Spatiotemporal Regulation of CRISPR/Cas9 Enables Efficient, Precise, and Heritable Edits in Plant Genomes
- 1Department of Biological Sciences, Birla Institute of Technology and Science (BITS), Pilani, India
- 2Department of Molecular and Cellular Biology, University of California, Davis, Davis, CA, United States
CRISPR/Cas-mediated editing has revolutionized crop engineering. Due to the broad scope and potential of this technology, many studies have been carried out in the past decade towards optimizing genome editing constructs. Clearly, the choice of the promoter used to drive gRNA and Cas9 expression is critical to achieving high editing efficiency, precision, and heritability. While some important considerations for choosing a promoter include the number and nature of targets, host organism, mode of transformation and goal of the experiment, spatiotemporal regulation of Cas9 expression using tissue-specific or inducible promoters enables higher heritability and efficiency of targeted mutagenesis with reduced off-target effects. In this review, we discuss specific studies that highlight the prospects and trade-offs associated with the choice of promoters on genome editing and emphasize the need for inductive exploration and discovery to further advance this area of research in crop plants.
1 Introduction
The Clustered, Regularly Interspaced, Short Palindromic Repeats (CRISPR)/CRISPR-associated protein (Cas) system is a powerful system for programmable editing of gene structure, expression, and epigenetics (Jinek et al., 2012; Adli, 2018). Unlike other genome editing tools such as meganucleases (MGN), zinc finger nucleases (ZFNs), and transcription activator-like effector-based nucleases (TALENs), the level of precision, flexibility, and ease of application and design of the CRISPR/Cas system is unparalleled (Huang et al., 2015; Arora and Narula, 2017). Further, due to its ability to target multiple genes, the CRISPR/Cas system can address the challenge of high redundancy in plant genes while establishing gene-function relationships (Xing et al., 2014; Abdelrahman et al., 2021).
The major components of the CRISPR/Cas9-mediated genome editing toolkit are Cas9 endonuclease and guide RNA (gRNA), which form a complex and cleave the target DNA adjacent to the protospacer adjacent motif (PAM). The gRNA acts as a guide for the Cas9 protein to make the double-strand breaks (DSBs) at the target site, which may either be repaired through an error-prone non-homologous end-joining (NHEJ) or the high-fidelity homologous recombination (HR) pathway (Iliakis et al., 2004; Wyman and Kanaar, 2006; Heyer et al., 2010). The optimization of the construct design, especially codon optimization of Cas9, gRNA design, and regulatory regions (promoters) used for driving gRNA and Cas9 expression, are critical to achieving high precision, targeting efficiency and heritability (Li et al., 2013; Johnson et al., 2015; Hassan et al., 2021). The efficiency is usually reported as the percentage of transgenic plants with a mutation at the intended target, while precision is assessed based on the frequency of off-target mutations. Heritability is estimated based on the number of plants that inherit the targeted mutation from one generation to another. Here, we highlight the impact of spatiotemporal regulation of Cas9/gRNA expression on mutagenesis efficiency, precision, and heritability of genome edits in plants.
1.1 Different Architectures of Clustered, Regularly Interspaced, Short Palindromic Repeats Constructs
Several architectures of the expression cassette for Cas9 and gRNA expression are being used in plants (Figure 1A) (Montecillo et al., 2020). A mixed dual promoter system has most frequently been used where a Pol II promoter drives Cas9 expression while a Pol III promoter is used for gRNA expression (Lowder et al., 2015). The use of two distinct Pol II promoters for Cas9 and sgRNA expression has also been successfully demonstrated (Čermák et al., 2017). Pol II promoters are especially useful for transcribing multiple gRNAs as a single transcriptional unit to facilitate multiplex editing when coupled with either self-cleaving hammerhead (HH) and hepatitis delta virus (HDV) ribozymes, bacterial CRISPR-associated RNA endoribonuclease Csy4, or endogenous tRNA processing enzymes to facilitate gRNA processing (Čermák et al., 2017). Since increasing the construct size by using two distinct Pol II promoters for Cas9 and sgRNA expression puts an additional constraint on transformation in plants, a single transcript unit (STU) system where a single promoter is used to express both sgRNAs and Cas9 is also effective (Tang et al., 2019). Insertion of sgRNA in the intron of the Cas9 further enhances the system’s efficiency (Figure 1A) (Ding et al., 2018). However, repeated use of the same Pol II promoter to express the gRNA and Cas9 risks homology-dependent gene silencing (Rajeevkumar et al., 2015). Recently, the use of bidirectional promoters to regulate Cas9 and gRNA expression has also been successfully demonstrated in plants (Ren et al., 2019). It facilitates coordinated expression of Cas9 and gRNA while shortened vector would be advantageous during plant transformation. However, due to a limited number of bidirectional promoters characterized to date, this system needs further investigations for wider applicability. Below we encapsulate the wide range of promoters that have been leveraged for genome editing in plants so far (Figure 1B).
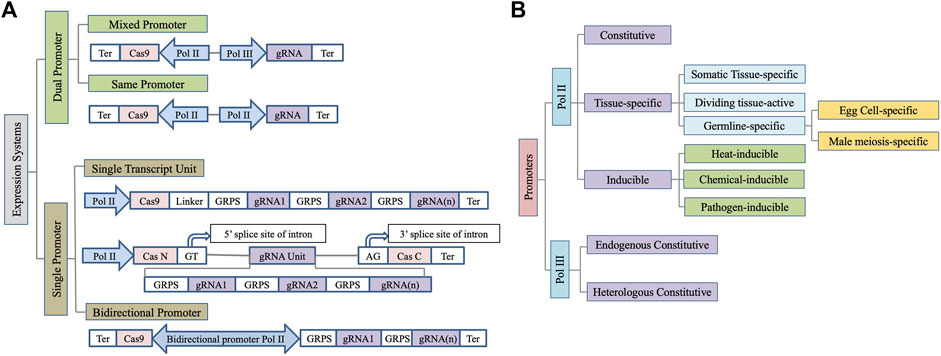
FIGURE 1. Schematic depiction of wide range of (A) expression systems and (B) promoters used for Cas9/gRNA expression in plants. Ter, Terminator; GRPS, Guide RNA processing system.
