The LAMMER Kinase, LkhA, Affects Aspergillus fumigatus Pathogenicity by Modulating Reproduction and Biosynthesis of Cell Wall PAMPs
- 1Laboratory of Cellular Differentiation, Department of Microbiology and Molecular Biology, College of Bioscience and Biotechnology, Chungnam National University, Daejeon, South Korea
- 2Institute of Biotechnology, Chungnam National University, Daejeon, South Korea
- 3Laboratory of Developmental Genetics, Department of Biology, College of Bioscience and Biotechnology, Chungnam National University, Daejeon, South Korea
The LAMMER kinase in eukaryotes is a well-conserved dual-specificity kinase. Aspergillus species cause a wide spectrum of diseases called aspergillosis in humans, depending on the underlying immune status of the host, such as allergy, aspergilloma, and invasive aspergillosis. Aspergillus fumigatus is the most common opportunistic fungal pathogen that causes invasive aspergillosis. Although LAMMER kinase has various functions in morphology, development, and cell cycle regulation in yeast and filamentous fungi, its function in A. fumigatus is not known. We performed molecular studies on the function of the A. fumigatus LAMMER kinase, AfLkhA, and reported its involvement in multiple cellular processes, including development and virulence. Deletion of AflkhA resulted in defects in colonial growth, production of conidia, and sexual development. Transcription and genetic analyses indicated that AfLkhA modulates the expression of key developmental regulatory genes. The AflkhA-deletion strain showed increased production of gliotoxins and protease activity. When conidia were challenged with alveolar macrophages, enodocytosis of conidia by macrophages was increased in the AflkhA-deletion strain, resulting from changes in expression of the cell wall genes and thus content of cell wall pathogen-associated molecular patterns, including β-1,3-glucan and GM. While T cell-deficient zebrafish larvae were significantly susceptible to wild-type A. fumigatus infection, AflkhA-deletion conidia infection reduced host mortality. A. fumigatus AfLkhA is required for the establishment of virulence factors, including conidial production, mycotoxin synthesis, protease activity, and interaction with macrophages, which ultimately affect pathogenicity at the organismal level.
Introduction
Aspergillus fumigatus is an opportunistic fungal pathogen that causes aspergillosis in immunocompromised individuals. Human infections caused by Aspergillus can range from allergenic disease to severe invasive aspergillosis (IA). A. fumigatus can grow and reproduce in soil, air dust, and decompose organic matter as a saprotroph. Conidia (2–3 μm), which are asexual spores, are sufficient to bypass mucociliary clearance and reach the alveoli in the lungs (Abad et al., 2010). While clearance of inhaled conidia occurs as a result of a highly coordinated immune response in immunocompetent hosts, conidia can efficiently adapt their physiology to the altered host environment in immunocompromised individuals. Epithelial barriers and innate immune cells, such as neutrophils, macrophages, and dendritic cells, play important roles in the removal of fungal invaders (Osherov, 2012). To recognize a pathogen, host cells contain pattern recognition receptors (PRRs), including Toll-like receptors (TLRs), C-type lectin receptors (CLRs), and NOD-like receptors (NLRs) (Van De Veerdonk et al., 2017). PRRs interact with pathogen-associated molecular patterns (PAMPs), which are usually absent from the host as essential structural components of pathogens (Hatinguais et al., 2020). Fungal cell wall components in conidia and hyphae are characteristic PAMPs, including glucan, chitin, and galactomannan (GM) (Marcos et al., 2016). Conidia have a protective outer layer comprised of hydrophobins and melanin and shed this outer layer during germination, exposing PAMPs to PRRs (Aimanianda and Latgé, 2010; Perez-Cuesta et al., 2020). The innate immune system induced by PAMP-PRR interaction causes phagocytosis, killing of fungi, and cytokine signaling for activation of the adaptive immune response. Stimulated macrophages cause phosphorylation and activation of mitogen-activated protein kinases (MAPKs) and produce pro-inflammatory cytokines and chemokines, such as tumor necrosis factor-α (TNF‐α) and interleukins (IL-1α, IL-6, and IL-8) (Rao, 2001; Dubourdeau et al., 2006; Dagenais and Keller, 2009). TNF-α enhances the host immune response by augmenting the phagocytic potential of phagocytes and by regulating the growth and differentiation of a wide range of cells.
Fungal products may contribute to fungal pathogenicity in immunocompromised hosts by evading or modulating host defenses. Several molecules secreted by A. fumigatus can inhibit the phagocytic activity of the host, including galactosaminogalactan (GAG), mycotoxins, such as fumagillin and gliotoxin (GT), and hydrolytic enzymes. GAG is a specific carbohydrate polymer that consists of galactose, galactosamine, and N-acetylgalactosamine, and is expressed by hyphae and secreted (Van De Veerdonk et al., 2017). GAG plays crucial roles in the adherence of hyphae to surfaces and formation of biofilms, can induce neutrophil apoptosis and increase resistance to neutrophil extracellular traps (NETs) (Fontaine et al., 2011; Lee et al., 2015). Mycotoxin fumagillin and GT can decrease the function of neutrophils and macrophages (Fallon et al., 2010; Schlam et al., 2016). While less than 20% of the environmental isolates produced GT, approximately 93% of A. fumigatus strains recovered from cancer patients with IA produced GT (Lewis et al., 2005; Sugui et al., 2007). A. fumigatus produces a wide range of proteases and hydrolytic enzymes to catalyze macromolecules, which enables the fungus to grow and survive in distinct environments (de Vries and Visser, 2001). The A. fumigatus genome contains more than 100 proteases (Watson et al., 2011). Alkaline protease 1 (Alp1) is the most abundant protein secreted by A. fumigatus and is detected in the airways of patients with asthma, but not in healthy individuals (Balenga et al., 2015).
LAMMER kinase is a dual-specificity kinase that phosphorylates serine/threonine and tyrosine residues (Yun et al., 1994). LAMMER kinases containing a conserved motif, ‘EHLAMMERILG,’ in sub-domain X of the kinase catalytic domain, are highly conserved in eukaryotes and have multiple functions in various physiological processes: growth, cell wall biogenesis, stress response, cell division, and differentiation (Lim and Park, 2019b). In contrast to the bacterial pathogen, A. fumigatus undergoes morphological transition in the host environment during infection: inert conidia swelling, germination, and growth into lung tissue, until finally undergoing dissemination. Various signaling pathways are involved in the morphogenesis and development of fungi. We investigated the multifunctional role of LAMMER kinase in morphogenetic plasticity and development, which are intimately coupled with virulence in pathogenic fungi.
To investigate the cellular function of LAMMER kinase in A. fumigatus, we cloned AflkhA, which is a homolog of Aspergillus nidulans AnlkhA. Deletion of AflkhA resulted in defects in colonial growth, conidia production, sexual development, a decreased ability to form biofilms, and increased protease activity and GT production. Additionally, deletion of AflkhA resulted in increased susceptibility to phagocytosis by alveolar macrophages and increased survival rate in a T cell development-deficient zebrafish infection assay. Here, we present, for the first time, that LAMMER kinase is involved in the development and virulence of the fungal pathogen A. fumigatus.
Results
AfLkhA Is Required for Colonial Growth and Asexual Development in A. fumigatus
We performed BLASTP analysis of the A. fumigatus genome database using A. nidulans AnLkhA as a query and identified AfLkhA (AFUA_1G16780) as the best hit (identity = 81%). AfLkhA is the only LAMMER kinase ortholog in A. fumigatus and contains 664 amino acids (73 kDa). Most of the LAMMER kinase orthologs in Aspergillus, except Aspergillus glaucus and Aspergillus terreus, include “EHLAMMEAVIG” as a LAMMER motif in the catalytic domain (Supplementary Figure 1). The catalytic domain of AfLkhA comprises 3rd–6th exons. With a high identity, AfLkhA might have functions similar to those of AnLkhA.
To characterize the functions of AfLkhA, we generated the AflkhA-deletion (ΔAflkhA) strain by replacing its open reading frame (ORF) with the ptrA marker in the AFIR928 strain (MAT1-2), which is a supermater strain. The ΔAflkhA strain was viable but showed reduced growth on agar medium (Figures 1A, B). There was no remarkable difference in the dry weight of liquid cultures among the wild type (WT), ΔAflkhA, and C’AflkhA strains (Figure 1C).
