- Workman Lab Stowers Institute for Medical Research, Kansas City, MO, United States
The mechanisms of epigenetic gene regulation—histone modifications, chromatin remodeling, DNA methylation, and noncoding RNA—use metabolites as enzymatic cofactors and substrates in reactions that allow chromatin formation, nucleotide biogenesis, transcription, RNA processing, and translation. Gene expression responds to demands from cellular processes that use specific metabolites and alters or maintains cellular metabolic status. However, the roles of metabolites—particularly nucleotides—as regulatory molecules in epigenetic regulation and biological processes remain largely unknown. Here we review the crosstalk between gene expression, nucleotide metabolism, and cellular processes, and explore the role of metabolism in epigenetics as a critical regulator of biological events.
Introduction
One-carbon metabolism governs cellular nutritional status by sensing input metabolites and generating and redistributing output metabolites (1, 2). The input metabolites of one-carbon metabolism are usually serine and glycine reacting in the folate cycle; output metabolites include S-adenosyl-L-methionine (SAMe), glutathione, nucleotides, and polyamines. This one-carbon metabolic network generates energy and coordinates intracellular redox status. One-carbon metabolism has been targeted in cancer therapy (1, 3), for example by using pyrimidine analogues such as fluorouracil, which inhibits thymidylate synthase in the folate metabolic pathway (3). Outputs of one-carbon metabolism play key roles in post-translational modification of histones, DNA methylation, chromatin remodeling, and RNA biogenesis including tRNA modifications (2, 4).
Chromatin modifications regulate transcription and gene expression to modify or maintain the cellular status. Metabolites are essential cofactors and substrates in the epigenetic marking of histones including those in nucleosomes. For example, histone/lysine acetyltransferases use acetyl-CoA to catalyze the addition of an acetyl group to an epsilon amino group of a lysine sidechain in the N-terminal “tail” domains of histones (5). Lysine acylation reactions also include propionylation, butyrylation, crotonylation, β-hydroxybutyrylation, succinylation, malonylation, and glutarylation (6). These different acylation reactions use different short-chain acyl intermediates, suggesting that the type of acylation is determined by the cell metabolic status. Histone deacetylases (HDACs) catalyze the hydrolysis of an N-ε-acetyl-L-lysine sidechain to generate acetate and L-lysine (7). Class I and II HDACs catalyze this hydrolysis in a Zn+-dependent manner; Class III HDACs (sirtuins) use the oxidized form of nicotinamide adenine dinucleotide, NAD+, as a co-substrate and transfer the acetyl group onto adenosine diphosphate ribose (ADPR) to form a 2’-O-acetyl-ADPR (8, 9). Therefore, sirtuins may be sensitive to cellular NAD/energy levels in their regulation of transcription. Methylation of histones and DNA requires SAMe (which is synthesized from ATP and methionine by methionine adenosyltransferase) as a cofactor. Histone demethylations are catalyzed by two major classes of lysine demethylases: lysine-specific histone demethylases (LSDs) and JmjC-domain-containing histone demethylases (JMJCs) (2). LSDs require flavin adenine dinucleotide (FAD), a redox-active coenzyme that is synthesized de novo from riboflavin (vitamin B2) (10). Demethylation by JMJCs requires Fe(II), consumes α-ketoglutaric acid/2-oxoglutaric acid and O2, and produces succinate, CO2, and formaldehyde (11); these demethylation reactions are directly involved in redox reactions. These reversible histone modifications enable crosstalk of metabolism with epigenetic regulation via coordination of nucleosome modifiers (12).
Nucleosome modifiers may sense cellular status by using metabolites in reactions that regulate gene expression. For example, acetylation and methylation can occur on the same lysine substrate, such as in histone H3 lysine 9 (H3K9) methylation and acetylation in mammals. H3K9 methylation is seen in silenced chromatin; however, H3K9 acetylation is a mark for transcription activity. Notably, a nontranscriptional function of histone methylation plays an important role in mediating gene expression and metabolism in Saccharomyces cerevisiae. Excess methyl group formed by abolishing phospholipid methylation is stored in core histones, leading to increased H3K36, K79, and K4 methylation (13). This feature of histones as methyl sinks adjusts the intracellular levels of toxic sulfides and reactive oxygen species (ROS) which are regulated in transsulfuration and sulfur amino acid catabolism (13). Metabolic enzymes not only catalyze metabolite generation, but also participate in chromatin modification. For example, in S. cerevisiae, Pyk1, a homolog of human pyruvate kinase M2 (PKM2), is a component of the serine-responsive SAM-containing metabolic enzyme (SESAME) complex, which contains serine metabolic enzymes (Ser33, Ser3, and Shm2), SAMe synthetases (Sam1 and Sam2), and an acetyl-CoA synthase (Acs2) (14). Pyk1 in SESAME phosphorylates H3T11 by utilizing phosphoenolpyruvate as the phosphate donor instead of ATP (14). The interaction of SESAME with the Set1 H3K4 methyltransferase complex enables crosstalk between H3K4 methylation and H3T11 phosphorylation in response to glycolysis and glucose-dependent serine metabolism (14). Remarkably, loss of phosphorylation on H3T11 extends the chronological lifespan of S. cerevisiae (15). Thus, histone and chromatin modifications use metabolites to regulate gene expression and cellular metabolism.
