Ostreococcus tauri Luminescent Reporter Lines as Biosensors for Detecting Pollution From Copper-Mine Tailing Effluents in Coastal Environments
- 1Departamento de Genética Molecular y Microbiología, Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile, Santiago, Chile
- 2Departamento de Oceanografía, Facultad de Ciencias Naturales y Oceanográficas, Universidad de Concepción, Concepción, Chile
- 3Instituto Milenio de Oceanografía, Concepción, Chile
- 4Sorbonne Universités, UPMC Univ Paris 06 & Centre National pour la Recherche Scientifique, UMR 7621, Laboratoire d'Océanographie Microbienne, Observatoire Océanologique, Banyuls-sur-Mer, France
- 5Programa de Doctorado en Ingeniería de Sistemas Complejos, Universidad Adolfo Ibáñez, Santiago, Chile
- 6Centre National de la Recherche Scientifique, Institut Jacques Monod, Université Paris Diderot (Paris 07), Paris, France
- 7Departamento de Ecología, Center of Applied Ecology and Sustainability, Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile, Santiago, Chile
- 8Centro GEMA- Genómica, Ecología y Medio Ambiente, Facultad de Ciencias, Universidad Mayor, Santiago, Chile
Phytoplankton cells are excellent biosensors for environmental monitoring and toxicity assessments in different natural systems. Green algae, in particular, appear to be more responsive to copper (Cu) disturbances. This is interesting considering that Cu pollution in coastal environments has increased over the last century, with enormous repercussions to marine ecosystems. Unfortunately, no high-throughput method exists for the environmental monitoring of Cu toxicity in seawater. To assess potential uses as biosensors of Cu pollution, high-throughput screening was performed on five luminescence reporter lines constructed in the green algae Ostreococcus tauri RCC745. The reporter line expressing the iron storage ferritin protein fused to luciferase (Fer-Luc) was the most sensitive, responding to Cu concentrations in the μM range. Fer-Luc was also the most sensitive reporter line for detecting toxicity in mining-derived polluted seawater predominantly contaminated by soluble Cu. Nevertheless, the Cyclin-Dependent-Kinase A (CDKA) reporter was most suitable for detecting the toxicity of copper-mine tailing effluents containing other metals (e.g., iron). These results highlight that Ostreococcus biosensors can serve as a reliable, inexpensive, and automated, high-throughput laboratory approach for performing seawater analyses of coastal areas subjected to metal disturbances. When challenged with Cu, O. tauri not only evidenced a rapid, transcriptional response for the tested genes, but also showed changes in a broad range of genes, especially as related to the stress response. Overall, the obtained results reinforce that a single biosensor is insufficient when dealing with complex mixtures of toxic compounds in natural environments.
Introduction
Biosensors contain biological components (e.g., cells) that react to target substances (e.g., pollutants), and these reactions generate an easily readable signal (e.g., photon emission) proportional to the concentration of the compound of interest or of associated by-products (D'Souza, 2001; Balootaki and Hassanshahian, 2014; Kaur et al., 2015). In recent years, phytoplankton cells have proven to be excellent biosensors for environmental monitoring and toxicity assessments (Gutiérrez et al., 2015; Martín-Betancor et al., 2015). This is because phytoplankton cells quickly respond to disturbances in marine environments (Campanella et al., 2001). In fact, whole-cell biosensors based on phytoplankton cells are used to ecologically screen for pesticides and trace metals (Chouteau et al., 2005; Payne, 2013), and the available evidence reveals a broad tolerance range to trace metals in this group (Davis et al., 2006; Wang et al., 2012).
Green algae, in particular, appear to be more responsive to copper (Cu) disturbances than other phytoplankton taxa (Henríquez-Castillo et al., 2015). Therefore, this group of organisms has potential applications in the design of Cu biosensors. Green algae from the genus Ostreococcus (Chlorophyta, Mamiellophyceae) are cosmopolitan marine primary producers that dominate eukaryotic pico-phytoplankton communities (i.e., cells < 3 μm in diameter) in several coastal ecosystems (O'Kelly et al., 2003; Collado-Fabbri et al., 2011; Ottesen et al., 2011). These microalgae include different clades (Monier et al., 2016) that are genetically adapted to specific environmental conditions (Rodríguez et al., 2005; Botebol et al., 2017). In particular, Ostreococcus tauri is a model photosynthetic organism used to study biological processes (Corellou et al., 2009; Lozano et al., 2014), including cell division (Moulager et al., 2010), circadian clock architecture (O'Neill et al., 2011), iron metabolism (Botebol et al., 2015; Lelandais et al., 2016), and vitamin assimilation (Paerl et al., 2017). These applications as a model organism arise due to unique characteristics, including a small size, compact sequenced genome, and ease of genetic manipulation. The genetically engineered O. tauri RCC745, which expresses the cell cycle gene Cyclin-Dependent-Kinase (CDKA) fused to firefly luciferase (CDKA-Luc), is a very sensitive biosensor in high-throughput screenings for herbicides and antifouling compounds frequently found in the marine environment (Moulager et al., 2010; Sanchez-Ferandin et al., 2013). However, the application of O. tauri reporter lines as biosensors for Cu has not yet been tested in the laboratory or in natural seawater conditions. Genomic analyses in Ostreococcus spp. suggest that Cu is an essential cofactor in cellular processes, such as in photosynthesis, where cytochrome b is replaced by the Cu protein plastocyanin (Piganeau et al., 2011), and in iron acquisition, where multicopper oxidase is the only transporting component in the O. tauri genome (Palenik et al., 2007).
While Cu is an essential trace metal for marine organisms, toxicity occurs at high levels, with consequent deleterious effects on ecosystems and for public health (Castilla, 1996; Liu et al., 2011; Zhuang et al., 2013; Vásquez et al., 2015). Over the past century, Cu concentrations have increased in several coastal zones worldwide, with notable cases of Cu pollution from mine tailings (Paytan et al., 2009). Such cases include the open-cut Ok Tedi mine in Papua New Guinea (Fallon et al., 2002; Kirsch, 2007), the Mole River mine in Australia (Ashley and Lottermoser, 1999), the Dabaoshan mining region in China (Lin et al., 2005), and the El Salvador mine in Chañaral Bay, Chile (Andrade et al., 2006). Current protocols for environmental analyses of Cu are based on spectroscopy, including atomic absorption and inductively coupled plasma-mass instruments. Despite being very sensitive and accurate, these methods are time-consuming, require elaborate sample preparation, and, most importantly, are expensive, thus limiting the number of samples that can be analyzed (Liu et al., 2011). There is, therefore, a need to develop low-cost, highly-sensitive, and high-throughput methods for determining Cu toxicity in coastal environments (Feng et al., 2016).