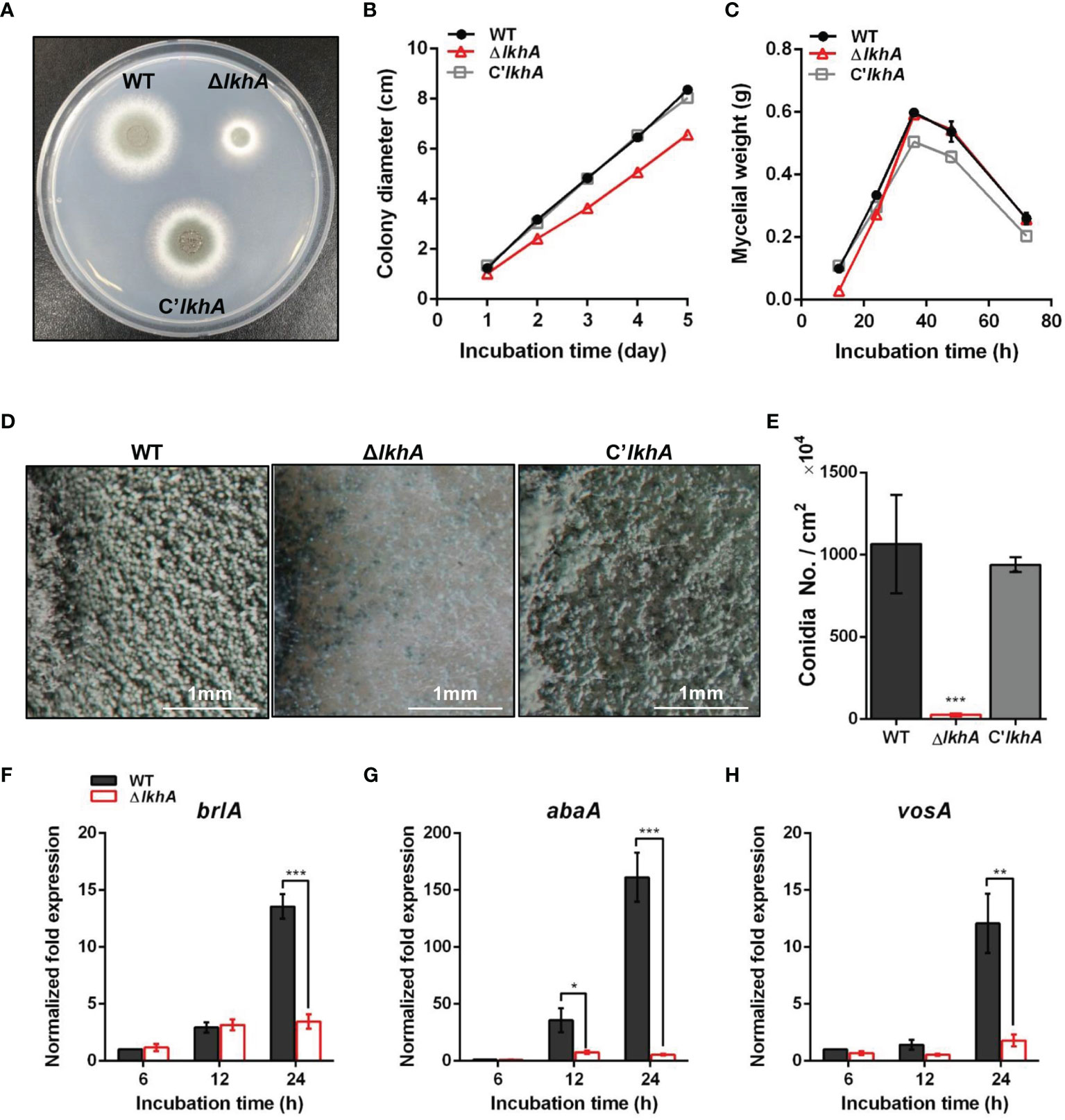
Figure 1 Vegetative growth and asexual development of ΔAflkhA strain. (A) Colony morphology of WT, ΔAflkhA, and C’AflkhA strains. Spores were inoculated onto GMM agar and incubated at 37°C for two days. (B) Radial growth of WT, ΔAflkhA, and C’AflkhA strains. Colony diameter of each strain was measured daily from point of inoculation over five days. When error bars are not shown, they are smaller than symbols (n = 5). (C) Mycelial production on liquid YCGMM. Dry weight of each strain was measured. When error bars are not shown, they are smaller than symbols (n = 3). (D) Conidiophore heads on GMM agar. (E) Quantification of conidia production (n = 4). *P < 0.05. (F-H) Expression patterns of brlA (F), abaA (G) and vosA (H) genes. Mycelial balls of strains obtained from YCGMM liquid culture were transferred to GMM agar and incubated for indicated hrs. Total RNA was extracted and RT-qPCR analysis was performed using 18S rRNA gene as an internal control. **P < 0.01, ***P < 0.001.
AfLkhA affects both vegetative growth and asexual development. Under a stereomicroscope, there was a reduction in the formation of conidiophores on the ΔAflkhA colony, compared to those of the WT (Figure 1D). In agreement with the phenotypic data, statistical analysis revealed an approximately 98% decrease in conidiospore production in the ΔAflkhA strain (Figure 1E). BrlA, AbaA, and WetA constitute the backbone of the central regulatory pathway (CRP) and regulate asexual development in Aspergillus (Etxebeste et al., 2010; Yu, 2010). Mycelia balls produced in liquid YCGMM were shifted to solid GMM to induce asexual development, and total RNA was prepared from the cultures at the indicated time post-induction. When we investigated the expression levels of CRP genes brlA and abaA, the ΔAflkhA strain showed significantly reduced transcript levels of brlA at 24 hr, and abaA at 12 and 24 hrs (Figures 1F, G). VosA is a Velvet family protein, which plays an important role in spore viability (Park et al., 2012). The level of vosA expression in the ΔAflkhA strain was reduced (Figure 1H). Taken together, these results indicate that AflkhA plays a pivotal role in the production of conidiophores. To elucidate the cellular function of LAMMER kinase in stress response, spotting analysis was performed on agar containing several stress chemicals. No significant changes were observed in the sensitivity to cell wall stress (calcofluor white and Congo red), oxidative stress (H2O2 and menadione), osmotic stress (KCl and NaCl), ion depletion stress (EDTA and EGTA), and endoplasmic reticulum stress (tunicamycin and dithiothreitol) (Supplementary Figure 2).
AfLkhA Is Required for Sexual Development
LAMMER kinase is involved in both asexual and sexual development in A. nidulans (Kang et al., 2013). To investigate the effect of AflkhA-deletion on sexual development in A. fumigatus, crosses were induced by the VeM method on oatmeal agar under sealed and dark conditions (Lim and Park, 2019a). Crossing of AFB62 strains with AFIR928 (WT × WT) and with C’AflkhA (WT × C’lkhA) produced cleistothecia, sexual fruiting bodies, containing a cloud of ascospores (Figure 2A, arrow). The crossing of the WT strain with the ΔAflkhA strain (WT × ΔlkhA) produced similar sizes of cleistothecium (Supplementary Figure 3). The number of cleistothecia (4 ± 2 cleistothecia/mm2) was significantly lower than that in the WT × WT cross (189 ± 17 cleistothecia/mm2) and the WT × C’lkhA cross (120 ± 20 cleistothecia/mm2) (Figure 2B). To investigate the formation of ascospores, we picked up cleistothecia and crushed them on glass slides. While the WT × WT cross produced mature ascospores with normal equatorial crests, the WT × ΔlkhA cross produced abnormal ascospores with no equatorial crests (Figure 2C).
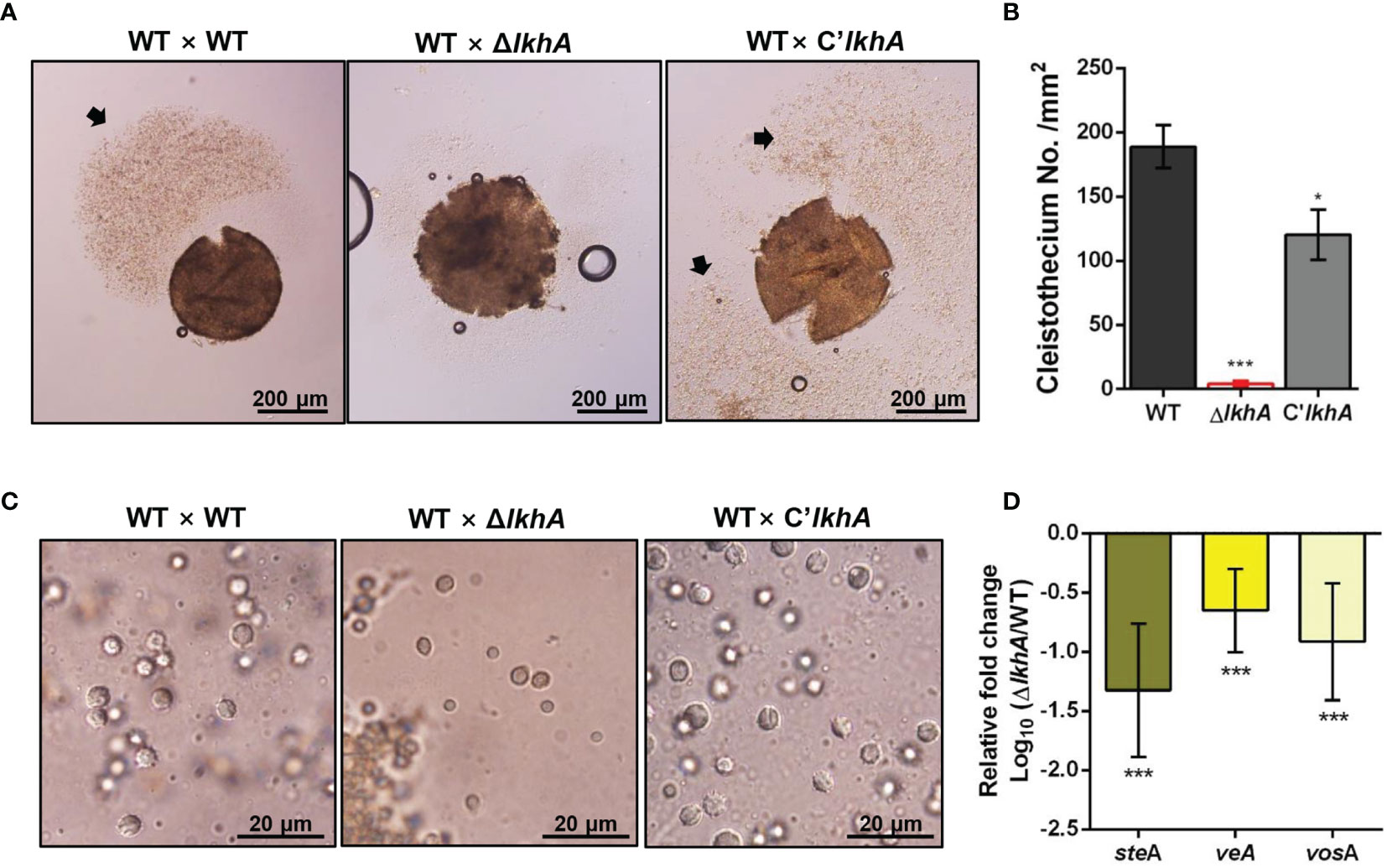
Figure 2 Role of AfLkhA in sexual development. (A) Morphology of the sexual fruiting body, cleistothecium, in WT, ΔAflkhA, and C’AflkhA strains. Mycelial balls were moved to oatmeal agar and incubated at 30°C for 14 days under dark conditions to induce sexual development. Arrow indicates a cloud of ascospores sprayed from cleistothecium. (B) Quantification of cleistothecia production (n = 5). Asterisk represents significant differences: *P < 0.05, ***P < 0.001. (C) Morphology of ascospores. Cleistothecium was ruptured on a glass slide to observe ascospores. (D) Relative expression patterns of steA, veA and vosA. Total RNA was extracted from cleistothecia. RT-qPCR analysis was performed using 18S rRNA gene as an internal control. The x-axis shows a list of genes. The y-axis indicate the relative mRNA abundance of the genes in ΔAflkhA strain compared to WT. The values were transformed and presented as Log10. ***P < 0.001.
