Combining bioinformatics and machine learning to identify common mechanisms and biomarkers of chronic obstructive pulmonary disease and atrial fibrillation
- 1Guang’anmen Hospital, China Academy of Chinese Medical Sciences, Beijing, China
- 2Graduate School, Beijing University of Chinese Medicine, Beijing, China
- 3Graduate School, China Academy of Chinese Medical Sciences, Beijing, China
- 4Graduate School, Hebei University of Chinese Medicine, Shijiazhuang, China
- 5Eye Hospital China Academy of Chinese Medical Sciences, China Academy of Chinese Medical Sciences, Beijing, China
Background: Patients with chronic obstructive pulmonary disease (COPD) often present with atrial fibrillation (AF), but the common pathophysiological mechanisms between the two are unclear. This study aimed to investigate the common biological mechanisms of COPD and AF and to search for important biomarkers through bioinformatic analysis of public RNA sequencing databases.
Methods: Four datasets of COPD and AF were downloaded from the Gene Expression Omnibus (GEO) database. The overlapping genes common to both diseases were screened by WGCNA analysis, followed by protein-protein interaction network construction and functional enrichment analysis to elucidate the common mechanisms of COPD and AF. Machine learning algorithms were also used to identify key biomarkers. Co-expression analysis, “transcription factor (TF)-mRNA-microRNA (miRNA)” regulatory networks and drug prediction were performed for key biomarkers. Finally, immune cell infiltration analysis was performed to evaluate further the immune cell changes in the COPD dataset and the correlation between key biomarkers and immune cells.
Results: A total of 133 overlapping genes for COPD and AF were obtained, and the enrichment was mainly focused on pathways associated with the inflammatory immune response. A key biomarker, cyclin dependent kinase 8 (CDK8), was identified through screening by machine learning algorithms and validated in the validation dataset. Twenty potential drugs capable of targeting CDK8 were obtained. Immune cell infiltration analysis revealed the presence of multiple immune cell dysregulation in COPD. Correlation analysis showed that CDK8 expression was significantly associated with CD8+ T cells, resting dendritic cell, macrophage M2, and monocytes.
Conclusions: This study highlights the role of the inflammatory immune response in COPD combined with AF. The prominent link between CDK8 and the inflammatory immune response and its characteristic of not affecting the basal expression level of nuclear factor kappa B (NF-kB) make it a possible promising therapeutic target for COPD combined with AF.
Introduction
Chronic obstructive pulmonary disease (COPD) is characterized by progressive airway obstruction. It is estimated that COPD will be the third most deadly disease in the world by 2030 (1). In clinical practice, COPD is often combined with multiple cardiovascular diseases, including heart failure, coronary atherosclerotic heart disease, and atrial fibrillation (AF) (2). AF, the most common type of arrhythmia, has been shown in epidemiological surveys to affect at least 33.5 million people worldwide (3). Heart failure and stroke are severe complications of AF, and the risk of heart failure in AF patients is about twice as high as normal, and the risk of stroke is 4–5 times higher (4, 5).
There is a strong association between COPD and AF, with studies showing a 2.23-fold increased risk of AF in COPD patients compared to non-COPD patients (6). Similarly, the prevalence of COPD in patients with AF reached 25% (7). In addition, combined COPD increases the recurrence rate and the incidence of adverse events after catheter ablation in patients with AF (8, 9). A meta-analysis that included 46 studies involving 4,232,784 AF patients showed that AF patients with comorbid COPD had a significantly increased risk of bleeding, cardiovascular event death, and all-cause mortality compared to AF patients without COPD (10). It is currently thought that enhanced sympathetic activity, altered cardiac structure, immune dysfunction, inflammation, and oxidative stress may be involved in the development of AF in patients with COPD. However, the exact mechanisms have not been fully elucidated (11). Meanwhile, drugs commonly used to treat COPD, such as beta-blockers, theophylline, and glucocorticoids, have been linked to an increased risk of AF development (12, 13). Therefore, it is of practical clinical significance to explore the potential mechanisms of COPD and AF co-morbidity at the genetic level and to find promising therapeutic targets for application.
The field of bioinformatics is developing rapidly, and large amounts of genetic data are publicly available to uncover many unknown pathophysiological mechanisms in the development of diseases and potential connections between diseases. Machine learning, an essential artificial intelligence component, has also been widely applied to bioinformatics research and has become an important tool (14). Based on this, this study integrates COPD and AF mRNA datasets in public databases and attempts to reveal the common biological mechanisms of COPD and AF co-morbidity through weighted gene co-expression network analysis (WGCNA), protein-protein interaction (PPI) network construction, and enrichment analysis. Random forest (RF), support vector machine (SVM), extreme gradient boosting (XGBoost), and generalized linear model (GLM) were used to screen potential biomarkers. A comprehensive analysis of key biomarkers was performed, including co-expression analysis, construction of “transcription factor (TF)-mRNA-microRNA (miRNA)” regulatory networks, and drug prediction. An examination of immune cell infiltration on the COPD dataset was also carried out. Figure 1 depicts the study flowchart.
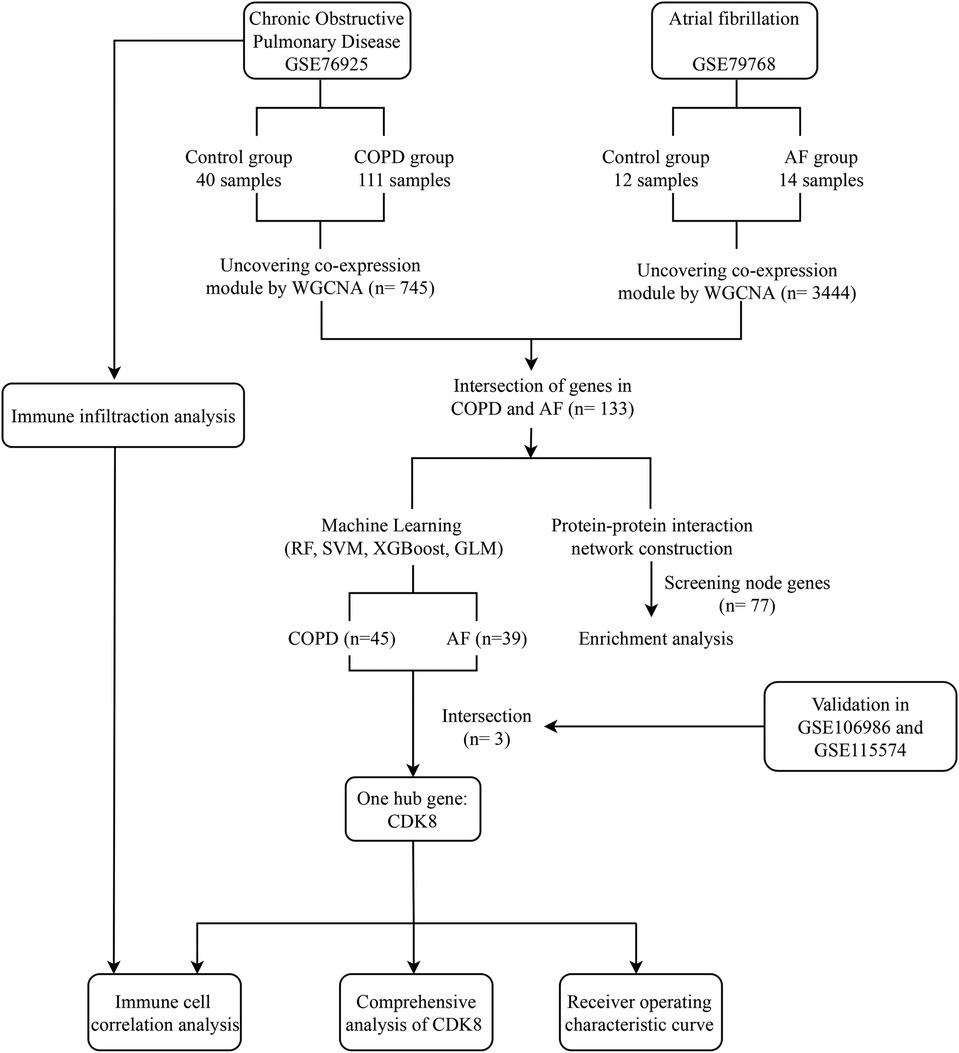
Figure 1. Study flowchart. GSE, gene expression omnibus series; WGCNA, weighted gene co-expression network analysis; RF, Random forest support vector machine; SVM, support vector machine; XGBoost, extreme gradient boosting; GLM, generalized linear model.
Materials and methods
Data source
We applied the Gene Expression Omnibus (GEO) database (15) (http://www.ncbi.nlm.nih.gov/geo/) to filter gene expression datasets for microarrays by qualifying the keywords “COPD” and “AF” with the filter criteria “Homo Sapiens” and “tissues.” Four datasets were finally obtained. In the COPD group, GSE76925 (lung tissue samples from 111 COPD patients and 40 control patients) (16) and GSE106986 (lung tissue samples from 14 COPD patients and 5 control patients) were selected. In the AF group, GSE79768 (left and right atrial tissue samples from 7 AF patients and 6 control patients) (17) and GSE115574 (left and right atrial tissue samples from 15 AF patients and 15 control patients) (18) were selected. GSE76925 and GSE79768 were used as training sets, GSE106986 and GSE115574 were used as the external validation set.