Cu in seawater can be detected in association with suspended particulate material (> 0.45 μm) or as dissolved Cu (< 0.45 μm). Dissolved Cu in seawater can exist in multiple states, including as hydrated ions, in colloidal forms, and complexed with inorganic and organic ligands, the latter of which being the predominant state in aquatic environments (i.e., 99%) (Coale and Bruland, 1988). Free Cu ions and labile complex forms are the most toxic to the biota (Meador, 1991). In contrast, Cu complexation by organic ligands can decrease bioavailability and, hence, potential Cu toxicity (Stauber and Florence, 1985; Gledhill et al., 1997). Discriminating between these Cu forms is analytically complex. Therefore, an appropriate Cu sensor needs to be responsive to the bioavailable fraction of Cu in seawater, which makes any laboratory-based approach difficult because of the inherent difficulties in determining the labile fraction.
In this work, a micro plate-based automated laboratory approach was used to screen five luminescent O. tauri RCC745 lines reporting several cellular and physiological processes. The aim of this was to establish potential uses as biosensors of Cu toxicity in the laboratory and in Cu-contaminated seawater samples from the Chañaral area (i.e., south-eastern South Pacific). This coastal area of northern Chile has been the discharge site for thousands of tons of Cu mine-derived waste for the last 60 years (Correa et al., 1999). Available evidence from this highly polluted site indicates that the labile fraction of Cu accounts for most of the toxicity in macro- and microorganisms (Medina et al., 2005; Andrade et al., 2006; Morán et al., 2008; De la Iglesia et al., 2012). After initial biosensor selection, the transcriptional responses of O. tauri to a Cu challenge were measured. O. tauri evidenced a rapid expression of the tested genes and changes in a broad range of genes related to the stress response. Overall, the obtained results highlight that Ostreococcus biosensors can be used as a reliable, inexpensive, and automated high-throughput laboratory approach for analyzing seawater from coastal areas subjected to metal disturbances.
Materials and Methods
Test Conditions for Ostreococcus Growth Inhibition
Wild-type O. tauri RCC745 (clade C) was grown in T25 aerated flasks (Sarstedt) in artificial seawater media supplemented with Keller nutrients (0.88 mM nitrate, 22 μM phosphate, and f/2 vitamins) (Keller et al., 1987). Trace metals were added without EDTA (Botebol et al., 2015). Copper was added as a Cu sulfate (20 nM final concentration). Exponential phase cultures were transferred onto 24-well plates at a cell density of 1 million cells mL−1 and incubated with 20 nM, 40 nM, 0.2 μM, 0.4 μM, 2 μM, 4 μM, and 40 μM of Cu sulfate. Each concentration was tested in triplicate using independent wells. Subsamples (20 μL) were taken, diluted in 180 μL of Keller media with glutaraldehyde (0.25% vol/vol final concentration), and stored at −20°C. Cells were counted by flow cytometry (Accuri C6 flow cytometer, BD) using a 488 nm excitation laser with detection through side scatter and red fluorescence, using log-scale amplifiers. After 24 h, Cu toxicity was expressed as the concentration of Cu that induced a 50% reduction in cellular growth, relative to the Cu-free control (50% effective concentration [EC50]). For each Cu treatment, cell abundances were normalized against the control samples. The percentages were then plotted against the logarithm of Cu concentrations. Regression analysis was used to determine EC50 values (Prism 6.0, GraphPad).
Test Conditions for Ostreococcus Luminescence Inhibition
O. tauri RCC745 lines engineered with a luciferase gene reporter system were grown until the exponential phase, as described in Sanchez-Ferandin et al. (2013). The following O. tauri RCC745 lines were used: (1) pHAPT::Luc, a high-affinity phosphate transporter transcriptional reporter (promoter of the gene fused to luciferase); (2) CCA1-Luc, a circadian clock gene translational reporter (full gene in frame fused to luciferase); (3) CDKA-Luc, the cell cycle controller Cyclin-Dependent Kinase A translational reporter; (4) Fer-RI-Luc, an iron-storage ferritin protein translational reporter (random insertion at the ferritin locus of the Fer-Luc transgene); and (5) Fer-RH-Luc, an iron-storage ferritin protein translational reporter (homologous recombination at the ferritin locus of the Fer-Luc transgene) (Corellou et al., 2009; Moulager et al., 2010; Djouani-Tahri et al., 2011; Lozano et al., 2014; Botebol et al., 2015). Exponential phase cells were transferred into a 96-well microplate at final densities of 5 × 106 cells mL−1. Luciferin (10 μM final concentration) was added to the microplates, and then Cu was added at the following final concentrations: 0.4, 1, 2, 3, 4, 6.5, 10, 20, and 40 μM. Each Cu concentration was tested in triplicate using independent wells. Control samples containing 20 nM of Cu (i.e., minimum concentration not affecting growth kinetics) were also included in triplicate. Luminescence linked to the expression of the constructs was monitored every hour for 30 h using a high-throughput luminometer (Berthold Technologies). After 24 h, Cu toxicity was expressed as the Cu concentration that induced a 50% reduction in luminescence relative to the control (i.e., EC50). Luminescence curves of the treatments were normalized against control samples. The percentages were then plotted against the logarithm of Cu concentrations. Regression analysis was used to determine EC50 values (Prism 6.0, GraphPad).
Ecotoxicology Test Using Natural Seawater From a Mine-Tailing Polluted Area
Seawater samples were collected in March 2013 from the Chañaral area (Figure S1) using metal-free Kemmerer bottles placed at a 1 m depth. The Chañaral area is heavily disturbed due to Cu mine tailings, with Cu levels being well-characterized (Andrade et al., 2006). Six sites were sampled, including one directly affected by riverine Cu discharge (i.e., Chañaral, Palito 200, Playa Palito, Canal Palito, and La Lancha) (Figure S1). Due to low Cu levels, the Playa Blanca site (15 km north of the Canal Palito site) was used as a reference site (Andrade et al., 2006; Henríquez-Castillo et al., 2015).
For ecotoxicology tests, the O. tauri CDKA-Luc and Fer-RI-Luc luminescent reporter lines were pre-acclimated in seawater supplemented with Keller nutrients. Late exponential phase cells were transferred onto 96-well microplates containing seawater dilutions for the six sample sites (5 x 106 cells mL−1 final densities). For this, seawater from each polluted site was diluted using seawater from the reference site (i.e., Playa Blanca). The following dilutions were used: 25 mL of polluted seawater with 150 mL of reference seawater (25/100), 50/125, 75/100, 87.5/87.5, 100/75, 125/50, 150/25, and 175/0. In the case of the seawater from Canal Palito, salinity was previously adjusted down using MiliQ water to values for the reference seawater (Playa Blanca). All seawater dilutions used in treatments were enriched with Keller nutrients plus Cu sulfate (20 nM) and iron-citrate (0.1 μM). Luciferine (10 μM final concentration) was added directly to the plates. Luminescence of the reporter lines was measured every hour for 60 h with a high-throughput luminometer (Berthold Technologies). After 48 h, seawater toxicity was expressed as the dilution of polluted seawater in reference seawater that induced a 50% reduction in luminescence relative to the control (i.e., with no polluted seawater). Luminescence curves of the treatments were normalized against control samples. Percentages were then plotted against the logarithm of the dilution.