An important production of one-carbon metabolism is nucleotide biosynthesis. Nucleotides are fundamental components of nucleosomes and RNA transcripts. However, it remains unclear how nucleotide/nucleoside metabolism influences chromatin modifications and epigenetics. Therefore, in this review we focus our discussion on nucleotide metabolism in epigenetic regulation and biological processes.
Chromatin Modifiers and Nucleotide Biosynthesis
Transcription is a mechanism to regulate metabolism and is regulated by chromatin modifiers. Chromatin modifiers often form protein complexes that modify chromatin structure and/or place epigenetic marks. ATP-dependent chromatin remodeling by the Switch/sucrose non-fermentable (SWI/SNF) family plays important function in gene activation and displaces acetylated histones (12). BAF60a, a subunit of SWI/SNF promotes chromatin accessibility at PPARγ and EBF2 transcription factor binding sites on thermogenic genes in brown and beige adipogenesis and thermogenesis (16). However, adipocyte-specific BAF60a knockout mice show more pronounced cold-induced browning of inguinal white adipose tissue induced by MC2R, a receptor for the adrenocorticotropic hormone (16). Thus, unexplored roles of SWI/SNF in energy generation is under consideration.
Some chromatin modifiers contain nucleotide metabolic enzymes. In Drosophila and mammals, the nucleotide biosynthetic enzyme guanosine 5’-monophosphate synthase (GMPS) forms a heteromeric complex with ubiquitin protease 7 (USP7) (17, 18). This complex is recruited to polycomb response elements and promotes polycomb silencing of homeotic genes through methylation of histone H3K27 (17). The suggested mechanism is that USP7-GMPS removes ubiquitin from ubiquitylated H2B which is associated with active chromatin. In mammalian cells, GMPS promotes p53 stabilization in the presence of etoposide, which induces DNA damage (19). GMPS is critical for expression of the p53 target genes p21 and bax, and for p53-dependent transcriptional suppression of cdc6 and mcm6 following etoposide treatment (19). GMPS also promotes de novo synthesis of guanine (20) (Figure 1). GMPS is highly expressed in ovary cancer tissue, in which p21 expression is reduced (22). A remaining question is whether recruitment of GMPS to the polycomb complex suppresses GMPS nucleoside synthase activity. Interestingly, the polycomb group gene super sex combs (sxc) encodes O-linked N-acetylglucosamine (GlcNAc) transferase (Ogt) in Drosophila (23). Ogt modifies serine and threonine residues of target proteins by adding a single N-acetylglucosamine in