Emergence of Ceftazidime/Avibactam and Tigecycline Resistance in Carbapenem-Resistant Klebsiella pneumoniae Due to In-Host Microevolution
- 1Department of Infectious Diseases, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, China
- 2Key Laboratory of Microbial Technology and Bioinformatics of Zhejiang Province, Hangzhou, China
- 3Regional Medical Center for National Institute of Respiratory Diseases, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 4Department of Clinical Laboratory, The Children’s Hospital, Zhejiang University School of Medicine, National Clinical Research Center for Child Health, Hangzhou, China
Klebsiella pneumoniae can cause both hospital- and community-acquired clinical infections. Last-line antibiotics against carbapenem-resistant K. pneumoniae (CRKP), such as ceftazidime/avibactam (CZA) and tigecycline (TGC), remain limited as treatment choices. This study aimed to investigate the mechanisms by which CRKP acquires CZA and TGC resistance in vivo under β-lactam antibiotic and TGC exposure. Three CRKP strains (XDX16, XDX31 and XDX51) were consecutively isolated from an inpatient with a urinary tract infection in two months. PFGE and MLST showed that these strains were closely related and belonged to sequence type (ST) 4496, which is a novel ST closely related to ST11. Compared to XDX16 and XDX31, XDX51 developed CZA and TGC resistance. Sequencing showed that double copies of blaKPC-2 were located on a 108 kb IncFII plasmid, increasing blaKPC-2 expression in XDX51. In addition, ramR was interrupted by Insertion sequence (IS) Kpn14 in XDX51, with this strain exhibiting upregulation of ramA, acrA and acrB expression compared with XDX16 and XDX31. Furthermore, LPS analysis suggested that the O-antigen in XDX51 was defective due to ISKpn26 insertion in the rhamnosyl transferase gene wbbL, which slightly reduced TGC susceptibility. In brief, CZA resistance was caused mainly by blaKPC-2 duplication, and TGC resistance was caused by ramR inactivation with additional LPS changes due to IS element insertion in wbbL. Notably, CRKP developed TGC and CZA resistance within one month under TGC and β-lactam treatment without exposure to CZA. The CRKP clone ST4496 has the ability to evolve CZA and TGC resistance rapidly, posing a potential threat to inpatients during antibiotic treatment.
Introduction
Klebsiella pneumoniae, is a major pathogen that can cause both nosocomial- and community-acquired infections, such as urinary tract infections, bacteremia, respiratory infections and soft tissue infections, especially in immunocompromised individuals (Ullmann, 1998; Holt et al., 2015). The emergence of carbapenem-resistant K. pneumoniae (CRKP) has limited antibiotic therapeutic choices and led to serious challenges in clinical treatment and infection control (Rice, 2008; Livermore et al., 2015a; Moradigaravand et al., 2017). The World Health Organization (WHO) considers carbapenem-resistant Enterobacteriaceae to be critical priority pathogens that require new antibiotics (Shrivastava et al., 2018). Ceftazidime/avibactam (CZA) and tigecycline (TGC) are last-line choices for the treatment of CRKP infections.
CZA is a novel β-lactam-β-lactamase inhibitor combination with the ability to inhibit the activity of AmpC, extended-spectrum β-lactamases (ESBLs), class A carbapenemases such as KPC and class D carbapenemases such as OXA-48 but not class B carbapenemases such as NDM, IMP and VIM (Shirley, 2018). CZA was approved for the treatment of complicated intra-abdominal infections and urinary tract infections by the FDA on February 25, 2015. A multicenter, observational study reported that CZA could be a reasonable substitute for colistin for the treatment of CRKP infections with reduced mortality (Van Duin et al., 2018). In the International Network For Optimal Resistance Monitoring (INFORM) surveillance programme, the in vitro susceptibility of carbapenemase-positive and MBL-negative isolates to CZA was 99.8% between 2015 and 2017 (Spiliopoulou et al., 2020). However, acquisition of CZA resistance has been reported after CZA therapy in recent years. Resistance to CZA has been observed in strains with mutations in AmpC, blaKPC-2 and blaKPC-3, and the mutation point was mostly in Ω-loop in blaKPC genes (Shirley, 2018; Zhang et al., 2020). In addition, high expression of KPC-3 was reported to be associated with CZA resistance (Humphries and Hemarajata, 2017).
The emergence of TGC resistance after exposure to TGC has also been reported (Duin et al., 2014; Lin et al., 2016; Ye et al., 2017). Overall, the primary TGC resistance mechanisms in K. pneumoniae include overexpression of efflux pumps, such as AcrAB and OqxAB, which can be mediated by regulators (RamR-RamA and OqxR-RarA) and global activators (MarA, SoxS and Rob) (Veleba and Schneiders, 2012). There also exist other known mechanisms, conferred by the Lon protease, Tet(A) protein and ribosomal protein S10, which is encoded by rpsJ (Villa et al., 2014). In addition, Linkevicius M et al. reported that lipopolysaccharide (LPS) defects could lead to reduced susceptibility to TGC (Linkevicius et al., 2016).
Among all the antibiotics tested by the INFORM surveillance programme (2015–2017) against 1,460 carbapenem-nonsusceptible Enterobacteriaceae isolates, CZA and TGC showed the highest susceptibility rates (73.0% and 78.1%, respectively) (Spiliopoulou et al., 2020). Strains resistant to these last-line antibiotics could pose a great threat to public health.
Here, we report a series of CRKP strains isolated in 2017 that have evolved resistance to both CZA and TGC during exposure to β-lactam antibiotics and TGC. The CRKP isolates resistant to CZA were isolated before CZA was approved in China. The aim of this study was to describe the phenotypic and genotypic adaption characteristics of these strains during in-host evolution and explain the molecular mechanisms of CZA and TGC resistance.
