Antimicrobial Drimane Sesquiterpenes Contribute to Balanced Antagonism but Do Not Structure Bacterial and Fungal Endophytes in the African Pepper Bark Tree Warburgia ugandensis
- 1Division of Terrestrial Ecosystem Research, Department of Microbiology and Ecosystem Science, Faculty of Life Sciences, University of Vienna, Vienna, Austria
- 2Bioresources Unit, Austrian Institute of Technology GmbH, Tulln, Austria
- 3Division of Molecular Systems Biology, Department of Ecogenomics and System Biology, Faculty of Life Sciences, University of Vienna, Vienna, Austria
- 4World Agroforestry Centre (ICRAF), Nairobi, Kenya
- 5Department for Plant Biochemistry, Albrecht-von-Haller-Institute, University of Goettingen, Goettingen, Germany
The African pepper bark tree, Warburgia ugandensis, accumulates antimicrobial drimane sesquiterpenes in all of its organs. One hypothesis states that plant defense compounds determine endophyte community structure. Another hypothesis suggests that they just facilitate the endophytic lifestyle by exerting a balanced antagonism. To explore this, a representative selection of endophytic bacterial and fungal isolates from this tree species was assayed together with six non-endophytic strains to determine their tolerance and susceptibility to the root and leaf extract fraction containing high and low drimane sesquiterpene amounts respectively. Inhibitory effects were explored by assessing both growth and growth efficiency, the latter of which relates respiratory activity to growth. The susceptibility of the tested strains showed considerable variation and the obtained patterns did not allow a clear distinction between root and leaf endophytes as well as endophytes and non-endophytes. In addition, all strains were also assayed against juglone, an antimicrobial and redox-active aromatic naphthoquinone. A comparison of differential pulse voltammograms and efficacy in variants of the deoxyribose degradation assay revealed that drimane sesquiterpenes possess anti- and pro-oxidant activities that compare to those of juglone. Leaf endophytes showed higher resistance to oxidative stress than root endophytes, quite contrary to the actual exposure. The obtained results support the notion that structural diverse plant defense compounds can contribute to a balanced antagonism against but not to structuring of endophyte communities. Oxidative stress seems to be involved in generating this effect albeit it cannot explain it alone.
Introduction
The term endophyte refers to organisms that at least spend part of their life cycle within a host plant. It includes taxa from archaea, bacteria, fungi and protists (Hardoim et al., 2015). Extensive culture-based and -independent analysis of endophytic bacteria and fungi in the last decades led to the identification of endophytes in both monocotyledonous and dicotyledonous plants, ranging from mosses to woody tree species. Nearly all plant species are assumed to represent potential hosts to one or more endophytic species (Rodriguez et al., 2009). Endophytes have to be considered as an important part of the plant microbiome and, in some instances, can even increase stress tolerance levels of the host plant (Redman et al., 2002; Sessitsch et al., 2012; Hardoim et al., 2015). In attempts to maintain a “stable relationship” with the host plant, endophytes produce various metabolites and enzymes, both of which are regarded as lucrative biotechnological resources (Strobel et al., 2004; Wang and Dai, 2011; Hardoim et al., 2015).
Besides various physiological defense mechanisms, plants are especially known to produce antimicrobial secondary metabolites, some of which can be induced by the colonization event of the micro-organism. The majority, however, is constitutive and supposed to help controlling the development of potential pathogens (Bednarek and Osbourn, 2009). Many of those metabolites show pronounced antibacterial or antifungal effects in in vitro assays (Grayer and Harborne, 1994; Cowan, 1999). Likewise, endophytic micro-organisms, especially fungi, have also been identified to produce diverse secondary metabolites with toxic activities against a wide range of organisms (Strobel et al., 2004).
The above described “stable relationship” may thus be considered as a balanced antagonism scenario that can either develop between host plant and the endophyte (Schulz et al., 1999) or, as suggested later, between the endophyte and other co-occurring micro-organisms in the host plant tissue (Schulz et al., 2015). In both cases, secondary metabolites can be involved in interactions that lead to a “balanced antagonism,” which could explain the symptomless development of endophytes in plant tissues. One concern with these hypotheses is that we do not know about the quality of metabolites that bacteria or fungi produce in the apoplast in which bacteria and fungi develop before they attack the cells. What we know has been gained from studies in which endophytes were cultured on artificial media in axenic cultures in the laboratory. Likewise, we do not know the actual amounts of plant secondary metabolites to which endophytic micro-organisms are exposed to in plant tissues. In comparison to other organisms, plants can accumulate rather large amounts, which is facilitated by evolving efficient compartmentalization structures within their tissues, such as resin canals, oil ducts, the vacuole for more polar compounds, or glandular hairs for more volatile derivatives, amongst others (Gershenzon, 2002; Hadacek, 2002). In the late stage of an endophyte's life cycle, when many of them start to develop saprophytic behavior (Schulz and Boyle, 2005), they can come into contact with the host's secondary metabolites as a consequence of tissue decomposition processes that destroy the compartmental barriers.
In a previous paper (Drage et al., 2014), we described the fungal and bacterial endophytes that could be detected in the roots, fruits, and leaves of the African pepper bark tree, Warburgia ugandensis Sprague, syn. W. salutaris (Bertol.f.) Chiov., which is classified into the rather primitive plant family Canellaceae. This family is known to occur as trees in the paleo- and neotropics. Its organs contain drimane sesquiterpenes (DS) that can cause a hot taste, similar to chili pepper. Various biological activities have been documented for drimane sesquiterpenes, such as antimicrobial, antifungal, antiprotozoal, insect antifeedant, cytotoxic, and molluscicidal activities (Jansen and de Groot, 2004). As a consequence, considerable interest exists from traditional medicine. A culture-independent approach, T-RFLP (terminal restriction fragment length polymorphism) combined with PCR cloning, was used to characterize the endophyte communities in the leaves and roots from ten trees growing in two different sites along the Rift valley in Kenya, Eastern Africa (Drage et al., 2014). Contrary to initial expectations, no correlations between the variable accumulation patterns of drimane sesquiterpenes in the roots and leaves of ten individuals and their fungal and bacterial endophyte communities could be found.
This study reports the culture-dependent recovery of bacterial and fungal endophytes from identical trees from the same accession sites from roots, fruits, leaves, leaf litter. The recovered strains were identified on the basis of sequence comparison of ribosomal marker gene regions. A representative subset of these isolates, five root endophytes and six leaf endophytes were compared to six randomly selected strains from sources, three bacteria and three fungi. All were assayed in liquid serial dilutions to determine their respective susceptibility or tolerance of drimane sesquiterpenes. Root and leaf extract fractions containing high and low amounts of drimane sesquiterpenes were incorporated into the assays. The growth in the dilution series was curve-fitted to a log-logistic growth model. Additionally, growth efficiency was determined to obtain information to what extent microbial respiration was affected (Drage et al., 2012).
Endophytes are thought to preferentially colonize the apoplast in plant tissues (McCully, 2001) and can be exposed to high concentrations of ROS (reactive oxygen species), a scenario of oxidative stress (Sharova and Medvedev, 2017). Their levels can rise as a consequence of stress to the host plant (Torres, 2010) or tissue differentiation (Swanson and Gilroy, 2010). Increased ROS concentrations are considered as toxic. In attempts to obtain some insights about a potential mode of action, the drimane sesquiterpene fraction was compared to juglone, a well-known redox-active plant metabolite (Chobot and Hadacek, 2009). Juglone was included into growth susceptibility and growth efficiency assays. In addition, we compared the drimane sesquiterpene fraction and juglone in the deoxyribose degradation assay, which informs about the potential of a compound to scavenge or generate ROS (Chobot, 2010), and an routine electrochemical method, differential pulse voltammetry.
The main objectives of this study were
(1) To isolate representative endophyte strains from roots and aerial organs of Warburgia and identify them on basis of rDNA marker genes.
(2) To explore if host endophytic bacteria and fungi tolerate drimane sesquiterpenes more or less compared to other, non-associated strains.
(3) To determine if drimane sesquiterpenes can affect ROS levels as a potential mode of action.
These objectives aim at testing the hypothesis that drimane sesquiterpenes can affect the endophyte community structure by antagonistic interactions. Higher amounts of drimane sesquiterpenes in root tissues should result in more pronounced effects. So far, two published studies provide support for such an assumption. The first one (Carter et al., 1999) showed that oat-colonizing fungi can degrade the saponin avenacin A-1 more efficiently, which could affect the structure of the root-associated fungal community. The second (Saunders and Kohn, 2009) found that fungi, which tolerated benzoxazines, were more abundant in producing maize plants than in non-producing mutant individuals. In a previous study, we explored if bacterial and fungal endophyte community structures in the pepper bark tree W. ugandensis correlated with drimane sesquiterpene patters, which not only varied between organs but also between individuals (Drage et al., 2014). None, however, could be found. In the former study, the community analysis was performed culture-independently by T-RFLP, the present study explores the same scenario by assaying selected endophytic isolates and non-associated strains against drimane sesquiterpenes.
Materials and Methods
Chemicals
All chemicals, if not stated otherwise, were obtained from Sigma Aldrich (Schnelldorf, Germany) and standard microbiology media from Merck (Darmstadt, Germany). Ultrapure water (18.2 MΩ cm) was prepared using a Millipore Milli-Q 185 Plus system (Millipore Corp., Billerica, MS). DNA polymerase was purchased from Solis BioDyne, (Tartu, Estonia), endonucleases HaeIII and AluI and Go Taq® Green Master kit from Promega GmbH (Mannheim, Germany).
Plant Material
In autumn 2007, leaves, leaf litter and young roots of W. ugandensis trees were sampled in Kenya from two sites, one east (near Rumuruti town, 0°19′ N/36°30′ E) and one west (near Kitale town, 01°00′ N/35°01′ E) of the Great Rift Valley in Kenya, Africa (Drage et al., 2014).