1.2 Constitutive Promoters
Constitutive promoters with sustained high expression in all cell types have been extensively used in transgenic systems. Both constitutive RNA Polymerase II (Pol II) and III (Pol III) promoters have been leveraged for genome editing applications in plants.
1.2.1 Polymerase II Constitutive Promoters
Constitutive Pol II promoters derived from plant pathogens (e.g., CaMV, NOS) or housekeeping genes (e.g., Ubiquitin, Actin, EF1A2) have been most widely used for driving Cas9 expression in plants with endogenous promoters exhibiting higher efficiency over heterologous promoters (Li et al., 2015). Besides, Cauliflower mosaic virus (CaMV) 35S promoter exhibits greater efficiency in dicots while maize Ubiquitin promoter (pZmUbi) exhibits higher mutagenesis rate in monocots (Feng et al., 2013). However, Cas9 driven by constitutive promoter generally leads to the formation of chimeras requiring screening of a large number of edited lines for several generations to obtain homozygous mutants (Feng et al., 2014). Especially in Arabidopsis, which is transformed using the floral dip method, constitutive promoters result in mosaics in T1 generation as mutations mainly occur after the first embryonic cell division (Fauser et al., 2014; Feng et al., 2014). Also, despite the high mutagenesis efficiency reported with these promoters, the heritability of mutations remains low due to the limited activity of constitutive promoters in germline cells. Similar results have been reported in soybean, where plant transformation depends on organogenesis and regeneration. Most edits are somatic and non-transmissible in soybean when Cas9 is expressed using a constitutive promoter (Zheng et al., 2020).
1.2.2 Polymerase III Constitutive Promoters
Due to high transcriptional efficiency with short transcripts, Pol III promoters have been extensively used for regulating gRNA expression. As is the case for Pol II promoters, endogenous Pol III promoters exhibit higher editing efficiency (Sun et al., 2015; Long et al., 2018; Qi et al., 2018; Ren et al., 2021). Due to limited characterization of Pol III promoters in less-studied systems, U6 and U3 promoters of Arabidopsis and rice have been most extensively used for gRNA expression in dicots and monocots, respectively (Ma et al., 2015; Lowder et al., 2016; Montecillo et al., 2020). Trimmed U3 and U6 promoters, with only essential elements for transcriptional initiation, have been used to further shorten the construct (Hao et al., 2020). However, there are some obvious limitations associated with Pol III promoters. For example, earlier U6 and U3 promoters were believed to have a highly conserved transcription start site at +1. Instead, the transcription start site of the U6 promoter seems to vary, which can have undesirable consequences for gRNA specificity (Ma et al., 2014). Some of the studies have also demonstrated Pol II activity of Pol III promoters with varying strength which can have adverse implications on localization and stability of gRNAs (Gao et al., 2018). The dual property (Pol II and Pol III) of the H1 promoter of humans has recently been utilized to drive both guide RNA and Cas9 expression (Gao et al., 2019). Further, Pol III promoters obviously lack spatiotemporal control and tunability, and due to their limited ability to regulate longer transcripts, they are not suitable for multiplex editing applications.
1.3 Tissue-Specific Promoters
Since sustained expression of Cas9 using constitutive promoter provides a wider opportunity for off-target effects, spatiotemporal control of Cas9 expression using tissue-specific promoters would be ideal for restricting undesirable Cas9 expression. Based on the experiment’s goal, researchers have employed several germline-specific, cell division-active, and somatic tissue-specific promoters for genome editing in plants (Table 1). We refer to promoters with preferential high expression in meristematic and germline cells as cell division-active as they may have some activity in other cells as well.
TABLE 1. List of spatiotemporally regulated promoters used for driving Cas9/gRNA expression in plants.
1.3.1 Cell Division-Active Promoters
Promoters of several genes active in proliferating cells of shoot apical meristem, root meristem, young leaves, anthers, pollen, embryo sac, embryo, and endosperm such as YAO (YAOZHE), ICU (INCURVATA), EFα1 (Elongation factor alpha 1), H4 (Histone 4), RPS5A (Ribosomal Protein S5A), CDC45 (Cell division control protein 45) have been leveraged for driving Cas9 expression in Arabidopsis. All these studies report higher editing efficiency and heritability of homozygous mutations with dividing tissue-specific promoters than constitutive promoters (Hyun et al., 2015; Yan et al., 2015; Osakabe et al., 2016; Tsutsui and Higashiyama, 2017; Feng Z. et al., 2018). Some of the genes even exhibited homozygous mutations in T1 generation itself with 100% heritability in T2 plants with RPS5A promoter-driven Cas9 (Tsutsui and Higashiyama, 2017). Later, Feng and co-workers (2018b) demonstrated higher editing efficiency, heritability, and frequency of homozygous mutants in T1 plants of Arabidopsis for multiplex editing by using YAO and CDC45 promoters compared to 35S promoter. Similar results were reported in tomato as well where three constitutive promoters, including CaMV35S, Pcubi4 (Polyubiquitin 4), and Slp16 (Ribosomal protein p16) were compared with the promoter of Elongation Factor-1alpha (SIEF1α) which is primarily active in meristematic cells of shoot tips and shoot apical meristems. The authors observed the highest mutation efficiency with SIEFα with the reduced number of mosaic mutations in T0 plants (Hashimoto et al., 2018). These results demonstrate the superiority of dividing cell-active promoters over constitutive promoters for individual loci and multiplex editing in plants.