Cu Concentration in Seawater From Polluted Sites
For determinations of total dissolved metals, seawater samples were acidified to pH < 2 with bidistilled ultra-pure grade in a class-100 HEPA laminar flowhood. Concentrations of metals (i.e., Ag, Cd, Co, Cu, Fe, Mo, Ni, Pb, V, and Zn) were determined by inductively coupled plasma mass spectrometry (XSERIES 2 spectrophotometer, Thermo Fisher Scientific) after pre-concentration using the APDC/DDDC organic extraction method (Bruland et al., 1979; Tovar-Sánchez, 2012). Furthermore, labile Cu concentrations (Cu_ASV) were determined by square wave anodic stripping voltammetry (SW-ASV) as described in Andrade et al. (2006).
Transcriptomic Analyses of Ostreococcus
O. tauri RCC745 cultures were grown until the exponential phase (15 × 106 cells/mL) in T175 aerated flasks under a 12:12 light-dark cycle (20 μmol photons cm−2 s), transferred on 27 T75 flasks (80 mL each), and incubated with a final Cu concentration of 0.4 or 2 μM. Control samples containing 20 nM of Cu were also included in triplicate. To obtain cell counts, 20 μL triplicate samples were collected 1, 4, and 12 h after the addition of Cu. These samples were diluted in 180 μL of Keller medium with glutaraldehyde (0.25% vol/vol final concentration) and stored at −20°C. Cells were counted by flow cytometry (Accuri C6 flow cytometer, BD) using a 488 nm excitation laser with detection through side scatter and red fluorescence, using log-scale amplifiers. For RNA extraction, the remnant culture volume was split into two 50 mL polypropylene tubes. Pluronic F68 was added (0.01% vol/vol final concentration) and centrifuged for 30 min at 12,000 × g. Cell pellets were frozen in liquid nitrogen and stored at −20°C until RNA extraction.
Total RNA was extracted from frozen cell pellets using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA concentrations were determined by absorption at 260 nm, and purity was evaluated by the 260/280 nm absorption ratio. Total RNA integrity was assessed using an Agilent 2100 Bioanalyzer. RNAseq libraries were constructed using the TruSeq Stranded mRNA Library Preparation Kit (Illumina) by the NGS Service FASTERIS (Switzerland). Libraries were multiplexed in one channel of 1 × 50 bp run on a MiSeq run for titration and quality control. Finally, the libraries were sequenced using an Illumina HiSeq 2000 sequencer. Raw image data were converted into sequence data by base-calling, defined as raw reads. Pandaseq was used to assemble the paired-end sequences with default values. All sequences were searched for homologies against the KEGG database and against the most recent O. tauri genome annotation (Blanc-Mathieu et al., 2014) using the DIAMOND protein aligner (Buchfink et al., 2014). The DEseq2 routine (Love et al., 2014) was applied for comparisons between treatments. Genes with absolute Log2FC values over 0.3 and p-adjusted values below 0.1 were selected for further analysis. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE114058 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114058).
Statistical Analyses
All statistical analyses were performed in R v.3.4.1. Dissimilarity matrices were created using Euclidian distances. Non-metric multidimensional scaling was performed using the isoMDS function in the MASS package. Heatmaps were generated using the Heatmap, Vegan, and gplots packages.
Results
Responses of Different Ostreococcus Luminescent Reporters to Cu Amendment
Five O. tauri luciferase reporter lines were tested using an automated microplate approach to determine potential applications as biosensors for Cu toxicity. These lines were pHAPT::Luc, CCA1-Luc, CDKA-Luc, Fer-RI-Luc, and Fer-RH-Luc. The addition of Cu resulted in a differential response of the reporter lines, with an initial increase in luminescence for first 6 h post-exposure, followed by dose-dependent decreases (Figure 1). For all lines, excepting pHAPT:Luc, this initial luminescence increase occurred at the highest Cu concentration tested. Clear dose-dependent kinetic responses were observed, with a maximal decrease in luminescence after 24 h of Cu exposure (Figure 1). EC50 values for Cu were calculated for each line 24 h after Cu addition (Table S1). EC50 values varied among the different lines, but ranged between 4 and 10 μM. Fer-RI-Luc was the most Cu-sensitive luminescent line, with an EC50 of 3.83 μM after 24 h.
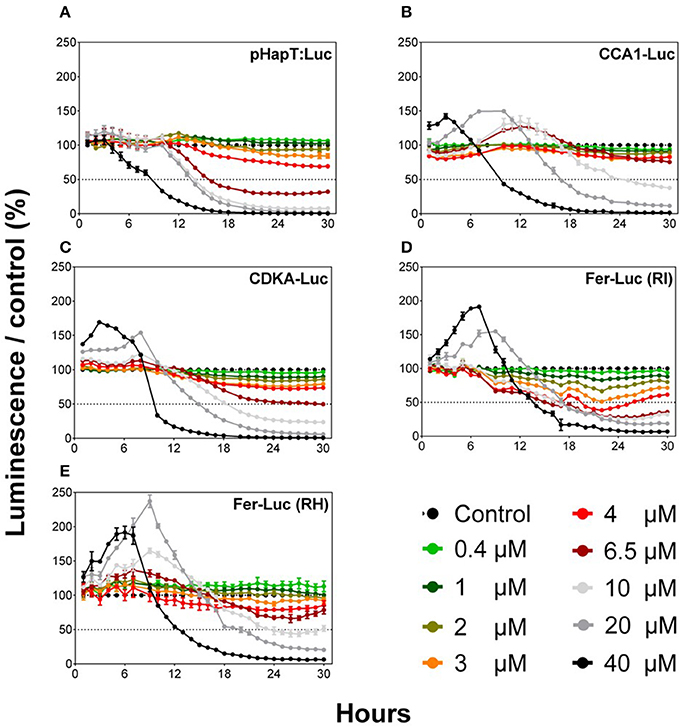
Figure 1. Comparison of the different O. tauri reporter luminescent lines after exposure to different Cu concentrations. Graphs represent normalized data compared to the control (KASW, 20 nM of Cu). Values represent the means ± SEM for triplicate wells.
Response of Fer-RI-Luc and CDKA-Luc Lines to Cu-Polluted Seawater
The sensitivities of Fer-RI-Luc and CDKA-Luc were tested using natural seawater collected from five Cu-mining polluted sites in the Chañaral area (i.e., Chañaral, Palito 200, Playa Palito, Canal Palito, and La Lancha), as well as from an unpolluted reference site (Playa Blanca) (Figure S1). The seawater from all polluted sites showed high levels of both total dissolved Cu (> 0.1 μM) and labile Cu (> 0.01 μM) (Table S2). Variable concentrations of Cu, Fe, Zn, Pb, V, Ni, and Co were found among sites (Figure 2A), with a clear difference between polluted sites and the reference site, as revealed by non-metric multidimensional scaling analysis of metal concentrations (Figure 2B). Similar concentrations of trace metals, other than the two Cu fractions, were found between La Lancha and the reference site (paired t-test, Cu, labile Cu; P < 0.001).