Bacterial and Fungal Endophytes: Isolation and Strain Identification
One cm2 leaf or fruit tissue, or 1 cm fine root pieces were dipped into 70% ethanol and flame treated. The surface-sterilized plant material was embedded into MS agar to facilitate recovery of endophytes after transport to the laboratory. The plates were prepared in Kenya, but further processing only commenced 10 days later in Austria.
The procedures of isolation and bacterial and fungal endophytes have been described in detail elsewhere (Drage et al., 2012). In brief, bacteria were isolated from macerated surface-sterilized plant tissue. After culturing 10−1 dilutions on TSA (tryptic soy broth agar) and R2A (Reasoner's 2A agar) plates, 96 randomly chosen colonies were investigated further. Fungi were transferred as they emerged from the MS medium-embedded plant tissue and repeatedly cultured on MEA (malt extract agar) until purity was obtained. The choice was based on morphological similarity and repeated occurrence on the same tissue type. Altogether, 42 isolates were chosen for ITS and partial 28S rRNA sequence analyses.
Procedures of DNA isolation and PCR targeting bacterial 16S rDNA and fungal ITS and partial 28S rRNA genes were standard and have been described previously (Drage et al., 2012, 2014). The nucleotide sequences determined in this study have been deposited in the GenBank database; accession numbers JF836819-45 and HQ130661-721.
For isolation of genomic DNA, bacteria were grown over night in tryptic soy broth. DNA was isolated as described (Reiter and Sessitsch, 2006). 16S rDNA PCR was carried out using the primers 8f (5′-AGAGTTTGATCCTGGCTCAG-3′), and 1520r (5′-AAGGAGGTGATCCAGCCGCA-3′) (Edwards et al., 1989), pHr (5′-TGCGGCTGGATCACCTCCTT-3′) and P23SR01 (5′-GGCTGCTTCTAAGCCAAC-3′) (Massol-Deya et al., 1995). PCRs were performed adding 1 μL extracted DNA to 1 μL PCR reaction buffer (Invitrogen Corp, Carlbad, CA), 2.5 mM MgCl2, 0.15 μM of each primer, 0.2 mM of deoxynucleoside triphosphate, and 2.5 U FIREPOL DNA polymerase (Solis BioDyne, Tartu, Estonia); cycler conditions: 5 min denaturation at 95°C, 30 cycles consisting of denaturation for 30 s at 95°C, primer annealing for 1 min at 53°C, polymerization for 2 min at 72°C, and final extension for 10 min at 72°C. Aliquots of the PCR products containing 200 ng of amplified DNA were digested with 5 U of endonuclease HaeIII and subsequently with 5 U of AluI for 3 h at 37°C. The digest was analyzed by 2.5% agarose gel electrophoresis. One isolate of each ribotype was identified by 16S rDNA sequencing with the primer 8f making use of the sequencing service of the company AGOWA (Berlin, Germany). Sequences visualization was done with the sequence alignment editor package BioEdit (Ibis Biosciences, Carlsbad, CA). The aligned sequences were identified by BLAST analysis.
Fungi were grown on MEA (malt extract agar) at room temperature. Fungal mycelium was washed from the plates with 1–2 ml of 0.1% Triton X100 buffer and collected by centrifugation. DNA isolation procedures were identical to bacteria. PCR was carried out by using primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) TW13 (5′-GGTCCGTGTTTCAAGACG-3′) to amplify a 1.5 kb PCR product containing the ITS and part of LSU sequence. For PCR, 1 μL of undiluted DNA was added to 14 μL Go Taq® Green Mastermix containing 1 μM of each primer. Cycler conditions were as following: 2.5 min following: 2.5 min denaturation at 95°C, 35 cycles consisting of denaturation for 30 s at 94°C, primer annealing for 30 s at 54°C, polymerization for 2 min at 72°C, and final extension for 5 min at 72°C. Sequencing, sequence alignment and identification was carried out as for bacteria.
Other Randomly Selected Strains
These included the bacterial strains Bacillus simplex DSM 1312, a human pathogen, Paenibacillus amylolyticus DSM 11730, a soil bacterium, Paraburkholderia phytofirmans PsJN, a plant growth promoting bacterium from onions. The chosen fungi were Penicillium expansum VIAM MA2811, an air-borne isolate, Fusarium avenaceum VIAM MA1512, an endophyte from tubers of the Apiaceae Cicuta virosa, and Fusarium oxysporum f. sp. lycopersici Fol 007, a tomato plant pathogen.
Drimane Sesquiterpenes
Leaves and roots were oven-dried at 35°C for up to 3 days. In Kenya, air-drying was not possible due to high humidity. Dried plant material was stored in paper bags at ambient temperature until further processing. To obtain extracts for bioassays, the whole available material from all 20 accessions was pooled and pulverized. The powder (no total weight determined) was extracted twice with MeOH for 24 h (250 mL MeOH/10 g pulverized dried plant material). Portions of the combined extract (~200 mg) were dissolved in 50 mL water and subjected to a one-way solid-phase extraction on 20 g of the resin Amberlite XAD-1180. The drimane sesquiterpenes were eluted by absolute ethanol. The procedure also removed all insoluble particles. The obtained fractions were analyzed by GC–MS after chemical derivatization into trimethylsilyl derivatives to determine their drimane sesquiterpene composition (MSTFA, Thermo Scientific, Waltham, MS). One hundred microgram of the dried ethanolic eluate were dissolved in 100 μL N-methyl.N-TMS.trifluoroacetamide (MSTFA, Thermo Scientific Inc., Waltham, MS). The silylation reaction was allowed to run for 1 h at room temperature. The instrument was a Autosystem XL gas chromatograph linked to a Turbomass quadrupole mass analyzer (Perkin Elmer, Waltham, MS), the column a Zebron 5 ms (Phenomenex, Torrance, CA), 18 m × 0.18 mm × 0.18 μm. The splitless injector was set to 250°C. One microliter was injected. The temperature gradient started from 70 to 300°C, 3°C min−1. The transfer line was set to 280°C, the ion source to 300°C). The procedures are analog to previously described ones (Drage et al., 2014).
Bacterial and Fungal Inoculum Preparation
Bacterial endophyte isolates were cultured on TSB medium (tryptic soy broth) prepared in a buffer (3 g TSB, 1 L 25 mM NaOH/KH2PO4 buffer, pH = 7.4). Fungal endophyte isolates were cultured on mannitol supplemented malt extract agar (10 g malt extract, 20 g mannitol, 3 g peptone, 15 g agar, 1 L water). Conidia formation was monitored by microscopic observation. Conidia were harvested using 0.9% aqueous NaCl supplemented with 5% DMSO. To 1 mL of the concentrated, harvested conidia suspension 1 mL of aqueous 14% sucrose and 1 % peptone was added. Bacterial inoculum was used immediately, fungal inoculum, was stored at −20°C until use. Before use, colony forming units (CFU) were determined by dilution plating and counting. For the assays, the stock solution was adjusted to 105 CFU mL−1. The choice of test bacterial and fungal strains was limited by sufficient performance of the strain in the assay system. The majority of strains that are listed in Table 1 were explored for assay suitability but only a small portion showed sufficient development that allowed proper scoring.
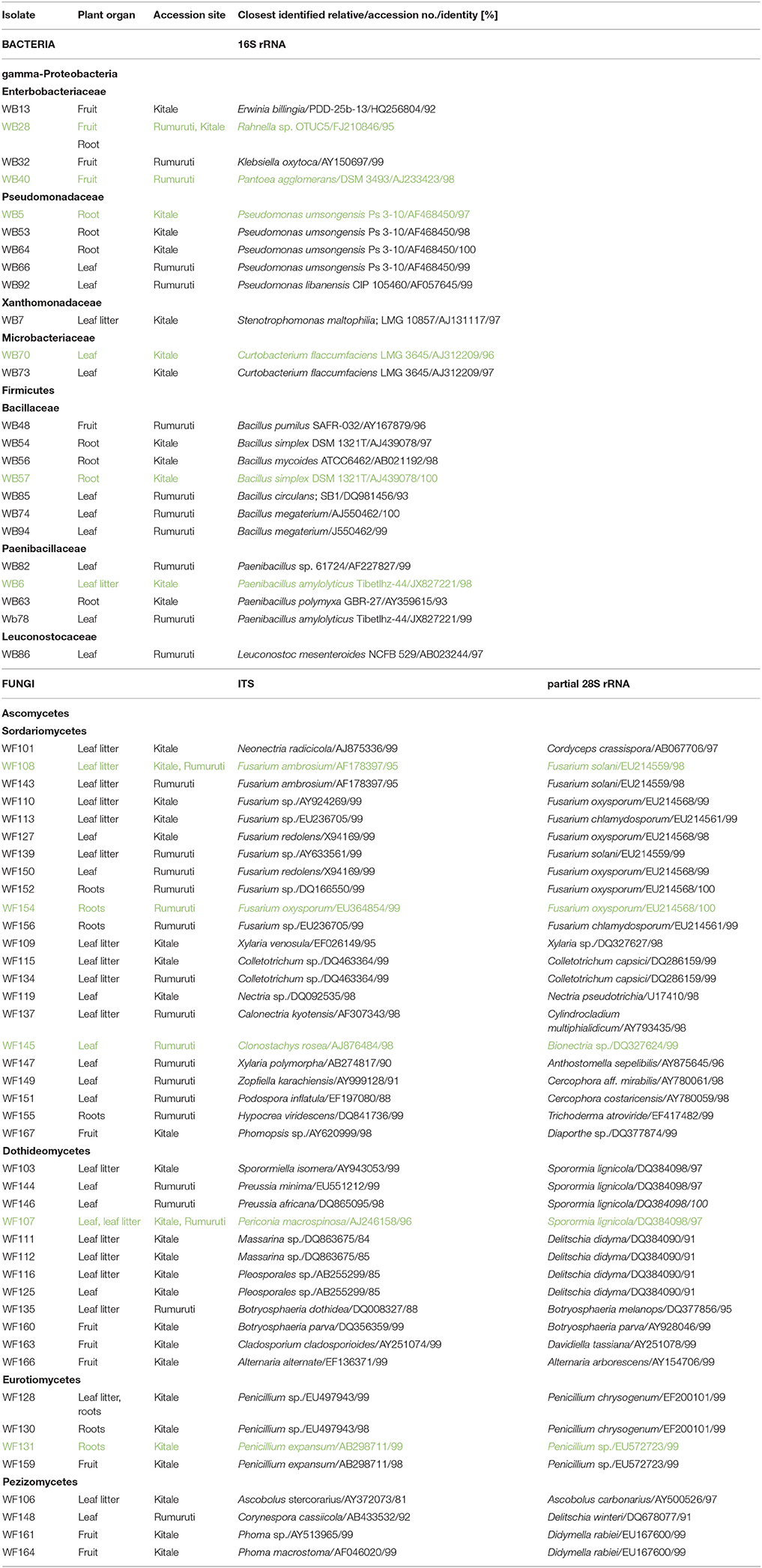
Table 1. Endophytic bacteria and fungi recovered from pepper bark tree (Warburgia ugandensis) roots, leaves, leaf litter and fruits (isolates marked in green color were chosen for susceptibility assays).