Next, we investigated the gene expression patterns in cleistothecia and sexual fruiting bodies. When A. fumigatus sexual development is induced by the VeM method, both asexual and sexual developmental reproductive organs are observed on oatmeal agar (Lim and Park, 2019a). To investigate changes in gene expression for only sexual development, we picked up ten cleistothecia from each crossing and extracted total RNAs from these sexual fruiting bodies. SteA is a transcription factor that controls filamentation and sexual development in A. nidulans (Vallim et al., 2000). VeA is a key developmental regulator in numerous fungal species, particularly Ascomycetes (Myung et al., 2012). VeA has been demonstrated to positively regulate sexual development in various Aspergillus species, including A. nidulans, Aspergillus cristatus, Aspergillus parasiticus, and Aspergillus flavus (Kim et al., 2002; Calvo et al., 2004; Duran et al., 2006; Liu et al., 2018a). VosA is required for the integrity of both asexual and sexual spores. Levels of both gene expression and proteins levels of vosA are high in conidia and ascospores (Ni and Yu, 2007). The expression of steA, veA, and vosA genes were downregulated in the WT × ΔlkhA cross (Figure 2D). Taken together, our data suggests that AfLkhA plays an essential role in sexual development.
AfLkhA Is Required for Negative Regulation of GT
To test whether the absence of AfLkhA would affect the biosynthesis of GT, a mycotoxin considered as a putative virulence factor, the production of GT and expression of corresponding genes were investigated. Thin-layer chromatography (TLC) images showed that the ΔAflkhA strain produced an increased amount of GT compared to the WT and C’AflkhA (Figure 3A). GT is synthesized by biosynthetic gene clusters composed of 12 genes (Wang et al., 2014). We examined the expression of genes encoding GT biosynthetic genes (gliP, gliZ, and gliA). GliZ is a Zn2-Cys6 transcription factor that controls gene expression of other genes involved in GT biosynthesis (Bok et al., 2006). GliP is a nonribosomal peptide synthase that catalyzes the first step in the GT biosynthetic pathway (Sugui et al., 2007). By exporting the GT, GliA is involved in protection from extracellular GT, including its own produced toxin (Wang et al., 2014). Expression levels of gliP, gliZ, and gliA were significantly higher in the ΔAflkhA strain than those in the WT strain (Figures 3B–D). A zinc-responsive transcriptional activator ZafA binds to ZafA-binding motifs in the upstream region of gliZ (Seo et al., 2019). The expression of zafA was upregulated in the ΔAflkhA strain (Figure 3E). Taken together, the ΔAflkhA strain increases GT production by regulating expression of zafA gene and GT cluster genes, suggesting that AfLkhA negatively controls GT production.
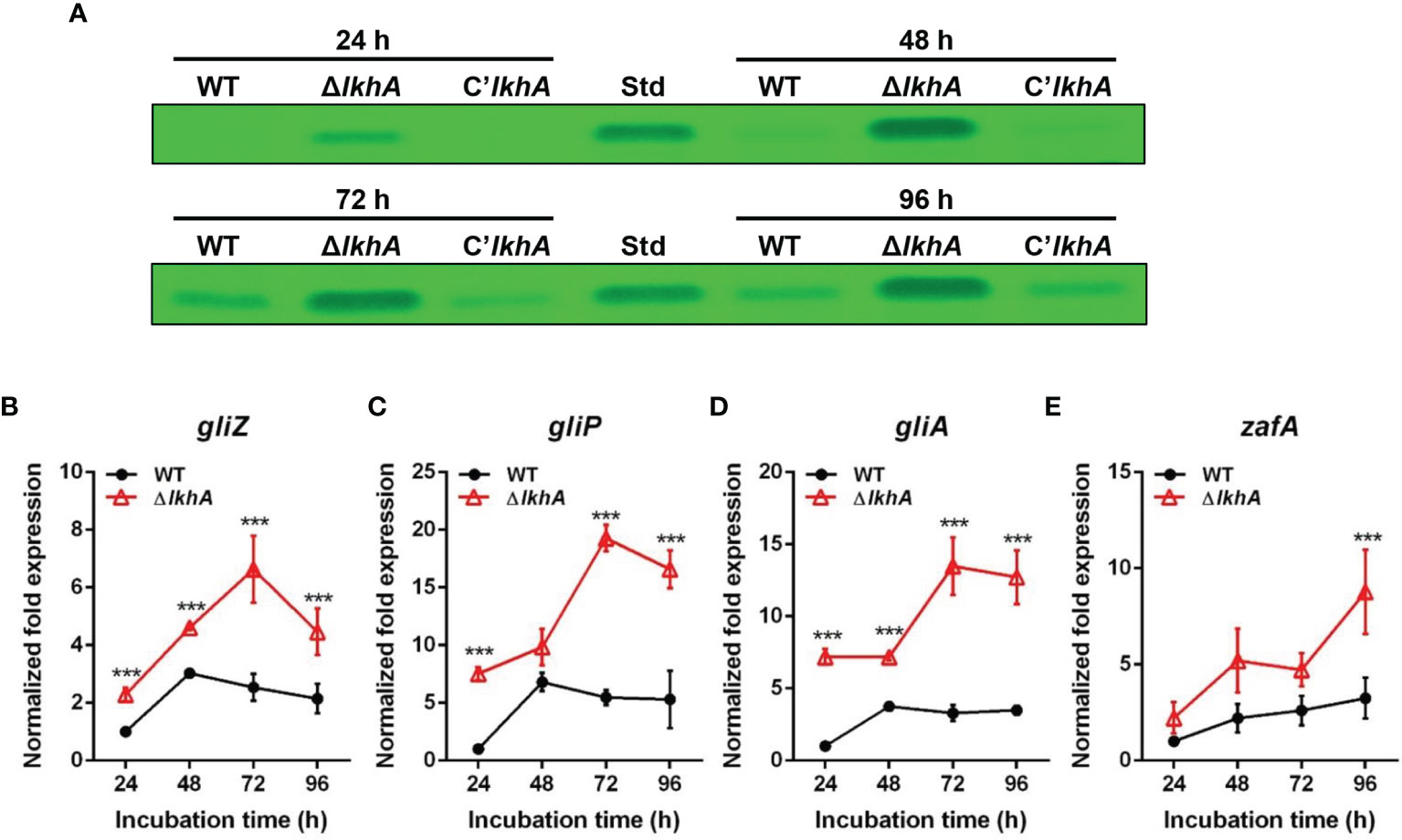
Figure 3 Role of AfLkhA in gliotoxin production. (A) Determination of GT production in WT, ΔAflkhA, and C’AflkhA strains. GT were extracted with chloroform from culture supernatant of each strain and subjected to TLC. Std: GT standard (Simga-Aldrich). (B–E) Total RNA was extracted from mycelial balls incubated in Czapek Dox broth. RT-qPCR analysis was performed using 18S rRNA gene as an internal control. Expression patterns of GT cluster genes, transcription factor gliZ (B), nonribosomal peptide synthetase gliP (C), transporter gliA (D), and transcriptional activator zafA (E). ***P < 0.001.
AfLkhA Is Involved in Protease Activity and Biofilm Formation
Hydrolytic enzymes are virulence factors that allow A. fumigatus to degrade molecules and obtain energy sources from the host environment (Richie et al., 2009). Various allergens show protease activity and cause protease-dependent release of the pro-inflammatory cytokines, IL-6, and IL-8 (Kauffman et al., 2000). We examined the possible role of AfLkhA in controlling protease activity in A. fumigatus.
When the cultures were grown on 1% skim milk agar, an increase in the proteolytic activity of the ΔAflkhA strain was visualized by degradation halos at the edge of the colonies (Figure 4A). Quantitative analysis using azocasein as a substrate detected a 37% increase in alkaline protease activity in the ΔAflkhA cultures with respect to the WT and C’AflkhA strains (Figure 4B). A. fumigatus-associated serine protease, Alp1, is the most powerful alkaline protease essential for both carbon and nitrogen acquisition (Sriranganadane et al., 2010). When the expression of the alp1 gene was determined using reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) at 72 hrs, the expression level of alp1 was higher in the ΔAflkhA strain than that in the WT strain (Figure 4C). These results suggest that AfLkhA is involved in alkaline protease activity by affecting transcriptional regulation.