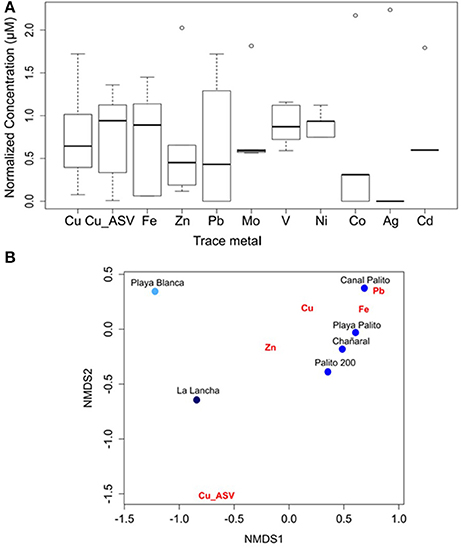
Figure 2. (A) Boxplot for trace-metal concentration at the study sites. Values were log(x+1) transformed and standardized. (B) Non-metric multidimensional scaling analysis according to metal concentrations that strongly varied between sites.
Cu-polluted seawater had negative, dose-dependent effects on the Fer-RI-Luc and CDKA-Luc reporter lines (Figure 3). The Fer-RI-Luc line showed a dual response to Cu additions, with an initial increase in luminescence followed by a dose-dependent decline for almost all treatments (Figures 3A,E). In contrast, CDKA-Luc luminescence decreased immediately after the addition of Cu amendments (Figures 3B,D,F).
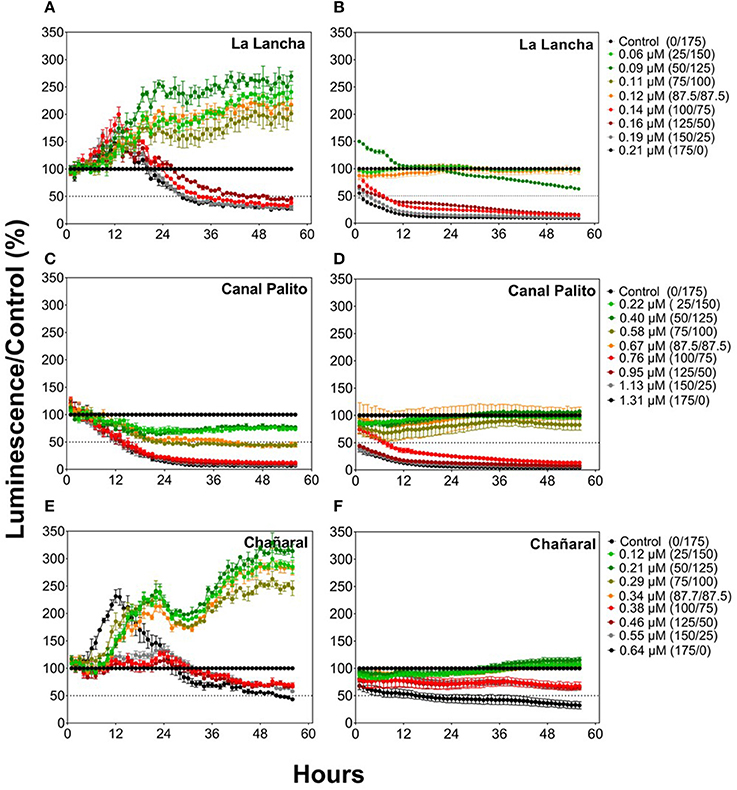
Figure 3. Responses of the O. tauri Fer-RI-Luc (A,C,E) and CDKA-Luc (B,D,F) reporter luminescent lines after exposure to seawater with different Cu concentrations. Graphs represent normalized data compared to the control lines (0.04 μM of Cu). Theoretical Cu concentrations were estimated based on initial concentrations in polluted sweater and on dilutions in reference seawater, as displayed in descriptions for the curves.
Transcriptional Response of Ostreococcus tauri to Cu Challenge
To study the effect of increased Cu concentrations on gene expressions in the luminescent reporter lines, transcriptional responses were studied through an RNAseq-based approach. Growth-curve analysis showed that the addition of 0.4 and 2 μM of Cu inhibited growth by 30 and 60%, respectively, at 24 h post-Cu amendment addition (Figure S2). Based on this, high-throughput transcriptomic analysis was performed at 1, 4, and 12 h post-Cu addition. General patterns of gene expression were found (Figure 4; Table S3). Significant changes in gene expression were observed 4 h post-Cu exposure, especially for the 2 μM treatment (Figure 4, cluster 2).
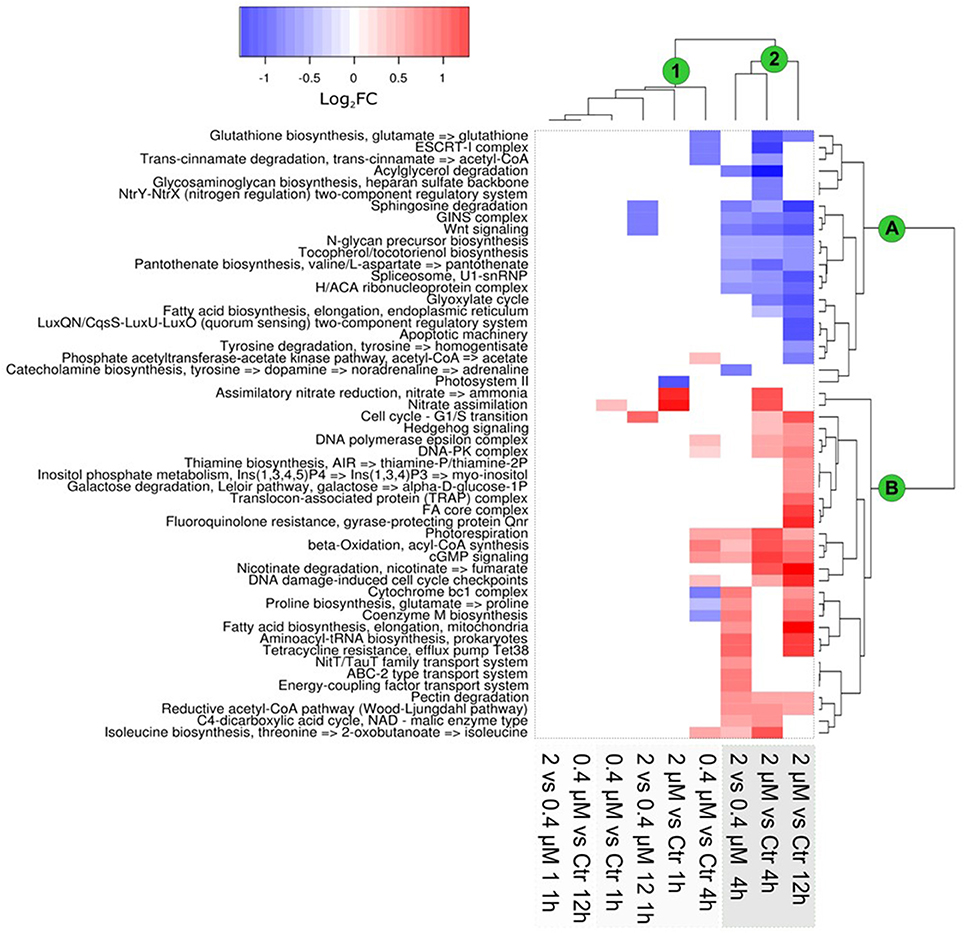
Figure 4. Transcriptional response of Ostreococcus tauri to Cu exposure, based on functional KEGG modules. Clustering of the data showed a differential expression of KEGG modules, as related to the time of Cu exposure and Cu concentration. Results are expressed (according to the color scale) in log base 2, i.e., as log2FC [+Cu]/[Ctrl]. The main clusters of modules and samples are marked with green circles. Clustering was based on Ward's method.