Growth Susceptibility (G)
Broth microdilution method (Hadacek and Greger, 2000; Engelmeier and Hadacek, 2006) was performed to determine the minimal inhibitory concentrations (MIC). DSs were tested in two-fold serial dilutions from 0.5 to 1,000 μg/mL and juglone from 0.01 to 350 μg/mL. Dilutions were performed in sterile 96-well U-shaped PS-microplates with lids (Greiner Bio-One, Kremsmünster, Austria). Stock solutions of the extracts were prepared in the specific medium for fungi and bacteria. The stock solution contained 5% MeOH (v/v) that had to be added to facilitate solubility. Controls were prepared with the same solvents and serially diluted as well. To each well, 50 μL inoculum was added. Microplates were sealed with parafilm to avoid evaporation and incubated at room temperature on a horizontal shaker with 120 rpm. Growth was scored turbidimetrically (570 nm) with a TECAN infinite M200 plate Reader (Tecan, Männedorf, Switzerland) after 2–4 days, depending on the growth rate of the tested strain. Assays were performed in triplicates and repeated once. Growth was expressed as percentage of the growth of the respective control in the serial dilution.
Growth Efficiency (GE)
The additional measurement of respiration (R) besides of growth (G) informs about the efficacy of the transfer of metabolic activity into biomass production. This enabled the calculation of a growth efficiency factor (GE), GE = G/G+R (Del Giorgio et al., 1997). Serial dilutions were performed similarly as for susceptibility determination. Microbial growth was scored by turbidity measurements at 570 nm with a Tecan Infinite M200 plate Reader (Tecan, Austria) similarly as in growth susceptibility assays. Respiration was measured immediately afterwards by using the same plates as part of a MicroResp™ analysis system (Macaulay Land Use Research Institute, Craigiebuckler, Aberdeen, UK) in a modified version (Drage et al., 2012). The additional incubation time for respiration measurements was 6 h for bacteria and 12 h for fungi. Absorbance of the MicroResp™ detection plates containing cresol red as indicator was measured at 570 nm at the beginning and the end of incubation period with a Tecan Infinite M200 plate Reader (Tecan, Männedorf, Switzerlan). All assays were performed in triplicate and repeated once. The obtained data were normalized before calculating GE (100% = growth or respiration of the respective strain on medium containing only the organic solvent).
Redox Chemistry
Deoxyribose degradation assays were performed as described (Chobot, 2010). In brief, samples were dissolved in aqueous 30 mM KH2PO4/KOH buffer solution and serially diluted (juglone 2–500 μM, drimane sesquiterpene leaf fractions 0.02–5 mg/mL). To 125 μL of the tested solution, 25 μL 2-deoxy-d-ribose in the same buffer and either 50 μL 50 μM FeCl3 or Fe(III)–EDTA complex were added. Twenty-five microliter 10.0 mM H2O2 and 25 μL aqueous 1.0 mM ascorbic acid were added then to start the Fenton reaction. The classic version utilizes ascorbic acid to reduce iron(III) to iron(II), which then reduces hydrogen peroxide (H2O2) to hydroxyl radical (•OH). The latter is a strong oxidant and attacks 2-deoxyribose, the oxidation products of which were quantified as thiobarbituric acid-reactive species (TBARS).
In the variants, ascorbic acid and H2O2 equivalents were replaced by buffer. In order to obtain sufficient H2O2 concentrations, the assay duration was extended from 1 to 16 h. The arising degradation products of deoxyribose reacted with thiobarbituric acid (dissolved in 3% trichloroacetic acid) to a red pigment that was detected photometrically at 532 nm after butanol (600 μL) extraction with a Tecan Infinite M200 plate Reader (Tecan, Männedorf, Switzerland).
Differential pulse voltammetry was performed as described (Kubicova et al., 2013). In brief, measurements were performed with a three-electrode system, μAutolab PGSTS type III (EcoChemie Inc., Utrecht, Netherlands); working electrode, 3 mm glassy carbon; reference electrode Ag/AgCl; counter electrode, platinum wire. Samples were prepared by dissolving 1 mL aqueous solution in 9 mL 0.1 M phosphate buffer (pH = 7.4). The scan potential was from −0.3 to +1.2 V.
Statistics
Dose-dependent effects on growth and respiration were curve fitted to a log-logistic model using R (R Core Team, 2017) and the package drc (Ritz et al., 2015).
Conc designates the concentration of the tested compound or extract; LL is the lower limit of the effect, UL the upper limit; the slope reflects the relation of effect in increase to dosage increase—in case of stimulation the values become negative; the EC50 value reflects the estimated concentration that causes 50 % inhibition (or stimulation). In addition, a lack-of-fit test was performed to check if the obtained data from 3 replicates of two repeats fit the obtained model. Missing values indicate either that no model fit was possible or that some variables could not be calculated.
For a total comparison of the effects of the leaf and root drimane sesquiterpene fraction and juglone on all tested bacterial and fungal strains, endophytes and non-endophytes,: EC50, LL (lower limit), UL (upper limit), slope (of the sigmoidal dose–response curve) and p (lack-of-fit test) were chosen to obtain a χ2-distance matrix that was visualized by non-metric multidimensional scaling (nMDS). If no model fit data were available, the values were set to zero. These tasks were performed with Primer 6 (Primer-E, Plymouth, UK). ANOSIM (Clarke, 1993) was used to test if leaf endophytes, root endophytes and the other strains form significantly different groups on basis of the obtained distance matrix of their dose–response model variables. The test statistics R can yield values between −1 and 1. Positive values suggest that groups are characterized by high similarity within them, values close to zero indicate that no differences exist between the groups and negative values point out that the groups share similarities between them.
The deoxyribose degradation assay was performed in triplicate and repeated once. The data were analyzed by ANOVA with a 95% Duncan multiple range test as post-hoc test to detect significant differences in effect levels. Kruskal-Wallis tests were performed to compare model parameters. The low n numbers caused data to lack normal distribution and to be heteroskedastic. These tasks was performed with Statgraphics Centurion XVII (Statpoint Technologies Inc., VA).
Results
Drimane Sesquiterpenes
Figure 1 exemplifies the difference in the drimane sesquiterpene composition between the leaf- and the root fraction of the pooled extracts that were used in the assays. The two illustrated GC analyses (Figure 1A) were carried out with identical sample amount injection. The MS spectra indicated that the majority of the peaks that show in the time window of the illustrated total ion chromatogram are possible drimane sesquiterpenes. Some of the more prominent peaks have been identified tentatively (Figure 1B) in a previous study (Drage et al., 2014). The chromatogram from the root fraction shows definitely more prominent peaks and many more minor peaks than the leaf fraction, proving some evidence for the presence of higher drimane sesquiterpene concentrations.
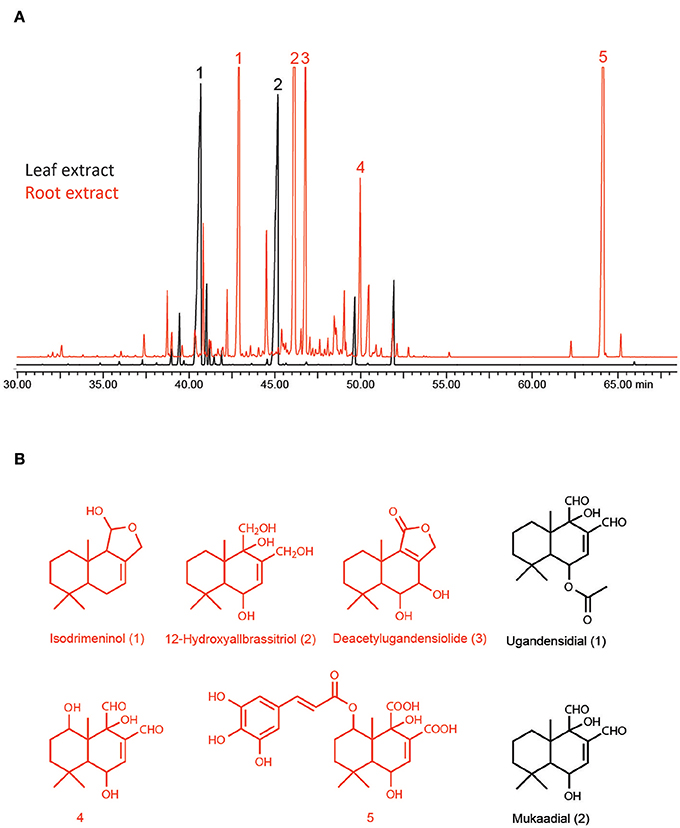
Figure 1. (A) GC–MS chromatograms of identical amounts of pepper bark tree root extract (brown) and leaf extract (black); (B) tentative structures of identified drimane sesquiterpenes.