Weighted gene co-expression network analysis
WGCNA analysis was performed on GSE76925 and GSE79768, respectively, to obtain modules closely associated with COPD and AF. WGCNA analysis was constructed using the “WGCNA” package in R (19). The genes were ranked based on the standard deviation of gene expression. The top 25% of genes with the largest fluctuations were selected for subsequent analysis, and outlier samples were excluded by hierarchical clustering. The R2 was set greater than 0.9, and a suitable soft threshold (β) was calculated to make the network conform to the scale-free distribution. The co-expression modules are identified by hierarchical clustering to obtain a hierarchical clustering tree. Finally, the module feature values and the correlation between module feature values and clinical features are calculated to obtain the expression spectrum of each module, which is expressed by the correlation coefficient as well as the p-value. Finally, we select the genes in the modules closely related to the disease for subsequent analysis.
Identification of overlapping genes and PPI network analyses
The genes in the modules closely related to disease in GSE76925 and GSE79768 obtained by WGCNA analysis were taken to intersect. The Venn diagram was used to visualize the overlapping genes. After that, the overlapping genes were imported into the “Search Tool for Interacting Genes” (STRING) online platform (https://cn.string-db.org/) (20). The species was limited to “Homo sapiens,” with the confidence score set to an intermediate value (confidence score > 0.4) to construct the PPI network. The results were exported to Cytoscape 3.7.2 for visualization (21).
Functional enrichment analysis
To further understand the common biological mechanisms between the two diseases, the protein information in the PPI network was enriched for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using the “clusterProfiler” package in R (22). The GO includes biological process (BP), cellular component (CC), and molecular function (MF) (23). The screening condition was set at adjust -P < 0.05, and the visualization was presented using the Sangerbox platform (http://vip.sangerbox.com/). Meanwhile, Gene set enrichment analysis (GSEA) enrichment analysis was performed on the GSE76925 and GSE79768 datasets to comprehensively analyze the key pathways associated with COPD and AF pathogenesis. The reference dataset was “c5.kegg.v7.4.symbols.gmt” from the MSigDB database (24). The significantly enriched pathways were identified with the screening criteria of P < 0.05 and FDR < 0.25, and the “enrichplot” package was used for visualization.
Identification of candidate genes based on machine learning algorithms
Machine learning is now widely used to identify characteristic genes. To identify key genes associated with COPD and AF, four machine learning algorithms, including RF, SVM, XGBoost, and GLM, were used to screen for key genes in COPD and AF, respectively. In both training datasets, the response variable was set to whether the diagnosis was COPD and AF, and the overlapping genes were set as explanatory variables. Use 70% of the data for model construction and 30% for model validation. Models for RF, SVM, XGBoost, and GLM were constructed separately using the “caret” R package (25). It is well known that while building a machine learning model with good results, it is equally important to evaluate the interpretation of the model, as only an interpretable machine learning model is likely to be more widely understood and adopted. The “DALEX” R package is a model interpretation package that has been developed to help understand the links between input variables and model outputs (26). It uses the size of the residuals to assess the quality of the model (smaller residuals mean better model quality) and the root mean square error (RMSE) to assess the importance of the variables (defined as how much the absence of a variable affects the predicted value of the response variable). Once modelling was completed, residual box plots were drawn for the four models using the “DALEX” R package and the RMSE was used to assess the importance of each gene in the model. Also, we use the “predict” function in R to verify the accuracy of the predictions of the model constructed by “caret”. Receiver operating characteristic (ROC) curves were then plotted using the “pROC” R package (27) and the area under the curve (AUC) was reported to assess the predictive effectiveness of models. Finally, we selected models with high predictive accuracy based on the quality of the model (assessed by the size of the residuals) and the area of the AUC. According to the gene importance score, select the top 20 genes from the constructed models for COPD and AF respectively, and then perform the intersection for these genes. Afterwards, we compared the differential expression of intersecting genes between the two datasets, with P < 0.05 considered to be significantly different, and used the “ggpubr” R package to plot boxplots. Finally, genes that were differentially expressed in the disease and control groups in both sets of data and that showed the same expression trend in both microarrays were identified as candidate hub genes.
Validation of hub genes and evaluation of prediction accuracy
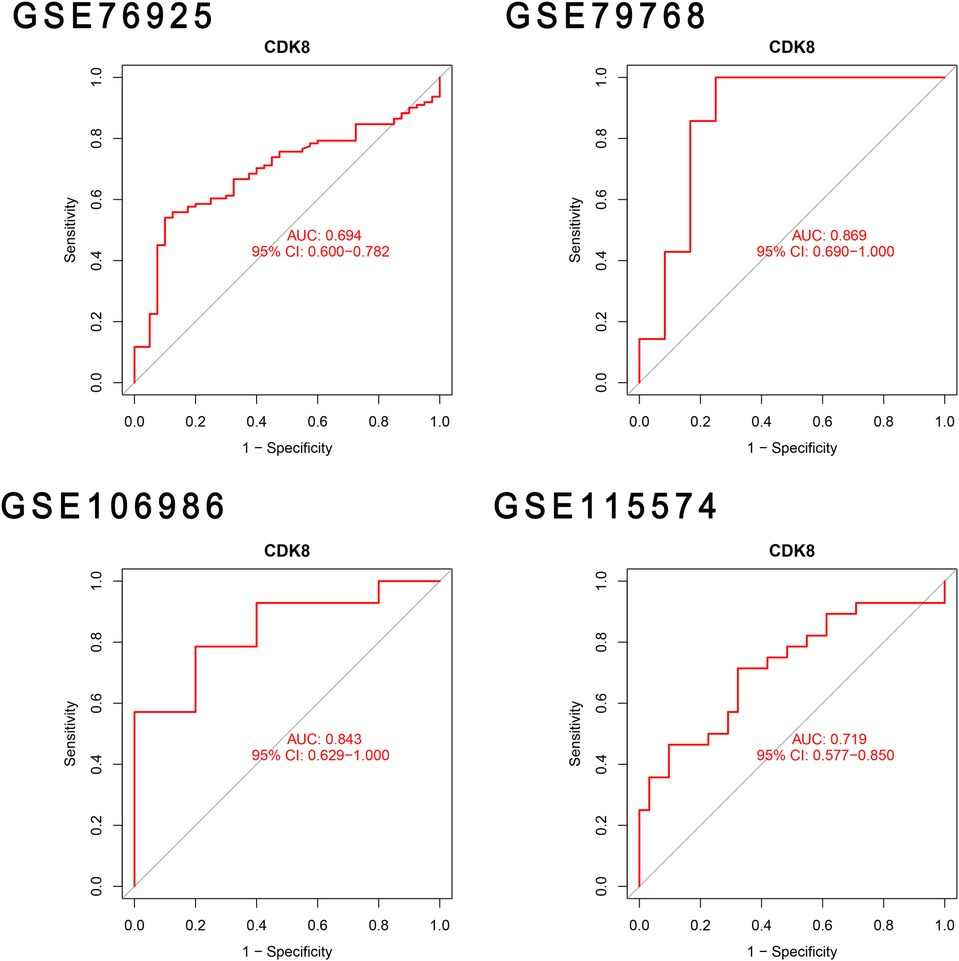
The identified candidate genes were validated in the validation sets for COPD and AF, respectively. The comparison of gene expression between disease group and control group with P < 0.05 was considered to be significantly different. The candidate genes with significant differences were finally considered to be hub genes and the boxplot was drawn for visualization. Afterwards, ROC curves of the diagnostic value of hub genes in the training set and validation set were plotted using “pROC” R package, and AUC was calculated to evaluate the accuracy of hub gene prediction.
Comprehensive analysis of hub genes
Hub genes were entered into the GeneMANIA online website (28) (http://genemania.org) for co-expression and functional enrichment analyses. The JASPAR database (29) (http://jaspar.genereg.net/) were used to predict TFs regulating hub genes. Prediction of miRNAs regulated by hub genes using the miRTarBase database (30) (https://mirtarbase.cuhk.edu.cn/), and experimentally validated miRNAs were selected. The results were visualized using Cytoscape to demonstrate the “TF-mRNA-miRNA” regulatory network. In addition, we used the DGIDB 3.0 database (31) (http://www.dgidb.org/) to predict potential drugs that could target the hub gene.
Immune cell infiltration analysis
CIBERSORT can calculate the proportion of different immune cells in the gene expression profile through a deconvolution algorithm (32). We performed an immune cell infiltration analysis of the COPD gene expression matrix (GSE76925) using the CIBERSORT algorithm. The “barplot” and “vioplot” packages were used to show the relative proportions and differences of immune cell types in the expression profile between the control and COPD groups. The “corrplot” package was used to show the correlation heat map of immune cells in the expression profile. The hub gene's expression was then taken from the expression profile. The correlation between the hub gene and immune cells was analyzed by Spearman correlation analysis, with P < 0.05 as the screening condition, and visualized by the “ggplot” package.
Results
Construction of co-expressed gene modules
WGCNA analysis was performed on the GSE76925 and GSE79768 datasets to identify co-expression modules associated with COPD and AF, respectively. The β selection analysis of GSE76925 showed that the network was closer to the scale-free network when the β = 4 (Figure 2A). Six modules were also identified, of which the yellow module was positively associated with COPD (correlation coefficient = 0.35, P = 9e-06) and contained 745 genes (Figures 2B,C). The β selection analysis of GSE79768 showed that the network was closer to the scale-free network when the β = 10 (Figure 2D). Eleven modules were identified, among which blue (correlation coefficient = 0.55, P = 0.004), pink (correlation coefficient = 0.42, P = 0.03), turquoise (correlation coefficient = 0.73, P = 3e-05), and yellow (correlation coefficient = 0.61, P = 0.001) were positively correlated with AF and contained 3,444 genes (Figures 2E,F). Among them, 952 were blue modules, 287 were pink modules, 1,526 were turquoise modules, and 705 were yellow modules. Detailed gene information is listed in Supplementary Table S1.