Material and Methods
Strains and Patient Characteristics
Three CRKP strains (XDX16, XDX31 and XDX51) were isolated from an 85-year-old male patient hospitalized in Sir Run Run Shaw Hospital, Zhejiang Province, China, in 2017. The patient was admitted for a urinary tract infection with multiple organ dysfunction. After admission, he received urological surgeries, including partial cystectomy, prostatic enucleation surgery and ureteric reimplantation. The patient suffered urinary tract infection with prolonged fever. Piperacillin-tazobactam (TZP, 4.5 g every 8 h) and cefoperazone-sulbactam (SCF, 2 g every 8 h) were alternately used as empirical therapies. Then, TGC (50 mg every 12 h) was used as a definitive therapy. During this period, three CRKP strains, named XDX16, XDX31 and XDX51, were isolated from the patient’s urine. In addition, XDX51 exhibited resistance to both CZA and TGC. The patient chose to be discharged against medical advice. The timeline of CRKP infection and antibiotic usage was shown in Figure 1. This study was approved by the ethics committees of the participating hospital (20170301-3)
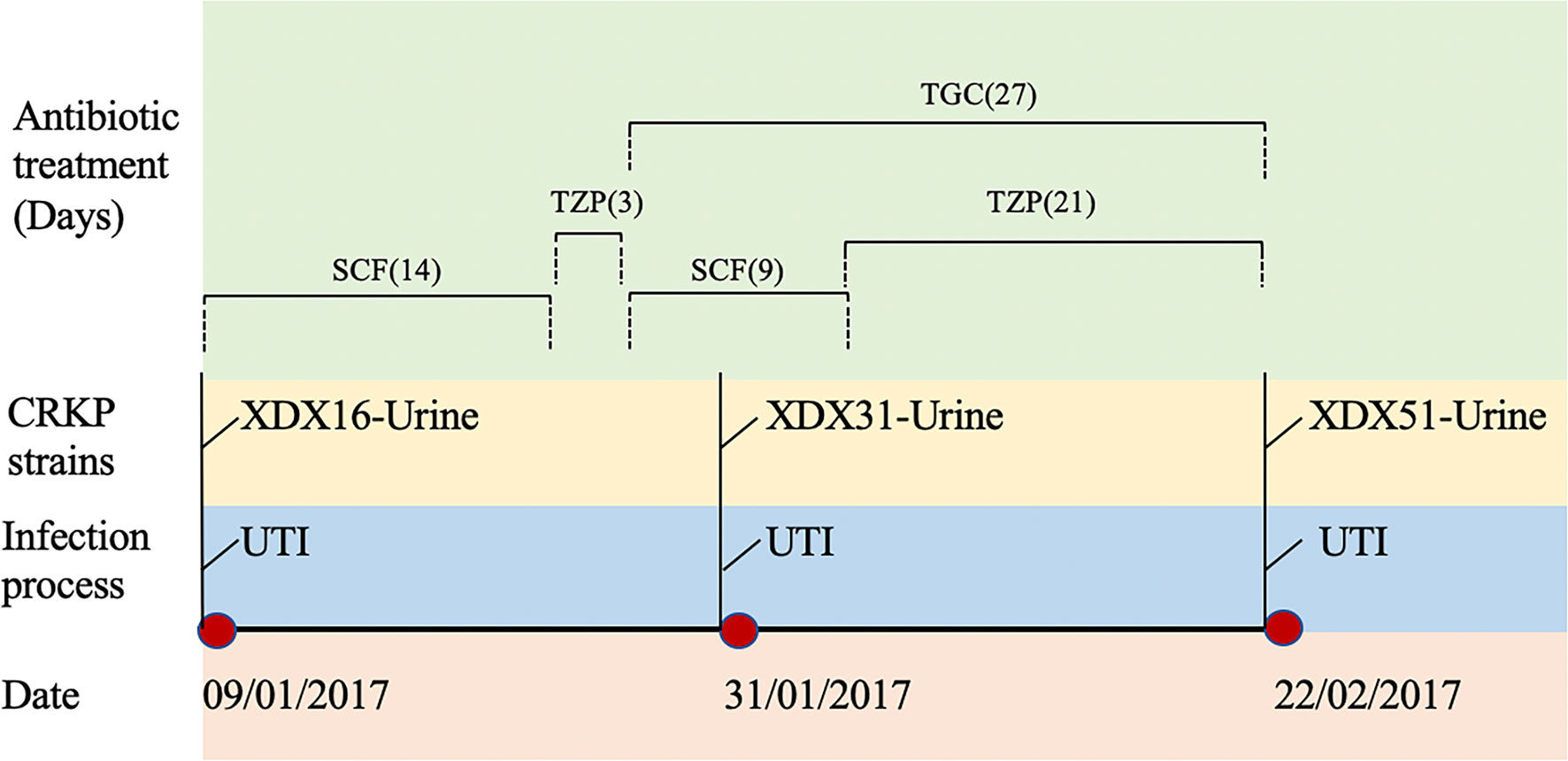
Figure 1 Timeline of CRKP infection and treatment during the patient’s hospitalization. XDX16, XDX31 and XDX51 are carbapenem-resistant Klebsiella pneumoniae strains. UTI, urinary tract infection; SCF, Cefoperazone-sulbactam; TZP, Piperacillin-tazobactam; TGC, Tigecycline.
Antimicrobial Susceptibility Testing
The antibiotics tested in this study were TGC, CZA, meropenem, imipenem, ertapenem, ceftazidime, amikacin, levofloxacin, fosfomycin, aztreonam and colistin. The antimicrobial susceptibility tests and minimum inhibitory concentrations (MICs) were based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST, Version 10.0, 2020) guidelines and breakpoints. The MICs of carbapenem and fosfomycin (with supplementation of glucose-6-phosphate in the agar) were determined by the agar dilution method, and the others were determined by the broth microdilution method with fresh cation-adjusted Mueller-Hinton broth. The MICs of TGC were also determined in the presence of Phe-Arg-β-naphthylamide (PAβN) at a concentration of 50 μg/mL to verify the function of the efflux pump in TGC resistance (Salehi et al., 2021).
Whole-Genome Sequencing and Analysis
The clonality of the strain series was confirmed by pulsed-field gel electrophoresis (PFGE) with a previously described protocol (Barton et al., 1995). Briefly, genomic DNA was digested with the restriction endonuclease XbaI and electrophoresed at 14°C for 20 h.
XDX16 and XDX51 were subjected to both Illumina paired-end sequencing (Illumina Inc., San Diego, CA) and long-read nanopore sequencing (Oxford Nanopore Technologies, Oxford, UK). For Illumina sequencing, genomic DNA was extracted using a QIAamp DNA MiniKit (Qiagen, New York, USA). The whole genome was assembled by canu (Koren et al., 2017), and the de novo assemblies were subsequently annotated with the Prokka pipeline (Seemann, 2014). Resistance genes were detected by ResFinder 3.2 with a 90% threshold for gene identification and a 60% minimum length coverage (Zankari et al., 2012). Multilocus sequence typing (MLST) was performed with the Center for Genomic Epidemiology guidelines (http://cge.cbs.dtu.dk/services/MLST/). Breseq (version 0.27.1) was used to find mutations in XDX51, with XDX16 as the reference strain (Barrick and Deatherage, 2014). All the mutations in XDX16, XDX31 and XDX51 were confirmed by PCR and Sanger sequencing (the primers are listed in Table S1).
The nucleotide sequences of XDX16 and XDX51 have been submitted to the NCBI database under the accession JAIWPV000000000-JAIWPW000000000.
qRT-PCR for Gene Expression Analysis
qRT-PCR was used to measure the expression of blaKPC-2, acrA, acrB and ramR of XDX16, XDX31 and XDX51. XDX16 was used as the reference strain, and the rpoB gene was used as the internal reference (the primers are listed in Table S1). RNA was extracted using the PureLink RNA Mini Kit (Invitrogen, Carlsbad, CA) in the exponential growth period of bacterial cells. Then, cDNA was obtained using a PrimeScript™ RT Reagent Kit (Takara, Kyoto, Japan). The expression level was assessed using TB Green™ Premix Ex Taq™ (Takara, Kyoto, Japan) in a LightCycler 480 system (Roche, Rotkreuz, Switzerland) with triplicate samples for each isolate, replicating three times independently using the comparative CT method. Genes were considered to be differentially expressed when the |log2 fold change| was greater than 1.5 (Livak and Schmittgen, 2001). The log2 fold change in blaKPC-2 expression was analyzed by an unpaired t test on GraphPad Prism, and p<0.05 was considered significant.