1.3.2 Germlines-Specific Promoters
With Cas9 expression restricted to germline cells, male meiosis and egg cell-specific promoters have the potential to enhance the heritability of mutations and reduce the frequency of somatic mutations. Both male and female germlines-specific promoters have been leveraged for this purpose.
1.3.2.1 Male Meiosis-Specific Promoters
Eid and co-workers (2016) tested three meiosis-specific promoters for driving Cas9 expression in Arabidopsis. MGE1p is specific to meiosis I, MGE2p is specific to meiosis I and II, while MGE3p is specific to meiosis II. All three promoters mediated mutagenesis in T1 plants, with MGE1p exhibiting the highest efficiency, while MGE3p had the lowest efficiency. MGE1p even outperformed the egg-cell-specific promoter of EC1.2p. Similarly, Xu and co-workers (2018) used meiocyte-specific promoter DMC1 (DNA Meiotic Recombinase 1), which is active in both male and female meiocytes, to drive Cas9 expression in Arabidopsis and reported 64% T1 plants as heterozygous while 37% of the T2 plants were homozygous for the targeted mutation. Conversely, the use of the DMC1 promoter for regulating Cas9 expression in maize led to 66% homozygous mutants in T0 generation itself which were stably inherited in T1 plants (Feng C. et al., 2018). Moreover, no off-target mutations were detected in the predicted loci. Since transgenic plants were regenerated using tissue culture procedure in maize, high expression of DMC1 in calli as well as germline cells likely contributed to the high mutagenesis efficiency with this promoter which can be leveraged for other grain crops as well.
1.3.2.2 Egg Cell-Specific Promoters
Cas9 expression driven by EC1.2 (Egg Cell 1.2)/DD45/(Downregulated in dif1 45) promoter that exhibits specific expression in the egg cells and one-cell stage embryos led to homozygous mutants in T1 Arabidopsis plants (Wang et al., 2015). Although its molecular basis is still unclear, a fusion promoter developed using the EC1.1 promoter and EC1.2 enhancer further improved the mutation efficiency. A similar observation was made in another study with Arabidopsis and soybean where four different promoters, including egg cell-specific promoters, AtP5p and AtEC1.2e1.1 of Arabidopsis, and GmEC1.1p and GmEC1.2p of soybean, were utilised for Cas9 expression (Zheng et al., 2020). The AtEC1.2e1.1 promoter resulting from the fusion of cis-regulatory elements of the EC1.1 and EC1.2 promoters exhibited higher mutation frequency than the other three promoters in T2 plants of Arabidopsis. In soybean, AtEC1.2e1.1 led to a mutation rate of 26.8%, while no mutations could be detected with the other three promoters in T0 soybean plants. Although the number of transgenic soybean lines was too low to draw a clear conclusion, both the studies clearly suggest that an optimized combination of egg cell-specific promoters can enhance the mutation efficiency and proportion of homozygous mutations.
Mao and co-workers (2016) compared the efficiency of DD45 promoter with male germline-specific promoters, SPL (SPOROCYTELESS) and LAT52 in Arabidopsis (Mao et al., 2016). SPL predominantly expresses in early microsporocytes while LAT52 expresses during late pollen development. Though all germ-line specific promoters led to heritable heterozygous mutations in the T2 generation, only DD45 promoter led to heritable homozygous mutations in the T1 generation. The authors speculate that since the primary target in the floral dip transformation method in Arabidopsis is ovules, the expression of DD45 promoter in zygotes might be synchronized with Agrobacterium infection of egg cells resulting in homozygotes in the T1 generation. Later, Miki and co-workers (2018) compared the DD45 and LAT52 promoters with dividing tissue-active YAO and CDC45 promoters for highly efficient RNA-targeted gene knock-in (targeted insertions of external genes) in Arabidopsis. The authors developed “all-in-one” constructs with HDR (Homology-directed Repair) donor sequence and sgRNA targeting genomic locus of interest for GFP knock-in into the ROS1 and DME (DEMETER) gene loci. Yet again, while precise knock-ins that could produce ROS1-GFP, DME-GFP, or GFP-DME fusions were accomplished with other promoters, only DD45 promoter led to heritable and efficient gene targeting in T2 and T3 plants, indicating that HDR may be more efficient in egg cells and/or early embryos. Wolter and co-workers (2018) compared the mutation efficiency of promoters of stem-cell identity regulator CLV3 (CLAVATA3) with YAO and EC1.1 for gene targeting by homologous recombination (HR) in Arabidopsis. While CLV3 and YAO promoters exhibited very low GT frequency like constitutive ubiquitin promoter, the egg cell-specific fusion promoter EC1.1 exhibited high genome targeting frequency with heritable events in 74% of the T1 lines (Wolter et al., 2018). Comparative analysis of editing efficiencies of RPS5a and DD45 promoters with several constitutive and tissue-specific promoters including p35S, pPcUbi, pAP1, pICU2, pGILT, and pALB to target Lhcb1 genes, which is present in five copies at two loci in the Arabidopsis genome also confirmed the superior performance of pRPS5a and DD45 among all the tested promoters (Ordon et al., 2020). These studies demonstrate the application of egg cell-specific promoters for wide range of genome editing applications.