an O-glycosidic linkage (Figure 2). Lack of sxc/Ogt results in failure to maintain polycomb transcriptional repression. O-GlcNAcylation of polycomb complex subunits is necessary for proper targeting of Hox repression in human (25), the role of the polycomb complex in glycosylation is unknown (Figure 2). Monitoring the expression of polycomb target genes upon adding supplemental UDP to sxc/Ogt-null mutants may help address the question of whether polycomb suppresses its target genes by suppressing glycosylation or whether UDP is required as a cofactor for the repression of polycomb target genes.
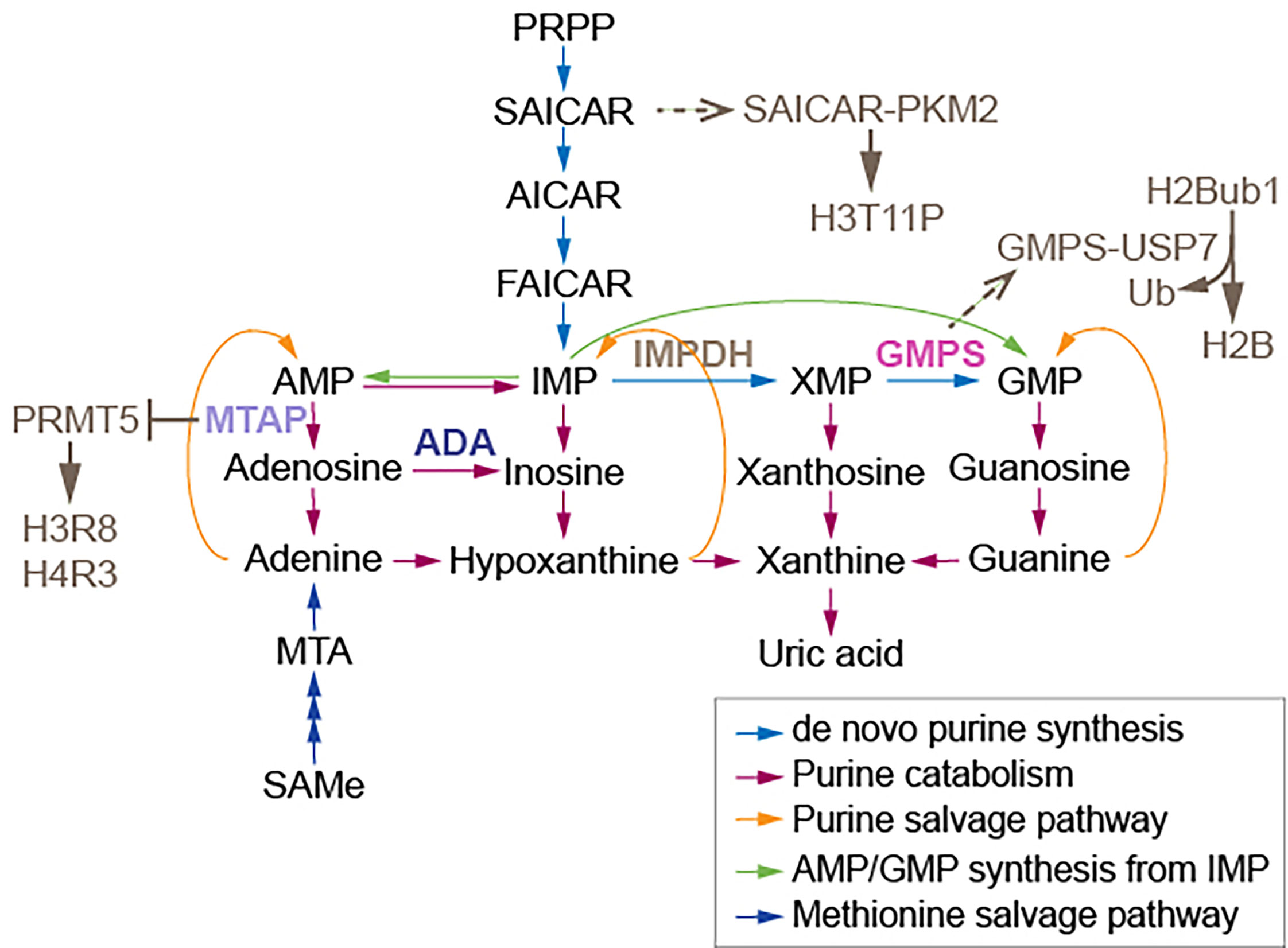
Figure 1 Enzymes and a synthase relaying on purine metabolism modify nucleosomes. The association of GMPS with USP7 removes monoubiquitylation of histone H2B. The association of SAICAR with PKM2 facilitates H3T11 phosphorylation. MTAP suppresses PRMT5, which methylates H3 arginine (R) 8 and H4R3. AMP, adenosine monophosphate; MTA, 5’-methylthioadenosine; PRPP, 5-phosphoribosyl-1-pyrophosphate; AICAR, 5-amino-1-[3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]imidazole-4-carboxamide; FAICAR, 5-formamidoimidazole-4-carboxamide ribotide; IMP, inosine monophosphate; XMP, xanthosine monophosphate; GMP, guanosine monophosphate; IMPDH, inosine monophosphate dehydrogenase; GMPS, guanine monophosphate synthase. Figure adapted from (21).
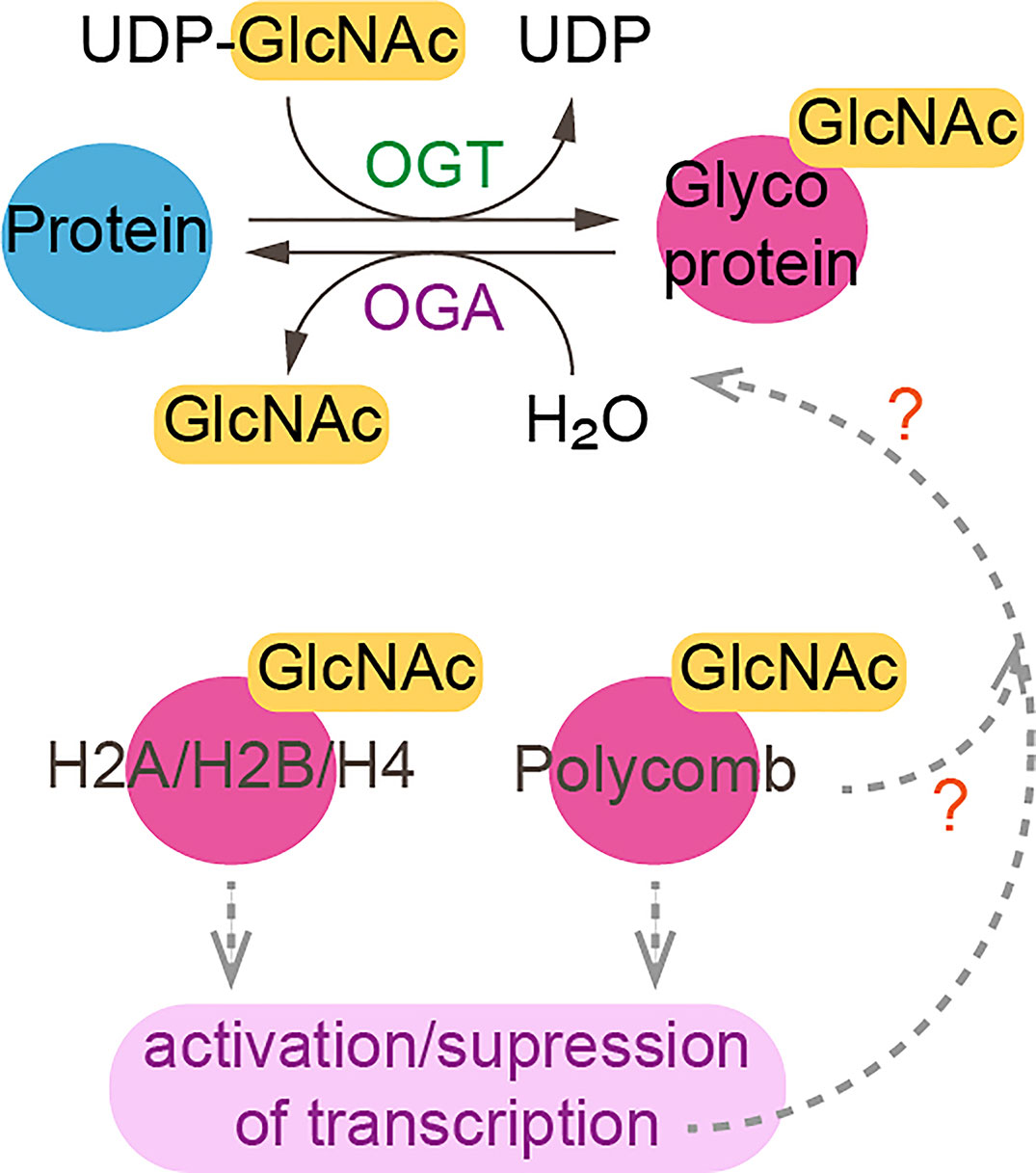
Figure 2 O-linked-N-acetylglucosaminylation. O-linked-N-acetylglucosaminylation (O-GlcNAcylation) occurs when O-GlcNAc is added to serine or threonine residues of nuclear or cytoplasmic proteins by O-GlcNAc transferase (OGT) (24). This reaction is reversible as O-GlcNAc can be removed by O-GlcNAcase (OGA). Linkage of GlcNac to histones and polycomb subunits affects gene expression. ?, unknown.
Nucleotides and DNA Damage
Nucleotide metabolic enzymes also have been found to directly modify nucleosomes. The interaction of SAICAR (phosphoribosylaminoimidazolesuccinocarboxamide) with PKM2 facilitates PKM2 activity and the phosphorylation and activation of Erk1/2, which is necessary for mitogen-induced cell proliferation in Hela cells (26, 27). SAICAR is an intermediate of de novo purine synthesis (Figure 1). SAICAR also stimulates histone H3T11 phosphorylation in vitro (27). Promotion of cell proliferation by SAICAR-PKM2 presumably involves cell cycle control and DNA replication because DNA replication is coordinated with nucleotide synthesis. Deoxynucleotide triphosphates (dNTPs) used in DNA replication are obtained from corresponding ribonucleotides through reduction