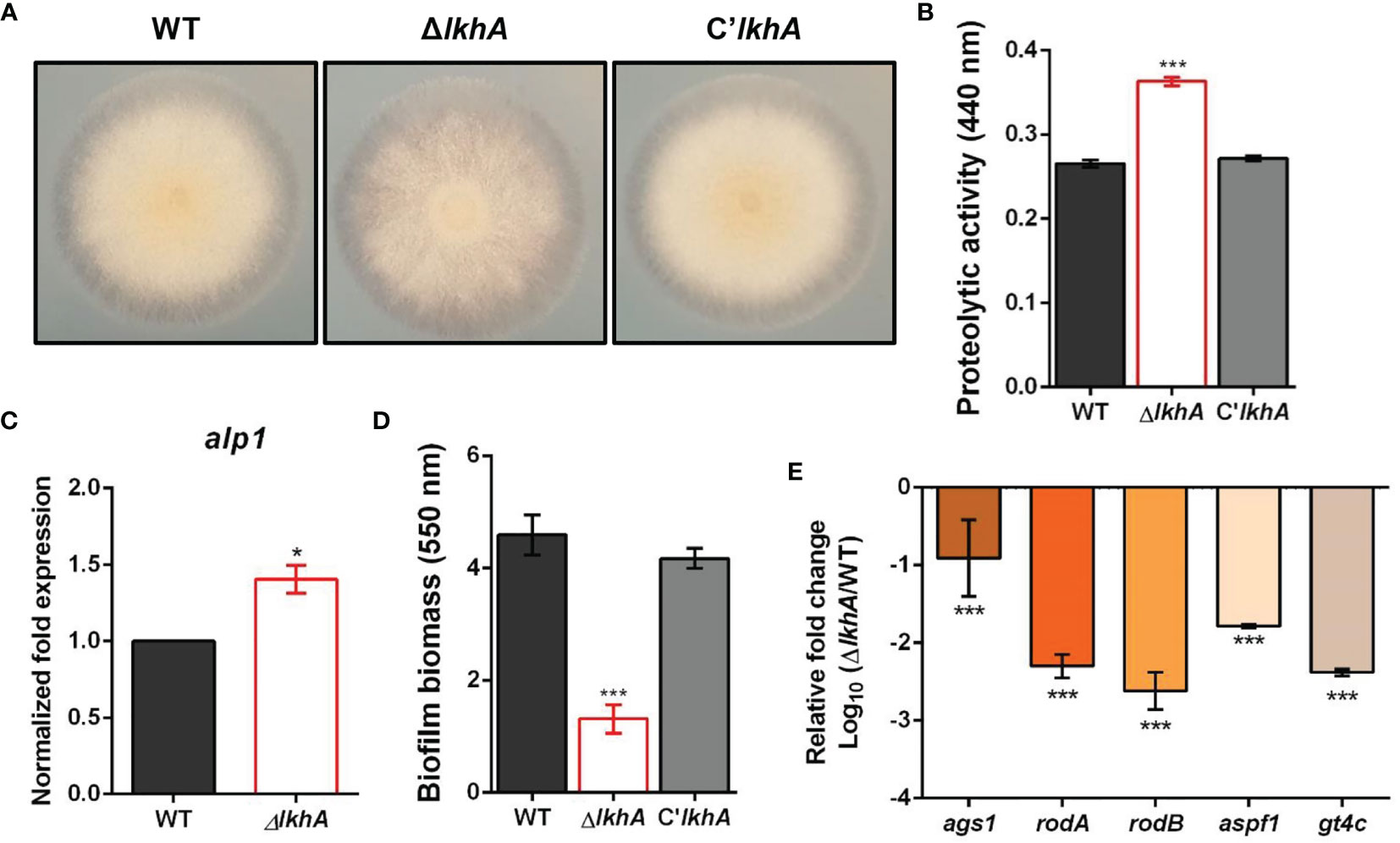
Figure 4 Roles of AfLkhA in protease production and biofilm formation. (A) Halo formed at edge of colonies of WT, ΔAflkhA, and C’AflkhA strains. Each strains were point-inoculated on Czapek Dox medium containing 1% skim milk and incubated at 30°C. (B) Quantification of proteolytic activity. Supernatants were obtained from Czapek Dox broth (pH 7.3) containing 1% skim milk powder for 3 days at 30°C. Protein concentrations were calculated with a BCA Protein Assay Kit. Protease activity was measured using an azocasein assay. The experiments included six replicates (n = 6). ***P < 0.001. (C) Expression pattern of alkaline protease alp1 gene. Total RNA was extracted from mycelial balls incubated in Czapek Dox broth. RT-qPCR analysis was performed using 18S rRNA gene as an internal control. *P < 0.05. (D) Crystal violet staining assay. Biofilm formation was measured after 16 hrs of growth in GMM. The biofilms were stained with 0.01% crystal violet and dissolved in 30% acetic acid solution. The amounts of dye were measured by spectrophotometry at 550 nm (n = 10). ***P < 0.001. (E) Relative expression patterns of α-1,3-glucan synthase (ags1), hydrophobins rodA, rodB, a ribonuclease (aspf1), and a GAG synthase (gt4c). Total RNA was extracted from biofilm cultures. RT-qPCR analysis was performed using 18S rRNA gene as an internal control. The x-axis shows a list of genes. The y-axis indicate the relative mRNA abundance of the genes in ΔAflkhA strain compared to WT. The values were transformed and presented as Log10. ***P < 0.001.
Another important feature of fungi is their ability to form biofilms, which are microbial communities attached to the surface of organic or inorganic matters. The extracellular matrix (ECM) functions in the linkage of fungal cells with themselves or the substratum during biofilm growth (Gibbons et al., 2012). Biofilm formation was determined using crystal violet, which stained the total biofilm biomass. Biofilm formation by the ΔAflkhA strain was reduced by 70% compared to that of the WT and C’lkhA strains (Figure 4D). The ECM in Aspergillus biofilm is composed of polysaccharides, pigments, and proteins, and its formation is affected by the activity of α-1,3-glucan synthases (Ags1) and hydrophobins (RodA and RodB) (Beauvais et al., 2005; Beauvais et al., 2007; Valsecchi et al., 2018). Under biofilm conditions, rodB genes are highly expressed, followed by rodA, among the seven genes for hydrophobins (RodA–RodG) (Beauvais and Latgé, 2015). Polysaccharide GAG has a role in the adherence of hyphae to surfaces, affecting biofilm formation (Fontaine et al., 2011). The gt4c gene, which encodes GAG synthase, is essential for GAG production (Briard et al., 2020). Ribonuclease Aspf1 is a secreted protein that is an antigen in the biofilm of A. fumigatus (Gastebois et al., 2013). While α-1,3-glucans and GAGs were detected in both in vivo and in vitro biofilms, the rodA, rodB, and aspf1 genes were detected only in the ECM in vitro biofilm condition (Beauvais and Latgé, 2015; Liu et al., 2018b). When the expression patterns of these genes in static biofilm conditions were analyzed by RT-qPCR analysis, the ΔAflkhA strain showed significantly decreased expression of ags1, rodA, rodB, aspf1, and gt4c (Figure 4E). These results suggest that LkhA plays a role in biofilm formation by regulating the transcription of genes that encode ECM and secreted proteins that are involved in adherence.
The Conidia of ΔAflkhA Were Susceptible to Phagocytosis by Alveolar Macrophages
To investigate whether AfLkhA is involved in the phagocytic process, macrophages were incubated with WT and ΔAflkhA conidia. ΔAflkhA conidia were more susceptible to phagocytosis than WT conidia (Figure 5A). More conidia of the ΔAflkhA strain (55%) were endocytosed by macrophages than those of the WT strain (31%) (Figure 5B). The phagocytic index (the average number of conidia per macrophage) was also greater for macrophages containing ΔAflkhA conidia (4.1 vs 1.9 c/m) (Figure 5C). Macrophages prefer to ingest ΔAflkhA conidia, suggesting that AfLkhA causes a possible change in the cell wall components of the conidia surface, which affects cell wall PAMP-PRR interactions between A. fumigatus conidia and host immune cells.
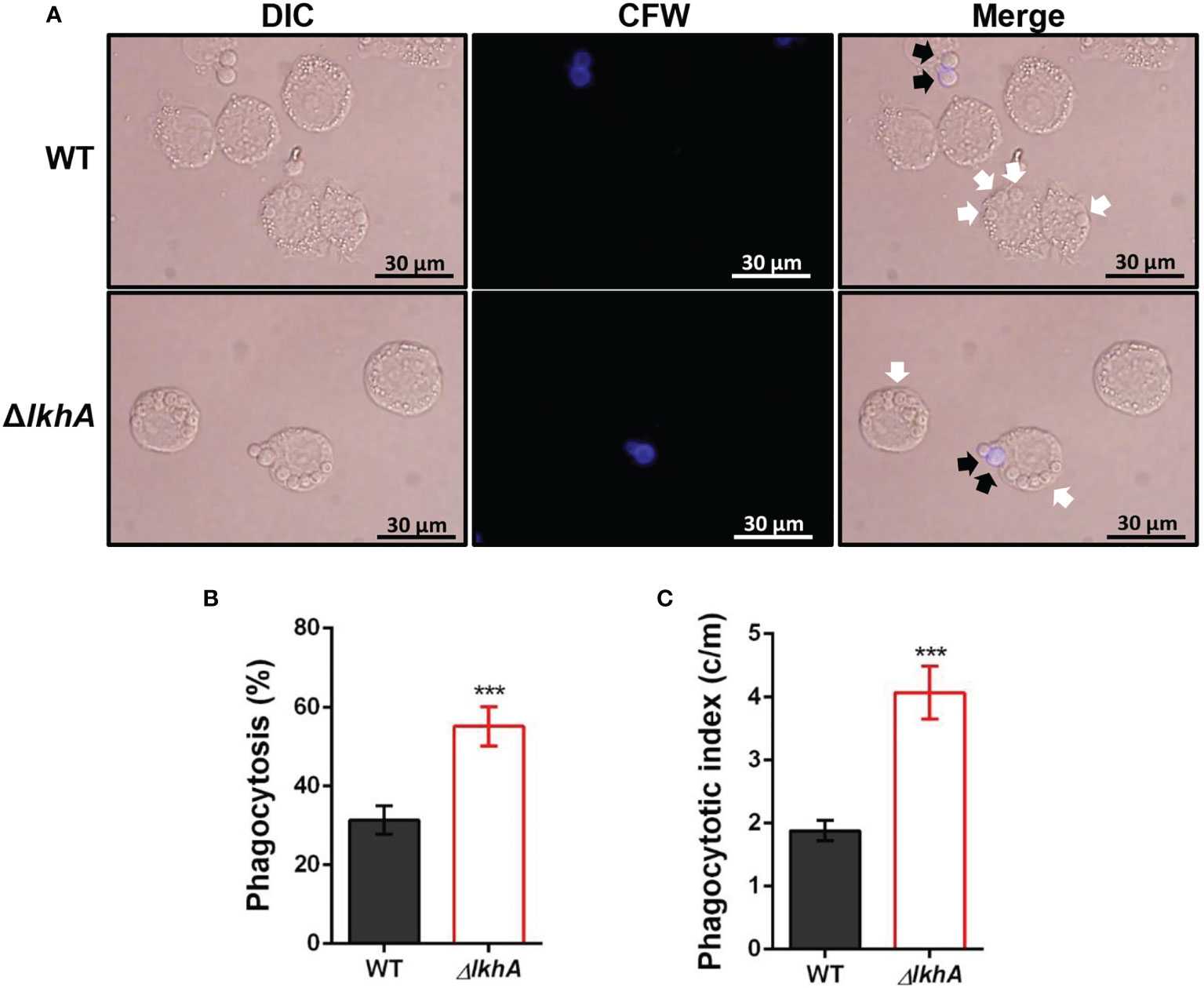
Figure 5 Alveolar macrophage response to ΔAflkhA conidia. MH-S murine alveolar macrophages were challenged with a three-fold concentration of WT and ΔAflkhA conidia and then incubated for 4 hrs at 37°C in an atmosphere of 5% CO2. (A) Microscopic analysis of the uptake of the fungal conidia by MH-S macrophages. External conidia (black arrows) were stained by calcofluor white (1 μg/mL in DPBS). White arrows indicate conidia endocytosed by macrophage cells. (B) Phagocytosis of conidia. The percentage of macrophages containing more than one ingested conidia was counted (n = 18). (C) Phagocytosis index. The average number of indigested conidia per macrophage (c/m) was calculated (n = 40). Statistical analysis was performed using the student’s t-test. ***P < 0.001.