1.3.3 Somatic Tissues-Specific Promoters
In situations where gene editing can be detrimental to the organism’s survival or lead to pleiotropic effects, hindering the further experimental investigation, somatic tissue-specific editing can be performed. For instance, to analyze the function of Hydroxycinnamoyl transferase (HCT), which is required for lignin biosynthesis in Arabidopsis, Liang and co-workers (2019) used a fibre-specific promoter pNSD3/SND1 to drive Cas9 expression. With a 90% decrease in HCT activity, the chimera plants exhibited a normal growth phenotype allowing biochemical analysis of the plants while the cell-specific editing was maintained in the T2 generation. Another fascinating example of using tissue-specific promoters for editing essential genes comes from a study done by Feder and co-workers (2020), where the authors demonstrated the use of fruit-specific phosphoenolpyruvate carboxylase 2 (PPC2) promoter in tomato for editing the SET-domain containing polycomb gene, SIEZ2, which earlier yielded pleiotropic phenotypes when targeted using RNA interference (Boureau et al., 2016). Although PPC2 promoter exhibited leakiness in seeds, the study allowed investigation of additional roles of SIEZ2 in fruit maturation. More recently, Bollier and co-workers (2021) used this approach for multigene editing of fundamentally important genes using root cap-specific promoter pSMB (SOMBRERO). The authors confirmed the editing of six genes simultaneously with high efficiency in T1 and T2 Arabidopsis plants. This system allows the knockdown of multiple genes with redundant or synergistic functions to dissect cell/tissue-specific genetic networks while maintaining the sequence integrity in germline cells.
1.4 Inducible Promoters
While germline-specific promoters help deal with the challenge of lower heritability of mutations, the inducible promoters may further reduce the off-target effects due to transient expression of Cas9. The inducible systems respond to either chemical inducers or biotic/abiotic stimuli (Qiu and Yu, 2009; Misra and Ganesan, 2021).
1.4.1 Chemical-Inducible Promoters
Several chemicals such as estradiol, ethanol, ecdysone, glucocorticoid, etc., have been used for developing inducible systems in plants. However, some of these can lead to untended growth defects (Kang et al., 1999; Roslan et al., 2001; Amirsadeghi et al., 2007). The estrogen-inducible chimeric transcription activator (XVE) system has been successfully used in several plant species without any undesirable impact on plant growth and morphology. Tang and workers (2019) used the XVE system to demonstrate the application of an STU system where Cas9, sgRNA, and ribozyme were expressed using a single estradiol-inducible promoter for regulated expression of Cas9 and sgRNA in the rice protoplast system. The authors observed leaky expression of Cas9 in some samples with low level of mutagenesis detected without estradiol treatment.
Recently Wang and co-workers (2020) demonstrated the application of an inducible editing system for creating gene knockouts in Arabidopsis. The authors tested four inducible promoters viz., pWOL: XVE, pWOX5:XVE, pSCR: XVE, and pWER: XVE, which drive expression in specific cell types of root meristem to target the PLT2 (PLETHORA2) gene. The PLT2 editing was assessed using YFP fluorescence as an indicator. Further, the cell-type specificity of their system in root meristems was demonstrated by targeting AP2/EREBP family transcription factor genes, PLT1 and PLT2. The double mutant plt1,2 phenotype was rescued using gPLT2-3xYFP, where 3xYFP was used to restrict the mobility of PLT2, thereby allowing tracking of cell-specific editing of PLT2. Results suggest that the cell-type-specific inducible system is a promising strategy for generating cell-type-specific high-precision gene knockouts.
1.4.2 Heat-Inducible Promoters
A heat-inducible promoter is induced by a high-temperature stimulus. There have been three studies so far demonstrating heat-inducible promoters for editing applications. Nandy and co-workers (2019) used promoter of soybean heat shock protein 17.5E (HSP17.5E) for driving Cas9 expression for targeted editing of endogenous phytoene desaturase (PDS) gene and β-Glucuronidase transgene in rice. The heat-shock treatment of calli/seedlings led to heritable heterozygous and homozygous mutations in T0 plants with more than 3 times mutation frequency and significantly reduced off-target effects compared to constitutive rice ubiquitin promoter. Conversely, Barone and co-workers (2020) demonstrated the use of heat shock inducible promoter Hsp26 to facilitate intragenomic gene targeting in maize. After transformation, induction of Cas9 in undifferentiated cells led to DSBs with simultaneous mobilization of donor template from pre-integrated T-DNA. High-frequency intragenomic HR led to non-chimeric heritable gene targeting with 4.7% target insertion in T0 plants. More recently, Hanania and colleagues (2020) used 18.2 Arabidopsis heat shock promoter to drive sgRNA expression, specially designed to excise the target T-DNA boundaries for excision of the transgene from Nicotiana tabacum BY2 (Bright Yellow 2) cells after confirmation of successful editing. Together these studies demonstrate wide applications of heat-inducible promoters in plants provided the duration and cycles of heat treatment are optimised.
1.4.3 Pathogen-Inducible Promoters
Pathogen-inducible promoters are a promising strategy to engineer disease resistance in crop plants. Ji and co-workers (2018) demonstrated the use of a virus-inducible CRISPR/Cas system to engineer resistance against beet curly top virus (BSCTV) in tobacco transient and Arabidopsis transgenic systems. After observing off-target effects in T2 plants using constitutive promoters, the authors tested BSCTV-inducible pV86 and pC86 promoters for Cas9 expression. Since the pathogen itself induces pathogen inducible systems, these are ideal for engineering disease resistance without needing an inducer and worrying about the mode of application. Also, as no other factors have been shown to induce these promoters, no leaky expression and off-target effects were detected in the absence of the pathogen.