reactions catalyzed by ribonucleotide reductase (RNR) (28). Imbalances in the dNTP pool increase mutation rates and replication anomalies; therefore, RNR activity and dNTP pool sizes are strictly regulated (29). At low ROS levels in mammalian cells, increased oligomerization of peroxiredoxin 2 (PRDX2) results in formation of a replisome-associated complex with the replication fork accelerator TIMELESS, which associates with chromatin (30). However, at high ROS levels induced by hydroxyurea inhibition of RNR, a smaller PRDX2 oligomer displaces TIMELESS from replisomes, resulting in attenuation of DNA replication (30). It remains unknown whether elevated ROS from different sources select different nucleotide synthetic pathways, and whether the pathways that generate ROS attenuate DNA replication.
Methylthioadenosine phosphorylase (MTAP) is required for the salvage of adenine and methionine (31) (Figure 1). Deletion of MTAP is frequently found in human tumors, including 53% of glioblastomas and 26% of pancreatic cancers (32). MTAP-deleted cancer cells accumulate methylthioadenosine (MTA), which inhibits the methyltransferase activity of protein arginine methyltransferase 5 (PRMT5) (32). The viability of MTAP-deleted cancer cells is diminished by depletion of PRMT5 (32), suggesting that lowed SAMe levels influence cancer cell viability, as PRMT5 is required for maintenance of cellular SAMe levels (33). It has been proposed that depletion of PRMT5 may be useful for cancer therapy (32); however, it is cautioned that PRMT5-dependent SAMe is required for proper mRNA splicing (33). It also remains unknown whether tumor suppressor genes, which may induce MTAP, are critically regulated by methyltransferase activity of PRMT5 in healthy cells. Inhibition of methionine adenosyltransferase 2A (MAT2A), which catalyzes the production of SAMe from ATP, reduces SAMe levels and inhibits the proliferation of MTAP-deleted HCT116 cells (34). However, MAT2A inhibition using AGI-24512 induces greater DNA damage in MTAP-deleted HCT116 cells than in wild type HCT116 cells (34). Another MAT2A inhibitor, AG-270, may be a stronger drug candidate than AGI-24512 as AG-270 reduces proliferation and DNA damage repair in MTAP-deleted cells (34). These observations suggest that loss of adenosine induces DNA damage in MTAP-deleted cells; however, no measurements of adenosine levels in MTAP-deleted cancer cells and HCT116 cells were performed. Such measurements of metabolites including nucleotides which require MTAP are needed to be understand the results of these preclinical studies of MAT2A inhibitors.