Expressional changes were evaluated for genes used in the ecotoxicology tests and for genes involved in the regulation of Cu homeostasis (Figure 5). HapT (OT_ostta02g02460) and CCA1 (OT_ostta02g04817) were upregulated in all conditions in response to Cu, especially in the 0.2 μM Cu treatment. While ferritin transcripts (OT_ostta02g03400) were significantly upregulated at 4 and 12 h after the addition of the 2 μM Cu treatment, CDKA transcripts (OT_ostta15g00670) were upregulated only at 0.2 μM (Figure 5). Overall, CDK transcript levels decreased in response to the addition of Cu. Genes related to Cu homeostasis, such as Superoxide Dismutase Cu/Zn-dependent (SOD1), the respective SOD1 Chaperone (CCS), and Cu-exporting ATPase (copB), displayed various expression patterns. Some were downregulated (e.g., copA), while others appeared upregulated (e.g., SOD1).
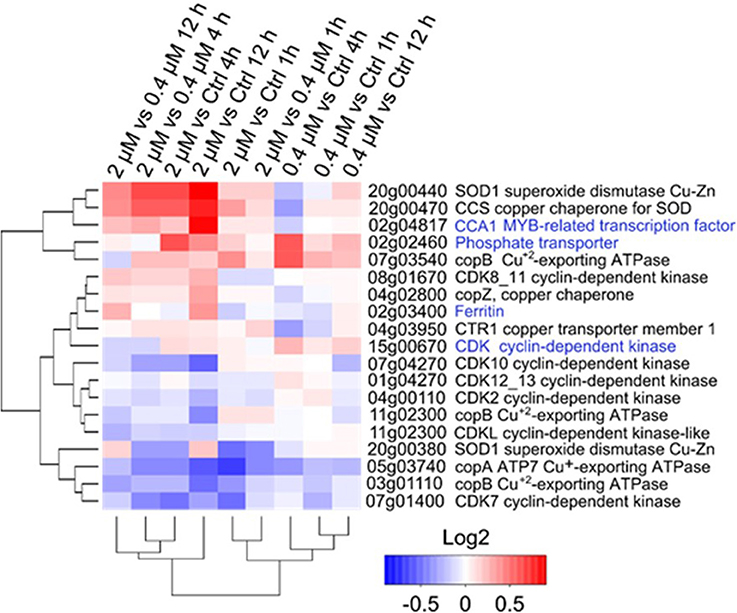
Figure 5. Transcriptional changes of the tested genes after Cu exposure in O. tauri RCC745. Cluster analysis is based on Euclidian distances of the Log2 fold-change values of genes for each treatment. Gene function for each transcript according to KEGG annotation is indicated at the right of the genomic location. Genes tested in the ecotoxicology analysis are highlighted in blue.
Discussion
Five different O. tauri luminescent lines were analyzed to determine applicability as a high-throughput system for Cu monitoring in coastal environments. Luminescent lines included a broad spectrum of genes and genetic constructions involved in different biological processes, including the circadian clock (CCA1), cell division (CDKA), metal homeostasis regulation (Fer), and phosphate transport (HapT) (Moulager et al., 2010; Sanchez-Ferandin et al., 2013). The obtained results indicate that increases in Cu exert global effects on gene expression, as reflected by the physiology of O. tauri. Impacts to gene expression were particularly notable for the Fer-RI-Luc iron-related luminescent line, which was the most effective in sensing toxicity in natural seawater samples predominantly contaminated by labile Cu. The differences in sensitivity between the Fer-RI-Luc and the CDKA-Luc lines highlight the need to use different biosensors when environmental pollutants are mixed, as usually occurs. Importantly, the conducted assays required small amounts of polluted seawater and non-complex sample preparation, thus facilitating the combination of multiple luminescence reporter lines to accurately detect metal pollution. The feasibility of assays, in terms of cost and time, represents a valuable contribution to the environmental monitoring of coastal areas. The presently tested luminescent biosensors showed major advantages compared to traditional assessment tests based on cell-growth inhibition. Specifically, the monitoring process is fully automated and can provide rapid, low-cost responses while using a high-throughput approach applicable to routine coastal pollution studies.
All luciferase reporter lines displayed dose-dependent growth inhibition in response to Cu amendments. These results suggest that Cu has a global toxic effect on the physiology and growth of O. tauri, a finding similar to prior reports for other phytoplankton species (Davis et al., 2006; Stuart et al., 2009; Jordi et al., 2012). Of the lines tested, Fer-RI-Luc was the most sensitive to Cu. This is in line with the nutritional coupling between Cu and Fe reported for other microbial species (Crichton and Pierre, 2001; Peers et al., 2005; Maldonado et al., 2006; Sunda, 2012), a coupling that can lead to nutritional imbalance at high Cu concentrations. Fer-RI-Luc construction was also highly sensitive when O. tauri was challenged with Cu-polluted seawater. Indeed, the minimum detection level was 0.06 μM Cu L−1, one of the lowest detection limits reported to date for a Cu whole-cell biosensor (Tag et al., 2007; Jarque et al., 2016). For the La Lancha site, where Cu was the only metal to evidence significant differences with the reference site, soluble copper, in a dose-dependent manner, accounted for seawater toxicity. The Fer-RI-Luc line could, therefore, be a sensitive biosensor for Cu toxicity when Cu is the sole source of pollution. However, when this luminescent line was tested with the riverine discharge water, luminescence inhibition was observed for all of the tested concentrations. Therefore, the recorded luminescence decrease cannot be attributed to only dose-dependent Cu effects. Water coming from the riverine discharge point contained a complex mixture of metals, including Mo, Ni, Co, Ag, Zn, Cd, and Pb. Metal interactions may account for the increased water toxicity at the riverine discharge site (Franklin et al., 2002). The CDKA-Luc reporter showed decreased luminescence for all tested polluted seawater samples, with dose-dependent decreases aligning with earlier data showing that the CDKA-Luc line is a sensitive biosensor of general toxicity (Sanchez-Ferandin et al., 2013). These results indicate that, when dealing with complex metal complexes such as those near the riverine discharge site, the CDKA-Luc line is more suitable to detect seawater toxicity. Overall, the obtained results reinforce the idea of combining several reporter lines, such as CDKA-Luc and Fer-RI-Luc, to discriminate between general and specific Cu toxicity in polluted seawater.