AfLkhA Affects Content of Cell Wall PAMPs
Immune cells detect PAMPs, which are conserved molecular signatures found on the cell surface of many pathogens. To investigate the role of LAMMER kinase in the formation of the conidial cell wall, the synthesis of β-1,3-glucan was investigated. β-1,3-glucan, one of the major fungal cell wall PAMPs, is detected by Dectin-1, a C-type lectin receptor of monocytes and macrophages (Faro-Trindade et al., 2012). In A. fumigatus, the expression of fks1 (encoding β-1,3-glucan synthase) was detected in swollen conidia and was detected during mycelial growth (Gastebois et al., 2013). While the fks1 gene was consistently expressed during vegetative growth in WT, fks1 mRNA was upregulated in the ΔAflkhA strain (Figure 6A). Increased expression of fks1 gene was also detected in the ΔAflkhA conidia (Figure 6B). In accordance with the increased expression of the fks1 gene, the amount of β-1,3-glucan in the ΔAflkhA conidia was significantly higher than that observed in the WT and C’AflkhA conidia (~2-fold higher in the ΔAflkhA strain compared to the WT) (Figure 6C). Next, we investigated the expression of ags1 (encoding α-1,3-glucan synthase) and chsG (encoding chitin synthase), which are required for fungal cell wall synthesis (Mellado et al., 1996; Beauvais et al., 2013). The ΔAflkhA strain expressed increased levels of ags1 and chsG (Supplementary Figure 4).
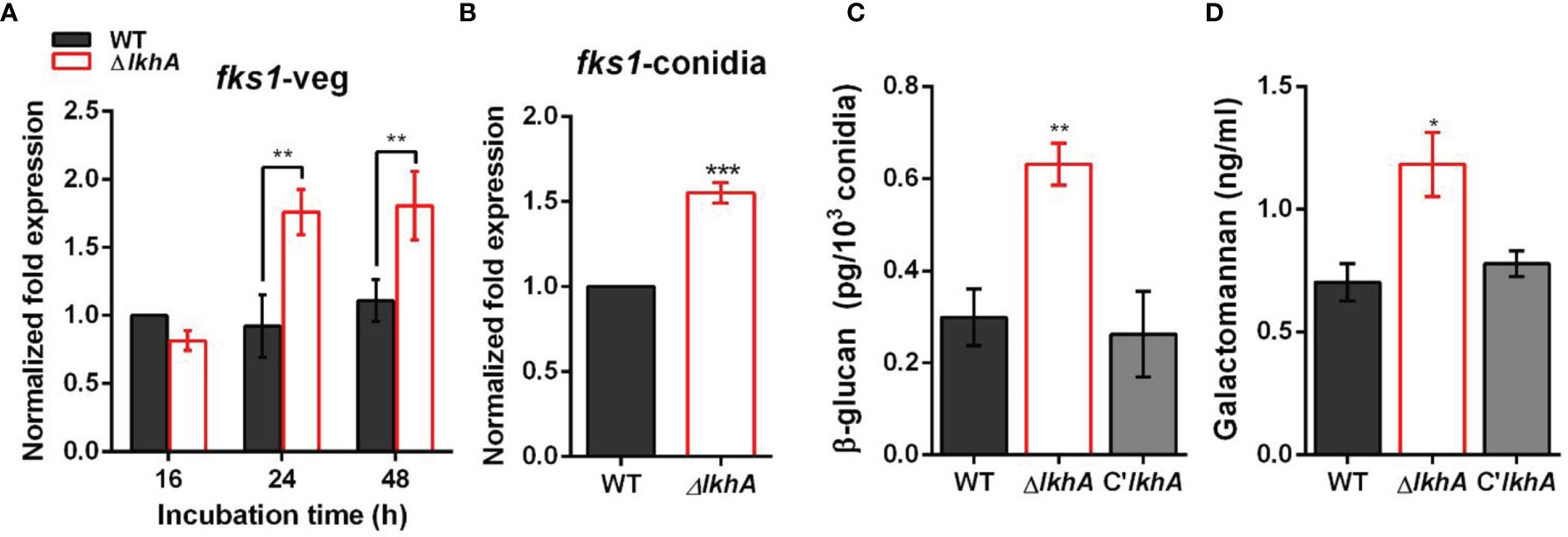
Figure 6 Role of AflkhA in biosynthesis of conidial cell walls. Expression level of β-1,3-glucan synthase fks1 in vegetative cells (A) and conidia (B). Spores were inoculated in MM liquid culture and incubated for the indicated number of hrs. Total RNA was extracted and RT-qPCR analysis was performed using 18S rRNA gene as an internal control. **P < 0.01, ***P < 0.001. (C) Amount of β-glucan (pg) per 103 conidia in WT and ΔAflkhA strains (n = 4). **P < 0.01 (D) Concentration (ng/mL) of GM. Conidia was inoculated in modified Brian medium and cultured at 37°C for 24 hrs. Extracellular GM content in the culture supernatant was assayed by ELISA (n = 4). *P < 0.05.
Next, we investigated the abundance of GM in vegetative cell cultures grown in Brian’s medium by an enzyme-linked immunosorbent assay (ELISA). GM mediates the binding of A. fumigatus conidia to DC-SIGN, a type II membrane C-type lectin in lung dendritic cells (DCs) and alveolar macrophages (Serrano-Gómez et al., 2004). We found that the ΔAflkhA strain produced more GM than the WT and C’AflkhA strains (~1.7 fold increase in ΔAflkhA compared to WT) (Figure 6D). Collectively, these data suggest that AflkhA affects fungal cell wall content, which leads to alterations in the innate immune recognition of PAMPs.
Macrophages Challenged With ΔAflkhA Conidia Induced Early ERK Activation and Produced More Cytokine TNF- α
Following pathogen recognition, the function of alveolar macrophages is dependent in part on MAPKs phosphorylation. Extracellular signal-regulated kinase (ERK) and p38 are MAPKs that are activated and subsequently translocate to the nucleus for transcriptional activation of various target genes in macrophage cells (Rao, 2001). We studied the activation of ERK and p38 in MH-S cells after infection with WT and ΔAflkhA conidia (Figure 7A). When macrophages were stimulated with WT conidia, ERK and p38 were strongly activated, as they displayed a significant mean fold of phosphorylation of ~3.5 for ERK (Figure 7B, WT) and ~4.1 for p38 (Figure 7C, WT) after 6 hrs as compared with non-infected cells. We found a strong response of ERK (phosphorylation fold of 3.0) at an early time point (2 hrs) after infection with ΔAflkhA conidia (Figure 7B, ΔlkhA), but no significant difference in phosphorylation patterns of p38 compared to cells challenged with WT conidia (Figure 7C, ΔlkhA).
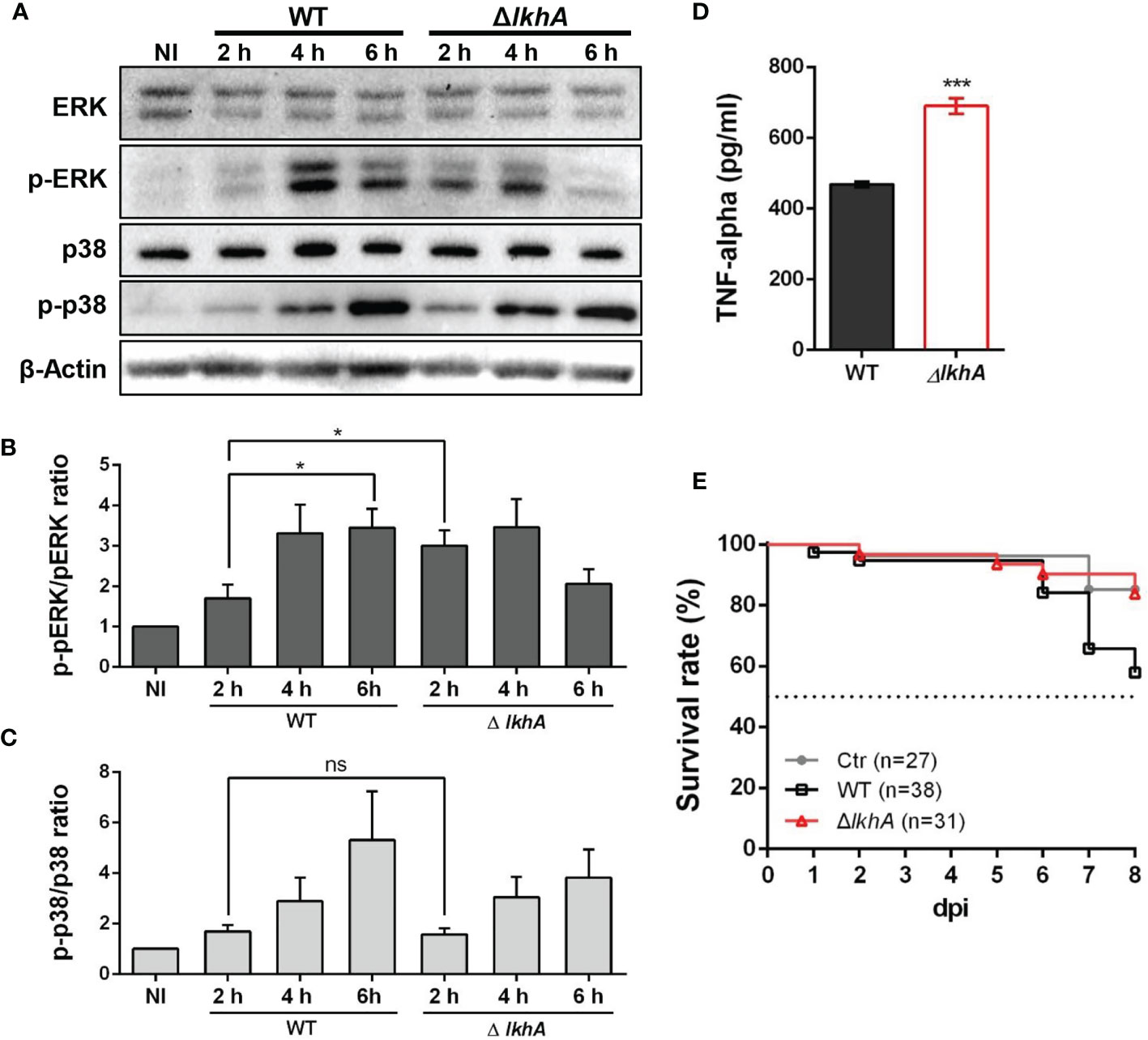
Figure 7 Effect of MAPK phosphorylation and cytokine release from alveolar macrophages treated with AflkhA-null conidia. MH-S cells were simulated with 10-fold of WT and ΔAflkhA conidia and then incubated for the indicated time points. (A) Phosphorylation of ERK and p38 after stimulation of MH-S cells with conidia was analyzed by western blotting. The level of phosphorylation was quantified after normalization to the levels of total ERK (B) or p38 (C) using ImageJ software. *P < 0.05. ns, not significant. (D) Release of TNF-α. MH-S cells were challenged with 10-fold WT and ΔAflkhA conidia. The release of TNF-α was measured by ELISA 6 hrs after infection. ***P < 0.001. (E) Virulence of A. fumigatus ΔAflkhA in T cell-deficient zebrafish larvae (foxn1 morphant). Conidia diluted in fluorescence dye were injected at 3 dpf (0 dpi). Survival was monitored at > 8 dpi. Ctr represents the control. Data are mean values.