Nicotinamide Adenine Dinucleotide
A metabolic cofactor consisting of nucleosides plays roles in redox reactions and in enzymatic reactions as a substrate. Nicotinamide adenine dinucleotide (NAD) is a central metabolic coenzyme due to its involvement in redox reactions. NAD consists of two nucleosides connected by a pyrophosphate. NAD exists in oxidized (NAD+) and reduced (NADH) forms. In mammalian cells, NAD+ is used by class III HDACs, including SIRT1 and SIRT2 (35). In yeast, Sir2 is homologous to human SIRT1 and functions in transcriptional silencing at the silent mating type loci HML and HMR and at telomeres as part of the SIR complex, which consists of Sir2, Sir3, and Sir4 (36). Sir2 deacetylates histones in an NAD+-dependent reaction. The deacetylation activity of Sir2 and other sirtuins is inhibited by nicotinamide, which inhibits silencing at the telomeres, mating type loci, and rDNA (37, 38). Mammalian SIRT1 and SIRT6 transcriptionally drive the circadian rhythm in distinct manners (39). Whereas SIRT1 establishes a repressive chromatin state by contributing to H3K9me2 and H3K27me3 at circadian clock-controlled genes by deacetylation of histone H3K9, BMAL1, and PER2 (40), SIRT6 regulates recruitment of another circadian activator, sterol regulatory element binding transcription factor 1 (SREBF1/SREBP1), to the promoter of the circadian clock-controlled gene Fasn (39). Removal of SIRT6 disrupts hepatic lipid metabolism and circadian rhythm (39). SIRT6 also regulates lysine fatty acylation, which removes long-chain fatty acyl group, such as myristoyl, from proteins (41). It is unknown whether circadian clock regulators are controlled by fatty acylation. Since fatty acid beta-oxidation is used to synthesize acetyl-CoA, circadian clock-controlled genes may sense NAD+ (redox carrier) levels or redox status via SIRT1 recruitment and connects to fatty acid metabolism via SIRT6 recruitment to control acetyl-CoA-levels. Aging involves greater risk of circadian rhythm disruption due to alteration of transcriptional regulation of clock-controlled genes (42). In a comparison of young (10 month) and old (22 month) mice, the addition of 3.2 g/L of the NAD+ precursor nicotinamide riboside reprogrammed and improved age-dependent disruption of circadian clock-controlled transcription in the liver, as monitored by BMAL1 ChIP sequencing (42).
ADP-Ribosylation
NAD also plays roles in the transfer reaction of adenosine diphosphate (ADP). In ADP-ribosylation, one or more ADP-riboses from NAD+ are transferred to a protein by mono (ADP-ribose) or poly(ADP-ribose) polymerases (PARPs). Poly ADP-ribosylation (PARylation) regulates a variety of cellular processes including chromatin decondensation, transcription, DNA damage response, and mitosis (43). PARP1 binds the DNA damage site and recruits DNA repair molecules through PARylation (44). Inhibitors of PARP and poly(ADP-ribose) glycohydrolase (PARG) are used in cancer therapy (43). Interestingly, progestin promotes PARP1-mediated PAR generation, and PAR and its degradation to ADP-ribose (ADPR) are essential for increases in nuclear ATP levels in response to progestin in human breast cancer cells (45). Upon progestin treatment, ATP diffuses from mitochondria into the nucleus for around 10 minutes; meanwhile, enough NAD+ accumulates for the PAR synthesis reaction that occurs 10 minutes after supplying progestin. ADP-ribose from PAR degradation is used by ADP-sugar pyrophosphate nudix hydrolase 5 (NUDIX5), which synthesizes and maintains nuclear ATP used in chromatin remodeling to maintain progestin-responsive transcription (45). PARG also regulates chromatin remodeling of genes that respond to retinoic acid (46). Gene expression in response to nuclear hormones may use ATP and PAR to transiently obtain large amounts of energy.
Nucleosides and RNA Editing
To escape from genome instability, damaged nucleotides are fixed by DNA repair pathways. Nucleosides in RNA are also edited. The editing of adenosine to inosine (A-to-I) in transcripts is catalyzed by adenosine deaminases (ADAs) (Figure 1) (47). This process frequently occurs at introns and 3’ untranslated regions (3’ UTRs). A greater frequency of A-to-I editing is found in micro RNA (miRNA) binding sites of target genes in tumor cells than in normal cells (47). The frequency of A-to-I editing by ADAs within miRNA is also greater in tumor cells than in normal cells (47). A single nucleotide polymorphism in the 3’ UTR of MDM4, which enables the binding of miR-191, leads to attenuation of tumor progression (48). However, pleural effusion ADA levels are significantly higher in malignant pleural mesothelioma patients than in healthy doners (49). Hence, changed expression of ADAs and miRNAs may be the consequences of tumorgenesis. Adenosine and inosine are products of de novo purine synthesis; inosine is also an intermediate of the purine salvage pathway (Figure 1). Alteration of A-to-I editing frequency may result from an imbalance in purine metabolism. ADA may adjust nucleoside pools by controlling inosine and miRNA. Measuring levels of other deaminases such as AMP deaminase and guanosine deaminase in addition to ADA levels may reveal that the nucleotide metabolic pathway is directly downstream of the tumor target. These measurements may also indicate the sensitivity of miRNA synthesis to each nucleotide. The 3’ UTR contains a poly(A) tail; a study showing how purine metabolism influences the poly(A) tail is also needed. The 5’ cap in mRNA consists of N7-methylguanine (N7meG), which connects to the first mRNA nucleotide and prevents mRNA degradation (50, 51). Since methylation of N7G requires SAMe, RNA processing may be affected by SAMe metabolism. Additional studies are needed to link pyrimidine metabolism to RNA synthesis.