Gene Knockout and Complementation Experiment
We used the lambda-Red knockout system described previously for wbbL gene knockout (Huang et al., 2014). Briefly, the FRT-flanked apramycin resistance cassette was amplified from the pIJ773 plasmid using a homologous region primer. The pACBSR-Hyg plasmid was introduced into XDX16 by electroporation for recombination. The knockout cassette was transformed into XDX16-pACBSR-Hyg. Correct transformants were screened using LBApra at 37°C and verified by PCR and Sanger sequencing. Colonies were screened for the loss of pACBSR-Hyg by streaking onto LBApra with or without hygromycin (100 μg/mL) and low-salt LB + hygromycin plates at 37°C overnight. The resistance marker was removed by pFLP-Hyg plasmid.
The amplified target gene (wbbL) and its promoter were cloned into the pCR2.1-Hyg plasmid, which was digested with the Xba1 FastDigest enzyme (Thermo Scientific, Waltham, the USA). The recombinant plasmids were introduced to resistant and knockout strains (XDX51 and XDX16∆wbbL) via chemical transformation. The empty vector (pCR2.1-Hyg) was introduced to the same strains as a blank control. The complemented sequence was confirmed by Sanger sequencing (the primers are listed in Table S1).
LPS Analysis
Extraction of XDX16, XDX16∆wbbL and XDX51 LPS was performed according to Michael R. Davis’s protocol (Davis and Goldberg, 2012). Then, 5 μl of LPS was electrophoresed on 15% Tris-glycine (Fdbio Science, Hangzhou, China) and directly stained using a fast silver staining kit (Beyotime Biotechnology, Shanghai, China). O-antigen typing analysis was performed using Kaptive (http://kaptive.holtlab.net/).
Results
Strains and Antibiotic Susceptibility
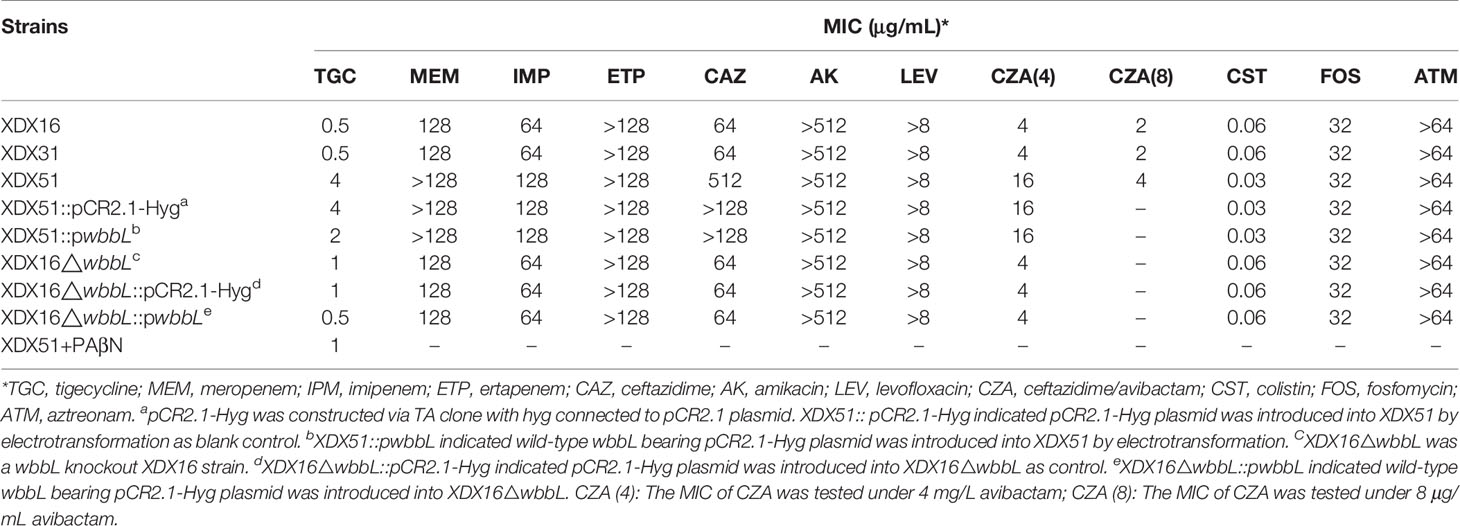
XDX16 and XDX31 were considered the index strains. Three strains were all identified to be resistant to meropenem, imipenem, ertapenem, ceftazidime, amikacin, levofloxacin and aztreonam. Significant differences in MICs were observed for CZA and TGC between XDX51 and the index strains. Specifically, for the index strains, TGC had an MIC of 0.5 μg/mL, while for XDX51, the MIC of TGC was 8-fold higher. Additionally, the MIC of CZA for XDX51 (16 μg/mL) was 4-fold higher than that for the index strains (4 μg/mL). In addition, the MICs of meropenem, imipenem and ceftazidime for XDX51 were also higher than those for the index strains (Table 1).
Genomic Characteristics
The PFGE results confirmed that the three strains were closely related (Figure S1). XDX16 and XDX51 were chosen for whole-genomic sequencing since XDX31 seemed to be the same passage as XDX16, with which it shared an identical PFGE typing result and resistance phenotype. According to genome-based MLST analysis, XDX16 and XDX51 belonged to the same novel ST, ST4496, which is closely related to ST11 with one single-locus variant on mdh.
XDX16 and XDX51 shared the same resistance gene distribution, including the aminoglycoside resistance genes aadA2 and rmtB, the sulfonamide resistance gene sul1, the beta-lactam resistance genes blaKPC-2 and blaSHV-182, the fosfomycin resistance gene fosA and the quinolone resistance genes oqxA and oqxB. Both of them had identical mutations in acrR and ompK35.
Identification of Plasmid Differences in XDX51
Both XDX16 and XDX51 had the IncFII plasmid harboring the antibiotic resistance genes blaKPC-2 and rmtB. Furthermore, there was duplication of blaKPC-2 on the plasmid of XDX51 (Figure 2A). The two copies were linked together and shared the same Insertion sequence (IS) 26. The surrounding structure of blaKPC-2 in our study was similar to the classic blaKPC-2 genetic environment in pKP048 in China reported in a previous study, with the gene order IS26, Tn3-resolvase, ISKpn8, blaKPC-2, ISKpn6, hypothetical protein and IS26 (Shen et al., 2009) (Figure 2B). In our study, there were two IS26 elements around the Tn3-based blaKPC-2 structure, with one IS26 replacing the Tn3 transposase with pKP048.