The proinflammatory cytokine TNF-α is produced in response to A. fumigatus conidia both in vivo and in vitro (Hohl et al., 2005; Dubey et al., 2014). The production of TNF-α was assessed in alveolar macrophage cells co-cultured with conidia of WT and ΔAflkhA strains for 6 hrs. Cells challenged with ΔAflkhA conidia contained substantially more TNF-α (690 ± 21) than cells with WT conidia (469 ± 8) (Figure 7D).
A Lack of AfLkhA Attenuates Virulence of A. fumigatus in the T Cell-Deficient Zebrafish Larvae Model
Our results showed that macrophages responded differently to WT and ΔAflkhA conidia (Figure 5). A previous report showed that the lack of an adaptive immune system in a T cell-deficient zebrafish model is suitable for studying the role of the innate immune system in pathogenicity (Rosowski et al., 2018a). To investigate the effects of innate immunity against WT and ΔAflkhA conidia in an in vivo model system, virulence tests were performed using T cell-deficient zebrafish larvae (foxn1 morphants) as an immunocompromised model. Forkhead box protein N1 (foxn1) encodes a wing-helix forkhead transcription factor that is required for thymic epithelial cell development (Lv et al., 2020). Nude mice with Foxn1 mutations have been used as xenograft models (Fogh et al., 1977). T cell-deficient larvae were highly susceptible to WT conidia infection, with ~42% mortality at 8 days post-infection (dpi), whereas the ΔAflkhA conidia abrogated the effects on mortality by ~16% (Figure 7E). Control mock infection resulted in a mortality rate of ~15%. These data suggest that AfLkhA contributes to fungal pathogenicity.
Discussion
LAMMER kinase plays a diverse role in both the development and stress responses of fungi (Lim and Park, 2019b). In this study, we performed BLASTP analysis using the AnLkhA sequence, identified one LAMMER kinase ortholog, constructed a deletion mutant, and characterized its function in the opportunistic fungal pathogen A. fumigatus.
The ΔAflkhA strain showed defects in colonial growth, and in both asexual and sexual development. The reduced radial growth and sporulation in the ΔAflkhA strain agreed with previous findings in A. nidulans. The lkhA-deletion strain in A. nidulans showed defects in conidiophore morphology and in sporogenesis during asexual development (Kang et al., 2013). The infectious life cycle of A. fumigatus begins with the production of conidia, which are easily spread in the air. Reproducing asexually allows the pathogen to proliferate in the host environment. AfLkhA, which plays a role in conidia production, may affect the dispersal and invasion of A. fumigatus during inhalation and infection in the host.
AnLkhA affects the maturation of sexual reproductive organs (cleistothecia), regulating the COP9 signalosome (CSN) csnD and a psi factor ppoA genes (Kang et al., 2013). The cell cycle and sexual differentiation of fungi, such as the fission yeast Schizosaccharomyces pombe and the plant pathogen Ustilago maydis, are affected by the deletion of LAMMER kinase (Yu et al., 2013; de Sena-Tomás et al., 2015). In addition, the LAMMER kinases of higher eukaryotes (tobacco PK12 and Drosophila DOA) modulate development and gene expression (Yun et al., 1994; Savaldi-Goldstein, 2003). Mutations in DOA cause defects in various structures irrespective of sex, and is required for sex determination (Du et al., 1998). As shown in Figure 2, the cross with the ΔAflkhA strain produced a decreased number of cleistothecia with abnormal ascospores due to the decreased transcription of sexual genes, steA, veA, and vosA. These data suggest that AflkhA is a novel regulator required for the completion of sexual development in A. fumigatus. The ability to reproduce sexually allows fungal pathogens to adapt to hostile environments; therefore, the sexual cycle can influence virulence in some pathogenic fungi (Butler, 2010). In Crytococcus neoformans, the basidiospores are infectious propagules in capsules that protect fungi from various stresses (Botts et al., 2009). Aspergillus ascospores are resistant to environmental stresses, including azole drugs (Zhang et al., 2017), suggesting that sexual reproduction is involved in the generation of resistant spores. Therefore, the contribution of LkhA to both asexual and sexual development may affect the pathogenicity of the opportunistic human pathogen A. fumigatus in nature.
Mycotoxins and hydrolytic enzymes are associated with virulence in A. fumigatus. Among mycotoxins, GT has attracted the most attention and proven activity. GT exerts a broad spectrum of immunosuppressive effects and is present in the sera of patients with IA. One possible mechanism of GT is the suppressive effect on host immune cells: inhibition of cytokine production, apoptosis of immune cells, and reduced activity of cytotoxic T-cells (Kupfahl et al., 2006; Arias et al., 2018). GT production is regulated by ZafA, the activation of which is dependent on the availability of zinc. The ΔAflkhA strain produced increased amounts of GT and showed increased transcription of GT cluster genes and zafA gene compared to WT (Figure 3), suggesting that LkhA negatively regulates GT production by controlling transcription of GT cluster genes and zafA gene in A. fumigatus.
A. fumigatus lives as a saprotroph in the environment, and most degradative enzymes are specific to plant cell wall components. However, as an opportunistic pathogen, this fungus possesses various enzymes required for infection of human tissues. In this study, we showed increased alkaline protease activity in the ΔAflkhA strain (Figures 4A–C). Based on the fact that the ΔAflkhA strain showed increased GT production and protease activity, we predicted an increase in pathogenicity in the ΔAflkhA strain. Surprisingly, the virulence of the ΔAflkhA strain was reduced in the T cell-deficient zebrafish larva model (Figure 7E). It is noteworthy that GT is not an essential virulence factor in immunosuppressed mice (Kupfahl et al., 2008).
Cell adhesion and biofilm formation are related to cell wall components. Although no significant change was observed in the sensitivity test for cell wall damaging agents (Supplementary Figure 2A), biofilm formation was significantly reduced in the ΔAflkhA strain (Fig 4D). Deletion of AflkhA reduced the expression of genes (ags1, rodA, rodB, aspf1, and gt4c) that are highly expressed in biofilms (Figure 4E). The ECM of biofilms in vivo contains α-1,3-glucans, GM, and GAGs (Beauvais and Latgé, 2015). In A. fumigatus, α-1,3-glucans polysaccharides are synthesized by three α-1,3-glucan synthases: Ags1, Ags2, and Ags3. Only the ags1-deletion strain showed a reduction in α-1,3-glucan content (Beauvais et al., 2005). During conidia swelling, cell wall α-1,3-glucans are exposed at the cell surface to function as an adhesin for the induction of aggregation (Fontaine et al., 2010). GAG is present on the surface of hyphae and covers up β-1,3-glucans from recognition by Dectin-1, which leads to decreased pulmonary inflammation (Gravelat et al., 2013). The Δuge3 and Δgt4c A. fumigatus strains lacking UDP-glucose-4-epimerase and GAG synthase, respectively, cannot produce GAG and show decreased adherence capability and biofilm formation, suggesting that GAG acts as a major adhesin (Gravelat et al., 2013; Briard et al., 2020). It is also noteworthy that the involvement of LAMMER kinase in cell wall integrity has been reported in Candida albicans and A. nidulans (Choi et al., 2014; Lim et al., 2020b). These results suggest that AfLkhA affects the cell wall composition of hyphae, resulting in a decrease in biofilm formation.
The ΔAflkhA conidia were phagocytosed more than those of the WT by alveolar macrophage cells in vitro (Figures 5A–C). Although one of the important step in pathogenesis is germination, how germination affects immune activation and fungal clearance is unknown. Recently, it is reported that a fast-germinating strain is cleared better than a slow-germinating strain in zebrafish larvae model (Rosowski et al., 2018b). The conidia from both the WT and the ΔAflkhA revealed initiation of germination in 3 hrs, but the ΔAflkhA showed reduced germination rate thereafter than the WT (Supplementary Figure 5). Next, we hypothesized that cell wall PAMPs of the ΔAflkhA conidia differed in composition from those in WT. An increased amount of β-1,3-glucan and GM was detected in the ΔAflkhA strain (Figure 6). β-1,3-glucan and GM are recognized by PRRs of alveolar macrophages (Marcos et al., 2016). Therefore, it is possible that increased amount of PAMPs, including β-1,3-glucan and GM in the ΔAflkhA strain induces binding with PRRs of alveolar macrophages, which renders the ΔAflkhA strain susceptible to phagocytosis.
Conidia-infected bone marrow macrophages lacking Dectin-1 showed decreased TNF release and reduced phosphorylation of ERK, suggesting that Dectin-1 is required for interaction with conidia and inflammasome activation (Briard et al., 2019). When macrophages were challenged with conidia, earlier phosphorylation of ERK and increased TNF-α secretion were induced by the ΔAflkhA conidia (Figures 7A–D). Although further investigation is required, these results demonstrate that increased β-1,3-glucan in the ΔAflkhA conidia activates Dectin-1-mediated signal transduction, which in turn activates the adaptive immune system of the host. Dectin-1 signaling through Syk-CARD9-MALT1 activates NF-κB and induces the expression of cytokines and chemokines, including TNF-α, IL-6, IL-1α, IL-1β, G-CSF, GM-CSF, and MIP-1 (Werner et al., 2009; Faro-Trindade et al., 2012; Briard et al., 2019).
When in alveoli, resting conidia begin to swell, and fungal PAMPs are exposed to host immune cells after removal of the outer cell wall layers of the conidia. Following germination, mycelia grow and form biofilms, which protect microbes against hostile environments in the host immune system. Although the mycelia of ΔAflkhA produced increased GT and alkaline protease activity in vitro, the virulence of the ΔAflkhA strain was decreased in an in vivo T cell-deficient zebrafish infection model system. The reduced virulence of the ΔAflkhA strain could be explained by structurally or qualitatively changed PAMPs in conidia, which affects recognition by PRRs of immune cells in the innate immune response. Following the PAMPs-PRRs interaction, macrophages activate ERK to increase the secretion of the cytokine TNF-α, which mediates the transition from innate to adaptive immunity (Figure 8). Cytokines produced by macrophages enhance cellular immune responses to eliminate conidia by recruiting other immune cells for the release of further cytokines (Park and Mehrad, 2009). We suggest that the regulation of AfLkhA on the biosynthesis of fungal PAMPs may be one of the contributing factors for turning on the immune responses by stimulation of macrophages.