2 Conclusions and Future Perspectives
It is evident from the above studies that regardless of the system used, spatiotemporal regulation of Cas9 enables greater precision and heritability compared to constitutive promoters. The dividing tissue-active promoters enhance the mutation efficiency, heritability and proportion of homozygous mutants in Arabidopsis. The germline-specific promoters further decrease the chimerism by limiting Cas9 expression to specific cell types along with improved heritability and frequency of homozygous mutations. Further, egg cell-specific expression of Cas9 has been shown to stimulate CRISPR-dependent HDR more efficiently (Miki et al., 2018). Somatic tissue-specific promoters, on the other hand, are a boon for characterizing essential genes without disturbing the integrity of germline cells. However, in cases where target tissue types may not have promoters with high enough activity to be deployed for editing, synthetic/fusion promoters may be designed to fine-tune the regulatory behaviours of the known regulatory elements (Ali and Kim, 2019; Zhang et al., 2020).
Inducible promoters provide tighter spatial as well as temporal control and therefore, would be an ideal system for cell type-specific high precision edits for both essential and non-essential genes. Use of transient expression of inducible promoters compared to somatic tissue-specific promoters for characterizing essential genes would likely reduce the frequency of somatic mutations also. However, no studies have been conducted so far in plants to compare the performance of the inducible promoters with tissue-specific promoters. The leaky expression, speed of response and limited mobility of inducer may be of concern especially when the target tissue is deep seated like ovules and embryo sacs. Further research is needed to fully explore the potential of inducible promoters vis-à-vis tissue-specific promoters.
Another major lacuna is that most of the studies described here have been conducted in Arabidopsis which has a very different mode of transformation compared to the crop species that are dependent on tissue culture procedures for transgenic generation. Also, variable response of some of these promoters has been observed in different host systems and laboratories (Shockey, 2020). Some of the factors that likely contribute to this variability include variable length of regulatory regions used in different studies, host-specific factors, variability in sequence of gRNAs and target region, and impact of other regulatory elements in the vector used. Therefore, comparative assessment of spatiotemporally-regulated promoters across different plant species would be required to assess the wider applicability of tissue-specific and inducible promoters beyond Arabidopsis. Considering the exponential advancements this technology has witnessed in the past decade and its societal impact, the quest for enhanced efficiency, specificity and heritability of Cas9-mediated editing will hopefully lead to a wider assortment of promoters in the future which can then be used for developing recommender systems for specific host species and genome editing applications.
Author Contributions
AM, AG and FR collected the data from the literature and helped drafting the manuscript. RS conceptualized, prepared the framework and finalized the draft of the review. All the authors read and approved the article.
Funding Statement
FR and RS acknowledge the financial assistance in the form of a Core Research Grant (CRG/2020/003466) from the Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Government of India.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelrahman, M., Wei, Z., Rohila, J. S., and Zhao, K. (2021). Multiplex Genome-Editing Technologies for Revolutionizing Plant Biology and Crop Improvement. Front. Plant Sci. 12, 721203. doi:10.3389/fpls.2021.721203
Adli, M. (2018). The CRISPR Tool Kit for Genome Editing and beyond. Nat. Commun. 9 (1), 1911–1913. doi:10.1038/s41467-018-04252-2
Ali, S., and Kim, W.-C. (2019). A Fruitful Decade Using Synthetic Promoters in the Improvement of Transgenic Plants. Front. Plant Sci. 10, 1433. doi:10.3389/fpls.2019.01433
Amirsadeghi, S., Mcdonald, A. E., and Vanlerberghe, G. C. (2007). A Glucocorticoid-Inducible Gene Expression System Can Cause Growth Defects in Tobacco. Planta 226 (2), 453–463. doi:10.1007/s00425-007-0495-1
Arora, L., and Narula, A. (2017). Gene Editing and Crop Improvement Using CRISPR-Cas9 System. Front. Plant Sci. 8, 1932. doi:10.3389/fpls.2017.01932
