the secretory and late maturation stages of murine tooth development.
During early tooth development invagination of the dental epithelium and condensation of the mesenchyme requires fine control and modulation of both short and long-range cellular communication. Members of both fibroblast growth factor (FGF) and wingless-related MMTV integration site (Wnt) families are expressed in the dental epithelium and have been proposed to regulate gene expression in the underlying mesenchyme during early odontogenesis (Kettunen et al., 2005), e.g., Wif1 is known to bind through the WIF domain to several Wnts, e.g., Wint4 controlling and modulating gradients of secreted morphogens which in turn regulate cellular communication during early tooth formation. At post-natal stages these genes play an active role in cell differentiation, e.g., Wint6 facilitate mineralizing of the murine tooth (Wang et al., 2010). Bioinformatic analysis suggests that some molecules; Amelx, Ambn, Clu, Tgfb1, Serpinb5 have multiple functions, playing different roles during early murine tooth development and mineralization. Most of the genes resulting from Ambn, Amelx, and Enam profile search play an important role in cell proliferation, migration, adhesion during early tooth development (E11.5–E16.5) while during the differentiation and mineralization stages (E17.5 up to P7) these genes contribute to attachment, organization, polarity of either ameloblasts or odontoblasts (Karavanova et al., 1992; Aberg et al., 2004; Nakamura et al., 2008). This multifunctionality may be due to the fact that expression of many of the 87 genes are induced or regulated by different transcription factors and regulators, e.g., Runx2 (Ambn, Bglap, Ibsp, and Col1a2), Fosl2 (Bglap, Col1a2), and VitaminD3/-VDR-RXR (Calb1, CST6, FOXO1), triggering different cellular responses during tooth development (McMahon et al., 1990; Fukumoto et al., 2004; Rufini et al., 2011; Romano et al., 2012). Runx2 suppresses the expression of Ambn and Amel in cultured tooth germs (Kobayashi et al., 2006). Fos is associated with a variety of biological processes, e.g., proliferation and differentiation. Fosl2 (Fra-2) knockout mice show an aberrant tooth formation (Smeyne et al., 1993). Dietary deficiencies in Vitamin D3 affect Calbindin (Calb1) expression in ameloblasts and odontoblasts in rat incisors leading to hypocalcemia and dentin hypomineralization (Berdal et al., 1995).
The in situ results for Amelx, Ambn, and Enam mRNA showed similarities with immunohistochemical localization of Serpinb5, Clusterin, and Prnp (French et al., 1993; Davaadorj et al., 2010; Khan et al., 2010). Expression of enamel proteins in bone cells has previously been reported (Spahr et al., 2006; Tamburstuen et al., 2010; Tamburstuen et al., 2011) and is supported by the presented in situ data.
The combination of several techniques like microarrays, real-time RT-PCR, ISH, and immuno-blotting combined with bioinformatics may serve to provide a more comprehensive view of the cellular consequences of changes in gene expression occurring during murine tooth development. The major challenge is at the level of analysis of microarray data, i.e., isolation of DE genes which are functionally related. The profile search function, used in this study, has shown itself as a useful tool when searching for related genes in large sets of microarray gene expression data. This may also be useful to identify novel genes during murine tooth development.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Zunyi Zhang, Dr. Andre Carvalho, Cuong Khuu, and Professor Randi Furseth Klinge for helpful discussions. We are also grateful to Dr. Emenike Eribe and Dr. Luisa Rodrigues for critical reading of the manuscript. The skilled technical assistance of Mrs. Toril Woldene, Mrs. Bente Gehrken, Mr. Benedicto Geronimo, and Mr. Nikki Agabao was highly appreciated.
Footnotes
References
Aberg, T., Wang, X. P., Kim, J. H., Yamashiro, T., Bei, M., Rice, R., Ryoo, H. M., and Thesleff, I. (2004). Runx2 mediates FGF signaling from epithelium to mesenchyme during tooth morphogenesis. Dev. Biol. 270, 76–93.
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300.
Berdal, A., Papagerakis, P., Hotton, D., Bailleul-Forestier, I., and Davideau, J. L. (1995). Ameloblasts and odontoblasts, target-cells for 1,25-dihydroxyvitamin D3: a review. Int. J. Dev. Biol. 39, 257–262.