Bacterial Endophytes
The 29 ribotypes found classified into three divisions: Gammaproteobacteria (12 isolates from 6 genera), Actinobacteria (4 isolates from 1 genus) and Firmicutes (13 isolates from 3 genera) (Table 1). On basis of a comparison of amplified partial 16S rRNA gene fragments, the most abundant genera comprised Bacillus (8 isolates), Pseudomonas (5 isolates), Curtobacterium (4 isolates), Paenibacillus (4 isolates), Erwinia (2 isolates), and Rahnella (2 isolates). Leuconostoc, Klebsiella, Stenotrophomonas, and Pantoea were recovered only once. In total, 10 bacterial genera were isolated from W. ugandensis tissues. Both accession sites, Kitale and Rumuruti, were represented equally. Six of ten isolated genera appeared to be organ-specific: Curtobacterium and Leuconostoc in leaves, Klebsiella and Pantoea in fruits and Stenotrophomonas in leaf litter. Two genera were found in two plant organs respectively, Pseudomonas in roots and leaves und Rahnella in roots and fruits. The most abundant genera, Bacillus and Paenibacillus were isolated from three organs: leaves, roots and fruits for Bacillus and leaves, leaf litter and roots for Paenibacillus.
All identifications of OTUs are based on sequence similarity of the 16S rDNA to the closest identified relative in the BLAST search and have been deposited in the in-house culture collection of AIT, from which they are available under the strain numbers listed in Table 1.
Fungal Endophytes
The further analyzed 61 fungal strains affiliated into 4 classes: Sordariomycetes (28 isolates from 11 genera), Dothideomycetes (20 isolates from eight genera), Eurotiomycetes (8 isolates from one genus) and Pezizomycetes (5 isolates from three genera) (Table 1). The most abundant genera included Fusarium (15 isolates), Penicillium (8 isolates), Periconia (7 isolates), Phoma and Sporormiella (3 isolates, respectively). Xylaria, Colletotrichum, Masserina, Preussia, Pleosporales, Botryosphaeria and Neonectria, were identified twice, respectively. We isolated 38 strains from Kitale and 23 from Rumuruti. In terms of different origins of isolation, we obtained 25 isolates from leaf litter, 16 from leaves, 12 from roots and 8 from fruits.
Fusarium species in leaves, leaf litter and roots occurred in both accession sites, the genus Penicillium only in Kitale leaf litter, roots and fruits. Periconia and Xylaria were isolated from both sites and showed up in leaves and leaf litter. By contrast, Phoma and Phomopsis were found in Kitale fruits only. Sporormiella, Masserina, Neonectria, and Ascobolus were isolated only from leaf litter in Kitale. Colletotrichum was a leaf litter genus of both sites. Pleosporales, by contrast, was only found in leaf litter from Kitale. Preussia, Clonostachys, Zopfiella, and Podospora were isolated from Rumuruti leaves, Calonectria from Rumuruti leaf litter and Hypocrea from Rumuruti roots. Botryoshaeria originated from leaf litter and fruits of both sites. Nectria originated from Kitale leaves, and Cladosporium und Alternaria from Kitale fruits.
All identifications of OTUs are based on sequence similarity of the ITS gene region of the 18S rDNA to the closest identified relative in the BLAST search and have been deposited in the in-house culture collection of AIT, from which they are available under the strain numbers listed in Table 1. The difficulties in exact identification are reflected by different BLAST hits of the same isolated of partial 28S rDNA gene region. In the text, the taxa yielded by the ITS search are used exclusively.
Comparison of Growth (G) and Growth Efficiency (GE)
Table 2 summarizes the results of all performed assays and presents model fits of G and GE of all performed assays. The root drimanes sesquiterpene fraction and juglone inhibited the growth of all tested strains without exception. By contrast, the leaf drimane sesquiterpene fraction stimulated the growth of most assayed strains except that of the bacteria Pseudomonas umsonginensis, a root endophyte, Bacillus simplex, the human pathogen, and Paenibacillus amylolyticus, the soil strain, all of which were slightly inhibited. Growth efficiency (GE) EC50 values either compared to those of G, or they were higher or lower. Lower GE EC50 values indicate that respiration increased dramatically in the high concentration range. Higher GE EC50 values indicate that the tested compound or extract stimulated growth in the low concentration range. If both EC50s compare, respiration follows growth dynamics.
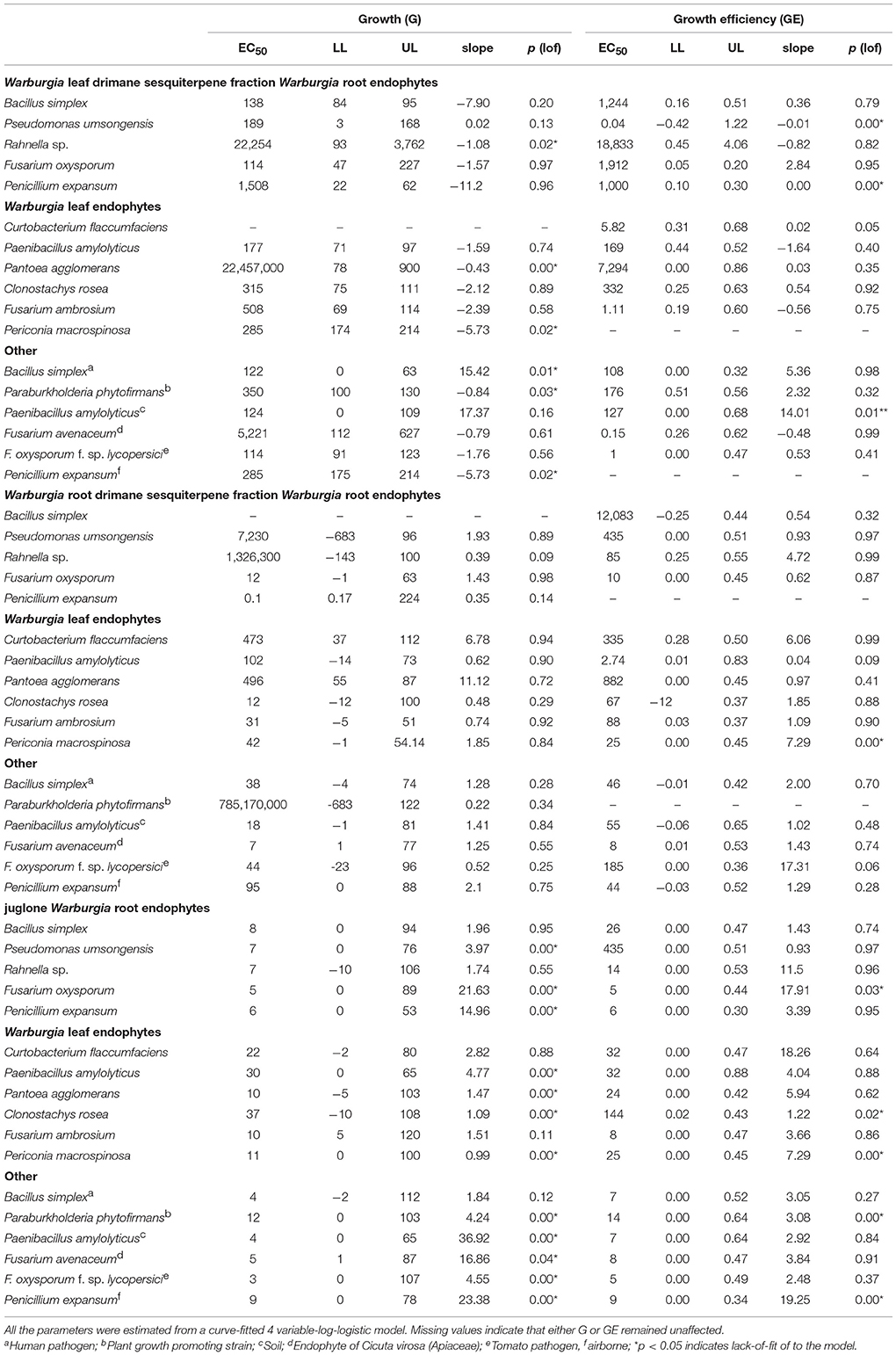
Table 2. Growth susceptibility (G) and growth efficiency (GE): EC50 (μg mL−1), LL (lower limit, μg mL−1), UL (upper limit, μg mL−1), slope (of sigmoidal dose–response curve), p (lack-of-fit test).
The drimane sesquiterpene root fraction inhibited the growth of all tested strains. Most affected were the fungal root endophytes Penicillium expansum and Fusarium oxysporum, the fungal leaf endophyte Clonostachys rosea, the Cicuta virosa tuber endophyte Fusarium avenaceum and the soil bacterium Paenibacillus amylolyticus. Unreasonably high EC50s in combination with very low slope values indicated extremely low dose–response relations within the tested concentration range. Root drimane sesquiterpene fraction inhibition of the plant growth-promoting bacterium Paraburkholderia phytofirmans and the root endophytic bacterium Rahnella sp. represent good examples for such effects. One endophytic bacterium, Bacillus simplex, remained nearly unaffected by the root drimane sesquiterpene fraction.