Barone, P., Wu, E., Lenderts, B., Anand, A., Gordon-Kamm, W., Svitashev, S., et al. (2020). Efficient Gene Targeting in maize Using Inducible CRISPR-Cas9 and Marker-free Donor Template. Mol. Plant 13 (8), 1219–1227. doi:10.1016/j.molp.2020.06.008
Bollier, N., Andrade Buono, R., Jacobs, T. B., and Nowack, M. K. (2021). Efficient Simultaneous Mutagenesis of Multiple Genes in Specific Plant Tissues by Multiplex CRISPR. Plant Biotechnol. J. 19 (4), 651–653. doi:10.1111/pbi.13525
Boureau, L., How-Kit, E., Drevensek, S., Rainieri, M., Joubès, J., Stammitti, L., et al. (2016). A CURLY LEAF Homologue Controls Both Vegetative and Reproductive Development of Tomato Plants. Plant Mol. Biol. 90 (4-5), 485–501. doi:10.1007/s11103-016-0436-0
Čermák, T., Curtin, S. J., Gil-Humanes, J., Čegan, R., Kono, T. J., Konečná, E., et al. (2017). A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants. The Plant Cell 29 (6), 1196–1217. doi:10.1105/tpc.16.00922
Ding, D., Chen, K., Chen, Y., Li, H., and Xie, K. (2018). Engineering Introns to Express RNA Guides for Cas9- and Cpf1-Mediated Multiplex Genome Editing. Mol. Plant 11 (4), 542–552. doi:10.1016/j.molp.2018.02.005
Eid, A., Ali, Z., and Mahfouz, M. M. (2016). High Efficiency of Targeted Mutagenesis in Arabidopsis via Meiotic Promoter-Driven Expression of Cas9 Endonuclease. Plant Cel Rep 35 (7), 1555–1558. doi:10.1007/s00299-016-2000-4
Fauser, F., Schiml, S., and Puchta, H. (2014). Both CRISPR/Cas-based Nucleases and Nickases Can Be Used Efficiently for Genome Engineering inArabidopsis Thaliana. Plant J. 79 (2), 348–359. doi:10.1111/tpj.12554
Feder, A., Jensen, S., Wang, A., Courtney, L., Middleton, L., Van Eck, J., et al. (2020). Tomato Fruit as a Model for Tissue-specific Gene Silencing in Crop Plants. Hortic. Res. 7 (1), 142–147. doi:10.1038/s41438-020-00363-4
Feng, C., Su, H., Bai, H., Wang, R., Liu, Y., Guo, X., et al. (2018a). High-efficiency Genome Editing Using a Dmc1 Promoter-Controlled CRISPR/Cas9 System in maize. Plant Biotechnol. J. 16 (11), 1848–1857. doi:10.1111/pbi.12920
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D.-L., et al. (2014). Multigeneration Analysis Reveals the Inheritance, Specificity, and Patterns of CRISPR/Cas-induced Gene Modifications in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111 (12), 4632–4637. doi:10.1073/pnas.1400822111
Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D.-L., Wei, P., et al. (2013). Efficient Genome Editing in Plants Using a CRISPR/Cas System. Cell Res 23 (10), 1229–1232. doi:10.1038/cr.2013.114
Feng, Z., Zhang, Z., Hua, K., Gao, X., Mao, Y., Botella, J., et al. (2018b). A Highly Efficient Cell Division-specific CRISPR/Cas9 System Generates Homozygous Mutants for Multiple Genes in Arabidopsis. Ijms 19 (12), 3925. doi:10.3390/ijms19123925
Gao, Z., Herrera-Carrillo, E., and Berkhout, B. (2019). A Single H1 Promoter Can Drive Both Guide RNA and Endonuclease Expression in the CRISPR-Cas9 System. Mol. Ther. - Nucleic Acids 14, 32–40. doi:10.1016/j.omtn.2018.10.016
Gao, Z., Herrera-Carrillo, E., and Berkhout, B. (2018). RNA Polymerase II Activity of Type 3 Pol III Promoters. Mol. Ther. - Nucleic Acids 12, 135–145. doi:10.1016/j.omtn.2018.05.001
Hanania, U., Ariel, T., Turbovski, A., Rathod, V. K. R., Oz, D., Tekoah, Y., et al. (2020). Sequential Genome Editing and Induced Excision of the Transgene in N. Tabacum BY2 Cells. Front. Plant Sci. 11, 607174. doi:10.3389/fpls.2020.607174
Hao, Y., Zong, W., Zeng, D., Han, J., Chen, S., Tang, J., et al. (2020). Shortened snRNA Promoters for Efficient CRISPR/Cas-based Multiplex Genome Editing in Monocot Plants. Sci. China Life Sci. 63, 933–935. doi:10.1007/s11427-019-1612-6
Hashimoto, R., Ueta, R., Abe, C., Osakabe, Y., and Osakabe, K. (2018). Efficient Multiplex Genome Editing Induces Precise, and Self-Ligated Type Mutations in Tomato Plants. Front. Plant Sci. 9, 916. doi:10.3389/fpls.2018.00916
Hassan, M. M., Zhang, Y., Yuan, G., De, K., Chen, J.-G., Muchero, W., et al. (2021). Construct Design for CRISPR/Cas-based Genome Editing in Plants. Trends Plant Sci. 26 (11), 1133–1152. doi:10.1016/j.tplants.2021.06.015
Heyer, W.-D., Ehmsen, K. T., and Liu, J. (2010). Regulation of Homologous Recombination in Eukaryotes. Annu. Rev. Genet. 44, 113–139. doi:10.1146/annurev-genet-051710-150955
Huang, H., Zheng, G., Jiang, W., Hu, H., and Lu, Y. (2015). One-step High-Efficiency CRISPR/Cas9-mediated Genome Editing in Streptomyces. Acta Biochim. Biophys. Sinica 47 (4), 231–243. doi:10.1093/abbs/gmv007
Hyun, Y., Kim, J., Cho, S. W., Choi, Y., Kim, J.-S., and Coupland, G. (2015). Site-directed Mutagenesis in Arabidopsis thaliana Using Dividing Tissue-Targeted RGEN of the CRISPR/Cas System to Generate Heritable Null Alleles. Planta 241 (1), 271–284. doi:10.1007/s00425-014-2180-5