O. tauri showed a strong transcriptional response to Cu amendments, as inferred from RNAseq data. Prior analyses on the main KEGG modules during the initial response to Cu amendments revealed a transcriptional inhibition of genes involved Photosystem II and nitrogen metabolism (Harrison et al., 1977; Harismendy et al., 2009). Later responses to Cu amendments included a well-defined group of overexpressed genes, some of which were related to the general stress response, such as the DNA damage-induced cell cycle checkpoint. Gene groups inhibited at the end of the experiments included genes related to cellular growth and biosynthesis. RNA-seq analysis confirmed the luminescence patterns for ferritin, which was induced soon after Cu addition before being downregulated later. For other genes, such as CCA1, hapT, and CDK, luminescence responses and expression patterns were quite different, and no clear dose-dependent response was observed. These differences may be due to the nature of the CDKA-Luc line, which reports CDKA protein levels vs. transcript levels in RNAseq data. Unlike RNAseq data, the luminescence curves established clear Cu dose-dependent responses of luminescence inhibition at 24 h. In addition to being low-cost, luminescent biosensors appear more sensitive, being able to detect toxicity in a dose-dependent manner.
Coastal pollution is increasing in step with expanding human activities. Despite the high impact of metallic pollutants on the environment, current tools for determining metal toxicity in coastal areas are scarce, expensive, and far from being routinely applied. The Chañaral area in northern Chile provides an excellent natural laboratory for understanding the complex repercussions of Cu pollution on aquatic and coastal ecosystems (Medina et al., 2005; Andrade et al., 2006; De la Iglesia et al., 2012). The northward current in this area generates a gradient in Cu concentrations, being highest at the riverine discharge site and decreasing in proximity to the northern reference site. A decrease in the complexity of the metal mixtures was also observed in a gradient from the discharge point to the reference site, thus permitting evaluations on the response of biosensors to different metal mixtures in natural marine samples. In this work, seawater from the most polluted site (i.e., La Lancha) was used to determine the effect of total dissolved Cu and labile dissolved Cu on two luciferase reporter lines, without interference from other metals present in seawater from the polluted sites located in the vicinity of the riverine discharge site.
Concluding Remarks
In this study, O. tauri biosensors were useful for studying Cu toxicity in seawater matrices. The presented data suggest that, when dealing with complex environmental samples, a combination of several reporter lines is required to assess seawater toxicity. Further interference and selectivity investigations are needed to fully understand responses to Cu by genetically engineered O. tauri biosensors and to develop more specific, sensitive biosensors based on the identification of Cu- responding genes, e.g., as derived from RNAseq approaches.
Author Contributions
RD, F-YB, and CH-C design the study. CH-C, RD, NT, and SA collected the samples. CH-C, AM, HB, J-CL, GL, and SA performed the experiments and data analysis. CH-C, RD, NT, SR-F, and GL perform bioinformatic analyses. All authors participates in wrote the paper.
Funding
This work has been funded by FONDECYT Grant No. 1171259 (to RD). Collaboration with France was funded through the CNRS International Research Network Diversity, Evolution and Biotechnology of Marine Algae (GDRI No. 0803).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2018.00022/full#supplementary-material
References
Andrade, S., Moffett, J., and Correa, J. A. (2006). Distribution of dissolved species and suspended particulate copper in an intertidal ecosystem affected by copper mine tailings in northern Chile. Mar. Chem. 101, 203–212. doi: 10.1016/j.marchem.2006.03.002
Ashley, P. M., and Lottermoser, B. G. (1999). Arsenic contamination at the Mole River mine, northern New South Wales. Aust. J. Earth Sci. 46, 861–874. doi: 10.1046/j.1440-0952.1999.00748.x
Balootaki, P. A., and Hassanshahian, M. (2014). Microbial biosensor for marine environments. Bull. Environ. Pharmacol. Life Sci. 3, 1–13.
Blanc-Mathieu, R., Verhelst, B., Derelle, E., Rombauts, S., Bouget, F.-Y., Carré, I., et al. (2014). An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies. BMC Genomics 15:1103. doi: 10.1186/1471-2164-15-1103
Botebol, H., Lelandais, G., Six, C., Lesuisse, E., Meng, A., Bittner, L., et al. (2017). Acclimation of a low iron adapted Ostreococcus strain to iron limitation through cell biomass lowering. Sci. Rep. 7:327. doi: 10.1038/s41598-017-00216-6
Botebol, H., Lesuisse, E., Sutak, R., Six, C., Lozano, J.-C., Schatt, P., et al. (2015). Central role for ferritin in the day/night regulation of iron homeostasis in marine phytoplankton. Proc. Natl. Acad. Sci. U.S.A. 112, 1–6. doi: 10.1073/pnas.1506074112
Bruland, K. W., Franks, R. P., Knauer, G. A., and Martin, J. H. (1979). Sampling and analytical methods for the determination of copper, cadmium, zinc, and nickel at the nanogram per liter level in sea water. Anal. Chim. Acta 105, 233–245.
Buchfink, B., Xie, C., and Huson, D. H. (2014). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Campanella, L., Cubadda, F., Sammartino, M. P., and Saoncella, A. (2001). An algal biosensor for the monitoring of water toxicity in estuarine environments. Water Res. 35, 69–76. doi: 10.1016/S0043-1354(00)00223-2
Castilla, J. C. (1996). Copper mine tailing disposal in northern Chile rocky shores: Enteromorpha compressa (Chlorophyta) as a sentinel species. Environ. Monit. Assess. 40, 171–184. doi: 10.1007/BF00414390
Chouteau, C., Dzyadevych, S., Durrieu, C., and Chovelon, J. M. (2005). A bi-enzymatic whole cell conductometric biosensor for heavy metal ions and pesticides detection in water samples. Biosens. Bioelectron. 21, 273–281. doi: 10.1016/j.bios.2004.09.032
Coale, K. H., and Bruland, K. W. (1988). Copper complexation in the Northeast Pacific. Limnol. Oceanogr. 33, 1084–1101.