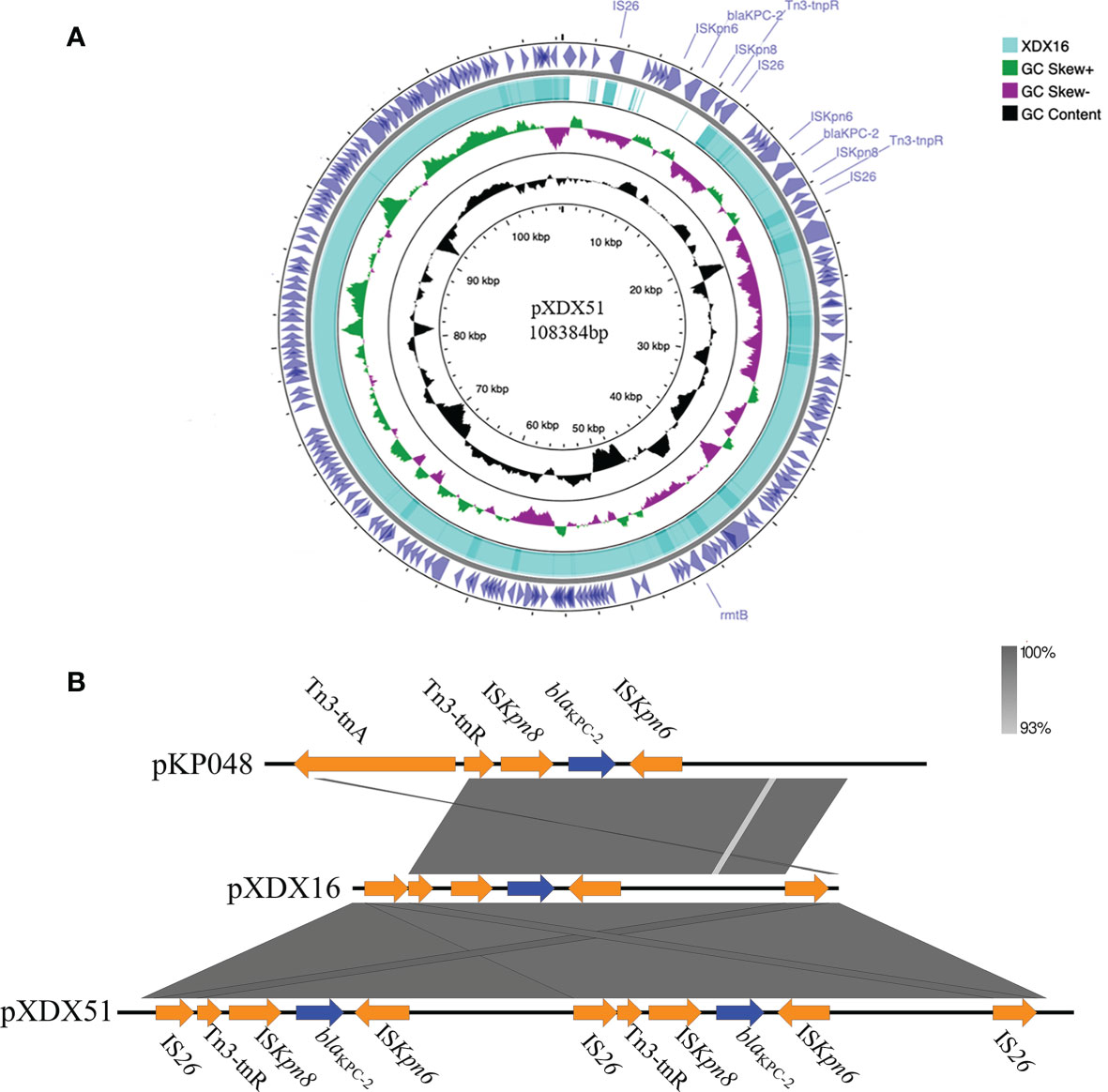
Figure 2 Characteristics of the blaKPC-2-bearing IncFII plasmid and the genetic environment of blaKPC-2. (A) Comparison of the IncFII plasmids in XDX51 and XDX16. There were double copies of blaKPC-2 in XDX51 compared to XDX16. (B) The blaKPC-2 environment in our study was compared with that in pKP048. The gene environment of blaKPC-2 in our study was IS26-Tn3-ISKpn8-blaKPC-2-ISKpn6-hypothetical protein-IS26. Blue arrows indicate resistance genes, and orange arrows indicate insertion sequences. The right inverted repeat (IRR) of IS26 was 5’-ggcactgttgcaaa-3’, the left inverted repeat (IRL) of IS26 was 5’-tttgcaacagtgcc-3’; the IRR of ISKpn8 was 5’-atgtcaagacccggctggttat-3’; and the IRL of ISKpn8 was 5’-atcccacgagtccagac-3’.
Identification of Whole-Genome Differences in XDX51
Comparison of the whole genomes of XDX16 and XDX51 also revealed that the wbbL gene (a rhamnosyl transferase gene) and ramR gene were interrupted by insertion sequences (Figure 3). A 769 bp ISKpn14 sequence was inserted at the 12 bp of ramR in XDX51. The insertion was responsible for interruption of the translation of the N-terminal amino acid sequence of RamR in XDX51. Similarly, wbbL was interrupted by a 1190 bp ISKpn26 sequence at 307 bp in XDX51. There were three SNPs detected in XDX51 compared to the prior wild type XDX16 using breseq. However, the three SNPs have occurred in the next wild type XDX31 isolate without TGC and CZA resistance phenotype. Hence, we inferred that these three SNPs were not responsible for TGC and CZA resistance (Table S2).
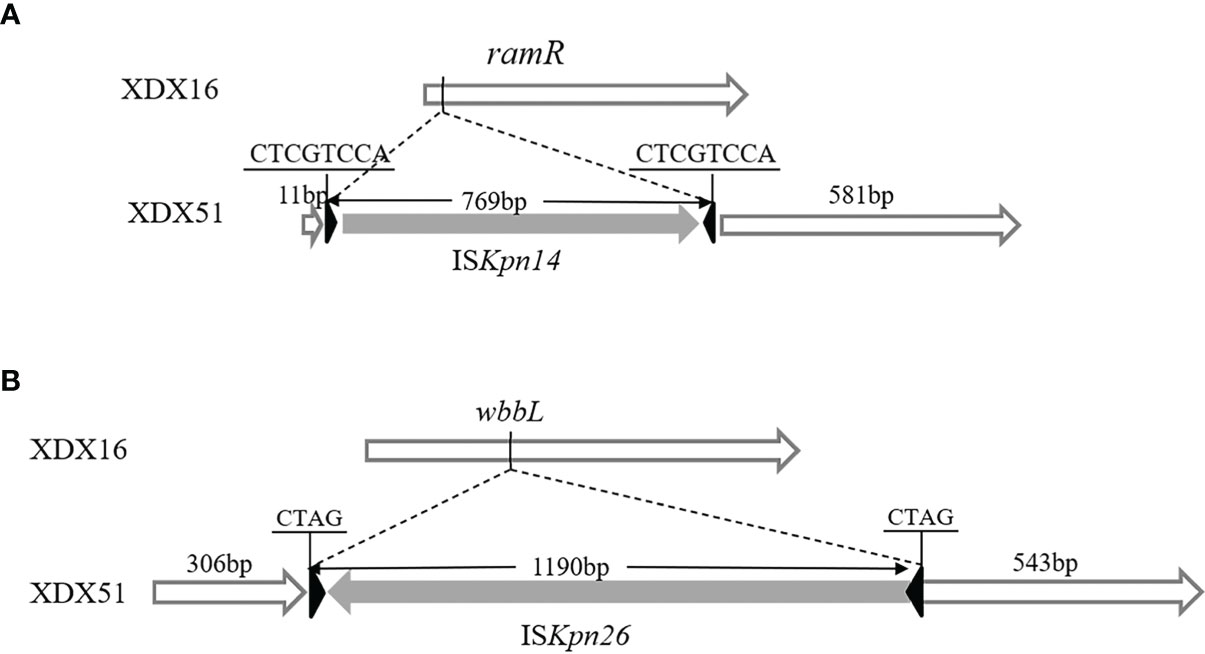
Figure 3 Schematic diagram of the gene structure. (A) The ramR gene was interrupted by ISKpn14 in XDX51; the IRL of ISKpn14 was 5’-GGTAATG-3’, and the IRR was 5’-CATTACC-3’. (B) The wbbL gene was interrupted by ISKpn26 in XDX51, the IRL of ISKpn26 was 5’-GGAAGGTGCGAA-3’, and the IRR was 5’-TTCGCACCTTCC-3’.