Leaf drimane sesquiterpene stimulation was most efficient for the root endophyte Fusarium oxysporum, the tomato pathogen Fus. oxysporum f. sp. lycopersici, and both the Bacillus simplex strains, the root endophyte and the human pathogen. The leaf endophytic bacterium Pantoea agglomerans and the root endophytic bacterium Rahnella sp. yielded very high EC50 values in terms of stimulation. This was caused by the fact that stimulation only showed at the highest tested concentrations. The leaf endophytic bacterium Curtobacterium flaccumfacies remained unaffected.
Juglone retarded the growth of all tested strains with EC50s from 3 to 37 μg mL−1. This is very narrow. By contrast, root drimane sesquiterpene fractions showed EC50s from 0.1 to >>1,000 μg mL−1. The most sensitive of the tested strains was the fungal tomato pathogen Fusarium oxysporum f. sp. lycopersici, and the most resistant the fungus Clonostachys rosea, which can also parasitize other fungi. In many strains, the detected lack-of-fit to the model is caused by the fact that slight or no inhibition changed to total inhibition within a few concentrations. As a result, the majority of the tested concentrations do not support the model in this statistical test.
Table 2 hints that the leaf and root drimane sesquiterpene fraction and the naphthoquinone juglone caused highly variable effects on the various endophytes and the other randomly selected strains. Figure 2 illustrates the dissimilarity of growth susceptibility (G, Figure 2A) and growth efficiency (GE, Figure 2B). An ANOSIM analysis yielded R-values that were close to zero and not significant. Consequently, the three strain groups do not differ in terms of growth susceptibility and efficiency in the performed assays.
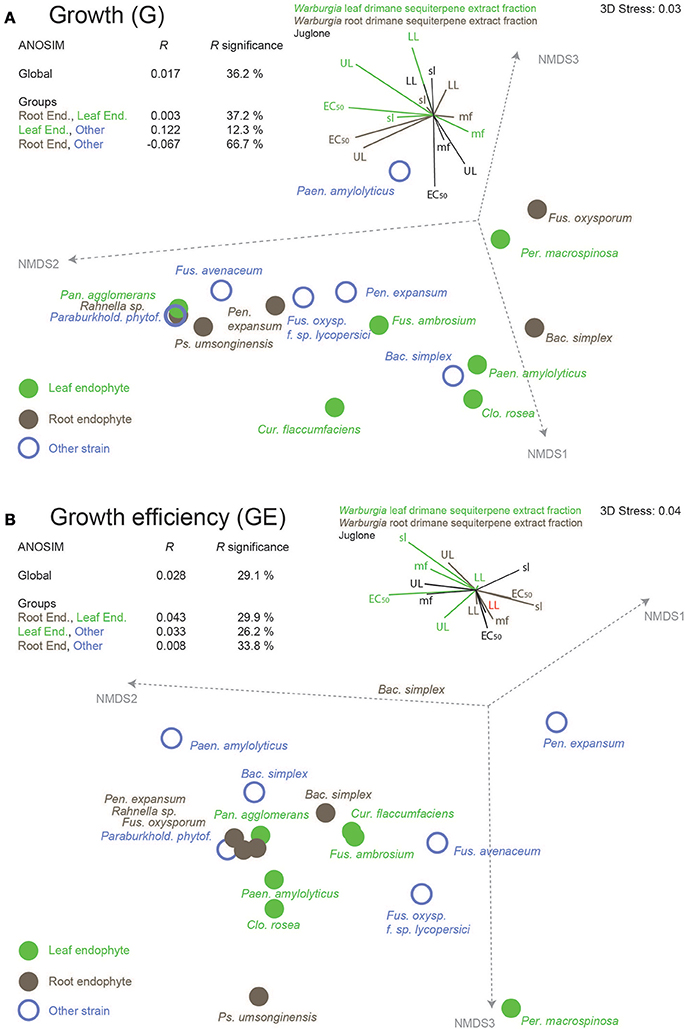
Figure 2. Dissimilarities of leaf and root endophytes of the African pepper bark tree, Warburgia ugandensis, and randomly selected other strains on basis of (A) growth susceptibility and (B) growth efficiency against the leaf and root drimane sesquiterpene fraction and the naphthoquinone juglone, a highly redox-active plant metabolite. Variable contributions of EC50, lower level (LL), upper level (UL), slope (of the sigmoidal dose–response curve) and lack-of-fit p (mf, model fit), all parameters from curve fitting of the original data to a log-logistic growth model, are illustrated as vector plots.
Figures 2A,B present dissimilarity plots of growth susceptibility (G) and growth efficiency (GE) of all isolates respectively. The variables are comprised of model parameters that were obtained from assaying the leaf and root drimane sesquiterpene fraction as well as the naphthoquinone juglone. The contributions of the single variables are indicated in a vector diagram that accompanies each plot. The analysis of similarity clearly documents that the three strain groups, leaf (and one fruit) endophytes, root endophytes, and randomly selected other strains, do not differ in terms of their assay results (R close to zero and not significant). This applies both to growth susceptibility (Figure 2A) and growth efficiency (Figure 2B). Both plots hint that leaf endophytes could be more resistant to the redox active juglone than root endophytes and the randomly selected strains. In fact, the growth susceptibility EC50s differ between leaf and root endophytes (Kruskal Wallis p = 0.006) and leaf endophytes and randomly selected isolates (Kruskal Wallis p = 0.02). The growth efficiency assay, however, did not follow this pattern.
Redox Chemistry
Figure 3 summarizes the results that were obtained in various variants (Figures 3A–C) of the deoxyribose degradation assay. Both the drimane sesquiterpenes (combined root extract, green bars) and juglone (red bars) decrease TBARS formation by their reductive power in the classical variant (Figure 3A). The effect was antioxidant. When no H2O2 was added (Figure 3B, variant 2), the assay setup tests if the drimane sesquiterpenes and juglone can reduce molecular oxygen to H2O2. Juglone was very efficient in doing this; higher concentrations caused a substantial pro-oxidant effect. Drimane sesquiterpenes, by contrast, induced only a weaker but still pro-oxidant effect. In the third variant (Figure 3C), the addition of ascorbic acid was skipped also. As a consequence, the test compound had to reduce iron(III) to iron(II) in addition. Again, juglone was the more efficient pro-oxidant, but effects were also visible for the drimane sesquiterpene extract fraction. A slight decrease in higher tested concentrations was probably caused by the onset of an antioxidant effect of a portion of the tested molecules in the mixture (Figure 3C). Figures 3A–C contain colored bars and white bars in the same graph. The colored bars indicate that iron was added as FeCl3 and thus available in free form. The white bars illustrate the chemical reactions in the identical setup with the only difference that iron was offered as EDTA complex. The differences clearly point to the fact that the redox effects are more pronounced if the test compound acted as ligand in a coordination complex, in which iron is the central atom. Without exception, this applied to the observed pro-oxidant effects (Figure 3C) but is less evident for the purely antioxidant effect in the classical setup of the deoxyribose degradation assay (Figure 3A).
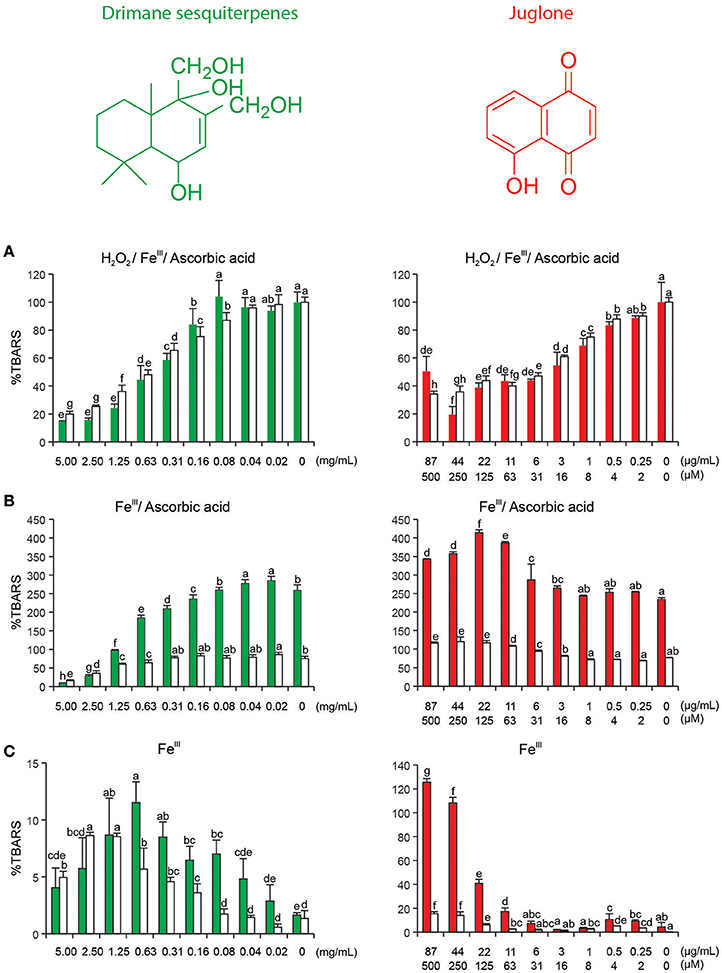
Figure 3. Drimane sesquiterpene root combined extract (representative structure) and naphthoquinone juglone in variants of deoxyribose degradation assay; TBARS, thiobarbituric acid reactive species that arise following deoxyribose degradation; (A) H2O2, ascorbic acid, and FeCl3, the classical setup; (B) ascorbic acid and FeCl3, testing if the assayed compound or extract can reduce O2 to H2O2; (C) FeCl3, testing testing if the assayed compound or extract can perform the complete reduction from O2 to OH; red bars, iron added as FeCl3 (iron can coordinated the test compound or extract components); white bars: iron added as Fe(III)–EDTA complex (iron remains in the complex with EDTA). All assays were performed in triplicates and replicated at least once. Bars represent mean + standard deviation, letters indicate 95% Duncan ANOVA.