Iliakis, G., Wang, H., Perrault, A. R., Boecker, W., Rosidi, B., Windhofer, F., et al. (2004). Mechanisms of DNA Double Strand Break Repair and Chromosome Aberration Formation. Cytogenet. Genome Res. 104 (1-4), 14–20. doi:10.1159/000077461
Ji, X., Si, X., Zhang, Y., Zhang, H., Zhang, F., and Gao, C. (2018). Conferring DNA Virus Resistance with High Specificity in Plants Using Virus-Inducible Genome-Editing System. Genome Biol. 19 (1), 197. doi:10.1186/s13059-018-1580-4
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337 (6096), 816–821. doi:10.1126/science.1225829
Johnson, R. A., Gurevich, V., Filler, S., Samach, A., and Levy, A. A. (2015). Comparative Assessments of CRISPR-Cas Nucleases' Cleavage Efficiency in Planta. Plant Mol. Biol. 87 (1), 143–156. doi:10.1007/s11103-014-0266-x
Kang, H.-G., Fang, Y., and Singh, K. B. (1999). A Glucocorticoid-Inducible Transcription System Causes Severe Growth Defects in Arabidopsis and Induces Defense-Related Genes. Plant J. 20 (1), 127–133. doi:10.1046/j.1365-313x.1999.00575.x
Li, J.-F., Norville, J. E., Aach, J., Mccormack, M., Zhang, D., Bush, J., et al. (2013). Multiplex and Homologous Recombination-Mediated Genome Editing in Arabidopsis and Nicotiana Benthamiana Using Guide RNA and Cas9. Nat. Biotechnol. 31 (8), 688–691. doi:10.1038/nbt.2654
Li, Z., Liu, Z.-B., Xing, A., Moon, B. P., Koellhoffer, J. P., Huang, L., et al. (2015). Cas9-Guide RNA Directed Genome Editing in Soybean. Plant Physiol. 169 (2), 960–970. doi:10.1104/pp.15.00783
Liang, Y., Eudes, A., Yogiswara, S., Jing, B., Benites, V. T., Yamanaka, R., et al. (2019). A Screening Method to Identify Efficient sgRNAs in Arabidopsis, Used in Conjunction with Cell-specific Lignin Reduction. Biotechnol. Biofuels 12 (1), 130–215. doi:10.1186/s13068-019-1467-y
Long, L., Guo, D. D., Gao, W., Yang, W. W., Hou, L. P., Ma, X. N., et al. (2018). Optimization of CRISPR/Cas9 Genome Editing in Cotton by Improved sgRNA Expression. Plant Methods 14 (1), 85–89. doi:10.1186/s13007-018-0353-0
Lowder, L. G., Zhang, D., Baltes, N. J., Paul, J. W., Tang, X., Zheng, X., et al. (2015). A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant Physiol. 169 (2), 971–985. doi:10.1104/pp.15.00636
Lowder, L., Malzahn, A., and Qi, Y. (2016). Rapid Evolution of Manifold CRISPR Systems for Plant Genome Editing. Front. Plant Sci. 7, 1683. doi:10.3389/fpls.2016.01683
Ma, H., Wu, Y., Dang, Y., Choi, J.-G., Zhang, J., and Wu, H. (2014). Pol III Promoters to Express Small RNAs: Delineation of Transcription Initiation. Mol. Ther. - Nucleic Acids 3, e161. doi:10.1038/mtna.2014.12
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 8 (8), 1274–1284. doi:10.1016/j.molp.2015.04.007
Mao, Y., Zhang, Z., Feng, Z., Wei, P., Zhang, H., Botella, J. R., et al. (2016). Development of Germ-line-specific CRISPR-Cas9 Systems to Improve the Production of Heritable Gene Modifications inArabidopsis. Plant Biotechnol. J. 14 (2), 519–532. doi:10.1111/pbi.12468
Miki, D., Zhang, W., Zeng, W., Feng, Z., and Zhu, J. K. (2018). CRISPR/Cas9-mediated Gene Targeting in Arabidopsis Using Sequential Transformation. Nat. Commun. 9 (1), 1967–1969. doi:10.1038/s41467-018-04416-0
Misra, S., and Ganesan, M. (2021). The Impact of Inducible Promoters in Transgenic Plant Production and Crop Improvement. Plant Gene 27, 100300. doi:10.1016/j.plgene.2021.100300
Montecillo, J. A. V., Chu, L. L., and Bae, H. (2020). CRISPR-Cas9 System for Plant Genome Editing: Current Approaches and Emerging Developments. Agronomy-Basel 10 (7). doi:10.3390/agronomy10071033
Nandy, S., Pathak, B., Zhao, S., and Srivastava, V. (2019). Heat-shock-inducible CRISPR/Cas9 System Generates Heritable Mutations in rice. Plant direct 3 (5), e00145. doi:10.1002/pld3.145
Ordon, J., Bressan, M., Kretschmer, C., Dall’Osto, L., Marillonnet, S., Bassi, R., et al. (2020). Optimized Cas9 Expression Systems for Highly Efficient Arabidopsis Genome Editing Facilitate Isolation of Complex Alleles in a Single Generation. Funct. Integr. Genomics 20 (1), 151–162. doi:10.1007/s10142-019-00665-4
Osakabe, Y., Watanabe, T., Sugano, S. S., Ueta, R., Ishihara, R., Shinozaki, K., et al. (2016). Optimization of CRISPR/Cas9 Genome Editing to Modify Abiotic Stress Responses in Plants. Sci. Rep. 6 (1), 26685–26710. doi:10.1038/srep26685
Qi, X., Dong, L., Liu, C., Mao, L., Liu, F., Zhang, X., et al. (2018). Systematic Identification of Endogenous RNA Polymerase III Promoters for Efficient RNA Guide-Based Genome Editing Technologies in maize. Crop J. 6 (3), 314–320. doi:10.1016/j.cj.2018.02.005