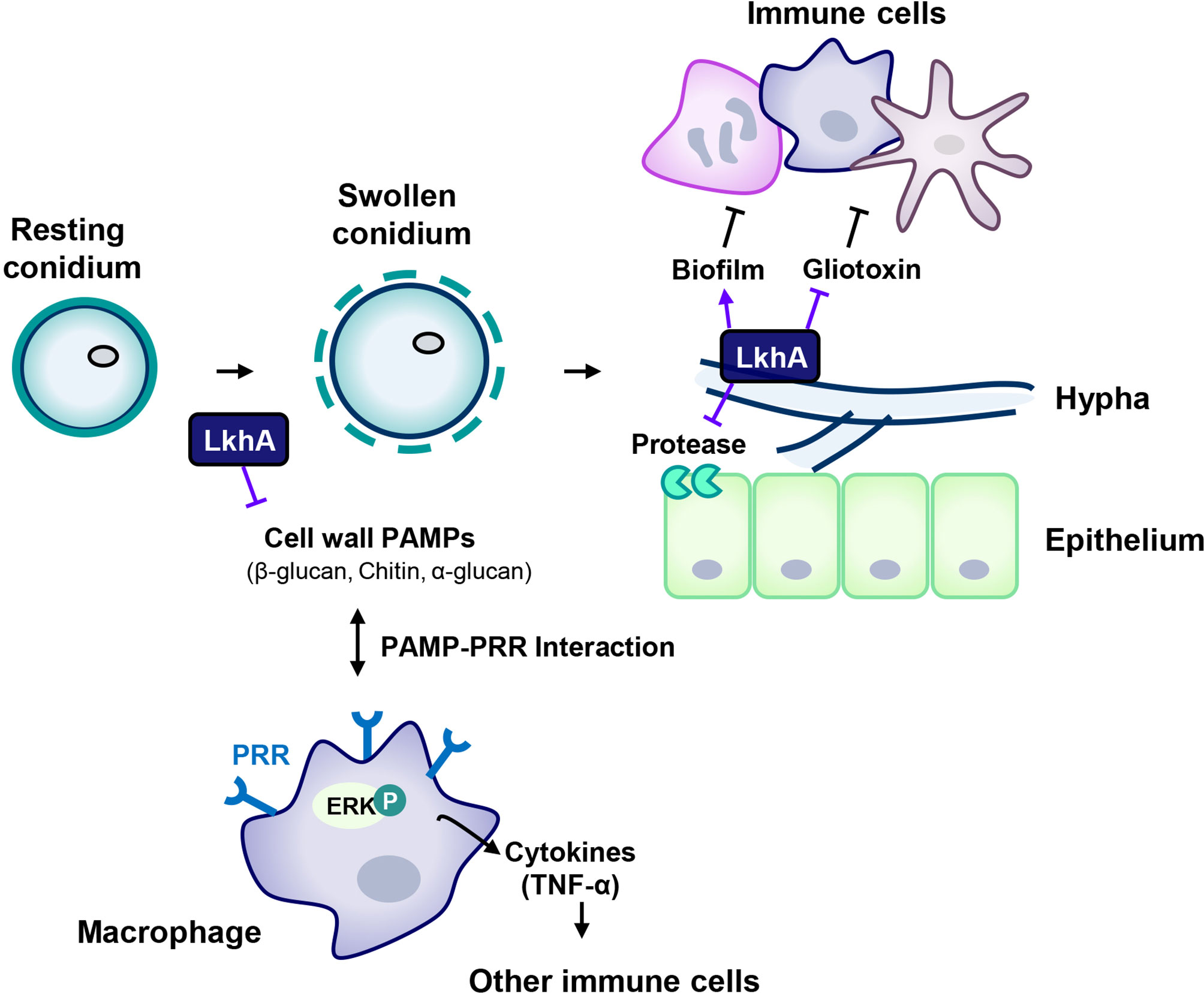
Figure 8 Proposed model for the involvement of LAMMER kinase in the interaction between A. fumigatus and immune cells. The composition of the cell wall continuously changes during the life cycle of A. fumigatus. The resting conidia first come into contact with host epithelial cells. When conidia swell, the outer layer (melanin and hydrophobins) is removed, uncovering the inner cell wall, which is mainly composed of glucan, chitin, and GM. LkhA regulates the transcription of genes for cell wall components and hydrophobins, which affects PAMP-PRR interactions in alveolar macrophages. This interaction mediates ERK signaling and stimulates the production of the proinflammatory cytokine TNF-α, which can promote the activation and proliferation of T cells. Hyphae differ in cell wall composition. The cell wall molecule GAG is secreted from hyphae and is present in the biofilm. The adhesion ability of A. fumigatus to the cell surface is important for virulence. LkhA is required for biofilm formation by modulating gene expression. During its growth, A. fumigatus produces GT, which is toxic to host cells. LkhA represses the expression of GT clustered genes and thus the production of GT. A. fumigatus also produces proteases to obtain nutrients by degrading macromolecules in the host environment. LkhA negatively regulates the expression of gene(s) for alkaline proteases and thus protease production. The virulence of A. fumigatus in the T cell-deficient zebrafish larva model was reduced by the loss of lkhA.
Cell walls are essential components of viability, morphology, and stress responses. The role of LAMMER kinase in cell wall integrity has been studied in fungi. AnLkhA is involved in cell wall synthesis by modulating the expression of cell wall synthase genes (fksA, chsC, and chsD) (Choi et al., 2014). The LAMMER kinase CaKns1 in C. albicans is required for cell wall integrity and adherence (Lim et al., 2020b). Although numerous studies have suggested that cell wall biosynthesis is a key factor in pathogenesis, the role of LAMMER kinase in fungal virulence has not been investigated in detail.
Our results provide insights into the role of AfLkhA in growth, asexual and sexual development, biofilm formation, alkaline protease activity, GT production, and cell wall PAMP biosynthesis. Here, we describe the involvement of A. fumigatus LAMMER kinase in virulence. AfLkhA is required for biosynthesis cell wall PAMPs, including β-1,3-glucan and GM. Changes in the PAMPs of fungal cell walls can affect cell wall PAMPs recognition by the PRRs located on the surface of host immune cells. This PRR-PAMP binding stimulates both the innate and adaptive immune systems that are required to fight fungal pathogens.
Materials and Methods
Growth Condition and Strains
The A. fumigatus strains employed in this study are listed in Supplementary Table 1. The supermater strains AFB62 (MAT1-1) and AFIR928 (MAT1-2) were obtained from the National Institutes of Health (NIH) (Sugui et al., 2011) and maintained in Aspergillus minimal medium with glucose (GMM) (Kong et al., 2013).
To construct the disruption cassette, fusion PCR was performed (Szewczyk et al., 2006). Using the genomic DNA of A. fumigatus as a template, 5’ and 3’ flanking regions of each AflkhA gene were amplified with 5’ UTR AflkhA F/R and 3’ UTR AflkhA F/R primer set, respectively. The pyrithiamine (PT) resistance gene (ptrA) product was amplified with the pTRI plasmid (Takara, Japan) as a template and ptrA F/R primer set (Kubodera et al., 2000). The three DNA fragments were then mixed, and fusion PCR was performed with “nested” primers (AflkhA nest F/R). The primers used in this study are listed in Supplementary Table 2. The disruption cassette was transformed into AFIR928 protoplasts generated using VinoTaste® Pro (Novozymes) [40]. Transformants were selected on GMM with PT, and correct recombination in genomic DNA was confirmed by PCR. To complement ΔAflkhA, the predicted promoter region and ORF of the AflkhA gene were amplified and transformed into the ΔAflkhA strain.
Phenotypic Analysis
For mycelial production, strains were cultivated in liquid complete medium (YCGMM), which was the GMM supplemented with 0.15% yeast extract and 0.15% casamino acid. To observe conidiophores, conidia were inoculated on solid YCGMM agar blocks and coverslip-cultured for 24 hrs (Lim et al., 2020a). To induce sexual development, the vegetative mass mating (VeM) method was used (Lim and Park, 2019a). Briefly, mycelial balls from isolates of opposite MAT1-1 and MAT1-2 mating types were mixed and transferred to oatmeal (Quaker Oats) agar plates, which were sealed with Parafilm and incubated at 30°C in the dark.
For susceptibility testing, conidial suspensions were resuspended to a concentration of 2×106 conidia per mL in distilled water and subjected to 10-fold serial dilution. After dilution, 5 μL of the solution was spotted onto GMM agar and incubated for two days at 37°C (control). Dithiothreitol (DTT) and tunicamycin (Tm), both of which induces UPR, was added to solid GMM to induce endoplasmic reticulum stress. To accomplish ion depletion stress, ethylenediaminetetraacetic acid (EDTA) and ethylene glycol tetraacetic acid (EGTA) were added to the GMM.
RNA Preparation, cDNA Synthesis, and Quantitative Real-Time PCR
Cells at each developmental stage were ground using liquid nitrogen with a mortar and pestle (Rocha et al., 2015). To extract RNA from cleistothecia, 20 cleistothecia were collected in a screw cap tube, frozen in liquid nitrogen, and vortexed with zirconida/silica beads (Biospec). Total RNA was extracted using TRIzol according to the manufacturer’s protocol (Invitrogen). Briefly, cell powder, 0.5 mm dia. zirconia/silica beads (BioSpec Products), Trizol reagent, and β-mercaptoethanol were added to a 2 mL centrifuge tube and homogenized by bead-beating at 4°C. Additional Trizol reagent and chloroform were added for 10 minutes and the supernatant was collected by centrifugation. RNA was precipitated with 100% ethanol and 8 M LiCl. Dried RNA pellets were dissolved in diethyl pyrocarbonate-treated water at 65°C. Synthesis of cDNA was achieved using 4 µg of extracted RNA, hexamer primers, and M-MLV reverse transcriptase (Enzynomics), as described in the manufacturer’s instructions. RT-qPCR was performed using the Bio-Rad CFX96 Real-Time PCR System (Bio-Rad) and a TOPrealTM qPCR 2X PreMIX Kit (Enzynomics). The transcription levels of target genes were normalized against those of the internal control gene 18S rRNA using the 2−ΔΔCt method as previously described (Park et al., 2015). The primers used for RT-qPCR are listed in Supplementary Table 2.