Figure S1 depicts a differential pulse voltammogram of the tested drimane sesquiterpene extract and juglone. The designated peaks indicate oxidation and reduction reactions. The voltammograms show clearly that both undergo redox reactions. The combined drimane sesquiterpene extract showed peaks at 0.10 and 0.47 V, juglone at −0.20 and 0.94 V (values are relative to Ag/AgCl electrode).
Discussion
The most notable finding of this study is that drimane sesquiterpenes, characteristic secondary metabolites of the African pepper bark tree, seem not to be able to cause community-structuring effects on associated bacterial and fungal endophytes. Both in terms of growth susceptibility and efficiency the selected respective isolates showed too much variation. Moreover, the randomly selected isolates also varied considerably. As a result, no differentiation of the three strain groups was possible.
The obtained results oppose those from two similarly targeted studies. The first one (Carter et al., 1999) investigated fungal endophytes from oat and wheat, both of which accumulate the antifungal saponin avenacin A-1 in their roots. Likewise, the authors identified strains that were either tolerant of or inhibited by avenacin A-1. The majority of them, however, was found out to be able to detoxify avenacin A-1 by deglucosylating it. The drimane sesquiterpenes of the pepper bark tree, by contrast, do not occur as glycosides. Moreover, the ability to induce hydrolytic reactions seems to be common among fungi (Oda et al., 2002). The second study compared recovered strains of fungal endophytes from benzoxazine producing and non-producing maize plants in terms of their susceptibility to 2-benzoxazolinone (Saunders and Kohn, 2009). The authors claim that their data support the hypothesis that plant secondary defense metabolites shape the community of fungal endophytes. This was more evident in 2-week than in 9-week old plants. By contrast, this study was (1) performed on a perennial tree species with tissues that were much older than 9 weeks; and (2) the quality of the culture-dependent obtained endophyte patterns in this study was regarded as too stochastic and unrepresentative to allow a statistical analysis of endophyte community structures.
The previously published culture-independent community structure analysis on Warburgia endophytes (Drage et al., 2014) attempted to find such correlations but failed to do so. When attempting to pinpoint host plant secondary metabolites as determinants of endophyte community structure, other factors might merit attention, for example, environmental factors. In tropical forests, rainfall amounts have been shown to affect the occurrence of endophyte lifestyles more than tree species composition. High precipitation appeared to favor endophytic host colonization and stimulated its growth within the host (Suryanarayanan et al., 2002). Furthermore, the high tree diversity in tropical forests does not reflect itself in endophyte diversity due to the occurrence of multi-host endophytes (Cannon and Simmons, 2002). High tree species diversity could also restrict endophyte diversity evolution because of low host tree individual apparency (May, 1991). The expansion of the host range is therefore regarded as useful adaptation for endophytes. The actual community structure could be the result of combined effects of abiotic factors, such as climate, topography, soil properties, and biotic, such as competing microbial species composition including their potential metabolites as well as those from the host plant (Suryanarayanan, 2011). Multi-host range patterns also occur in other functional fungal groups like wood-rotting fungi (Lindblad, 2000; Gilbert et al., 2002; Parfitt et al., 2010), ectomycorrhizal fungi (Diédhiou et al., 2010; Tedersoo et al., 2010) and arbuscular mycorrhizal fungi (Zhao et al., 2003). Leaf and leaf litter endophytic isolates were more numerous in this study than those that were discovered from root. Abiotic stimuli could induce a shift in their life strategy and they may become necrotrophic or pathogenic in their switch to a saprophytic lifestyle (Bahnweg et al., 2005; Promputtha et al., 2010). Several common degrader genera were detected among the endophytic isolates: Xylaria, Colletotrichum, Hypocrea, Preussia, Sporormiella, Ascobolus, Calonectria, Pleosporales, and Periconia.
Strains for the assays were selected in attempts to obtain a representative selection of the bacterial and fungal endophyte diversity in Warburgia. The selected bacterial isolates belonged to the abundant genera Bacillus, Pseudomonas, and Paenibacillus that have already been identified in the culture-independent exploration (Drage et al., 2014). These form a major group of endophytic bacteria that have been isolated as endophytes from various plant species, encompassing diverse plants such as trees and aquatic plants, but can occur also as saprophytes and pathogens (O'Neill et al., 1992; Shishido et al., 1995; Cankar et al., 2005; Ferreira et al., 2008; Izumi et al., 2008; Ulrich et al., 2008a,b; Khan and Doty, 2009; Pardatscher and Schweigkofler, 2009; Filteau et al., 2010; Chen et al., 2012). Bacillus simplex is not only a human pathogen but can also occur in soils and forms associations with arbuscular mycorrhizal fungi (Lecomte et al., 2011). Paenibacillus is known to promote plant growth (Rybakova et al., 2016). Pseudomonas umsongensis, to our knowledge, was never found as endophyte previously. The genus Curtobacterium was also isolated as endophyte from Eucalyptus, Populus, and Ulmus (Mocali et al., 2003; Ulrich et al., 2008b), but occurs also as a common plant pathogen; especially C. flaccumfaciens is well documented to occur world-wide (Osdaghi et al., 2016). Rahnella was isolated as endophyte from temperate trees, sweet potatoes and Polygonum and as rhizosphere bacterium with plant growth promoting activities (Cankar et al., 2005; Taghavi et al., 2009; Filteau et al., 2010). Pantoea belongs to an ubiquitously occurring opportunistic endophyte genus (Wang et al., 2006; Doty et al., 2009). The selected fungal isolates Periconia and Penicillium are common endophytes with a multi-host range (Gazis and Chaverri, 2010; Sanchez et al., 2010; Banerjee, 2011). Fusarium is well known as pathogen and endophyte (Gordon and Martyn, 1997; Banerjee, 2011). Clonostachys rosea occurs in various ecological niches, in above-ground plant organs and in soils, in which it can degrade phenolic acids (Schroers, 2001).
The root drimane sesquiterpene fraction inhibited most of the tested strains. Interestingly, one of the randomly selected strains, the plant growth-promoting bacterium Paraburkholderia phytofirmans (Sessitsch et al., 2005), which occurs on onions and probably never encountered drimane sesquiterpenes during its evolution, was one of the most resistant.
Rather unexpectedly, the majority of the assayed strains were stimulated by the leaf drimane sesquiterpene fraction. This could be due to hormetic effects of drimane sesquiterpenes. Low dosages could have caused stimulatory effects (Hadacek et al., 2011). Conversely, the extract fraction could contain further still unidentified compounds that can be utilized more efficiently as growth substrates than those present in the medium. This question cannot be answered on basis of the results that were obtained within this study. Still, they support the hypothesis that plant secondary metabolites can contribute to a balanced antagonism that facilitates and endophytic lifestyles of an microbe (Schulz et al., 1999; Schulz and Boyle, 2005).
The deoxyribose degradation assay and the voltammograms provide evidence that both juglone and drimane sesquiterpenes can undergo similar redox chemical reactions. They not only cause pro-oxidative but also antioxidative effects, always depending on the chemical milieu of their immediate environment. The chemistry is similar albeit not identical.
Likewise, tolerance and susceptibility to juglone and the drimane sesquiterpenes varies substantially between the tested strains. In interacting with oxygen and its reactive species on hand, and metals in form coordination complex formation, secondary metabolites of any origin participate in a specific but also highly complex systems chemistry (Hadacek and Bachmann, 2015) that can affect the biotic interactions of their producers. Bacteria and fungi are not only confronted by reactive oxygen or nitrogen species when they encounter host plant secondary metabolites such as juglone and drimane sesquiterpenes. They also have to cope with oxidative stress as saprophytes, a life style in which a similar redox chemistry is used to form hydroxyl radicals to start polymer (cellulose and lignin) degradation chain reactions in attempts to obtain nutrients (Baldrian and Valaskova, 2008; Arantes et al., 2012; Masai et al., 2014). It seems likely that microbes differ in their tolerance of oxidative stress scenarios, which can be caused by different agents and circumstances and occur at different stages of the life cycle. The sum of these interactions most likely constitutes only one of several components, which determine its successful establishment as an endophyte, pathogen or saprophyte. Many deterministic processes can remain hidden by an at first glance more neutral picture of a process, in which identifying a single component as determinant has to remain an unsuccessful attempt. This complexity reflects itself in comparison of growth susceptibility and growth efficiency. The management of stimulation and inhibition is variably managed by the different strains.
Despite the observed complexity, one finding merits further exploration. In this study, leaf endophytes appeared to be more resistant against redox activity causing oxidative stress as suggested by the lower susceptibility against juglone. Compared to root endophytes, they are exposed to higher levels of oxidative stress. Leaves contain chloroplasts, in which photosynthesis runs, which can add considerably to levels of exposure, especially when the host plants are exposed to abiotic stress (Foyer and Noctor, 2009). This might constitute a constraint for developing into a leaf endophyte or even later, into a successful leaf pathogen. How secondary metabolites affect these processes, might also remain ambiguous, simply because the chemistry into which they can enter is ambiguous too. They can cause both antioxidant and pro-oxidant effect in different chemical environments (Figures 3A–C) that can turn out as beneficial or harmful to both the producer and recipient.