Chai, Y., Jiang, X., Ito, Y., Bringas, P. Jr., Han, J., Rowitch, D. H., Soriano, P., Mcmahon, A. P., and Sucov, H. M. (2000). Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127, 1671–1679.
Cheadle, C., Vawter, M. P., Freed, W. J., and Becker, K. G. (2003). Analysis of microarray data using Z score transformation. J. Mol. Diagn. 5, 73–81.
Davaadorj, P., Tokuyama, R., Ide, S., Tadokoro, S., Kudoh, K., and Satomura, K. (2010). Possible involvement of maspin in tooth development. Histochem. Cell Biol. 134, 603–614.
Farquharson, M., Harvie, R., and Mcnicol, A. M. (1990). Detection of messenger RNA using a digoxigenin end labelled oligodeoxynucleotide probe. J. Clin. Pathol. 43, 424–428.
Feng, J., Mcdaniel, J. S., Chuang, H. H., Huang, O., Rakian, A., Xu, X., Steffensen, B., Donly, K. J., MacDougall, M., and Chen, S. (2012). Binding of amelogenin to MMP-9 and their co-expression in developing mouse teeth. J. Mol. Histol. doi:10.1007/s10735-012-9423-1
Fleischmannova, J., Matalova, E., Tucker, A. S., and Sharpe, P. T. (2008). Mouse models of tooth abnormalities. Eur. J. Oral Sci. 116, 1–10.
French, L. E., Chonn, A., Ducrest, D., Baumann, B., Belin, D., Wohlwend, A., Kiss, J. Z., Sappino, A. P., Tschopp, J., and Schifferli, J. A. (1993). Murine clusterin: molecular cloning and mRNA localization of a gene associated with epithelial differentiation processes during embryogenesis. J. Cell Biol. 122, 1119–1130.
Fukumoto, S., Kiba, T., Hall, B., Iehara, N., Nakamura, T., Longenecker, G., Krebsbach, P. H., Nanci, A., Kulkarni, A. B., and Yamada, Y. (2004). Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J. Cell Biol. 167, 973–983.
Hougaard, D. M., Hansen, H., and Larsson, L. I. (1997). Non-radioactive in situ hybridization for mRNA with emphasis on the use of oligodeoxynucleotide probes. Histochem. Cell Biol. 108, 335–344.
Hu, J. C., Lertlam, R., Richardson, A. S., Smith, C. E., McKee, M. D., and Simmer, J. P. (2011). Cell proliferation and apoptosis in enamelin null mice. Eur. J. Oral Sci. 119(Suppl 1), 329–337.
Karavanova, I., Vainio, S., and Thesleff, I. (1992). Transient and recurrent expression of the Egr-1 gene in epithelial and mesenchymal cells during tooth morphogenesis suggests involvement in tissue interactions and in determination of cell fate. Mech. Dev. 39, 41–50.
Kettunen, P., Loes, S., Furmanek, T., Fjeld, K., Kvinnsland, I. H., Behar, O., Yagi, T., Fujisawa, H., Vainio, S., Taniguchi, M., and Luukko, K. (2005). Coordination of trigeminal axon navigation and patterning with tooth organ formation: epithelial-mesenchymal interactions, and epithelial Wnt4 and Tgfbeta1 regulate semaphorin 3a expression in the dental mesenchyme. Development 132, 323–334.
Khan, Q. E., Press, C. M., Sehic, A., Landin, M. A., Risnes, S., and Osmundsen, H. (2010). Expression of prion protein gene and presence of prion protien during development of mouse molar tooth germ. Eur. J. Oral Sci. 118, 559–565.
Kim, K. M., Lim, J., Choi, Y. A., Kim, J. Y., Shin, H. I., and Park, E. K. (2012). Gene expression profiling of oral epithelium during tooth development. Arch. Oral Biol. doi: 10.1016/j.archoralbio.2012.02.019
Kobayashi, I., Kiyoshima, T., Wada, H., Matsuo, K., Nonaka, K., Honda, J. Y., Koyano, K., and Sakai, H. (2006). Type II/III Runx2/Cbfa1 is required for tooth germ development. Bone 38, 836–844.
McMahon, A. P., Champion, J. E., Mcmahon, J. A., and Sukhatme, V. P. (1990). Developmental expression of the putative transcription factor Egr-1 suggests that Egr-1 and c-fos are coregulated in some tissues. Development 108, 281–287.