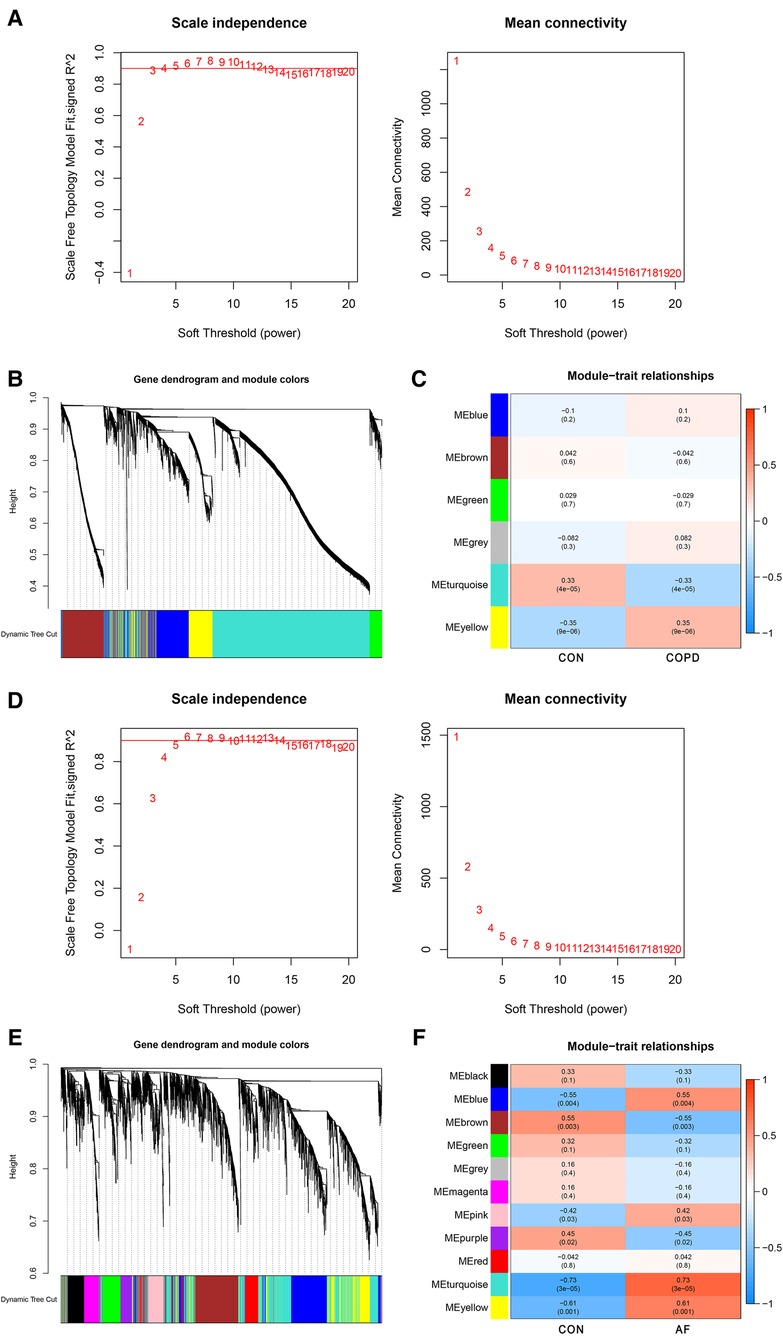
Figure 2. Identification of COPD and AF module genes via WGCNA. (A) The scale-free fit index for soft-thresholding powers and mean connectivity (GSE76925). (B) Dendrogram of the genes clustered (GSE76925). (C) Module-trait relationships heatmap (GSE76925). The numbers in each cell means the correlation coefficient and p-value. (D) The scale-free fit index for soft-thresholding powers and mean connectivity (GSE79768). (E) Dendrogram of the genes clustered (GSE79768). (F) Module-trait relationships heatmap (GSE79768).
Identification of overlapping genes and construction of PPI network
By taking the intersection of the genes of COPD and AF-related modules obtained by WGCNA, 133 genes common to COPD and AF were obtained (Figure 3A). After that, we constructed a PPI network of overlapping genes and excluded genes that did not interact. Finally, we obtained 77 interacting genes in the network (Figure 3B).
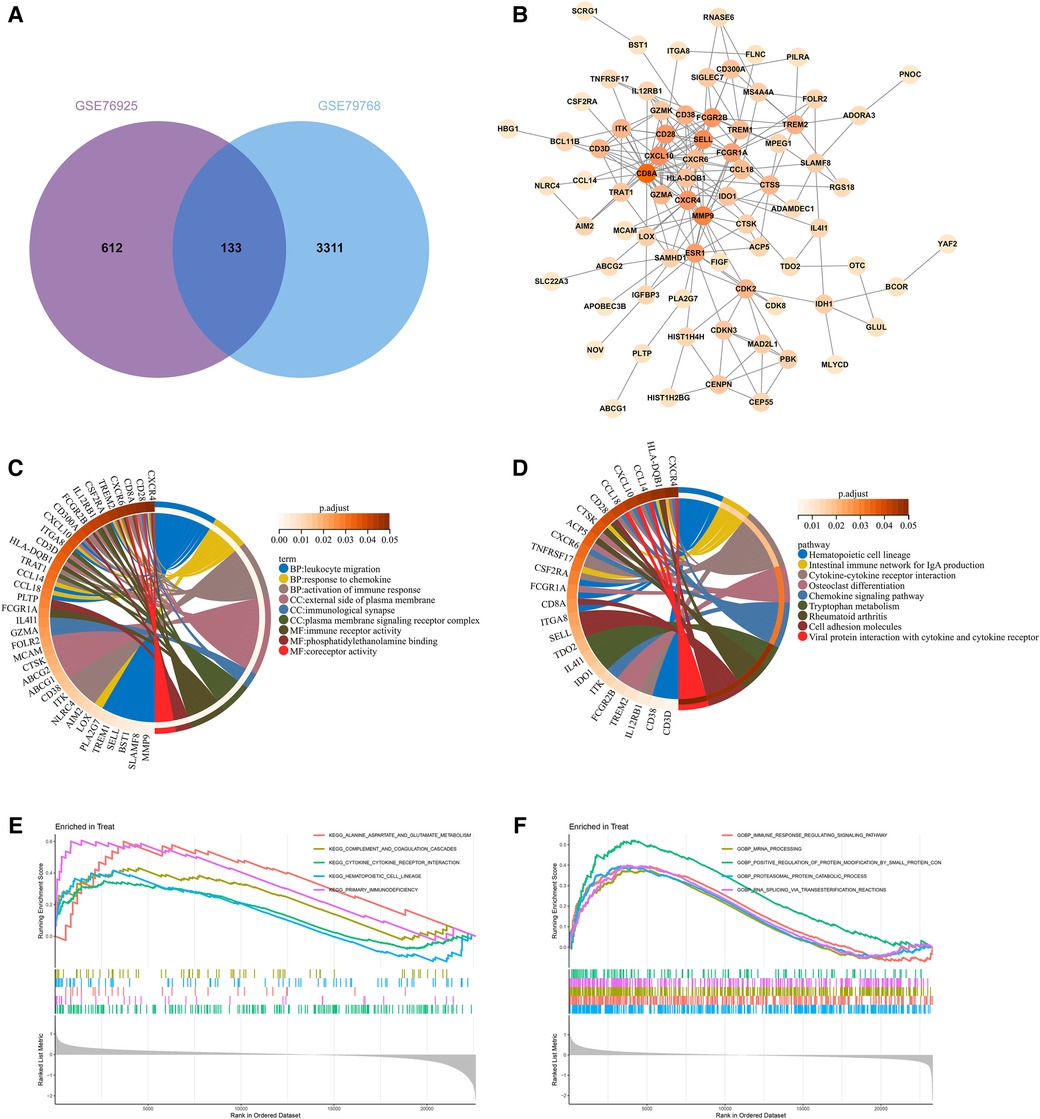
Figure 3. Construction of overlapping gene PPI networks and enrichment analysis. (A) Venn diagram of COPD and AF module genes. (B)PPI network. (C) KEGG pathway analysis of genes. Different colors represent various significant pathways and related enriched genes. (D) GO analysis of genes. Different colors represent various significant pathways and related enriched genes. (E) GSEA enrichment analysis of upregulated genes in the COPD dataset. (F) GSEA enrichment analysis of upregulated genes in the AF dataset.
Functional enrichment analysis
77 genes in the PPI network were analyzed for GO and KEGG enrichment to reveal the underlying molecular biological processes shared between COPD and AF. The GO enrichment analysis revealed that 77 genes were primarily enriched in the “Leukocyte migration,” “Response to chemokine,” “Cellular response to chemokine,” “Myeloid leukocyte migration,” “Activation of immune response,” “Regulation of inflammatory response”(BP); “External side of plasma membrane,” “Immunological synapse,” “Plasma membrane signaling receptor complex,” (CC); “Immune receptor activity,” “Phosphatidylethanolamine binding,” “Coreceptor activity,” (MF) (Figure 3C, Supplementary Table S2). Also, KEGG analysis showed that genes were enriched in “Hematopoietic cell lineage,” “Intestinal immune network for IgA production,” “Cytokine-cytokine receptor interaction,” “Chemokine signaling pathway,” “Cell adhesion molecules,” “T cell receptor signaling pathway” (Figure 3D, Supplementary Table S3). GSEA enrichment analysis revealed that genes upregulated in the GSE76925 dataset were mainly enriched in signaling pathways such as “Cytokine-cytokine receptor interaction,” “Primary immunodeficiency,” “Alanine aspartate and glutamate metabolism,” “Hematopoietic cell lineage,” “Complement and coagulation cascade” (Figure 3E). The genes upregulated in the GSE79768 dataset were mainly enriched in “RNA degradation,” “Ubiquitin-mediated proteolysis,” “Spliceosome,” “Leukocyte transendothelial migration,” and “Natural killer cell-mediated cytotoxicity” (Figure 3F).