Author Contributions
BM, AS, and FH devised the study and experiments. SD, BM, AS, FH, AM, and RJ collected material. SD, DE, BM, FH, and MG performed the experiments and analyzed data. VC provided additional methods and data. SD, BM and FH wrote the first manuscript draft. All authors read and commented on the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by grant P19851-B17 of Austrian Science Fund (FWF) to BM and FH. SD, DE, VC, and FH are grateful to Marianne Popp, former head of the Department of Chemical Ecology and Ecosystem Research of the Faculty of Life Science of the University of Vienna for providing laboratory space, equipment and support. BM and MG are grateful to Melanie Kuffner for skillful assistance. The Open Access Publication Fonds of the University of Göttingen is acknowledged for covering publications costs.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2017.00138/full#supplementary-material
References
Arantes, V., Jellison, J., and Goodell, B. (2012). Peculiarities of brown-rot fungi and biochemical Fenton reaction with regard to their potential as a model for bioprocessing biomass. Appl. Microbiol. Biotechnol. 94, 323–338. doi: 10.1007/s00253-012-3954-y
Bahnweg, G., Heller, W., Stich, S., Knappe, C., Betz, G., Heerdt, C., et al. (2005). Beech leaf colonization by the endophyte Apiognomonia errabunda dramatically depends on light exposure and climatic conditions. Plant Biol. 7, 659–669. doi: 10.1055/s-2005-872943
Baldrian, P., and Valaskova, V. (2008). Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol. Rev. 32, 501–521. doi: 10.1111/j.1574-6976.2008.00106.x
Banerjee, D. (2011). Endophytic fungal diversity in tropical and subtropical plants. Res. J. Microbiol. 6, 54–62. doi: 10.3923/jm.2011.54.62
Bednarek, P., and Osbourn, A. (2009). Plant-microbe interactions: chemical diversity in plant defense. Science 324, 746–748. doi: 10.1126/science.1171661
Cankar, K., Kraigher, H., Ravnikar, M., and Rupnik, M. (2005). Bacterial endophytes from seeds of Norway spruce (Picea abies L. Karst). FEMS Microbiol. Lett. 244, 341–345. doi: 10.1016/j.femsle.2005.02.008
Cannon, P. F., and Simmons, C. M. (2002). Diversity and host preference of leaf endophytic fungi in the Iwokrama forest reserve, Guyana. Mycologia 94:210. doi: 10.2307/3761797
Carter, J. P., Spink, J., Cannon, P. F., Daniels, M. J., and Osbourn, A. E. (1999). Isolation, characterization, and avenacin sensitivity of a diverse collection of cereal-root-colonizing fungi. Appl. Environ. Microbiol. 65, 3364–3372.
Chen, W. M., Tang, Y. Q., Mori, K., and Wu, X. L. (2012). Distribution of culturable endophytic bacteria in aquatic plants and their potential for bioremediation in polluted waters. Aquat. Biol. 15, 99–110. doi: 10.3354/ab00422
Chobot, V. (2010). Simultaneous detection of pro- and antioxidative effects in the variants of the deoxyribose degradation assay. J. Agric. Food Chem. 58, 2088–2094. doi: 10.1021/jf902395k
Chobot, V., and Hadacek, F. (2009). Milieu-dependent pro- and antioxidant activity of juglone may explain linear and nonlinear effects on seedling development. J. Chem. Ecol. 35, 383–390. doi: 10.1007/s10886-009-9609-5
Clarke, K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Austral. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Del Giorgio, P. A., Cole, J. J., and Cimbleris, A. (1997). Respiration rates in bacteria exceed phytoplankton production in unproductive aquatic systems. Nature 385, 148–151. doi: 10.1038/385148a0
Diédhiou, A. G., Selosse, M.-A., Galiana, A., Diabaté, M., Dreyfus, B., Bâ, A. M., et al. (2010). Multi-host ectomycorrhizal fungi are predominant in a Guinean tropical rainforest and shared between canopy trees and seedlings. Environ. Microbiol. 12, 2219–2232. doi: 10.1111/j.1462-2920.2010.02183.x
Doty, S. L., Oakley, B., Xin, G., Kang, J. W., Singleton, G., Khan, Z., et al. (2009). Diazotrophic endophytes of native black cottonwood and willow. Symbiosis 47, 23–33. doi: 10.1007/BF03179967
Drage, S., Engelmeier, D., Bachmann, G., Sessitsch, A., Mitter, B., and Hadacek, F. (2012). Combining microdilution with MicroResp™: microbial substrate utilization, antimicrobial susceptibility and respiration. J. Microbiol. Meth. 88, 399–412. doi: 10.1016/j.mimet.2012.01.006
Drage, S., Mitter, B., Tröls, C., Muchugi, A., Jamnadass, R. H., Sessitsch, A., et al. (2014). Antimicrobial drimane sesquiterpenes and their effect on endophyte communities in the medical tree Warburgia ugandensis. Front. Microbiol. 5:13. doi: 10.3389/fmicb.2014.00013
Edwards, U., Rogall, T., Blöcker, H., Emde, M., and Böttger, E. C. (1989). Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17, 7843–7853.
Engelmeier, D., and Hadacek, F. (2006). “Antifungal natural products. Assays and applications,” in Naturally Occurring Bioactive Compounds. Vol. 3, eds M. Rai and M. C. Carpinella (New York, NY: Elsevier (Advances in Phytomedicine)), 423–467.
Ferreira, A., Quecine, M. C., Lacava, P. T., Oda, S., Azevedo, J. L., and Araújo, W. L. (2008). Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol. Lett. 287, 8–14. doi: 10.1111/j.1574-6968.2008.01258.x
Filteau, M., Lagacé, L., LaPointe, G., and Roy, D. (2010). Seasonal and regional diversity of maple sap microbiota revealed using community PCR fingerprinting and 16S rRNA gene clone libraries. Syst. Appl. Microbiol. 33, 165–173. doi: 10.1016/j.syapm.2010.02.003.
Foyer, C. H., and Noctor, G. (2009). Redox regulation in photosynthetic organisms. signaling, acclimation, and practical implications. Antioxid. Redox Signal. 11, 861–905. doi: 10.1089/ars.2008.2177
Gazis, R., and Chaverri, P. (2010). Diversity of fungal endophytes in leaves and stems of wild rubber trees (Hevea brasiliensis) in Peru. Fungal Ecol. 3, 240–254. doi: 10.1016/j.funeco.2009.12.001
Gershenzon, J. (2002). “Secondary metabolites and plant defense,” in Plant Physiology. 3rd Edn., eds L. Taiz und E. Zeiger (Sunderland, MA: Sinauer), 283–308.
Gilbert, G. S., Ferrer, A., and Carranza, J. (2002). Polypore fungal diversity in a moist tropical forest. Biodivers. Conserv. 11, 947–957. doi: 10.1023/A:1015896204113
Gordon, T. R., and Martyn, R. D. (1997). The evolutionary biology of Fusarium oxysporum. Annu. Rev. Phytopathol. 35, 111–128. doi: 10.1146/annurev.phyto.35.1.111
Grayer, R. J., and Harborne, J. B. (1994). A survey of antifungal compounds from higher plants, 1982? Phytochemistry 37, 19–42. doi: 10.1016/0031-9422(94)85005-4
Hadacek, F. (2002). Secondary metabolites as plant traits. Current assessment and future perspectives. Crit. Rev. Plant Sci. 21, 273–322. doi: 10.1080/0735-260291044269
Hadacek, F., and Bachmann, G. (2015). Low-molecular-weight metabolite systems chemistry. Front. Environm. Sci. 3:12. doi: 10.3389/fenvs.2015.00012
Hadacek, F., Engelmeier, D., Bachmann, G., and Chobot, V. (2011). Hormesis and a chemical raison d'ětre for secondary plant metabolites. Dose Respon. 9, 79–116. doi: 10.2203/dose-response.09-028.Hadacek
Hadacek, F., and Greger, H. (2000). Testing of antifungal natural products: methodologies, comparability of results and assay choice. Phytochem. Anal. 11, 137–147. doi: 10.1002/(SICI)1099-1565(200005/06)11:3<137::AID-PCA514>3.0.CO;2-I
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mo.l Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
Izumi, H., Anderson, I. C., Killham, K., and Moore, E. R. B. (2008). Diversity of predominant endophytic bacteria in European deciduous and coniferous trees. Can. J. Microbiol. 54, 173–179. doi: 10.1139/w07-134
Jansen, B. J. M., and de Groot, A. (2004). Occurrence, biological activity and synthesis of drimane sesquiterpenoids. Nat. Prod. Rep. 21, 449–477. doi: 10.1039/b311170a
Khan, Z., and Doty, S. L. (2009). Characterization of bacterial endophytes of sweet potato plants. Plant Soil 322, 197–207. doi: 10.1007/s11104-009-9908-1
Kubicova, L., Hadacek, F., and Chobot, V. (2013). Quinolinic acid: neurotoxin or oxidative stress modulator? Int. J. Mol. Sci. 14, 21328–21338. doi: 10.3390/ijms141121328
Lecomte, J., St-Arnaud, M., and Hijri, M. (2011). Isolation and identification of soil bacteria growing at the expense of arbuscular mycorrhizal fungi. FEMS Microbiol. Lett. 317, 43–51. doi: 10.1111/j.1574-6968.2011.02209.x
Lindblad, I. (2000). Host specificity of some wood-inhabiting fungi in a tropical forest. Mycologia 92:399. doi: 10.2307/3761497
Masai, E., Katayama, Y., and Fukuda, M. (2014). Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci. Biotechnol. Biochem. 71, 1–15. doi: 10.1271/bbb.60437
Massol-Deya, A. A., Odelson, D. A., Hickey, R. F., and Tiedje, J. M. (1995). “Bacterial community fingerprinting of amplified 16S and 16–23S ribosomal DNA gene sequences and restriction endonuclease analysis(ARDRA),” in Molecular Microbial Ecology Manual, eds A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (Dordrecht: Springer Science), 289–296.