Collado-Fabbri, S., Vaulot, D., and Ulloa, O. (2011). Structure and seasonal dynamics of the eukaryotic picophytoplankton community in a wind-driven coastal upwelling ecosystem. Limnol. Oceanogr. 56, 2334–2346. doi: 10.4319/lo.2011.56.6.2334
Corellou, F., Schwartz, C., Motta, J., Djouani-Tahri, E. B., Sanchez, F., and Bouget, F.-Y. (2009). Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote Ostreococcus. Plant Cell 21, 3436–3449. doi: 10.1105/tpc.109.068825
Correa, J., Castilla, J., Ramírez, M., Varas, M., Lagos, N., Vergara, S., et al. (1999). Copper, copper mine tailings and their effect on marine algae in Northern Chile. J. Appl. Phycol. 11, 57–67. doi: 10.1046/j.1365-3040.2003.01073.x
Crichton, R. R., and Pierre, J. L. (2001). Old iron, young copper: from Mars to Venus. Biometals 14, 99–112. doi: 10.1023/A:1016710810701
D'Souza, S. F. (2001). Microbial biosensors. Biosens. Bioelectron. 16, 337–353. doi: 10.1016/S0956-5663(01)00125-7
Davis, A. K., Hildebrand, M., and Palenik, B. (2006). Gene expression induced by copper stress in the diatom Thalassiosira pseudonana. Eukaryotic Cell 5, 1157–1168. doi: 10.1128/EC.00042-06
De la Iglesia, R., Valenzuela-Heredia, D., Andrade, S., Correa, J., and González, B. (2012). Composition dynamics of epilithic intertidal bacterial communities exposed to high copper levels. FEMS Microbiol. Ecol. 79, 720–727. doi: 10.1111/j.1574-6941.2011.01254.x
Djouani-Tahri, E. B., Christie, J. M., Sanchez-Ferandin, S., Sanchez, F., Bouget, F. Y., and Corellou, F. (2011). A eukaryotic LOV-histidine kinase with circadian clock function in the picoalga Ostreococcus. Plant J. 65, 578–588. doi: 10.1111/j.1365-313X.2010.04444.x
Edgar, R., Domrachev, M., and Lash, A. E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. doi: 10.1093/nar/30.1.207
Fallon, S. J., White, J. C., and McCulloch, M. T. (2002). Porites corals as recorders of mining and environmental impacts: Misima Island, Papua New Guinea. Geochim. Cosmochim. Acta 66, 45–62. doi: 10.1016/S0016-7037(01)00715-3
Feng, C.-Y., Wei, J.-F., Li, Y.-J., Yang, Y.-S., Wang, Y.-H., Lu, L., et al. (2016). An on-chip pollutant toxicity determination based on marine microalgal swimming inhibition. Analyst 141, 1761–1771. doi: 10.1039/C5AN02384J
Franklin, N. M., Stauber, J. L., Lim, R. P., and Petocz, P. (2002). Toxicity of metal mixtures to a tropical freshwater alga (Chlorella sp): the effect of interactions between copper, cadmium, and zinc on metal cell binding and uptake. Environ. Toxicol. Chem. 21, 2412–2422. doi: 10.1002/etc.5620211121
Gledhill, M., Nimmo, M., Hill, S. J., and Brown, M. T. (1997). The toxicity of copper (II) species to marine algae, with particular reference to macroalgae. J. Phycol. 33, 2–11.
Gutiérrez, J. C., Amaro, F., and Martín-González, A. (2015). Heavy metal whole-cell biosensors using eukaryotic microorganisms: an updated critical review. Front. Microbiol. 6:48. doi: 10.3389/fmicb.2015.00048
Harismendy, O., Ng, P. C., Strausberg, R. L., Wang, X., Stockwell, T. B., Beeson, K. Y., et al. (2009). Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 10:R32. doi: 10.1186/gb-2009-10-3-r32
Harrison, W. G., Eppley, R. W., and Renger, E. H. (1977). Phytoplankton nitrogen metabolism, nitrogen budgets, and observations on copper toxicity: controlled ecosystem pollution experiment. Bull. Mar. Sci. 27, 44–57.
Henríquez-Castillo, C., Rodríguez-marconi, S., Rubio, F., Trefault, N., Andrade, S., and De la.Iglesia, R. (2015). Eukaryotic picophytoplankton community response to copper enrichment in a metal-perturbed coastal environment. Phycol. Res. 63, 189–196. doi: 10.1111/pre.12087
Jarque, S., Bittner, M., Blaha, L., and Hilscherova, K. (2016). Yeast biosensors for detection of environmental pollutants: current state and limitations. Trends Biotechnol. 34, 408–419. doi: 10.1016/j.tibtech.2016.01.007
Jordi, A., Basterretxea, G., Tovar-Sánchez, A., Alastuey, A., and Querol, X. (2012). Copper aerosols inhibit phytoplankton growth in the Mediterranean Sea. Proc. Natl. Acad. Sci. U.S.A. 109, 21246–21249. doi: 10.1073/pnas.1207567110
Kaur, H., Kumar, R., Babu, J. N., and Mittal, S. (2015). Advances in arsenic biosensor development - a comprehensive review. Biosens. Bioelectron. 63, 533–545. doi: 10.1016/j.bios.2014.08.003
Keller, M. D., Seluin, R. C., Claus, W., and Guillard, R. R. L. (1987). Media for the culture of oceanic ultraphytoplankton. J. Phycol. 23, 633–638. doi: 10.1016/0198-0254(88)92621-0
Kirsch, S. (2007). Indigenous movements and the risks of counterglobalization: tracking the campaign against Papua New Guinea's Ok Tedi mine. Am. Ethnol. 34, 303–321. doi: 10.1525/ae.2007.34.2.303
Lelandais, G., Scheiber, I., Paz-Yepes, J., Lozano, J. C., Botebol, H., Pilátová, J., et al. (2016). Ostreococcus tauri is a new model green alga for studying iron metabolism in eukaryotic phytoplankton. BMC Genomics 17:319. doi: 10.1186/s12864-016-2666-6
Lin, C., Tong, X., Lu, W., Yan, L., Wu, Y., Nie, C., et al. (2005). Environmental impacts of surface mining on mined lands, affected streams and agricultural lands in the Dabaoshan Mine region, southern China. Land Degrad. Dev. 16, 463–474. doi: 10.1002/ldr.675
Liu, M., Zhao, H., Chen, S., Yu, H., Zhang, Y., and Quan, X. (2011). A “turn-on” fluorescent copper biosensor based on DNA cleavage-dependent graphene-quenched DNAzyme. Biosens. Bioelectron. 26, 4111–4116. doi: 10.1016/j.bios.2011.04.006
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lozano, J. C., Schatt, P., Botebol, H., Vergé, V., Lesuisse, E., Blain, S., et al. (2014). Efficient gene targeting and removal of foreign DNA by homologous recombination in the picoeukaryote Ostreococcus. Plant J. 78, 1073–1083. doi: 10.1111/tpj.12530
Maldonado, M. T., Allen, A. E., Chong, J. S., Lin, K., Leus, D., Karpenko, N., et al. (2006). Copper-dependent iron transport in coastal and oceanic diatoms. Limnol. Oceanogr. 51, 1729–1743. doi: 10.4319/lo.2006.51.4.1729
Martín-Betancor, K., Rodea-Palomares, I., Muñoz-Martín, M. A., Leganés, F., and Fernández-Piñas, F. (2015). Construction of a self-luminescent cyanobacterial bioreporter that detects a broad range of bioavailable heavy metals in aquatic environments. Front. Microbiol. 6:186. doi: 10.3389/fmicb.2015.00186
Meador, J. P. (1991). The interaction of pH, dissolved organic carbon, and total copper in the determination of ionic copper and toxicity. Aquat. Toxicol. 19, 13–31.