Identification of hub genes based on machine learning algorithms
To identify key genes associated with COPD and AF, we constructed models using four machine learning methods and evaluated the models based on residuals and ROC. Box line plots of residuals and ROC are shown in Figures 4A–D. It can be seen that the SVM, XGB and RF models all exhibit similar excellent performance. Therefore, we selected the top 20 genes predicted by RF, SVM, and XGB in each of these two datasets based on the importance scores of the genes assessed by RMSE. Forty-five genes in total in the GSE76925 dataset and 39 in GSE79768, and 11 intersecting genes were obtained after taking the intersection. (Figure 4E, Supplementary Tables S4, S5). After verifying the differential expression and expression trends, three candidate genes, cyclin dependent kinase 8 (CDK8), solute carrier family 22 member 15 (SLC22A15), and TNF receptor superfamily member 17 (TNFRSF17), were finally obtained, and the box plots of differential expression are shown in Figure 5.
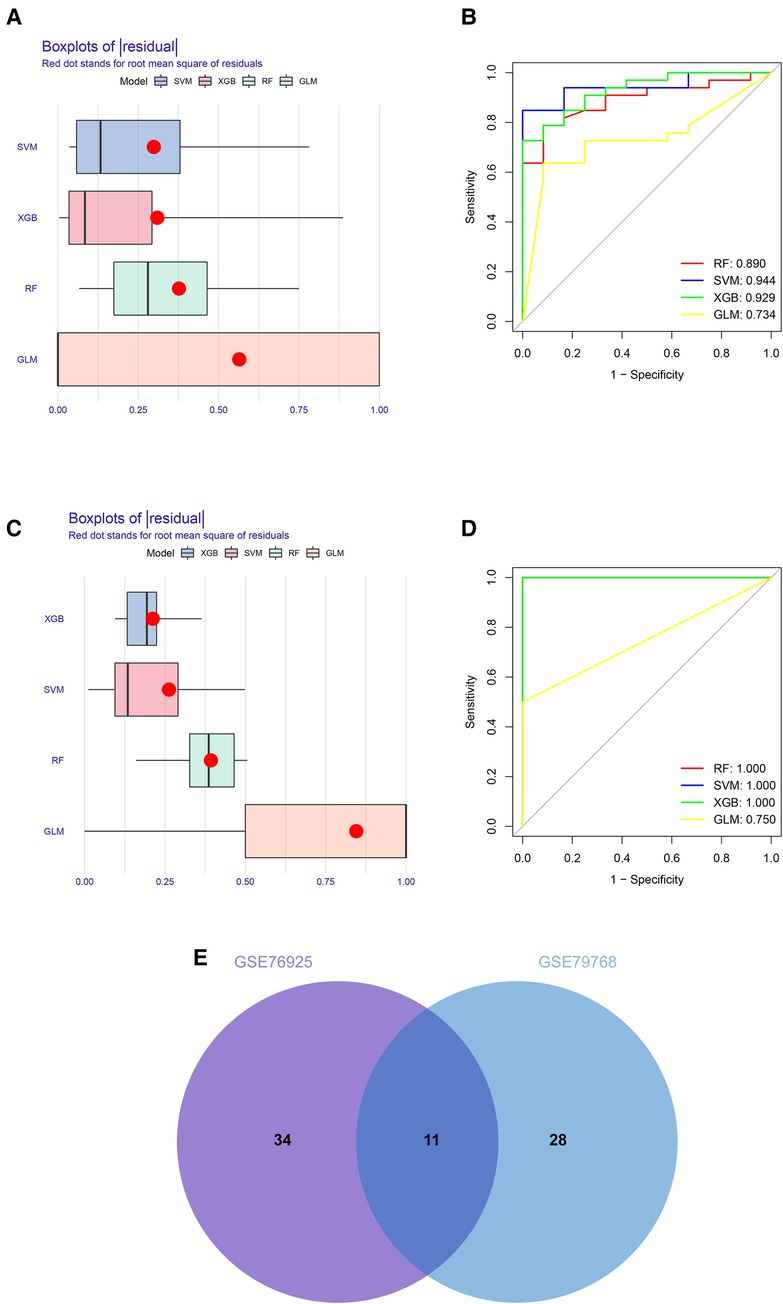
Figure 4. Construction and assessment of machine learning models for COPD and AF. (A) Boxplots of the residuals of the COPD. Red dot stands for root mean square of residuals. (B) ROC curves for model prediction accuracy in COPD dataset. (C) Boxplots of the residuals of the AF. (D) ROC curves for model prediction accuracy in AF dataset (E) Venn diagram of COPD and AF model prediction genes.
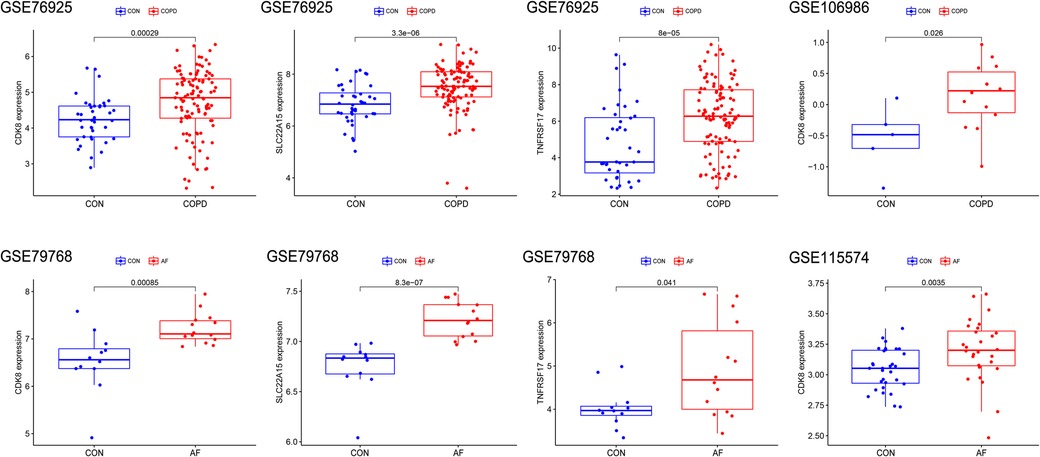
Figure 5. Validation of Hub gene expression levels in training and validation sets. The red box represents the disease group, and the blue represents the control group.
Validation of hub genes and assessment of predictive accuracy
The differential expression of CDK8, SLC22A15, and TNFRSF17 was confirmed in the external validation set. Only CDK8 showed a significant increase in expression in both the training and validation sets (P < 0.05) Figure 5. Thus, CDK8 was finally identified as a hub gene that may be related to AF and COPD. the CDK8′s ROC curves demonstrated that in all four data sets, the AUC was near to or greater than 0.7 (Figure 6). It is suggested that CDK8 may be effective for detecting COPD combined with AF.
Comprehensive analysis of hub genes
Twenty genes associated with hub genes were predicted by GeneMANIA, along with 1,326 interactions. This suggests a complex interaction between the hub gene and the remaining 20 genes. The functional enrichment results were mainly associated with DNA-templated transcription, positive regulation of DNA-templated transcription, and regulation of transcription initiation from the RNA polymerase II promoter (Figure 7A). The “TF-mRNA-miRNA” regulatory network contains 11 TFs that regulate CDK8, and five miRNAs (Figure 7B). DGIDB predicted 20 drugs targeting CDK8, including ALVOCIDIB, ACACETIN, and RONICICLIB (Figure 7C).
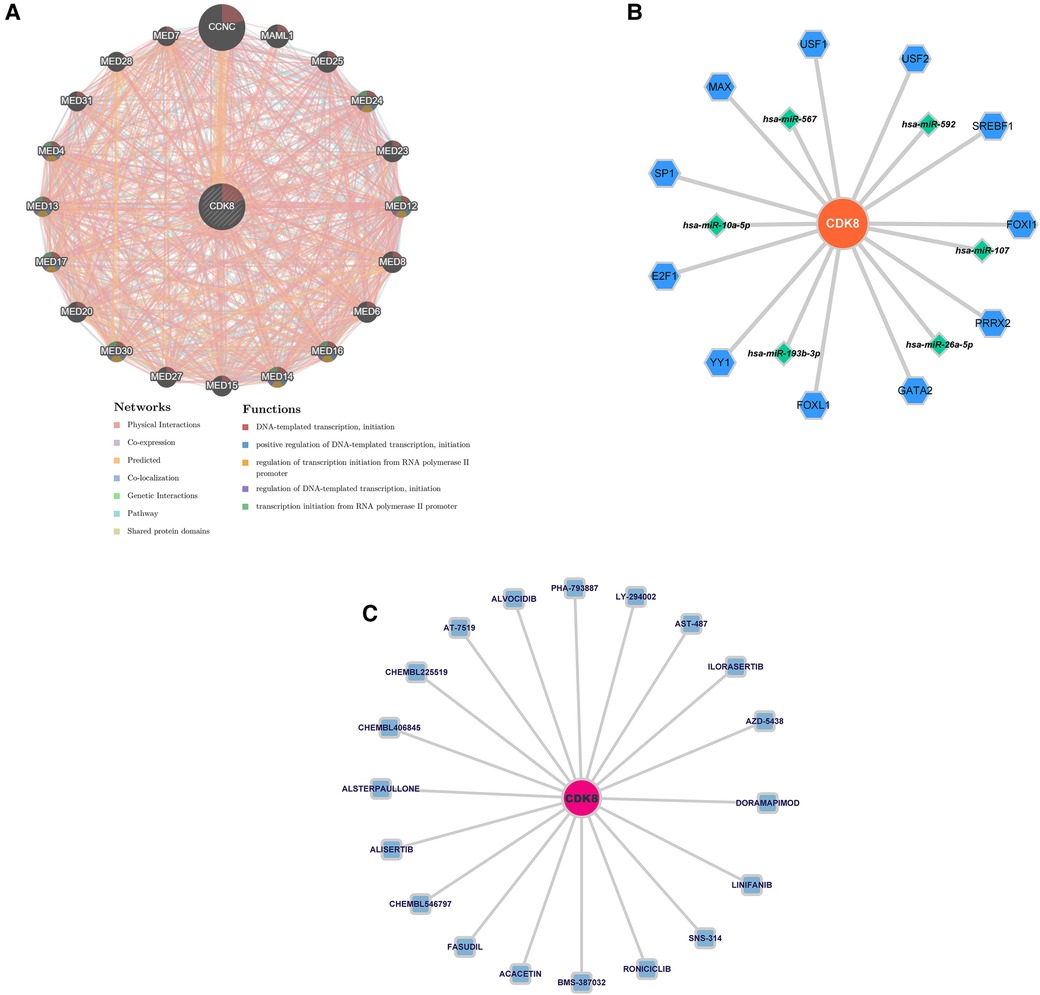
Figure 7. (A) Co-expression network of CDK8. (B) The “TF-mRNA-miRNA” regulatory network of CDK8. Blue hexagons are TFs; green diamonds are miRNAs. (C) Drug prediction for CDK8 based on DIGDB database. Blue squares are potential drugs.