McCully, M. E. (2001). Niches for bacterial endophytes in crop plants: a plant biologist's view. Funct. Plant Biol. 28:983. doi: 10.1071/PP01101
Mocali, S., Bertelli, E., Di Cello, F., Mengoni, A., Sfalanga, A., Viliani, F., et al. (2003). Fluctuation of bacteria isolated from elm tissues during different seasons and from different plant organs. Res. Microbiol. 154, 105–114. doi: 10.1016/S0923-2508(03)00031-7
Oda, Y., Saito, K., Ohara-Takada, A., and Mori, M. (2002). Hydrolysis of the potato glycoalkaloid α-chaconine by filamentous fungi. J. Biosci. Eng. 94, 321–325. doi: 10.1263/jbb.94.321
O'Neill, G. A., Chanway, C. P., Axwlrood, P. E., Radley, R. A., and Holl, F. B. (1992). An asessment of spruce growth response specificity after inoculation with co-existent rhizosphere bacteria. Can. J. Bot. 70, 2347–2353.
Osdaghi, E., Taghavi, S. M., Hamzehzarghani, H., Fazliarab, A., Harveson, R. M., and Lamichhane, J. R. (2016). Occurrence and characterization of a new red-pigmented variant of Curtobacterium flaccumfaciens, the causal agent of bacterial wilt of edible dry beans in Iran. Eur. J. Plant Pathol. 146, 129–145. doi: 10.1007/s10658-016-0900-3
Pardatscher, R., and Schweigkofler, W. (2009). Microbial biodiversity associated with the walnut Juglans regia L. in South Tyrol (Italy). Mitt. Klosterneubg. 59, 24–30.
Parfitt, D., Hunt, J., Dockrell, D., Rogers, H. J., and Boddy, L. (2010). Do all trees carry the seeds of their own destruction? PCR reveals numerous wood decay fungi latently present in sapwood of a wide range of angiosperm trees. Fungal Ecol. 3, 338–346. doi: 10.1016/j.funeco.2010.02.001
Promputtha, I., Hyde, K. D., McKenzie, E. H. C., Peberdy, J. F., and Lumyong, S. (2010). Can leaf degrading enzymes provide evidence that endophytic fungi becoming saprobes? Fungal Divers. 41, 89–99. doi: 10.1007/s13225-010-0024-6
R Core Team (2017). R. A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at https://www.R-project.org/
Redman, R. S., Sheehan, K. B., Stout, R. G., Rodriguez, R. J., and Henson, J. M. (2002). Thermotolerance generated by plant/fungal symbiosis. Science 298:1581. doi: 10.1126/science.1078055
Reiter, B., and Sessitsch, A. (2006). Bacterial endophytes of the wildflower Crocus albiflorus analyzed by characterization of isolates and by a cultivation-independent approach. Can. J. Microbiol. 52, 140–149. doi: 10.1139/w05-109
Ritz, C., Baty, F., Streibig, J. C., and Gerhard, D. (2015). Dose-response analysis using R. PLoS ONE 10:e0146021. doi: 10.1371/journal.pone.0146021
Rodriguez, R. J., White, J. F., Arnold, A. E., and Redman, R. S. (2009). Fungal endophytes. Diversity and functional roles. New Phytol. 182, 314–330. doi: 10.1111/j.1469-8137.2009.02773.x
Rybakova, D., Cernava, T., Köberl, M., Liebminger, S., Etemadi, M., and Berg, G. (2016). Endophytes-assisted biocontrol. Novel insights in ecology and the mode of action of Paenibacillus. Plant Soil 405, 125–140. doi: 10.1007/s11104-015-2526-1
Sanchez, M. S., Bills, G. F., Dominguez, A. L., and Zabalgogeazcoa, I. (2010). Endophytic mycobiota of leaves and roots of the grass Holcus lanatus. Fungal Divers 41, 115–123. doi: 10.1007/s13225-009-0015-7
Saunders, M., and Kohn, L. M. (2009). Evidence for alteration of fungal endophyte community assembly by host defense compounds. New Phytol. 182, 229–238. doi: 10.1111/j.1469-8137.2008.02746.x
Schroers, H. J. (2001). A monograph of Bionectria (Ascomycota, Hypocreales, Bionectriaceae) and its Clonostachys anamorphs. Stud. Mycol. 46, 1–214.
Schulz, B., and Boyle, C. (2005). The endophytic continuum. Mycol. Res. 109, 661–686. doi: 10.1017/s095375620500273x
Schulz, B., Haas, S., Junker, C., Andree, N., and Schobert, M. (2015). Fungal endophytes are involved in multiple balanced antagonisms. Curr. Sci. 109, 39–45.
Schulz, B., Römmert, A.-K., Dammann, U., Aust, H. R., and Strack, D. (1999). The endophyte-host interaction. A balanced antagonism? Mycol. Res. 103, 1275–1283. doi: 10.1017/S0953756299008540
Sessitsch, A., Coenye, T., Sturz, A. V., Vandamme, P., Ait Barka, E., Salles, J. F., et al. (2005). Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int. J. Syst. Evol. Microbiol. 55(Pt 3), 1187–1192. doi: 10.1099/ijs.0.63149-0
Sessitsch, A., Hardoim, P., Doring, J., Weilharter, A., Krause, A., Woyke, T., et al. (2012). Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol. Plant Microb. Interact. 25, 28–36. doi: 10.1094/mpmi-08-11-0204
Sharova, E. I., and Medvedev, S. S. (2017). Redox reactions in apoplast of growing cells. Russ. J. Plant Physiol. 64, 1–14. doi: 10.1134/S1021443717010149
Shishido, M., Loeb, B. M., and Chanway, C. P. (1995). External and internal root colonization of lodgepole pine seedlings by two growth-promoting Bacillus strains originated from different root microsites. Can. J. Microbiol. 41, 707–713.
Strobel, G., Daisy, B., Castillo, U., and Harper, J. (2004). Natural products from endophytic microorganisms. J. Natl. Prod. 67, 257–268. doi: 10.1021/np030397v
Suryanarayanan, T. S. (2011). “Diversity of fungal endophytes in tropical trees,” in Endophytes of Forest Trees: Biology and Applications, eds A. M. Pirttilä and A. C. Frank (Dordrecht: Springer), 67–80.
Suryanarayanan, T. S., Murali, T. S., and Venkatesan, G. (2002). Occurrence and distribution of fungal endophytes in tropical forests across a rainfall gradient. Can. J. Bot. 80, 818–826. doi: 10.1139/b02-069
Swanson, S., and Gilroy, S. (2010). ROS in plant development. Physiol. Plant. 138, 384–392. doi: 10.1111/j.1399-3054.2009.01313.x
Taghavi, S., Garafola, C., Monchy, S., Newman, L., Hoffman, A., Weyens, N., et al. (2009). Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl. Environ. Microbiol. 75, 748–757. doi: 10.1128/AEM.02239-08
Tedersoo, L., Sadam, A., Zambrano, M., Valencia, R., and Bahram, M. (2010). Low diversity and high host preference of ectomycorrhizal fungi in western Amazonia, a neotropical biodiversity hotspot. Isme J. 4, 465–471. doi: 10.1038/ismej.2009.131
Torres, M. A. (2010). ROS in biotic interactions. Physiol. Plant. 138, 414–429. doi: 10.1111/j.1399-3054.2009.01326.x
Ulrich, K., Stauber, T., and Ewald, D. (2008a). Paenibacillus? A predominant endophytic bacterium colonising tissue cultures of woody plants. Plant Cell Tiss. Organ Cult. 93, 347–351. doi: 10.1007/s11240-008-9367-z
Ulrich, K., Ulrich, A., and Ewald, D. (2008b). Diversity of endophytic bacterial communities in poplar grown under field conditions. FEMS Microbiol. Ecol. 63, 169–180. doi: 10.1111/j.1574-6941.2007.00419.x
Wang, E. T., Tan, Z. Y., Guo, X. W., Rodriguez-Duran, R., Boll, G., and Martinez-Romero, E. (2006). Diverse endophytic bacteria isolated from a leguminous tree Conzattia multiflora grown in Mexico. Arch. Microbiol. 186, 251–259. doi: 10.1007/s00203-006-0141-5
Wang, Y., and Dai, C.-C. (2011). Endophytes: a potential resource for biosynthesis, biotransformation, and biodegradation. Ann. Microbiol. 61, 207–215. doi: 10.1007/s13213-010-0120-6
Keywords: plant secondary metabolites, drimane sesquiterpenes, host plant defense, susceptibility, balanced antagonism, oxidative stress, endophytes
Citation: Drage S, Mitter B, Engelmeier D, Chobot V, Gorfer M, Muchugi A, Jamnadass RH, Sessitsch A and Hadacek F (2017) Antimicrobial Drimane Sesquiterpenes Contribute to Balanced Antagonism but Do Not Structure Bacterial and Fungal Endophytes in the African Pepper Bark Tree Warburgia ugandensis. Front. Ecol. Evol. 5:138. doi: 10.3389/fevo.2017.00138
Received: 29 June 2017; Accepted: 30 October 2017;
Published: 21 November 2017.
Edited by:
Thomas Seth Davis, Colorado State University, United StatesReviewed by:
Maria Carolina Blassioli Moraes, Embrapa Genetic Resources and Biotechnology, BrazilMaya Louise Evenden, University of Alberta, Canada
Copyright © 2017 Drage, Mitter, Engelmeier, Chobot, Gorfer, Muchugi, Jamnadass, Sessitsch and Hadacek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Franz Hadacek, franz.hadacek@biologie.uni-goettingen.de
 Sigrid Drage
Sigrid Drage Birgit Mitter
Birgit Mitter Doris Engelmeier
Doris Engelmeier Vladimir Chobot3
Vladimir Chobot3  Angela Sessitsch
Angela Sessitsch Franz Hadacek
Franz Hadacek