Expression Level of Antibiotic Resistance Genes
Compared to the copy numbers in XDX16, the copy numbers of blaKPC-2 in XDX51 determined by qRT-PCR (2.57 ± 0.3) were consistent with those found in the genome sequence. The blaKPC-2 expression level of XDX51 was 3.54 ± 0.5 times higher than that of XDX16 (Figure 4A). To verify the function of enhanced KPC expression on CZA resistance in XDX51, we performed CZA MIC test with avibactam at a higher concentration of 8 μg/mL, after which XDX51 could restore its susceptibility to CZA under sufficient avibactam.
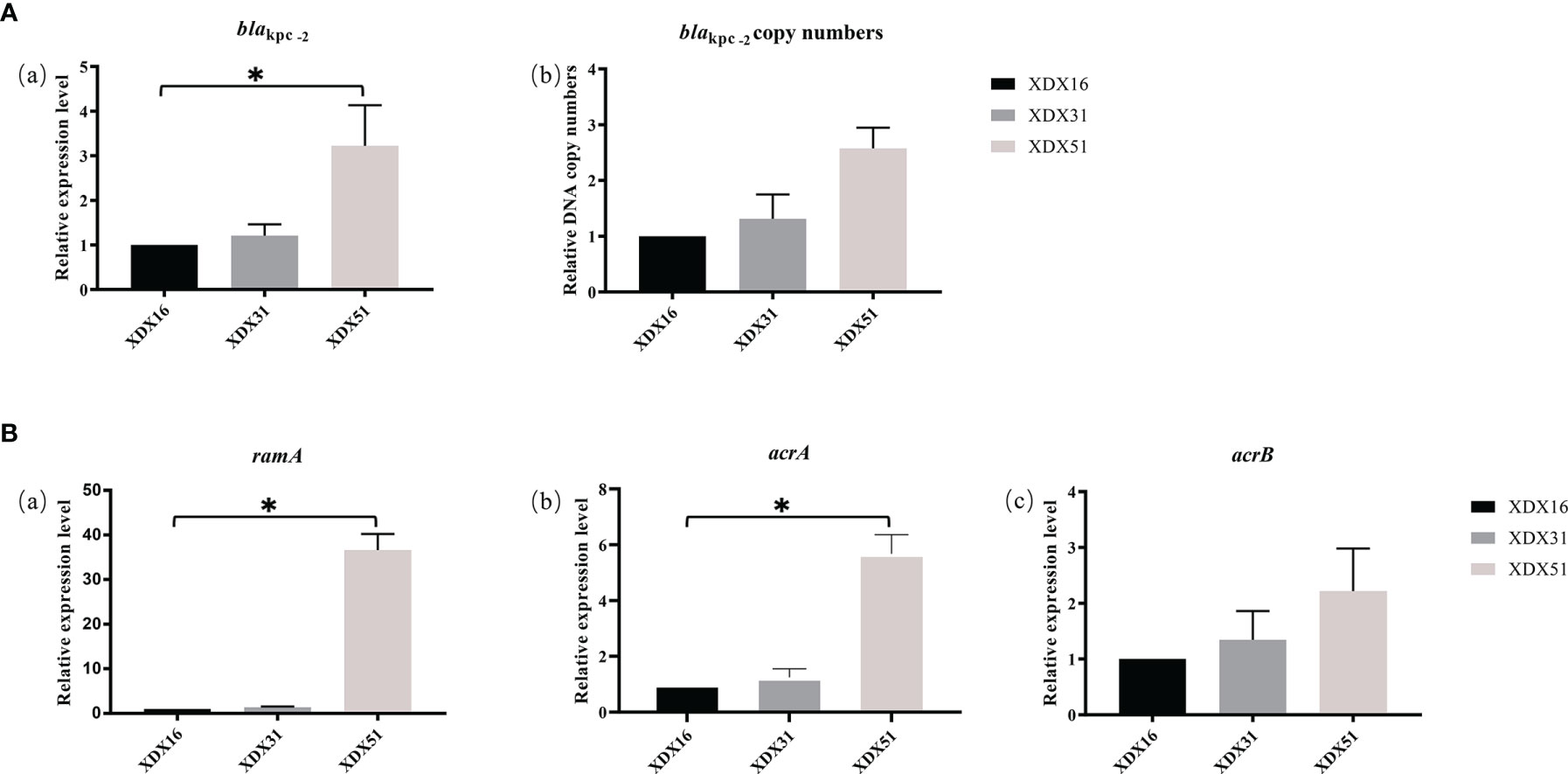
Figure 4 Relative expression level or DNA copy numbers of (A) blaKPC-2- and (B) AcrAB-TolC-related genes. XDX16 was used as the reference strain, and rpoB was used as the reference gene. The bars represent the mean ± standard deviation (SD) of triplicate biological repeats; the mean differences in log2 fold change were analyzed using an unpaired t test. *p < 0.05.
ramR is a regulator of the AcrA/B system. We evaluated the regulatory effects of ramR alteration in XDX51 by qRT-PCR. The results showed that insertion in ramR led to overexpression of ramA, which was 36.6 times higher in XDX51 than in XDX16. The acrA and acrB expression levels in XDX51 were also upregulated compared to those in the index strains (Figure 4B). These results suggested that insertion in ramR affected the expression level of AcrAB-TolC efflux pumps. To further confirm the effect of ramR alteration on efflux pumps, we performed an efflux pump suppression test with PAβN, and there was a 4-fold decrease in the MIC of TGC for XDX51.
Effect of the wbbL Gene on Phenotype
To further verify the effect of wbbL inactivation on antibiotic resistance, we constructed wbbL gene knockout and complementation strains. The MIC of TGC for the XDX16 wbbL gene knockout strain (XDX16∆wbbL) (1 μg/mL) increased 2-fold compared with that for XDX16 (0.5 μg/mL). The pCR2.1-Hyg vector was transformed into XDX16∆wbbL and into XDX51 as a control. Wild-type wbbL was cloned into the pCR2.1-Hyg vector (pwbbL) and introduced to the gene knockout strain (XDX16∆wbbL) and TGC-resistant strain with inactivated wbbL (XDX51). TGC sensitivity was restored in XDX16∆wbbL::pwbbL (0.5 μg/mL) and decreased in XDX51::pwbbL (2 μg/mL) but not in the empty vector-harboring strain. These results suggested that wbbL had a slight effect on the TGC resistance phenotype. The MICs of the isolates are presented in Table 1.
wbbL belongs to the O-antigen cluster. We hypothesized that wbbL could affect LPS biosynthesis in XDX51. Thus, LPS in XDX16∆wbbL and XDX51 was analyzed by SDS-PAGE and silver staining. Generally, the O-antigen bands were within 15–40 kD. There were fewer bands in XDX16∆wbbL and XDX51 within the 35–40 kD range, indicating that the O-antigen in XDX51 was shorter than that in the wbbL wild-type strain XDX16 (Figure S2).