Immune cell infiltration analysis
By observing the enrichment analysis results, we found that immune-related pathways were significantly enriched, suggesting that immune dysfunction may be involved in the development of AF in COPD patients. Therefore, we performed an immune cell infiltration analysis of gene expression profiles in COPD. Figure 8A shows the ratio of immune cells in the control group to the COPD group. Compared to the control group, the COPD group had higher levels of plasma cells, CD8+ T cells, T follicular helper cells, Gamma-delta (γδ) T cells, macrophage M0, and resting dendritic cells, and lower levels of monocytes, macrophage M1, and activated dendritic cell (Figure 8B). Positive correlations were found between activated mast cells and neutrophils (r = 0.60), T cells and plasma cells (r = 0.42), and γδ T cells and T follicular helper cells (r = 0.42). In contrast, Macrophage M1 and activated dendritic cells were negatively correlated (r = −0.54) (Figure 8C). This suggests that patients with COPD have a different immune pattern compared to normal patients and that there are interactions between different types of immune cells. Detailed results of the immune cell infiltration analysis are shown in Supplementary Table S6.
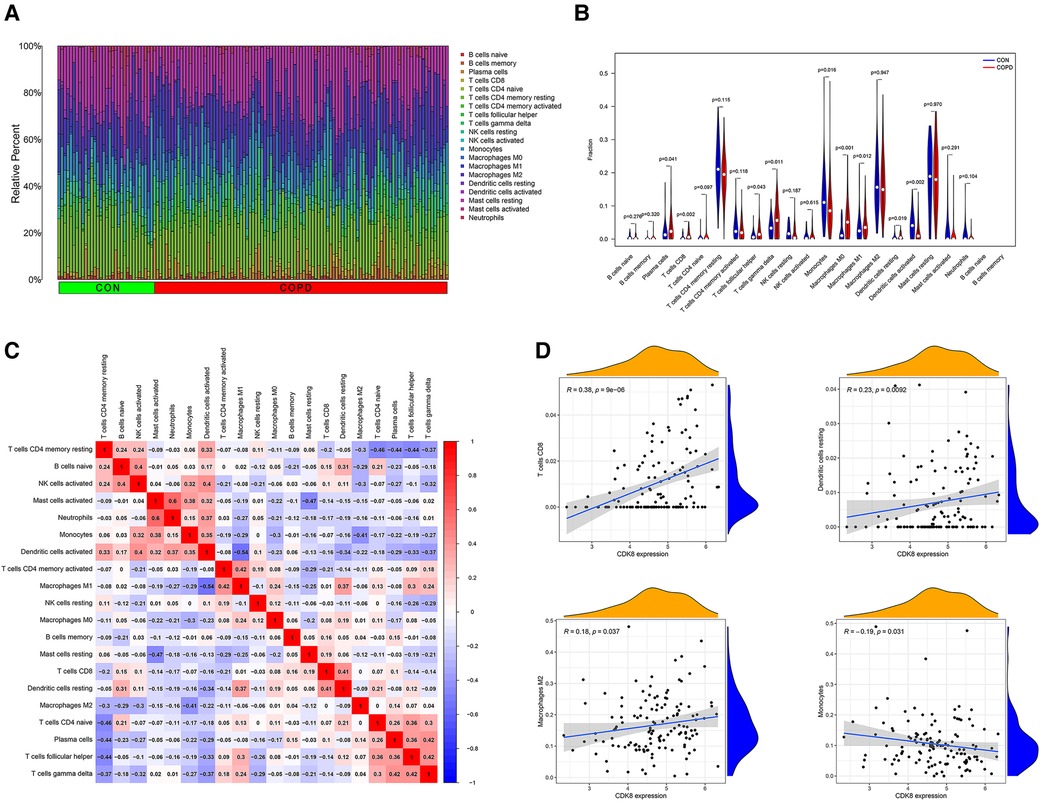
Figure 8. Immune cell infiltration analysis of GSE76925 and immune cell correlation analysis of CDK8. (A) The proportion of immune cells in different samples. (B) Comparison of immune cell ratios in the COPD and control groups. (C) Heat map of correlation analysis between immune cells. (D) Analysis of the correlation between CDK8 and immune cells.
Correlation analysis of CDK8 and immune cells showed that CDK8 expression correlated with four immune cell types, positively with CD8+ T cells, resting dendritic cells, and macrophage M2, and negatively with monocytes (Figure 8D).
Discussion
COPD affects more than 300 million people worldwide and causes approximately 3 million deaths yearly (33). COPD increases the incidence of AF and the risk of subsequent cardiovascular death. Similarly, comorbid AF increases the incidence of ischemic stroke, respiratory failure, and heart failure events in patients with COPD (34–37). The vicious circle relationship between COPD and AF, and the contradiction of pharmacological treatment, makes it urgent to explore the mechanisms of COPD and AF co-morbidity and to find potential therapeutic targets. It is generally believed that COPD-induced structural changes in the heart, increased sympathetic activity, and hypoxia-induced oxidative stress accelerates the development of AF. But recent studies have highlighted the role of immune dysfunction in the pathogenesis of both diseases, finding that immune dysfunction may play a prominent role in the subsequent inflammatory response, cardiac remodeling, structural remodeling, and neural remodeling (11, 38). However, the exact mechanisms have not been fully elucidated. In this context, we performed the first joint analysis of genetic datasets from both diseases to reveal common mechanisms and key biomarkers for the development of both diseases and to reveal changes in immune cells in COPD by immune cell infiltration analysis.
After screening of overlapping genes and the construction of PPI networks, 77 genes with interactions were obtained. They are mainly involved in biological processes such as activation of the immune response, regulation of inflammatory response, leukocyte migration, response to chemokines, immune receptor activity, and signaling pathways such as hematopoietic cell lineage, IgA-producing intestinal immune network, cytokine-cell receptor interactions, and T cell receptors. Combined with the results of GSEA enrichment analysis, we suggest that immune and inflammatory responses are the key mechanisms linking these two diseases, as confirmed by the results of previous studies (39–41). COPD is accompanied by a long-term and persistent chronic inflammatory response in the airways and lung parenchyma, leading to subsequent airway remodeling and destruction of the lung parenchyma (42). After inhalation of smoke or other toxic particles, the immune response is activated, after which macrophages release various cytokines and chemokines, including tumor necrosis factor-alpha (TNF-α), interleukin 1 beta (IL-1β), interleukin 6 (IL-6), C-X-C motif chemokine ligand 1 (CXCL1), and C-X-C motif chemokine ligand 8 (CXCL8), which attract circulating neutrophils, monocytes, and lymphocytes in the lungs leading to an inflammatory response (39, 43). At the same time, the activation of immune and inflammatory responses is not limited to the lungs, as studies have shown that COPD patients are accompanied by elevated circulating c-reactive protein (CRP), IL-6, CXCL8, and TNF-α (44). Changes in immune cells, especially macrophages, and increases in cytokines and chemokines such as TNF-α, IL-6, IL-1β, C-X-C motif chemokine ligand 1 (CXCL1), and C-X-C motif chemokine ligand 2 (CXCL2) have also been observed in AF (45, 46).