Qiu, Y., and Yu, D. (2009). Over-expression of the Stress-Induced OsWRKY45 Enhances Disease Resistance and Drought Tolerance in Arabidopsis. Environ. Exp. Bot. 65 (1), 35–47. doi:10.1016/j.envexpbot.2008.07.002
Rajeevkumar, S., Anunanthini, P., and Sathishkumar, R. (2015). Epigenetic Silencing in Transgenic Plants. Front. Plant Sci. 6, 693. doi:10.3389/fpls.2015.00693
Ren, C., Liu, Y., Guo, Y., Duan, W., Fan, P., Li, S., et al. (2021). Optimizing the CRISPR/Cas9 System for Genome Editing in Grape by Using Grape Promoters. Hortic. Res. 8 (1), 52–12. doi:10.1038/s41438-021-00489-z
Ren, Q., Zhong, Z., Wang, Y., You, Q., Li, Q., Yuan, M., et al. (2019). Bidirectional Promoter-Based CRISPR-Cas9 Systems for Plant Genome Editing. Front. Plant Sci. 10, 1173. doi:10.3389/fpls.2019.01173
Roslan, H. A., Salter, M. G., Wood, C. D., White, M. R. H., Croft, K. P., Robson, F., et al. (2001). Characterization of the Ethanol-Inducible Alc Gene-Expression System in Arabidopsis thaliana. Plant J. 28 (2), 225–235. doi:10.1046/j.1365-313x.2001.01146.x
Shockey, J. (2020). Gene Editing in Plants: Assessing the Variables through a Simplified Case Study. Plant Mol. Biol. 103 (1), 75–89. doi:10.1007/s11103-020-00976-2
Sun, X., Hu, Z., Chen, R., Jiang, Q., Song, G., Zhang, H., et al. (2015). Targeted Mutagenesis in Soybean Using the CRISPR-Cas9 System. Sci. Rep. 5 (1), 10342–10410. doi:10.1038/srep10342
Tang, X., Ren, Q., Yang, L., Bao, Y., Zhong, Z., He, Y., et al. (2019). Single Transcript Unit CRISPR 2.0 Systems for Robust Cas9 and Cas12a Mediated Plant Genome Editing. Plant Biotechnol. J. 17 (7), 1431–1445. doi:10.1111/pbi.13068
Tsutsui, H., and Higashiyama, T. (2017). pKAMA-ITACHI Vectors for Highly Efficient CRISPR/Cas9-mediated Gene Knockout in Arabidopsis thaliana. Plant Cel Physiol 58 (1), 46–56. doi:10.1093/pcp/pcw191
Wang, X., Ye, L., Lyu, M., Ursache, R., Löytynoja, A., and Mähönen, A. P. (2020). An Inducible Genome Editing System for Plants. Nat. Plants 6 (7), 766–772. doi:10.1038/s41477-020-0695-2
Wang, Z. P., Xing, H. L., Dong, L., Zhang, H. Y., Han, C. Y., Wang, X. C., et al. (2015). Egg Cell-specific Promoter-Controlled CRISPR/Cas9 Efficiently Generates Homozygous Mutants for Multiple Target Genes in Arabidopsis in a Single Generation. Genome Biol. 16 (1), 144–212. doi:10.1186/s13059-015-0715-0
Wolter, F., Klemm, J., and Puchta, H. (2018). Efficientin Plantagene Targeting in Arabidopsis Using Egg Cell‐specific Expression of the Cas9 Nuclease ofStaphylococcus Aureus. Plant J. 94 (4), 735–746. doi:10.1111/tpj.13893
Wyman, C., and Kanaar, R. (2006). DNA Double-Strand Break Repair: All's Well that Ends Well. Annu. Rev. Genet. 40, 363–383. doi:10.1146/annurev.genet.40.110405.090451
Xing, H. L., Dong, L., Wang, Z. P., Zhang, H. Y., Han, C. Y., Liu, B., et al. (2014). A CRISPR/Cas9 Toolkit for Multiplex Genome Editing in Plants. BMC Plant Biol. 14 (1), 327–412. doi:10.1186/s12870-014-0327-y
Xu, P., Su, H., Chen, W., and Lu, P. (2018). The Application of a Meiocyte-specific CRISPR/Cas9 (MSC) System and a Suicide-MSC System in Generating Inheritable and Stable Mutations in Arabidopsis. Front. Plant Sci. 9, 1007. doi:10.3389/fpls.2018.01007
Yan, L., Wei, S., Wu, Y., Hu, R., Li, H., Yang, W., et al. (2015). High-efficiency Genome Editing in Arabidopsis Using YAO Promoter-Driven CRISPR/Cas9 System. Mol. Plant 8 (12), 1820–1823. doi:10.1016/j.molp.2015.10.004
Zhang, S., Guo, F., Yan, W., Dai, Z., Dong, W., Zhou, J., et al. (2020). Recent Advances of CRISPR/Cas9-based Genetic Engineering and Transcriptional Regulation in Industrial Biology. Front. Bioeng. Biotechnol. 7, 459. doi:10.3389/fbioe.2019.00459
Keywords: Cas9, inducible promoter, tissue-specific promoter, genome editing, CRISPR
Citation: Rahman F, Mishra A, Gupta A and Sharma R (2022) Spatiotemporal Regulation of CRISPR/Cas9 Enables Efficient, Precise, and Heritable Edits in Plant Genomes. Front. Genome Ed. 4:870108. doi: 10.3389/fgeed.2022.870108
Received: 05 February 2022; Accepted: 24 March 2022;
Published: 26 April 2022.
Edited by:
Gözde S Demirer, California Institute of Technology, United StatesReviewed by:
Lily Cheung, Georgia Institute of Technology, United StatesTomáš Čermák, Ball Horticultural Company, United States
Jennifer Brophy, Stanford University, United States
Copyright © 2022 Rahman, Mishra, Gupta and Sharma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rita Sharma, rita.genomics@gmail.com, rita.sharma@pilani.bits-pilani.ac.in
 Farhanur Rahman
Farhanur Rahman Apurva Mishra
Apurva Mishra Archit Gupta
Archit Gupta Rita Sharma
Rita Sharma