Discussion
Chromatin consumes metabolites. Examining the connections between metabolism, cellular events, epigenetic marks, and transcription will further reveal how epigenetics mirrors cellular metabolism. Monitoring chromatin status in the condition where the status of nucleotide biogenesis is modified will increase our understanding of the roles of epigenetics in one-carbon metabolism. The molecular linkages between chromatin modifiers and different nucleotides need further study.
Author Contributions
TS wrote the manuscript. JW edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Stowers Institute for Medical Research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Yang M, Vousden KH. Serine and One-Carbon Metabolism in Cancer. Nat Rev Cancer (2016) 16(10):650–62. doi: 10.1038/nrc.2016.81
2. Suganuma T, Workman JL. Chromatin and Metabolism. Annu Rev Biochem (2018) 87:27–49. doi: 10.1146/annurev-biochem-062917-012634
3. Shuvalov O, Petukhov A, Daks A, Fedorova O, Vasileva E, Barlev NA. One-Carbon Metabolism and Nucleotide Biosynthesis as Attractive Targets for Anticancer Therapy. Oncotarget (2017) 8(14):23955–77. doi: 10.18632/oncotarget.15053
4. Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, et al. Sulfur Amino Acids Regulate Translational Capacity and Metabolic Homeostasis Through Modulation of tRNA Thiolation. Cell (2013) 154(2):416–29. doi: 10.1016/j.cell.2013.06.043
5. Christensen DG, Xie X, Basisty N, Byrnes J, McSweeney S, Schilling B, et al. Post-Translational Protein Acetylation: An Elegant Mechanism for Bacteria to Dynamically Regulate Metabolic Functions. Front Microbiol (2019) 10:1604. doi: 10.3389/fmicb.2019.01604
6. Dutta A, Abmayr SM, Workman JL. Diverse Activities of Histone Acylations Connect Metabolism to Chromatin Function. Mol Cell (2016) 63(4):547–52. doi: 10.1016/j.molcel.2016.06.038
7. Milazzo G, Mercatelli D, Di Muzio G, Triboli L, De Rosa P, Perini G, et al. Histone Deacetylases (HDACs): Evolution, Specificity, Role in Transcriptional Complexes, and Pharmacological Actionability. Genes (Basel) (2020) 11(5):1–49. doi: 10.3390/genes11050556
8. Gallinari P, Di Marco S, Jones P, Pallaoro M, Steinkuhler C. HDACs, Histone Deacetylation and Gene Transcription: From Molecular Biology to Cancer Therapeutics. Cell Res (2007) 17(3):195–211. doi: 10.1038/sj.cr.7310149
9. Feldman JL, Dittenhafer-Reed KE, Denu JM. Sirtuin Catalysis and Regulation. J Biol Chem (2012) 287(51):42419–27. doi: 10.1074/jbc.R112.378877
10. Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1. Cell (2004) 119(7):941–53. doi: 10.1016/j.cell.2004.12.012
11. Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone Demethylation by a Family of JmjC Domain-Containing Proteins. Nature (2006) 439(7078):811–6. doi: 10.1038/nature04433
12. Suganuma T, Workman JL. Crosstalk Among Histone Modifications. Cell (2008) 135(4):604–7. doi: 10.1016/j.cell.2008.10.036
13. Ye C, Sutter BM, Wang Y, Kuang Z, Tu BP. A Metabolic Function for Phospholipid and Histone Methylation. Mol Cell (2017) 66(2):180–93.e188. doi: 10.1016/j.molcel.2017.02.026
14. Li S, Swanson SK, Gogol M, Florens L, Washburn MP, Workman JL, et al. Serine and SAM Responsive Complex SESAME Regulates Histone Modification Crosstalk by Sensing Cellular Metabolism. Mol Cell (2015) 60(3):408–21. doi: 10.1016/j.molcel.2015.09.024
15. Oh S, Suganuma T, Gogol MM, Workman JL. Histone H3 Threonine 11 Phosphorylation by Sch9 and CK2 Regulates Chronological Lifespan by Controlling the Nutritional Stress Response. Elife (2018) 7:1–20. doi: 10.7554/eLife.36157
16. Liu T, Mi L, Xiong J, Orchard P, Yu Q, Yu L, et al. BAF60a Deficiency Uncouples Chromatin Accessibility and Cold Sensitivity From White Fat Browning. Nat Commun (2020) 11(1):2379. doi: 10.1038/s41467-020-16148-1