In response to these results, we performed an immune cell infiltration analysis in GSE76925, which showed a higher proportion of plasma cells, CD8+ T cells, T follicular helper cells, γδ T cells, macrophage M0, and resting dendritic cells, and a lower proportion of monocytes, macrophage M1, and activated dendritic cells in COPD lung tissue compared with controls. Studies have shown that CD8+ T lymphocytes increase in number and activity in COPD and produce many cytokines such as Interferon gamma (IFN-γ) and TNF-α (47). γδ T cells are nontraditional T cells, and despite their small number, a study by Murdoch (48) et al. found that γδ T cells increase interleukin 17A (IL-17) production during acute allergic airway disease and are involved in disease pathogenesis. Plasma cells are widely present in the connective tissue of the lamina propria of the respiratory tract and participate in the adaptive humoral immune response by synthesizing antibodies in response to the invasion of the respiratory tract (49). Studies have shown that the severity of COPD is positively correlated with the development of tertiary lymphoid organs (TLOs), and IL-21 T-follicular-helper (Tfh)-like cells have been observed in TLOs of COPD patients, suggesting that Tfh may be involved in the formation of TLOs (50, 51). Dendritic cells are critical antigen-presenting cells involved in adaptive immune activation in COPD. However, Givi (52) et al. found that chronic exposure to harmful particles impairs dendritic cells maturation and inhibits antigen-presenting capacity. Macrophages are differentiated from monocytes and play a key role in chronic inflammation in COPD patients. Naive macrophage M0 can be induced to differentiate into M1- and M2-type macrophages under different conditions, with M1-type mainly playing a pro-inflammatory role (53). Interestingly, our study found a lower proportion of M1-type macrophages in the COPD group compared with controls, the reason for which needs to be further investigated. Similarly, immune cells are the primary cell type in the heart, and one study showed that immune cells accounted for 10.4% of all cell types in atrial tissue (54). More dendritic cells were found in the left atrial myocardium of patients with AF compared to those with sinus rhythm. Increased numbers of neutrophils, lymphocytes, and macrophages were also observed in the atrial adipose tissue (55, 56). These findings suggest that immune cell changes in COPD may also be involved in developing AF.
A biomarker, CDK8, was identified by a machine learning algorithm and validated in the validation set, and it was significantly upregulated in both COPD and AF groups. CDK8 is a serine/threonine protein kinase that plays an important role in transcriptional regulation by binding to cell cycle protein C (57). Recent studies have shown that CDK8 is also involved in the inflammatory response, as Chen et al. (58) found that in response to TNF-α stimulation, nuclear factor kappa B (NF-kB) and CDK8 are jointly recruited to the promoters of response genes, driving the expression of NF-kB early response genes CXCL8, CXCL2, and C-X-C motif chemokine ligand 3 (CXCL3). In addition, CDK8 is involved in the activation of hypoxia inducible factor 1 subunit alpha (HIF1A) (59). lungs of COPD patients overexpress HIF1A, which is associated with hypoxia and inflammatory response (60). Elevated expression of HIF1A is also observed during AF, which may be involved in the structural remodeling of the left atrium (61). CDK8 also highlights potential advantages as a therapeutic target, and NF-kB plays an important role in activating immune inflammatory responses in COPD and AF by encoding chemokines and cytokines (62, 63). Transcription of NF-kB requires activation of CDK8. Studies have shown that reducing CDK8 activity inhibits NF-kB-driven transcription but has no effect on the basal expression of NF-kB-regulated genes or promoters (58). This avoids the detrimental effects of NF-kB blockers due to reduced NF-kB expression levels, making CDK8 a more promising therapeutic target. In addition, we performed a co-expression analysis of CDK8, “TF-mRNA-miRNA” network construction, and drug prediction. These CDK8-associated mRNAs, TFs, and miRNAs also contribute to understanding the CDK8 association network. At the same time, the results predicted by DGIDB may become new drugs for treating COPD combined with AF. Several studies have shown that inhibitors of CDK8 have an inhibitory effect on inflammatory immune responses (64, 65). Studies by Schmerwitz et al. (66) have found that ALVOCIDIB (also known as Flavopiridol) is able to against inflammation by effectively blocking the activation of endothelial cells through the inhibition of NF-kB consensus promoter activity and thereby disrupting the interaction between inflammatory-induced leukocytes and endothelial cells. This suggests that drugs targeting CDK8 may be promising for treating COPD combined with AF.
Analysis of the correlation between CDK8 and immune cell infiltration showed a positive correlation between CDK8 and CD8+ T cells, resting dendritic cells, and macrophage M2 and a negative correlation with monocytes. Patients with COPD have increased infiltration of CD8+ T cells in lung tissue and produce pro-inflammatory factors such as TNF-α (47). The involvement of TNF-α in AF involves multiple mechanisms, and TNF-α has been shown to disrupt intracellular calcium homeostasis in atrial myocytes by decreasing the expression of T-type calcium channel α1G subunit (TCCA 1G) and sarcoplasmic reticulum Ca-ATPase (SERCA2a) thereby participating in AF (67, 68). In addition, TNF-α directly reduces collagen synthesis in cardiomyocytes, enhances matrix metallopeptidase 2 (MMP-2) and matrix metallopeptidase 9 (MMP-9) activities, promotes collagen breakdown, and exacerbates myocardial fibrosis (69, 70). M2 macrophage infiltration in the lungs of patients with COPD is significantly increased. A study by Kaku (71) et al. showed that M2 macrophages are strongly associated with the severity of COPD and a predicted reduction in expiratory force volume in one second (FEV), suggesting the involvement of M2 macrophages in the development of COPD. Macrophages also secrete elastolytic enzymes, such as MMP-2 and MMP-9, which directly lead to the destruction of lung structures and fibrosis of the atria (72–74). Increased sympathetic nervous system activity triggered by COPD promotes the development of AF (11). Studies have revealed that high catecholamine levels induce sympathetic remodeling by acting on the β1-adrenergic receptors on macrophages to produce inflammatory factors such as TNF-α, nerve growth factor (NGF), and interleukin 1 alpha (IL-1). This process may cause the onset of AF (75). Dendritic cell function in AF and COPD is still being studied. Ravi (76) et al. found that monocyte migration capacity was reduced in COPD patients, which may partially explain the negative correlation between CDK8 and monocytes. However, the exact mechanism still needs further investigation. In conclusion, our findings provide a new perspective on the pathogenesis of COPD combined with AF from the viewpoint of the inflammatory immune response and suggest a biomarker CDK8 that could potentially be a therapeutic target.
This study also has several limitations. First, the data in this study were obtained based on the GEO database. Although a dataset containing more samples was selected and validated in an external dataset, the results may be biased due to the different platforms from which they were obtained. Additionally, due to COPD being a heterogeneous disease, there are various phenotypes of COPD, such as small airway-predominant disease, frequent exacerbators, and asthma-COPD overlap, which have different pathophysiological mechanisms that are not completely the same (77). Recent studies have shown that within 90 days of an acute exacerbation of COPD, patients are at a significantly increased risk of an emergency department visit or hospital admission related to AF (78). Another cohort study also found an increased risk of AF in asthmatic patients (79). However, our study was unable to analyze the association between the different phenotypes of COPD and AF separately, so further research is needed to investigate the relationship between these different COPD phenotypes and AF at the genetic level. Secondly, the AUC area of CDK8 in the COPD training set was less than 0.7, and although the AUC area was improved in the validation set, its value in practical clinical applications still needs further validation. Finally, the specific mechanisms of immune inflammatory response and CDK8 in COPD with AF and the association between CDK8 and immune cells need further proof from subsequent in vivo and in vitro experiments.
Conclusion
In this study, through bioinformatic analysis, we found that disturbances in immune regulation and subsequent activation of the inflammatory response may have a significant role in COPD combined with AF. Through machine learning algorithms, CDK8 was finally identified as a key biomarker, and inhibitors targeting CDK8 may be able to be promising therapeutic agents for COPD combined with AF by inhibiting NF-kB-induced immune inflammatory responses.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.'
Ethics statement
Our data comes from the GEO database, GEO belongs to public databases. The patients involved in the database have obtained ethical approval. Users can download relevant data for free for research and publish relevant articles. Our study is based on open source data, so there are no ethical issues or other conflicts of interest. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
KY, ZS and JL conceived of and designed the research plans. ZS, TZ and XS completed data processing and bioinformatics analyses. ZS, JL and TW drafted the manuscript. KY, ZS, and JD checked the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by grants from General Program of National Natural Science Foundation of China (81473466, 81873173) and the National Key Research and Development Program of China (2019YFC1708703).
Acknowledgments
We thank the Sangerbox for providing us with the platform as part of the data analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2023.1121102/full#supplementary-material.