Medina, M., Andrade, S., Faugeron, S., Lagos, N., Mella, D., and Correa, J. A. (2005). Biodiversity of rocky intertidal benthic communities associated with copper mine tailing discharges in northern Chile. Mar. Pollut. Bull. 50, 396–409. doi: 10.1016/j.marpolbul.2004.11.022
Monier, A., Worden, A. Z., and Richards, T. A. (2016). Phylogenetic diversity and biogeography of the Mamiellophyceae lineage of eukaryotic phytoplankton across the oceans. Environ. Microbiol. Rep. 8, 461–469. doi: 10.1111/1758-2229.12390
Morán, A. C., Hengst, M. B., De La Iglesia, R., Andrade, S., Correa, J. A., and González, B. (2008). Changes in bacterial community structure associated with coastal copper enrichment. Environ. Toxicol. 27, 2239–2245. doi: 10.1897/08-112.1
Moulager, M., Corellou, F., Vergé, V., Escande, M. L., and Bouget, F. Y. (2010). Integration of light signals by the retinoblastoma pathway in the control of S phase entry in the Picophytoplanktonic cell Ostreococcus. PLoS Genet. 6:e1000957. doi: 10.1371/journal.pgen.1000957
O'Kelly, C. J., Sieracki, M. E., Thier, E. C., and Hobson, I. C. (2003). A transient bloom of Ostreococcus (Chlorophyta, Prasinophyceae) in West Neck Bay, Long Island, New York. J. Phycol. 39, 850–854. doi: 10.1046/j.1529-8817.2003.02201.x
O'Neill, J. S., Van Ooijen, G., Dixon, L. E., Troein, C., Corellou, F., Bouget, F. Y., et al. (2011). Circadian rhythms persist without transcription in a eukaryote. Nature 469, 554–558. doi: 10.1038/nature09654
Ottesen, E. A., Marin, R., Preston, C. M., Young, C. R., Ryan, J. P., Scholin, C. A., et al. (2011). Metatranscriptomic analysis of autonomously collected and preserved marine bacterioplankton. ISME J. 5, 1881–1895. doi: 10.1038/ismej.2011.70
Paerl, R. W., Bouget, F. Y., Lozano, J. C., Vergé, V., Schatt, P., Allen, E. E., et al. (2017). Use of plankton-derived vitamin B1 precursors, especially thiazole-related precursor, by key marine picoeukaryotic phytoplankton. ISME J. 11, 753–765. doi: 10.1038/ismej.2016.145
Palenik, B., Grimwood, J., Aerts, A., Rouzé, P., Salamov, A., Putnam, N., et al. (2007). The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. U.S.A. 104, 7705–7710. doi: 10.1073/pnas.0611046104
Payne, R. J. (2013). Seven reasons why protists make useful bioindicators. Acta Protozool. 52, 105–113. doi: 10.4467/16890027AP.13.0011.1108
Paytan, A., Mackey, K. R., Chen, Y., Lima, I. D., Doney, S. C., Mahowald, N., et al. (2009). Toxicity of atmospheric aerosols on marine phytoplankton. Proc. Natl. Acad. Sci. U.S.A. 106, 4601–4605. doi: 10.1073/pnas.0811486106
Peers, G., Quesnel, S., and Price, N. M. (2005). Copper requirements for iron acquisition and growth of coastal and oceanic diatoms. Limnol. Oceanogr. 50, 1149–1158. doi: 10.4319/lo.2005.50.4.1149
Piganeau, G., Grimsley, N., and Moreau, H. (2011). Genome diversity in the smallest marine photosynthetic eukaryotes. Res. Microbiol. 162, 570–577. doi: 10.1016/j.resmic.2011.04.005
Rodríguez, F., Derelle, E., Guillou, L., Le Gall, F., Vaulot, D., and Moreau, H. (2005). Ecotype diversity in the marine picoeukaryote Ostreococcus (Chlorophyta, Prasinophyceae). Environ. Microbiol. 7, 853–859. doi: 10.1111/j.1462-2920.2005.00758.x
Sanchez-Ferandin, S., Leroy, F., Bouget, F. Y., and Joux, F. (2013). A new, sensitive marine microalgal recombinant biosensor using luminescence monitoring for toxicity testing of antifouling biocides. Appl. Environ. Microbiol. 79, 631–638. doi: 10.1128/AEM.02688-12
Stauber, J. L., and Florence, T. M. (1985). Interactions of copper and manganese: a mechanism by which manganese alleviates copper toxicity to the marine diatom, Nitzschia closterium (Ehrenberg) W. Smith. Aquat. Toxicol. 7, 241–254.
Stuart, R. K., Dupont, C. L., Johnson, D. A., Paulsen, I. T., and Palenik, B. (2009). Coastal strains of marine Synechococcus species exhibit increased tolerance to copper shock and a distinctive transcriptional response relative to those of open-ocean strains. Appl. Environ. Microbiol. 75, 5047–5057. doi: 10.1128/AEM.00271-09
Sunda, W. G. (2012). Feedback interactions between trace metal nutrients and phytoplankton in the ocean. Front. Microbiol. 3:204. doi: 10.3389/fmicb.2012.00204
Tag, K., Riedel, K., Bauer, H. J., Hanke, G., Baronian, K. H. R., and Kunze, G. (2007). Amperometric detection of Cu2+ by yeast biosensors using flow injection analysis (FIA). Sens. Actuators B Chem. 122, 403–409. doi: 10.1016/j.snb.2006.06.007
Tovar-Sánchez, A. (2012). “Sampling approaches for trace element determination in seawater,” in Comprehensive Sampling and Sample Preparation, ed J. Pawliszyn (Academic Press), 317–334.
Vásquez, K. Y., Orellana, S. A., Monsalve, S. M., Vergara, J. K., Zamora, C. S., Muñoz, D. V., et al. (2015). Exposure to fine particles by mine tailing and lung function effects in a panel of schoolchildren, chañaral, chile. J. Environ Prot. 118–128. doi: 10.4236/jep.2015.62014
Wang, B., Axe, L., Michalopoulou, Z.-H., and Wei, L. (2012). Effects of Cd, Cu, Ni, and Zn on brown tide alga Aureococcus anophagefferens growth and metal accumulation. Environ. Sci. Technol. 46, 517–524. doi: 10.1021/es202790p
Keywords: copper pollution, mine tailings, biosensors, Ostreococcus, ferritin, CDKA, luciferase reporter
Citation: Henríquez-Castillo C, Botebol H, Mouton A, Ramírez-Flandes S, Lozano J-C, Lelandais G, Andrade S, Trefault N, De la Iglesia R and Bouget F-Y (2018) Ostreococcus tauri Luminescent Reporter Lines as Biosensors for Detecting Pollution From Copper-Mine Tailing Effluents in Coastal Environments. Front. Environ. Sci. 6:22. doi: 10.3389/fenvs.2018.00022
Received: 03 December 2017; Accepted: 09 April 2018;
Published: 22 May 2018.
Edited by:
Jean Armengaud, Commissariat à l'Energie Atomique et aux Energies Alternatives (CEA), FranceReviewed by:
Joseph Alexander Christie-Oleza, University of Warwick, United KingdomRafael Bosch, Universidad de les Illes Balears, Spain
Copyright © 2018 Henríquez-Castillo, Botebol, Mouton, Ramírez-Flandes, Lozano, Lelandais, Andrade, Trefault, De la Iglesia and Bouget. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rodrigo De la Iglesia, rdelaiglesia@bio.puc.cl
François-Yves Bouget, francois-yves.bouget@obs-banyuls.fr
 Carlos Henríquez-Castillo
Carlos Henríquez-Castillo Hugo Botebol4
Hugo Botebol4  Salvador Ramírez-Flandes
Salvador Ramírez-Flandes Jean-Claude Lozano
Jean-Claude Lozano Nicole Trefault
Nicole Trefault Rodrigo De la Iglesia
Rodrigo De la Iglesia