17. van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, et al. GMP Synthetase Stimulates Histone H2B Deubiquitylation by the Epigenetic Silencer USP7. Mol Cell (2005) 17(5):695–707. doi: 10.1016/j.molcel.2005.02.013
18. Sarkari F, Sanchez-Alcaraz T, Wang S, Holowaty MN, Sheng Y, Frappier L. EBNA1-Mediated Recruitment of a Histone H2B Deubiquitylating Complex to the Epstein-Barr Virus Latent Origin of DNA Replication. PLoS Pathog (2009) 5(10):e1000624. doi: 10.1371/journal.ppat.1000624
19. Reddy BA, van der Knaap JA, Bot AG, Mohd-Sarip A, Dekkers DH, Timmermans MA, et al. Nucleotide Biosynthetic Enzyme GMP Synthase is a TRIM21-Controlled Relay of P53 Stabilization. Mol Cell (2014) 53(3):458–70. doi: 10.1016/j.molcel.2013.12.017
20. Bollee G, Harambat J, Bensman A, Knebelmann B, Daudon M, Ceballos-Picot I. Adenine Phosphoribosyltransferase Deficiency. Clin J Am Soc Nephrol (2012) 7(9):1521–7. doi: 10.2215/CJN.02320312
21. Donti TR, Cappuccio G, Hubert L, Neira J, Atwal PS, Miller MJ, et al. Diagnosis of Adenylosuccinate Lyase Deficiency by Metabolomic Profiling in Plasma Reveals a Phenotypic Spectrum. Mol Genet Metab Rep (2016) 8:61–6. doi: 10.1016/j.ymgmr.2016.07.007
22. Wang P, Zhang Z, Ma Y, Lu J, Zhao H, Wang S, et al. Prognostic Values of GMPS, PR, CD40, and P21 in Ovarian Cancer. PeerJ (2019) 7:e6301. doi: 10.7717/peerj.6301
23. Gambetta MC, Oktaba K, Muller J. Essential Role of the Glycosyltransferase Sxc/Ogt in Polycomb Repression. Science (2009) 325(5936):93–6. doi: 10.1126/science.1169727
24. Konzman D, Abramowitz LK, Steenackers A, Mukherjee MM, Na HJ, Hanover JA. O-GlcNAc: Regulator of Signaling and Epigenetics Linked to X-Linked Intellectual Disability. Front Genet (2020) 11:605263. doi: 10.3389/fgene.2020.605263
25. Maury JJ, El Farran CA, Ng D, Loh YH, Bi X, Bardor M, et al. RING1B O-GlcNAcylation Regulates Gene Targeting of Polycomb Repressive Complex 1 in Human Embryonic Stem Cells. Stem Cell Res (2015) 15(1):182–9. doi: 10.1016/j.scr.2015.06.007
26. Jurecka A, Zikanova M, Kmoch S, Tylki-Szymanska A. Adenylosuccinate Lyase Deficiency. J Inherit Metab Dis (2015) 38(2):231–42. doi: 10.1007/s10545-014-9755-y
27. Keller KE, Doctor ZM, Dwyer ZW, Lee YS. SAICAR Induces Protein Kinase Activity of PKM2 That Is Necessary for Sustained Proliferative Signaling of Cancer Cells. Mol Cell (2014) 53(5):700–9. doi: 10.1016/j.molcel.2014.02.015
28. Thelander L. Ribonucleotide Reductase and Mitochondrial DNA Synthesis. Nat Genet (2007) 39(6):703–4. doi: 10.1038/ng0607-703
29. Torrents E. Ribonucleotide Reductases: Essential Enzymes for Bacterial Life. Front Cell Infect Microbiol (2014) 4:52. doi: 10.3389/fcimb.2014.00052
30. Somyajit K, Gupta R, Sedlackova H, Neelsen KJ, Ochs F, Rask MB, et al. Redox-Sensitive Alteration of Replisome Architecture Safeguards Genome Integrity. Science (2017) 358(6364):797–802. doi: 10.1126/science.aao3172
31. Kryukov GV, Wilson FH, Ruth JR, Paulk J, Tsherniak A, Marlow SE, et al. MTAP Deletion Confers Enhanced Dependency on the PRMT5 Arginine Methyltransferase in Cancer Cells. Science (2016) 351(6278):1214–8. doi: 10.1126/science.aad5214
32. Mavrakis KJ, McDonald ER 3rd, Schlabach MR, Billy E, Hoffman GR, deWeck A, et al. Disordered Methionine Metabolism in MTAP/CDKN2A-Deleted Cancers Leads to Dependence on PRMT5. Science (2016) 351(6278):1208–13. doi: 10.1126/science.aad5944
33. Suganuma T, Swanson SK, Gogol M, Garrett TJ, Conkright-Fincham J, Florens L, et al. MPTAC Determines APP Fragmentation via Sensing Sulfur Amino Acid Catabolism. Cell Rep (2018) 24(6):1585–96. doi: 10.1016/j.celrep.2018.07.013
34. Kalev P, Hyer ML, Gross S, Konteatis Z, Chen CC, Fletcher M, et al. MAT2A Inhibition Blocks the Growth of MTAP-Deleted Cancer Cells by Reducing PRMT5-Dependent mRNA Splicing and Inducing DNA Damage. Cancer Cell (2021) 39(2):209–24.e211. doi: 10.1016/j.ccell.2020.12.010