References
1. Decramer M, Janssens W, Miravitlles M. Chronic obstructive pulmonary disease. Lancet. (2012) 379:1341–51. doi: 10.1016/S0140-6736(11)60968-9
2. Leong P, Macdonald M, Ko B, Bardin P. Coexisting chronic obstructive pulmonary disease and cardiovascular disease in clinical practice: a diagnostic and therapeutic challenge. Med J Aust. (2019) 210:417–23. doi: 10.5694/mja2.50120
3. Chugh S, Havmoeller R, Narayanan K, Singh D, Rienstra M, Benjamin EJ, et al. Worldwide epidemiology of atrial fibrillation: a global burden of disease 2010 study. Circulation. (2014) 129:837–47. doi: 10.1161/CIRCULATIONAHA.113.005119
4. Nicoli C, O’Neal W, Levitan E, Singleton M, Judd S, Howard G, et al. Atrial fibrillation and risk of incident heart failure with reduced versus preserved ejection fraction. Heart. (2022) 108:353–9. doi: 10.1136/heartjnl-2021-319122
5. Massera D, Wang D, Vorchheimer DA, Negassa A, Garcia M. Increased risk of stroke and mortality following new-onset atrial fibrillation during hospitalization. Europace. (2017) 19:929–36. doi: 10.1093/europace/euw110
6. Liao K, Chen C. Incidence and risk factors of atrial fibrillation in Asian COPD patients. Int J Chron Obstruct Pulmon Dis. (2017) 12:2523–30. doi: 10.2147/COPD.S143691
7. Konecny T, Park J, Somers K, Konecny D, Orban M, Soucek F, et al. Relation of chronic obstructive pulmonary disease to atrial and ventricular arrhythmias. Am J Cardiol. (2014) 114:272–7. doi: 10.1016/j.amjcard.2014.04.030
8. Proietti M, Laroche C, Drozd M, Vijgen J, Cozma D, Drozdz J, et al. Impact of chronic obstructive pulmonary disease on prognosis in atrial fibrillation: a report from the EURObservational research programme pilot survey on atrial fibrillation (EORP-AF) general registry. Am Heart J. (2016) 181:83–91. doi: 10.1016/j.ahj.2016.08.011
9. Gu J, Liu X, Tan H, Zhou L, Jiang W, Wang Y, et al. Impact of chronic obstructive pulmonary disease on procedural outcomes and quality of life in patients with atrial fibrillation undergoing catheter ablation. J Cardiovasc Electrophysiol. (2013) 24:148–54. doi: 10.1111/j.1540-8167.2012.02448.x
10. Romiti G, Corica B, Pipitone E, Vitolo M, Raparelli V, Basili S, et al. Prevalence, management and impact of chronic obstructive pulmonary disease in atrial fibrillation: a systematic review and meta-analysis of 4,200,000 patients. Eur Heart J. (2021) 42:3541–54. doi: 10.1093/eurheartj/ehab453
11. Simons S, Elliott A, Sastry M, Hendriks J, Arzt M, Rienstra M, et al. Chronic obstructive pulmonary disease and atrial fibrillation: an interdisciplinary perspective. Eur Heart J. (2021) 42:532–40. doi: 10.1093/eurheartj/ehaa822
12. Wilchesky M, Ernst P, Brophy J, Platt R, Suissa S. Bronchodilator use and the risk of arrhythmia in COPD: part 2: reassessment in the larger Quebec cohort. Chest. (2012) 142:305–11. doi: 10.1378/chest.11-1597
13. Huerta C, Lanes S, García Rodríguez L. Respiratory medications and the risk of cardiac arrhythmias. Epidemiology. (2005) 16:360–6. doi: 10.1097/01.ede.0000158743.90664.a7
14. Auslander N, Gussow A, Koonin E. Incorporating machine learning into established bioinformatics frameworks. Int J Mol Sci. (2021) 22:2903. doi: 10.3390/ijms22062903
15. Barrett T, Wilhite S, Ledoux P, Evangelista C, Kim I, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. (2013) 41:D991–995. doi: 10.1093/nar/gks1193
16. Morrow J, Zhou X, Lao T, Jiang Z, DeMeo D, Cho M, et al. Functional interactors of three genome-wide association study genes are differentially expressed in severe chronic obstructive pulmonary disease lung tissue. Sci Rep. (2017) 7:44232. doi: 10.1038/srep44232
17. Tsai F, Lin Y, Chang S, Chang G, Hsu Y, Lin Y, et al. Differential left-to-right atria gene expression ratio in human sinus rhythm and atrial fibrillation: implications for arrhythmogenesis and thrombogenesis. Int J Cardiol. (2016) 222:104–12. doi: 10.1016/j.ijcard.2016.07.103
18. Çubukçuoğlu D, Durdu S, Doğan Y, Erdemli E, Özdağ H, Akar A. Molecular signatures of human chronic atrial fibrillation in primary mitral regurgitation. Cardiovasc Ther. (2021) 2021. doi: 10.1155/2021/5516185
19. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. (2008) 9:559. doi: 10.1186/1471-2105-9-559
20. Szklarczyk D, Gable A, Nastou K, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. (2021) 49:D605–12. doi: 10.1093/nar/gkaa1074
21. Shannon P, Markiel A, Ozier O, Baliga N, Wang J, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. doi: 10.1101/gr.1239303
22. Yu G, Wang L, Han Y, He Q. Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS. (2012) 16:284–7. doi: 10.1089/omi.2011.0118
23. Gene Ontology Consortium. The gene ontology resource: enriching a GOld mine. Nucleic Acids Res. (2021) 49:D325–34. doi: 10.1093/nar/gkaa1113
24. Subramanian A, Tamayo P, Mootha V, Mukherjee S, Ebert B, Gillette M, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
25. Jiang H, Zhang X, Wu Y, Zhang B, Wei J, Li J, et al. Bioinformatics identification and validation of biomarkers and infiltrating immune cells in endometriosis. Front Immunol. (2022) 13:944683. doi: 10.3389/fimmu.2022.944683
26. Biecek P. DALEX: explainers for complex predictive models in R. J Mach Learn Res. (2018) 19:3245–9.
27. Grau J, Grosse I, Keilwagen J. PRROC: computing and visualizing precision-recall and receiver operating characteristic curves in R. Bioinformatics. (2015) 31:2595–7. doi: 10.1093/bioinformatics/btv153
28. Franz M, Rodriguez H, Lopes C, Zuberi K, Montojo J, Bader G, et al. GeneMANIA update 2018. Nucleic Acids Res. (2018) 46:W60–4. doi: 10.1093/nar/gky311
29. Castro-Mondragon J, Riudavets-Puig R, Rauluseviciute I, Lemma R, Turchi L, Blanc-Mathieu R, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. (2022) 50. doi: 10.1093/nar/gkab1113
30. Huang H, Lin Y, Cui S, Huang Y, Tang Y, Xu J, et al. miRTarBase update 2022: an informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. (2022) 50:D222–30. doi: 10.1093/nar/gkab1079
31. Cotto K, Wagner A, Feng Y, Kiwala S, Coffman A, Spies G, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. (2018) 46:D1068–73. doi: 10.1093/nar/gkx1143
32. Chen B, Khodadoust M, Liu C, Newman A, Alizadeh A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods in Mol Biol (Clifton, NJ). (2018) 1711. doi: 10.1007/978-1-4939-7493-1_12
33. GBD 2015 Chronic Respiratory Disease Collaborators. Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990-2015: a systematic analysis for the global burden of disease study 2015. Lancet Respir Med. (2017) 5:691–706. doi: 10.1016/S2213-2600(17)30293-X
34. Grymonprez M, Vakaet V, Kavousi M, Stricker B, Ikram M, Heeringa J, et al. Chronic obstructive pulmonary disease and the development of atrial fibrillation. Int J Cardiol. (2019) 276:118–24. doi: 10.1016/j.ijcard.2018.09.056
35. Huang B, Yang Y, Zhu J, Liang Y, Zhang H, Tian L, et al. Clinical characteristics and prognostic significance of chronic obstructive pulmonary disease in patients with atrial fibrillation: results from a multicenter atrial fibrillation registry study. J Am Med Dir Assoc. (2014) 15:576–81. doi: 10.1016/j.jamda.2014.04.009
36. Liu C, Chen Y, Chang Y, Chien W, Lin H, Cheng C, et al. New-Onset atrial fibrillation is a risk factor of ischemic stroke in chronic obstructive pulmonary disease. Healthcare (Basel). (2022) 10:381. doi: 10.3390/healthcare10020381
37. Chen C, Liao K. The impact of atrial fibrillation in patients with COPD during hospitalization. Int J Chron Obstruct Pulmon Dis. (2018) 13:2105–12. doi: 10.2147/COPD.S166534
38. Yao Y, Yang M, Liu D, Zhao Q. Immune remodeling and atrial fibrillation. Front Physiol. (2022) 13:927221. doi: 10.3389/fphys.2022.927221
39. Barnes P. Inflammatory mechanisms in patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. (2016) 138:16–27. doi: 10.1016/j.jaci.2016.05.011
40. Caramori G, Casolari P, Barczyk A, Durham A, Di S, Adcock I. COPD Immunopathology. Semin Immunopathol. (2016) 38:497–515. doi: 10.1007/s00281-016-0561-5
41. Hu Y, Chen Y, Lin Y, Chen S. Inflammation and the pathogenesis of atrial fibrillation. Nat Rev Cardiol. (2015) 12:230–43. doi: 10.1038/nrcardio.2015.2
42. Hogg J, Chu F, Utokaparch S, Woods R, Elliott W, Buzatu L, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. (2004) 350:2645–53. doi: 10.1056/NEJMoa032158
43. Saqib U, Sarkar S, Suk K, Mohammad O, Baig M, Savai R. Phytochemicals as modulators of M1-M2 macrophages in inflammation. Oncotarget. (2018) 9:17937–50. doi: 10.18632/oncotarget.24788
44. Agustí A, Edwards L, Rennard S, MacNee W, Tal-Singer R, Miller B, et al. Persistent systemic inflammation is associated with poor clinical outcomes in COPD: a novel phenotype. PLoS One. (2012) 7:e37483. doi: 10.1371/journal.pone.0037483