Discussion
Under antibiotic pressure, bacteria adapt to the host or environment with genomic and phenotypic changes (Linkevicius et al., 2016). Here, we described a series of closely related CRKP strains that evolved resistance to CZA and TGC in vivo in a hospitalized patient. ST analysis showed that the strains belonged to a new ST, ST4496, closely related to ST11. Before the CZA- and TGC-resistant strain XDX51 was isolated, TGC was used for 27 days, and β-lactam antibiotics (cefoperazone-sulbactam and piperacillin-tazobactam) were prescribed alternately, while there was no exposure to CZA. Compared to the index strains, three genomic differences were identified in XDX51, including duplication of blaKPC-2, ramR and wbbL insertions caused by IS elements. Ye et al. reported that deletion of the ramR ribosomal binding site could cause in vivo development of TGC resistance during TGC treatment (Ye et al., 2017). Long-term usage of antibiotics such as β-lactams and TGC could have led to the evolution of the series of CRKP strains in our study with gene mutations related to antibiotic resistance.
Increased KPC expression could enhance ceftazidime hydrolysis, which could not be completely inhibited by avibactam (Zhang et al., 2020). Both XDX16 and XDX51 carried an IncFII plasmid containing blaKPC-2 and rmtB. The MICs of CZA were 4-fold higher for XDX51 than for XDX16. There was duplication of blaKPC-2 on the XDX51 plasmid due to unequal crossover of the IS26 composite transposon causing enhanced KPC expression levels, as identified by qRT-PCR. KPC-23-producing K. pneumoniae has been reported to be resistant to CZA due to increased ceftazidime hydrolysis without prior exposure to CZA (Galani et al., 2019). Similarly, Humphries RM reported CZA resistance due to increased expression of KPC-3 in clinical isolates previously unexposed to CZA (Humphries and Hemarajata, 2017). Both situations above were combined with membrane porin deficiency. In addition, Ω-loop alterations caused by KPC mutations can prevent avibactam binding and lead to CZA resistance, such as mutations at position 179 in blaKPC-2 or blaKPC-3 (Livermore et al., 2015b; Barnes et al., 2017). CZA resistance caused by KPC point mutations tends to occur after the strains are exposed to CZA either in vivo or in vitro (Livermore et al., 2015b; Barnes et al., 2017; Shields et al., 2017). Additionally, Antinori E. reported that deletion in blaKPC-3 in clinical K. pneumoniae could also result in CZA resistance (Antinori et al., 2020). In our study, the CZA-resistant strain evolved double copies of blaKPC-2 compared with the index strains. An increase in the MICs of carbapenem and CAZ for XDX51 indicated enhanced CAZ hydrolysis due to overexpression of blaKPC-2. Our conclusion was confirmed by other studies as well. Shen Z. et al. reported that the expression level of blaKPC-2 in CRKP with CZA MIC 4-8 μg/mL group was 4.2-4.8-fold higher than that in CZA MIC 1-2 μg/mL and ≤0.5 μg/mL group, and hydrolysis activities of CAZ was 4-4.6-fold higher in CZA MIC 4-8 group than the other two groups, indicating that the enhanced expression of blaKPC-2 could result in the decrease of CZA susceptibility due to the increased hydrolysis activity of CAZ (Shen et al., 2017). Hence, we considered the enhanced KPC expression was responsible for CZA resistance in XDX51.
Nonfunctional RamR can lead to TGC resistance via regulation of efflux pumps. Mutations in ramR, including deletion, mutation and insertion, resulting in TGC resistance have been previously reported (Hentschke et al., 2010; Ye et al., 2017). In XDX51, ramR insertion caused by ISKpn14 was identified through whole-genome sequencing. It was interrupted at 12 bp, affecting the translation of N-terminal amino acids. The RamR N-terminus acts as a DNA-binding site (Yamasaki et al., 2013). Without its DNA binding function, RamR is not able to inhibit RamA expression, and as a result, the efflux pump AcrAB-TolC is upregulated. As we speculated, qRT-PCR showed upregulated expression levels of ramA, acrA and acrB compared to that in the baseline strains. Moreover, efflux pump inhibition experiments with PAβN demonstrated the role of AcrAB-TolC in the increase in the TGC MIC in XDX51. Briefly, in our study, the dysregulation of RamR caused by IS element insertion was a major molecular mechanism for TGC resistance in XDX51.
In addition to antibiotic resistance-related genomic differences, LPS phenotype-related gene mutations occurred during within-host evolution. The 307 bp wbbL gene in XDX51 was interrupted by ISKpn26, which shares 99% amino acid similarity with IS5. WbbL is a rhamnosyl transferase that transfers L-Rha residues to the O4 position of D-Glc or D-GlcNAc to obtain a complete O-antigen (Erbing et al., 1977; Izquierdo et al., 2003). O-antigen typing showed that XDX16 and XDX51 belonged to OL101 and that wbbL was an important component of the OL101 locus (Follador et al., 2016). LPS analysis suggested that the O-antigen in the XDX51 and wbbL knockout strains was shorter than that in the wbbL wild-type strain. In addition, the TGC MIC for the reconstructed wbbL defective mutant was 2-fold higher than that for the strain with the complete wbbl sequence. Leth K et al. reported that wbbL mutations could cause serotype shifts and MIC changes for mecillinam in combination with blaCTX-M mutations in E. coli. A similar study also reported that mutations in LPS-related genes could cause antibiotic resistance (Anton, 1995; Leth et al., 2019). In our study, the wbbL mutation had a slight effect on TGC susceptibility, with a twofold change in the MIC. Our result was consistent with Linkevicius’s report showing that defects in LPS-related genes (rfaC, rfaE, lpcA) could cause low levels of TGC resistance (Linkevicius et al., 2016).
In our study, the Insertion sequences played an important role in phenotypic changes in antibiotic resistance and genomic variation during within-host evolution, causing gene devitalization and duplication. Antibiotic resistance caused by ISs has been previously reported (Yang et al., 2020). Under long-term antibiotic stress, genomic variation caused by ISs could help the host overcome environmental challenges. Compared to XDX16 and XDX31, longer-term usage of the β-lactam-β-lactamase inhibitor and TGC led to the ISKpn14 insertion in wbbl, ISKpn26 insertion in ramR and replicative IS26 transposition with the blaKPC-2 transposon structure. The ISs above were widely distributed in the genomes of the baseline strains, and their movement in the genome led to key antibiotic resistance. Furthermore, composite transposons with two identical ISs, such as IS26 on the blaKPC-2-bearing plasmid in our study, can become mobilized and transferable among different strains. The dissemination of antibiotic resistance genes could cause great challenges to public health (Partridge et al., 2018). In the future, more clinical isolates would be studied to investigate the characteristic of CRKP that are prone to develop last line antibiotic resistance.
Conclusions
In brief, we tracked how a CRKP strain developed TGC and CZA resistance phenotypes during within-host antibiotic treatment. Three different genomic mutations were identified in XDX51, and all of them were caused by Insertion sequences. Double copies of blaKPC-2 contributed to the CZA MIC changes due to enhanced ceftazidime hydrolysis. CZA resistance occurred without previous exposure to CZA. In addition, ramR inactivation led to TGC resistance. Furthermore, inactivation of wbbL was identified as being associated with LPS O-antigen deficiency and slightly reduced the susceptibility to TGC. The ST4496 clone needs attention, as it has the potential to evolve TGC and CZA resistance rapidly under TGC and β-lactam antibiotic exposure.