35. Covarrubias AJ, Perrone R, Grozio A, Verdin E. NAD(+) Metabolism and Its Roles in Cellular Processes During Ageing. Nat Rev Mol Cell Biol (2021) 22(2):119–41. doi: 10.1038/s41580-020-00313-x
36. Ghidelli S, Donze D, Dhillon N, Kamakaka RT. Sir2p Exists in Two Nucleosome-Binding Complexes With Distinct Deacetylase Activities. EMBO J (2001) 20(16):4522–35. doi: 10.1093/emboj/20.16.4522
37. Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of Silencing and Accelerated Aging by Nicotinamide, a Putative Negative Regulator of Yeast Sir2 and Human SIRT1. J Biol Chem (2002) 277(47):45099–107. doi: 10.1074/jbc.M205670200
38. Wierman MB, Smith JS. Yeast Sirtuins and the Regulation of Aging. FEMS Yeast Res (2014) 14(1):73–88. doi: 10.1111/1567-1364.12115
39. Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, et al. Partitioning Circadian Transcription by SIRT6 Leads to Segregated Control of Cellular Metabolism. Cell (2014) 158(3):659–72. doi: 10.1016/j.cell.2014.06.050
40. Belden WJ, Dunlap JC. SIRT1 Is a Circadian Deacetylase for Core Clock Components. Cell (2008) 134(2):212–4. doi: 10.1016/j.cell.2008.07.010
41. Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, et al. SIRT6 Regulates TNF-Alpha Secretion Through Hydrolysis of Long-Chain Fatty Acyl Lysine. Nature (2013) 496(7443):110–3. doi: 10.1038/nature12038
42. Levine DC, Hong H, Weidemann BJ, Ramsey KM, Affinati AH, Schmidt MS, et al. NAD(+) Controls Circadian Reprogramming Through PER2 Nuclear Translocation to Counter Aging. Mol Cell (2020) 78(5):835–49.e837. doi: 10.1016/j.molcel.2020.04.010
43. Slade D. PARP and PARG Inhibitors in Cancer Treatment. Genes Dev (2020) 34(5-6):360–94. doi: 10.1101/gad.334516.119
44. Min A, Im SA. PARP Inhibitors as Therapeutics: Beyond Modulation of PARylation. Cancers (Basel) (2020) 12(2):1–16. doi: 10.3390/cancers12020394
45. Wright RH, Lioutas A, Le Dily F, Soronellas D, Pohl A, Bonet J, et al. ADP-Ribose-Derived Nuclear ATP Synthesis by NUDIX5 is Required for Chromatin Remodeling. Science (2016) 352(6290):1221–5. doi: 10.1126/science.aad9335
46. Le May N, Iltis I, Ame JC, Zhovmer A, Biard D, Egly JM, et al. Poly (ADP-Ribose) Glycohydrolase Regulates Retinoic Acid Receptor-Mediated Gene Expression. Mol Cell (2012) 48(5):785–98. doi: 10.1016/j.molcel.2012.09.021
47. Anastasiadou E, Jacob LS, Slack FJ. Non-Coding RNA Networks in Cancer. Nat Rev Cancer (2018) 18(1):5–18. doi: 10.1038/nrc.2017.99
48. Wynendaele J, Bohnke A, Leucci E, Nielsen SJ, Lambertz I, Hammer S, et al. An Illegitimate microRNA Target Site Within the 3' UTR of MDM4 Affects Ovarian Cancer Progression and Chemosensitivity. Cancer Res (2010) 70(23):9641–9. doi: 10.1158/0008-5472.CAN-10-0527
49. Nakajima Y, Kuribayashi K, Ishigaki H, Tada A, Negi Y, Minami T, et al. Adenosine Deaminase in Pleural Effusion and Its Relationship With Clinical Parameters in Patients With Malignant Pleural Mesothelioma. Cancer Invest (2020) 38(6):356–64. doi: 10.1080/07357907.2020.1776313
50. Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cocrystal Structure of the Messenger RNA 5' Cap-Binding Protein (Eif4e) Bound to 7-Methyl-GDP. Cell (1997) 89(6):951–61. doi: 10.1016/s0092-8674(00)80280-9
Keywords: nucleotide metabolism, chromatin modifiers, DNA damage, metabolism, histone modifications, NAD, ADP-ribosylation, RNA editing
Citation: Suganuma T and Workman JL (2021) Nucleotide Metabolism Behind Epigenetics. Front. Endocrinol. 12:731648. doi: 10.3389/fendo.2021.731648
Received: 27 June 2021; Accepted: 09 August 2021;
Published: 30 August 2021.
Edited by:
Francisco Verdeguer, University of Zurich, SwitzerlandReviewed by:
Kathryn Wellen, University of Pennsylvania, United StatesNaresh Chandra Bal, KIIT University, India
Copyright © 2021 Suganuma and Workman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tamaki Suganuma, tas@stowers.org; Jerry L. Workman, jlw@stowers.org
 Tamaki Suganuma
Tamaki Suganuma Jerry L. Workman
Jerry L. Workman