Nakamura, T., De Vega, S., Fukumoto, S., Jimenez, L., Unda, F., and Yamada, Y. (2008). Transcription factor epiprofin is essential for tooth morphogenesis by regulating epithelial cell fate and tooth number. J. Biol. Chem. 283, 4825–4833.
Osmundsen, H., Landin, M. A., From, S. H., Kolltveit, K. M., and Risnes, S. (2007). Changes in gene-expression during development of the murine molar tooth germ. Arch. Oral Biol. 52, 803–813.
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45.
Reiner, A., Yekutieli, D., and Benjamini, Y. (2003). Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375.
Romano, R. A., Solomon, L. W., and Sinha, S. (2012). Tp63 in oral development, neoplasia, and autoimmunity. J. Dent. Res. 91, 125–132.
Rufini, A., Barlattani, A., Docimo, R., Velletri, T., Niklison-Chirou, M. V., Agostini, M., and Melino, G. (2011). p63 in tooth development. Biochem. Pharmacol. 82, 1256–1261.
Smeyne, R. J., Vendrell, M., Hayward, M., Baker, S. J., Miao, G. G., Schilling, K., Robertson, L. M., Curran, T., and Morgan, J. I. (1993). Continuous c-fos expression precedes programmed cell death in vivo. Nature 363, 166–169.
Spahr, A., Lyngstadaas, S. P., Slaby, I., and Pezeshki, G. (2006). Ameloblastin expression during craniofacial bone formation in rats. Eur. J. Oral Sci. 114, 504–511.
Tamburstuen, M. V., Reppe, S., Spahr, A., Sabetrasekh, R., Kvalheim, G., Slaby, I., Syversen, U., Lyngstadaas, S. P., and Reseland, J. E. (2010). Ameloblastin promotes bone growth by enhancing proliferation of progenitor cells and by stimulating immunoregulators. Eur. J. Oral Sci. 118, 451–459.
Tamburstuen, M. V., Reseland, J. E., Spahr, A., Brookes, S. J., Kvalheim, G., Slaby, I., Snead, M. L., and Lyngstadaas, S. P. (2011). Ameloblastin expression and putative autoregulation in mesenchymal cells suggest a role in early bone formation and repair. Bone 48, 406–413.
Thesleff, I., and Hurmerinta, K. (1981). Tissue interactions in tooth development. Differentiation 18, 75–88.
Thesleff, I., and Mikkola, M. (2002). The role of growth factors in tooth development. Int. Rev. Cytol. 217, 93–135.
Tucker, A. S., and Sharpe, P. T. (1999). Molecular genetics of tooth morphogenesis and patterning: the right shape in the right place. J. Dent. Res. 78, 826–834.
Wang, C., Ren, L., Peng, L., Xu, P., Dong, G., and Ye, L. (2010). Effect of Wnt6 on human dental papilla cells in vitro. J. Endod. 36, 238–243.
Zhang, Y., Zhang, X., Lu, X., Atsawasuwan, P., and Luan, X. (2011). Ameloblastin regulates cell attachment and proliferation through RhoA and p27. Eur. J. Oral Sci. 119(Suppl 1), 280–285.
Keywords: tooth development, ameloblastin, amelogenin, enamelin
Citation: Landin MAdSS, Shabestari M, Babaie E, Reseland JE and Osmundsen H (2012) Gene expression profiling during murine tooth development. Front. Gene. 3:139. doi: 10.3389/fgene.2012.00139
Received: 23 April 2012; Accepted: 11 July 2012;
Published online: 31 July 2012.
Edited by:
Moisés Santillán, Centro de Investigación y Estudios Avanzados del IPN, MexicoReviewed by:
Moisés Santillán, Centro de Investigación y Estudios Avanzados del IPN, MexicoLev Guzman, Instituto Politecnico Nacional Unidad Profesional Interdisciplinaria de Ingeniería y Tecnologías Avanzadas, Mexico
Osbaldo Resendis-Antonio, Universidad Nacional Autónoma de México, Mexico
Copyright: © 2012 Landin, Shabestari, Babaie, Reseland and Osmundsen. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Maria A. dos Santos Silva Landin, Department of Biomaterials, Institute for Clinical Dentistry, PO Box 1109, Blindern, 0317 Oslo, Norway. e-mail: m.d.s.s.landin@odont.uio.no; dosantla@online.no