Data Availability Statement
The datasets presented in this study can be found in online repositories. This Whole Genome Shotgun project has been deposited at GenBank under the accession JAIWPV000000000-JAIWPW000000000.
Ethics Statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
XHa and QS conceived the idea and designed the experiments. XHa and YM performed the experiments. XHa analyzed the data. JQ, PZ, and PL helped with materials and reagents. XHa wrote the manuscript. YJ, DZ, XW, XHu, and YY reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the China National Natural Science Foundation grant (81830069).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.757470/full#supplementary-material
References
Antinori, E., Unali, I., Bertoncelli, A., Mazzariol, A. (2020). Klebsiella Pneumoniae Carbapenemase (KPC) Producer Resistant to Ceftazidime–Avibactam Due to a Deletion in the Blakpc3 Gene. Clin. Microbiol. Infect. 26, 946.e1–946.e3. doi: 10.1016/j.cmi.2020.02.007
Anton, D. N. (1995). Resistance to Mecillinam Produced by the Co-Operative Action of Mutations Affecting Lipopolysaccharide, Spot, and Cya or Crp Genes of Salmonella Typhimurium Typhimurium. Mol. Microbiol. 16, 587–595. doi: 10.1111/j.1365-2958.1995.tb02421.x
Barnes, M. D., Winkler, M. L., Taracila, M. A., Page, M. G., Desarbre, E., Kreiswirth, E. B. N., et al (2017). Klebsiella Pneumoniae Carbapenemase-2 (KPC-2), Substitutions at Ambler Position Asp179, and Resistance to Ceftazidime- Avibactam: Unique Antibiotic-Resistant Phenotypes Emerge From -Lactamase Protein Engineering Melissa. MBio 2, 1–17. doi: 10.1128/mBio.00528-17
Barrick, D. E., Deatherage, J. E. (2014). Identification of Mutations in Laboratory Evolved Microbes From Next-Generation Sequencing Data Using Breseq. Methods Mol. Biol. 1151, 1–22. doi: 10.1007/978-1-4939-0554-6
Barton, B. M., Harding, G. P., Zuccarelli, A. J. (1995). A General Method for Detecting and Sizing Large Plasmids. Anal. Biochem. 226, 235–240. doi: 10.1006/abio.1995.1220
Davis, M. R., Goldberg, J. B. (2012). Purification and Visualization of Lipopolysaccharide From Gram-Negative Bacteria by Hot Aqueous-Phenol Extraction. J. Vis. Exp. 1, 3196. doi: 10.3791/3916
Duin, C. D., Infectious, D., Carolina, N., Farm, M., Kaye, K. S., Kalayjian, R. C., et al. (2014). Tigecycline Therapy for Carbapenem-Resistant Klebsiella Pneumoniae ( CRKP ) Bacteriuria Leads to Tigecycline Resistance. Clin. Microbiol. Infect. 20, O1117–O1120. doi: 10.1111/1469-0691.12714
Erbing, C., Lindberg, B., Lönngren, J. (1977). Structural Studies on the Klebsiella O Group 12 Lipopolysaccharide. Carbohydr. Res. 56, 1–5.
Follador, R., Heinz, E., Wyres, K. L., Ellington, M. J., Kowarik, M., Holt, K. E., et al. (2016). The Diversity of Klebsiella Pneumoniae Surface Polysaccharides. Microb. Genomics 2, e000073. doi: 10.1099/mgen.0.000073
Galani, I., Antoniadou, A., Karaiskos, I., Kontopoulou, K., Giamarellou, H., Souli, M. (2019). Genomic Characterization of a KPC-23-Producing Klebsiella Pneumoniae ST258 Clinical Isolate Resistant to Ceftazidime-Avibactam. Clin. Microbiol. Infect. 25, 763.e5–763.e8. doi: 10.1016/j.cmi.2019.03.011
Hentschke, M., Wolters, M., Sobottka, I., Rohde, H., Aepfelbacher, M. (2010). ramR Mutations in Clinical Isolates of Klebsiella Pneumoniae With Reduced Susceptibility to Tigecycline. Antimicrob. Agents Chemother. 54, 2720–2723. doi: 10.1128/AAC.00085-10
Holt, K. E., Wertheim, H., Zadoks, R. N., Baker, S., Whitehouse, C. A., Dance, D. (2015). Genomic Analysis of Diversity, Population Structure, Virulence, and Antimicrobial Resistance in Klebsiella Pneumoniae, an Urgent Threat to Public Health. Proc. Natl. Acad. Sci. U.S.A. 112, E3574–E3581. doi: 10.1073/pnas.1501049112
Huang, T. W., Lam, I., Chang, H. Y., Tsai, S. F., Palsson, B. O., Charusanti, P. (2014). Capsule Deletion via a λ-Red Knockout System Perturbs Biofilm Formation and Fimbriae Expression in Klebsiella Pneumoniae MGH 78578. BMC Res. Notes 7, 1–13. doi: 10.1186/1756-0500-7-13
Humphries, R. M., Hemarajata, P. (2017). Resistance to Ceftazidime-Avibactam in Klebsiella Pneumoniae Due to Porin Mutations & the Increased Expression of KPC-3. Antimicrob. Agents Chemother. 61, e00537–e00517. doi: 10.1128/AAC.00537-17
Izquierdo, L., Merino, S., Regue, M., Rodriguez, F., Toma, J. M. (2003). Synthesis of a Klebsiella Pneumoniae O-Antigen Heteropolysaccharide ( O12 ) Requires an ABC 2 Transporter. J. Bacteriol. 185, 1634–1641. doi: 10.1128/JB.185.5.1634-1641.2003
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., Phillippy, A. M. (2017). Canu : Scalable and Accurate Long-Read Assembly via Adaptive K-Mer Weighting and Repeat Separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Leth, K., Katrine, N., Hansen, H., Boye, J., Jenny, N., Knudsen, D., et al. (2019). Mutational Change of CTX - M - 15 to CTX - M - 127 Resulting in Mecillinam Resistant Escherichia Coli During Pivmecillinam Treatment of a Patient. Microbiologyopen 8, e941. doi: 10.1002/mbo3.941
Lin, Y., Huang, Y., Huang, H., Yang, T., Wang, F. (2016). International Journal of Antimicrobial Agents In Vivo Evolution of Tigecycline-Non-Susceptible Klebsiella Pneumoniae Strains in Patients: Relationship Between Virulence and Resistance. Int. J. Antimicrob. Agents 48, 485–491. doi: 10.1016/j.ijantimicag.2016.07.008
Linkevicius, M., Anderssen, J. M., Sandegren, L., Andersson, D. I. (2016). Fitness of Escherichia Coli Mutants With Reduced Susceptibility to Tigecycline. J. Antimicrob. Chemother. 71, 1307–1313. doi: 10.1093/jac/dkv486
Livak, K. J., Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Livermore, D. M., Maya, J. J., Nordmann, P., Wang, H., Woodford, N., Quinn, J. P. (2015a). Clinical Epidemiology of the Global Expansion of Klebsiella Pneumoniae Carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/S1473-3099(13)70190-7.Clinical