45. Sun Z, Zhou D, Xie X, Wang S, Wang Z, Zhao W, et al. Cross-talk between macrophages and atrial myocytes in atrial fibrillation. Basic Res Cardiol. (2016) 111:63. doi: 10.1007/s00395-016-0584-z
46. Zhang Y, Cao H, Han X, Teng F, Chen C, Yang J, et al. Chemokine receptor CXCR-2 initiates atrial fibrillation by triggering monocyte mobilization in mice. Hypertension. (2020) 76:381–92. doi: 10.1161/HYPERTENSIONAHA.120.14698
47. Williams M, Todd I, Fairclough L. The role of CD8 + T lymphocytes in chronic obstructive pulmonary disease: a systematic review. Inflamm Res. (2021) 70:11–8. doi: 10.1007/s00011-020-01408-z
48. Murdoch J, Lloyd C. Resolution of allergic airway inflammation and airway hyperreactivity is mediated by IL-17-producing {gamma}{delta}T cells. Am J Respir Crit Care Med. (2010) 182:464–76. doi: 10.1164/rccm.200911-1775OC
49. Kato A, Hulse K, Tan B, Schleimer R. B-lymphocyte lineage cells and the respiratory system. J Allergy Clin Immunol. (2013) 131:933–57; quiz 958. doi: 10.1016/j.jaci.2013.02.023
50. Polverino F, Cosio B, Pons J, Laucho-Contreras M, Tejera P, Iglesias A, et al. B cell-activating factor. An orchestrator of lymphoid follicles in severe chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2015) 192:695–705. doi: 10.1164/rccm.201501-0107OC
51. Ladjemi M, Martin C, Lecocq M, Detry B, Nana F, Moulin C, et al. Increased IgA expression in lung lymphoid follicles in severe chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2019) 199:592–602. doi: 10.1164/rccm.201802-0352OC
52. Givi M, Folkerts G, Wagenaar G, Redegeld F, Mortaz E. Cigarette smoke differentially modulates dendritic cell maturation and function in time. Respir Res. (2015) 16:131. doi: 10.1186/s12931-015-0291-6
53. Akata K, van Eeden S. Lung macrophage functional properties in chronic obstructive pulmonary disease. Int J Mol Sci. (2020) 21:853. doi: 10.3390/ijms21030853
54. Litviňuková M, Talavera-López C, Maatz H, Reichart D, Worth C, Lindberg E, et al. Cells of the adult human heart. Nature. (2020) 588:466–72. doi: 10.1038/s41586-020-2797-4
55. Smorodinova N, Bláha M, Melenovský V, Rozsívalová K, Přidal J, Ďurišová M, et al. Analysis of immune cell populations in atrial myocardium of patients with atrial fibrillation or sinus rhythm. PLoS One. (2017) 12:e0172691. doi: 10.1371/journal.pone.0172691
56. Begieneman M, Rijvers L, Kubat B, Paulus W, Vonk A, van Rossum A, et al. Atrial fibrillation coincides with the advanced glycation end product N(ɛ)-(carboxymethyl)lysine in the atrium. Am J Pathol. (2015) 185:2096–104. doi: 10.1016/j.ajpath.2015.04.018
57. Wang X, Wang J, Ding Z, Ji J, Sun Q, Cai G. Structural flexibility and functional interaction of mediator Cdk8 module. Protein Cell. (2013) 4:911–20. doi: 10.1007/s13238-013-3069-y
58. Chen M, Liang J, Ji H, Yang Z, Altilia S, Hu B, et al. CDK8/19 Mediator kinases potentiate induction of transcription by NFκB. Proc Natl Acad Sci U S A. (2017) 114:10208–13. doi: 10.1073/pnas.1710467114
59. Perez-Perri J, Dengler V, Audetat K, Pandey A, Bonner E, Urh M, et al. The TIP60 Complex is a conserved coactivator of HIF1A. Cell Rep. (2016) 16:37–47. doi: 10.1016/j.celrep.2016.05.082
60. Shukla S, Walters E, Simpson J, Keely S, Wark P, O’Toole R, et al. Hypoxia-inducible factor and bacterial infections in chronic obstructive pulmonary disease. Respirology. (2020) 25:53–63. doi: 10.1111/resp.13722
61. Xu Y, Sharma D, Du F, Liu Y. The role of toll-like receptor 2 and hypoxia-induced transcription factor-1α in the atrial structural remodeling of non-valvular atrial fibrillation. Int J Cardiol. (2013) 168:2940–1. doi: 10.1016/j.ijcard.2013.03.174
62. Sun X, Feng X, Zheng D, Li A, Li C, Li S, et al. Ergosterol attenuates cigarette smoke extract-induced COPD by modulating inflammation, oxidative stress and apoptosis in vitro and in vivo. Clin Sci. (2019) 133:1523–36. doi: 10.1042/CS20190331
63. Liu M, Li W, Wang H, Yin L, Ye B, Tang Y, et al. CTRP9 Ameliorates atrial inflammation, fibrosis, and vulnerability to atrial fibrillation in post-myocardial infarction rats. J Am Heart Assoc. (2019) 8:e013133. doi: 10.1161/JAHA.119.013133
64. Chen X, Yan Y, Cheng X, Zhang Z, He C, Wu D, et al. A novel CDK8 inhibitor with poly-substituted pyridine core: discovery and anti-inflammatory activity evaluation in vivo. Bioorg Chem. (2023) 133:106402. doi: 10.1016/j.bioorg.2023.106402
65. Guo Z, Wang G, Lv Y, Wan Y, Zheng J. Inhibition of Cdk8/Cdk19 activity promotes treg cell differentiation and suppresses autoimmune diseases. Front Immunol. (2019) 10:1988. doi: 10.3389/fimmu.2019.01988
66. Schmerwitz U, Sass G, Khandoga A, Joore J, Mayer B, Berberich N, et al. Flavopiridol protects against inflammation by attenuating leukocyte-endothelial interaction via inhibition of cyclin-dependent kinase 9. Arterioscler Thromb Vasc Biol. (2011) 31:280–8. doi: 10.1161/ATVBAHA.110.213934
67. Rao F, Xue Y, Wei W, Yang H, Liu F, Chen S, et al. Role of tumour necrosis factor-a in the regulation of T-type calcium channel current in HL-1 cells. Clin Exp Pharmacol Physiol. (2016) 43:706–11. doi: 10.1111/1440-1681.12585
68. Kao Y, Chen Y, Cheng C, Lee T, Chen Y, Chen S. Tumor necrosis factor-alpha decreases sarcoplasmic reticulum Ca2+-ATPase expressions via the promoter methylation in cardiomyocytes. Crit Care Med. (2010) 38:217–22. doi: 10.1097/CCM.0b013e3181b4a854
69. Goudis C, Kallergis E, Vardas P. Extracellular matrix alterations in the atria: insights into the mechanisms and perpetuation of atrial fibrillation. Europace. (2012) 14:623–30. doi: 10.1093/europace/eur398
70. Siwik D, Chang D, Colucci W. Interleukin-1beta and tumor necrosis factor-alpha decrease collagen synthesis and increase matrix metalloproteinase activity in cardiac fibroblasts in vitro. Circ Res. (2000) 86:1259–65. doi: 10.1161/01.res.86.12.1259
71. Kaku Y, Imaoka H, Morimatsu Y, Komohara Y, Ohnishi K, Oda H, et al. Overexpression of CD163, CD204 and CD206 on alveolar macrophages in the lungs of patients with severe chronic obstructive pulmonary disease. PLoS One. (2014) 9:e87400. doi: 10.1371/journal.pone.0087400
72. Vlahos R, Bozinovski S. Role of alveolar macrophages in chronic obstructive pulmonary disease. Front Immunol. (2014) 5:435. doi: 10.3389/fimmu.2014.00435
73. Xu J, Cui G, Esmailian F, Plunkett M, Marelli D, Ardehali A, et al. Atrial extracellular matrix remodeling and the maintenance of atrial fibrillation. Circulation. (2004) 109:363–8. doi: 10.1161/01.CIR.0000109495.02213.52
74. Abe I, Teshima Y, Kondo H, Kaku H, Kira S, Ikebe Y, et al. Association of fibrotic remodeling and cytokines/chemokines content in epicardial adipose tissue with atrial myocardial fibrosis in patients with atrial fibrillation. Heart Rhythm. (2018) 15:1717–27. doi: 10.1016/j.hrthm.2018.06.025
75. Lyu J, Wang M, Kang X, Xu H, Cao Z, Yu T, et al. Macrophage-mediated regulation of catecholamines in sympathetic neural remodeling after myocardial infarction. Basic Res Cardiol. (2020) 115:56. doi: 10.1007/s00395-020-0813-3
76. Ravi A, Plumb J, Gaskell R, Mason S, Broome C, Booth G, et al. COPD Monocytes demonstrate impaired migratory ability. Respir Res. (2017) 18:90. doi: 10.1186/s12931-017-0569-y
77. Segal L, Martinez F. Chronic obstructive pulmonary disease subpopulations and phenotyping. J Allergy Clin Immunol. (2018) 141:1961–71. doi: 10.1016/j.jaci.2018.02.035
78. Hirayama A, Goto T, Shimada Y, Faridi M, Camargo C, Hasegawa K. Acute exacerbation of chronic obstructive pulmonary disease and subsequent risk of emergency department visits and hospitalizations for atrial fibrillation. Circ Arrhythm Electrophysiol. (2018) 11:e006322. doi: 10.1161/CIRCEP.118.006322
Keywords: chronic obstructive pulmonary disease, atrial fibrillation, microarray, machine learning, biomarker
Citation: Sun Z, Lin J, Zhang T, Sun X, Wang T, Duan J and Yao K (2023) Combining bioinformatics and machine learning to identify common mechanisms and biomarkers of chronic obstructive pulmonary disease and atrial fibrillation. Front. Cardiovasc. Med. 10:1121102. doi: 10.3389/fcvm.2023.1121102
Received: 11 December 2022; Accepted: 14 March 2023;
Published: 28 March 2023.
Edited by:
Miguel Hueso, Bellvitge University Hospital, SpainReviewed by:
Dmitry A. Sychev, Ministry of Health, RussiaSerghei Covantsev, S.P. Botkin Clinical Hospital, Russia
Xue He, Central South University, China
© 2023 Sun, Lin, Zhang, Sun, Wang, Duan and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kuiwu Yao yaokuiwu@126.com
These authors have contributed equally to this work and share first authorship
Specialty Section: This article was submitted to Cardiovascular Genetics and Systems Medicine, a section of the journal Frontiers in Cardiovascular Medicine
 Ziyi Sun
Ziyi Sun Jianguo Lin
Jianguo Lin Tianya Zhang4,†
Tianya Zhang4,†  Xiaoning Sun
Xiaoning Sun Kuiwu Yao
Kuiwu Yao