Livermore, D. M., Warner, M., Jamrozy, D., Mushtaq, S., Nichols, W. W., Mustafa, N., et al. (2015b). In Vitro Selection of Ceftazidime-Avibactam Resistance in Enterobacteriaceae With KPC-3 Carbapenemase. Antimicrob. Agents Chemother. 59, 5324–5330. doi: 10.1128/AAC.00678-15
Moradigaravand, D., Martin, V., Peacock, S. J., Parkhill, J. (2017). Evolution and Epidemiology of Multidrug-Resistant Klebsiella Pneumoniae in the United Kingdom and Ireland. MBio 8, 1–13. doi: 10.1128/mBio.01976-16
Partridge, S. R., Kwong, S. M., Firth, N., Jensen, S. O. (2018). Mobile Genetic Elements Associated With Antimicrobial Resistance. Clin. Microbiol. Rev. 31, e00088–e00017. doi: 10.1128/CMR.00088-17
Rice, L. B. (2008). Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens : No ESKAPE. J. Infect. Dis. 197, 1079–1081. doi: 10.1086/533452
Salehi, B., Ghalavand, Z., Yadegar, A., Eslami, G. (2021). Characteristics and Diversity of Mutations in Regulatory Genes of Resistance-Nodulation-Cell Division Efflux Pumps in Association With Drug-Resistant Clinical Isolates of Acinetobacter Baumannii. Antimicrob. Resist. Infect. Control 10, 1–12. doi: 10.1186/s13756-021-00924-9
Seemann, T. (2014). Prokka : Rapid Prokaryotic Genome Annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shen, Z., Ding, B., Ye, M., Wang, P., Bi, Y., Wu, S., et al. (2017). High Ceftazidime Hydrolysis Activity and Porin OmpK35 Deficiency Contribute to the Decreased Susceptibility to Ceftazidime/Avibactam in KPC-Producing Klebsiella Pneumoniae. J. Antimicrob. Chemother. 72, 1930–1936. doi: 10.1093/jac/dkx066
Shen, P., Wei, Z., Jiang, Y., Du, X., Ji, S., Yu, Y., et al. (2009). Novel Genetic Environment of the Carbapenem-Hydrolyzing Beta-Lactamase KPC-2 Among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53, 4333–4338. doi: 10.1128/AAC.00260-09
Shields, R. K., Chen, L., Cheng, S., Chavda, K. D., Press, E. G. (2017). Emergence of Ceftazidime-Avibactam Resistance Due to Plasmid-Borne blaKPC-3 Mutations During Treatment of Carbapenem-Resistant Klebsiella Pneumoniae Infections. Antimicrob. Agents Chemother. 61, 1–11.
Shirley, M. (2018). Ceftazidime-Avibactam: A Review in the Treatment of Serious Gram-Negative Bacterial Infections. Drugs 78, 675–692. doi: 10.1007/s40265-018-0902-x
Shrivastava, S. R., Shrivastava, P. S., Ramasamy, J. (2018). World Health Organization Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. J. Med. Soc. 32, 76–77. doi: 10.4103/jms.jms_25_17
Spiliopoulou, I., Kazmierczak, K., Stone, G. G. (2020). In Vitro Activity of Ceftazidime/Avibactam Against Isolates of Carbapenem-Non-Susceptible Enterobacteriaceae Collected During the INFORM Global Surveillance Programm-17. J. Antimicrob. Chemother. 75, 384–391. doi: 10.1093/jac/dkz456
Ullmann, U. (1998). Klebsiella Spp. As Nosocomial Pathogens : Epidemiology, Taxonomy, Typing Methods, and Pathogenicity Factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/CMR.11.4.589
Van Duin, D., Lok, J. J., Earley, M., Cober, E., Richter, S. S., Perez, F., et al. (2018). Colistin Versus Ceftazidime-Avibactam in the Treatment of Infections Due to Carbapenem-Resistant Enterobacteriaceae. Clin. Infect. Dis. 66, 163–171. doi: 10.1093/cid/cix783
Veleba, M., Schneiders, T. (2012). Tigecycline Resistance Can Occur Independently of the ramA Gene in Klebsiella Pneumoniae. Antimicrob. Agents Chemother. 56, 4466–4467. doi: 10.1128/AAC.06224-11
Villa, L., Feudi, C., Fortini, D., García-fernández, A., Carattoli, A. (2014). Genomics of KPC-Producing Klebsiella Pneumoniae Sequence Type 512 Clone Highlights the Role of RamR and Ribosomal S10 Protein Mutations in Conferring Tigecycline Resistance. Antimicrob. Agents Chemother. 58, 1707–1712. doi: 10.1128/AAC.01803-13
Yamasaki, S., Nikaido, E., Nakashima, R., Sakurai, K., Fujiwara, D., Fujii, I., et al. (2013). The Crystal Structure of Multidrug-Resistance Regulator RamR With Multiple Drugs. Nat. Commun. 4, 2078. doi: 10.1038/ncomms3078
Yang, T. Y., Wang, S. F., Lin, J. E., Griffith, B. T. S., Lian, S. H., Hong, Z., et al. (2020). Contributions of Insertion Sequences Conferring Colistin Resistance in Klebsiella Pneumoniae. Int. J. Antimicrob. Agents 55, 105894. doi: 10.1016/j.ijantimicag.2020.105894
Ye, M., Ding, B., Qian, H., Xu, Q., Jiang, J., Huang, J., et al. (2017). In Vivo Development of Tigecycline Resistance in Klebsiella Pneumoniae Owing to Deletion of the ramR Ribosomal Binding Site. Int. J. Antimicrob. Agents 50, 523–528. doi: 10.1016/J.IJANTIMICAG.2017.04.024
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: carbapenem-resistant K. pneumoniae, ceftazidime/avibactam, tigecycline, in vivo, insertion sequences
Citation: Han X, Shi Q, Mao Y, Quan J, Zhang P, Lan P, Jiang Y, Zhao D, Wu X, Hua X and Yu Y (2021) Emergence of Ceftazidime/Avibactam and Tigecycline Resistance in Carbapenem-Resistant Klebsiella pneumoniae Due to In-Host Microevolution. Front. Cell. Infect. Microbiol. 11:757470. doi: 10.3389/fcimb.2021.757470
Received: 12 August 2021; Accepted: 08 October 2021;
Published: 25 October 2021.
Edited by:
Ziad Daoud, Central Michigan University, United StatesReviewed by:
Kai Zhou, First Affiliated Hospital of Southern University of Science and Technology, ChinaThomas Jové, INSERM U1092 Anti-Infectieux Supports Moléculaires des Résistances et Innovations Thérapeutiques, France
Copyright © 2021 Han, Shi, Mao, Quan, Zhang, Lan, Jiang, Zhao, Wu, Hua and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunsong Yu, yvys119@zju.edu.cn
†These authors have contributed equally to this work
 Xinhong Han
Xinhong Han Qiucheng Shi
Qiucheng Shi Yihan Mao1,2,3
Yihan Mao1,2,3  Jingjing Quan
Jingjing Quan Peng Lan
Peng Lan Yan Jiang
Yan Jiang Dongdong Zhao
Dongdong Zhao Xueqing Wu
Xueqing Wu Xiaoting Hua
Xiaoting Hua Yunsong Yu
Yunsong Yu