Microscopy
For microcopy, the Olympus System microscope Model BX51 (Olympus) equipped with UPlanSApo 60X and UPlanFL 100X objective lenses (Olympus) and a stereomicroscope Model SMZ800 (Nikon) were used. Images were captured with a DP71 digital camera (Olympus) and processed using the DP manager imaging software (Olympus).
GT Analysis
GT production was analyzed in a time-course experiment. Spores (105 conidia/mL) were inoculated in 200 mL of Czapek Dox broth (pH 7.3) and incubated at 37°C. GT was extracted from the supernatant with chloroform, dried, and resuspended in 200 μl of methanol. Approximately 10 μL of each methanol solution and GT standard (Sigma-Aldrich) were loaded onto a TLC silica plate (Silica gel 60 F254; Merck) and developed in a mobile phase composed of chloroform: methanol (90: 10 v/v). The plate was dried at room temperature, and spots of GT were visualized under UV light (254 nm).
Protease Activity
Spores (5 × 103 conidia) were point inoculated on Czapek Dox medium containing 1% skim milk powder (Difco) instead of sodium nitrate and incubated for three days at 30°C. The degradation halos indicate protease activity. Supernatants were collected from Czapek Dox broth (pH 7.3) containing 1% skim milk powder for three days at 30 ◦C, and then precipitated with four volumes of ice-cold acetone and kept in a cold room overnight. Protein pellets were collected by centrifugation at 5,000 g for 15 minutes at 4°C and air-dried. Protein concentrations were calculated using the BCA Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. An azocasein assay was also performed as previously described, with some modifications (Shemesh et al., 2017). Azocasein (Sigma-Aldrich) was dissolved at a concentration of 5 mg/mL in an assay buffer containing 50 mM Tris (pH 8.0), 0.2 M NaCl, 5 mM CaCl2, and 0.05% Triton X-100. Each protein solution (200 µL) was mixed with 500 µL of azocasein solution and incubated for 90 minutes at 180 rpm at 30 ◦C. The reactions were stopped by adding 200 µL of 12% (v/v) trichloroacetic acid. The reaction mixtures were left at room temperature and centrifuged at 8,000 × g. The supernatant (200 µL) was mixed with 200 µL of 1 M NaOH. The absorbance of the released azo dye was determined at 436 nm.
Biofilm Formation Assay
Fungal biofilm assays were performed using 12-well plates, as described previously (Lohmar et al., 2019). Wells were inoculated with 1 mL of GMM medium containing 105 conidia and incubated for 16 hrs at 37°C. Biofilms were washed three times with PBS, stained with 1 mL of 0.01% (w/v) crystal violet solution, and incubated for 6 hrs. Destaining was conducted with 1 mL of 30% acetic acid. Adhesion capacity was quantified by measuring absorbance at 550 nm using a spectrophotometer and represented as the absorbance of crystal violet dye bound to the biofilm cells.
Culture of Macrophages and A. fumigatus Stimulation
The mouse alveolar macrophage MH-S cell line (ATCC CRL-2019) was cultured in RPMI-1640 medium (Welgene) supplemented with 0.05 mM 2-mercaptoethanol and 10% fetal bovine serum (FBS), and incubated at 37°C in 5% CO2 (Matsunaga et al., 2001).
MAPK phosphorylation was assayed using a previously described method (Dubourdeau et al., 2006). Briefly, MH-S cells were starved for 15–16 hrs in RPMI 1640 medium without FBS, stimulated with 10-fold WT or ΔAflkhA conidia, and washed with cold RPMI medium to remove any unbound conidia. After incubation at 37°C in 5% CO2 for different incubation times (2, 4, and 6 hrs), the cells were washed with cold DPBS (Welgene) and frozen at -70°C before analysis.
Phagocytosis Assay
MH-S cells (2 × 105 macrophages/well) were plated on 12-well cell culture plates with cover glass and incubated for 2 hrs at 37°C in 5% CO2. The cells were stimulated with WT or ΔAflkhA conidia (6 × 105 conidia/well) for 1 hr and washed three times with DPBS. Macrophage conidia were co-cultured for an additional 2 hrs. Wells were then washed with DPBS and stained with calcofluor white (1 μg/mL in DPBS) to label extracellular conidia. The number of A. fumigatus conidia phagocytosed by macrophages was quantified as the percentage of macrophages containing at least one ingested conidium. The phagocytic index was calculated as the average number of ingested conidia per phagocytosing macrophage.
Preparation of Protein Extracts and Immunoblot Analysis
Protein extracts were resuspended in radioimmunoprecipitation assay buffer (Elpis Biotech) with protease inhibitor cocktail (Calbiochem) and phosphatase inhibitor [0.1 M phenylmethylsulfonyl fluoride (PMSF), 0.5 M sodium fluoride, 0.1 M sodium orthovanadate] according to the manufacturer’s protocol. Lysates were collected using a cell scraper and cleared by centrifugation at 13,000 rpm for 15 minutes at 4°C. Equal amounts of total protein extracts were boiled for 5 minutes with 5X sample loading buffer (350 mM Tris-HCl pH 6.8, 600 mM dithiothreitol [DTT], 36% glycerol, 350 mM sodium dodecyl sulfate [SDS], 0.012% [w/v] bromophenol blue), and separated by 10% SDS-PAGE gels. Total proteins were electroblotted onto hybond-P polyvinylidene difluoride (PVDF) membranes (GE Healthcare) and blocked with 5% skim milk in TBST (20 mM Tris-HCl, pH 7.5, 30 mM NaCl, 0.05% Tween 20) for 2 hrs. The membranes were probed with anti-Erk1/2 antibody (1:10000; Cell Signaling Technology), anti-phospho-Erk1/2 antibody (1:10000; Cell Signaling Technology), anti-p38 MAPK antibody (1:10000; BioLegend), anti-phospho-p38 MAPK antibody (1:10000; Santa Cruz Biotechnology), and anti-β-actin antibody (1:10000; Cell Signaling Technology) as primary antibodies, and goat anti-rabbit IgG-HRP (1:10000; Enzo Life Sciences), and goat anti-mouse IgG-HRP (1:5000; Santa Cruz Biotechnology) as secondary antibodies. Immunological detection was performed using an ECL chemiluminescence system (Advansta). Phosphorylation levels were quantified after normalization to the levels of total ERK and p38 using densitometry scanning and ImageJ software.
Cytokine Measurements
For in vitro cytokine expression, co-culture of macrophage cells with A. fumigatus conidia was performed as described previously with some modifications (Dubourdeau et al., 2006). Briefly, MH-S cells were added to 96-well plates at a density of 5 × 105 cells. The cells were stimulated with WT or ΔAflkhA conidia at a conidium-to-macrophage ratio of 10:1 for 6 hrs and then frozen at -70°C. TNF-α was quantified in supernatants by ELISA using a mouse TNF-α detection kit (Invitrogen, Thermo Fisher Scientific). Data are presented as the average TNF-α value (pg/mL) for biological replicates.
Zebrafish Infection Assay by Conidial Microinjection
Zebrafish larvae (foxn1/Casper mutants) at three days post-fertilization (dpf) were obtained from the Zebrafish Center for Disease Modeling (Daejeon, Korea) (Lv et al., 2020). All experiments using zebrafish were conducted with protocols approved by the Animal Ethics Committee of Chungnam National University (CNU-00191). Conidial microinjection was performed as previously described (Bolcome et al., 2008; Knox et al., 2014), but modified to some extent. Conidia from two-day-old cultures at a concentration of 108 conidia/mL were mixed 1:1 with fluorescein isothiocyanate–dextran (Sigma-Aldrich) for clear visualization of injection success and injected into the common cardinal vein/duct of Cuvier. For survival analysis, infected larvae were monitored daily, and mortality was recorded.
Polysaccharide Analysis
The β-1,3-glucan amounts in conidia were determined using the enzymatic yeast beta-glucan Megazyme kit following the manufacturer’s protocol (Megazyme). Briefly, two-day-old conidia were collected, mixed with 2 M KOH for 30 minutes in an ice water bath, and incubated with 1.2 M sodium acetate buffer (pH 3.8) and Glucazyme™ for 16 hrs at 40°C. After incubation, 10 mL of water was added to each tube, and 100 μL aliquots were transferred to new tubes. GOPOD reagent was added and incubated for 20 minutes at 40°C. The optical density was determined at 510 nm. This test was performed in quadruplicate.
For galactomannan production, 50 mL of modified Brian medium (2% asparagine, 5% glucose, 2.4 g/L NH4NO3, 10 g/L KH2PO4, 2 g/L MgSO4-7H2O, 26 mg/L ZnSO4-7H2O, 2.6 mg/L CuSO4-5H2O, 1.3 mg/L Co(NO3)2- 6H2O, 65 mg/L CaCl2, pH 5.4) was inoculated with 5 × 107 conidia and incubated at 37°C at 180 rpm for 24 hrs (Brian et al., 1961; Fontaine et al., 2011; Gravelat et al., 2013). Culture supernatants were prepared by filtration of the culture on Miracloth. Extracellular GM content in the culture supernatant was assayed by ELISA using the GM Ag kit (San Diego, USA), following the manufacturer’s instructions.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: A FungiDB (https://fungidb.org/fungidb/. The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by The Animal Ethics Committee of Chungnam National University.
Author Contributions
J-YL, C-HK, and H-MP designed the experiments. J-YL, YK, SW, JJ, and Y-RL performed the experiments. J-YL, YK, SW, and JJ performed interpretation of the results and data analysis. C-HK and H-MP contributed resource. J-YL and H-MP wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the grants from the National Research Foundation of Korea (2020R1F1A1073075 to H-MP, 2020R1A6A3A01099306 to J-YL, and 2018M3A9B8021980 to C-HK).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Kyung J. Kwon-Chung of the NIAID/NIH for providing the supermater AFB62 and AFIR928 strains. We would like to thank Editage (www.editage.co.kr) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.756206/full#supplementary-material
References
Abad, A., Victoria Fernández-Molina, J., Bikandi, J., Ramírez, A., Margareto, J., Sendino, J., et al. (2010). What Makes Aspergillus Fumigatus a Successful Pathogen? Genes and Molecules Involved in Invasive Aspergillosis. Rev. Iberoam. Micol. 27, 155–182. doi: 10.1016/j.riam.2010.10.003
Aimanianda, V., Latgé, J. P. (2010). Fungal Hydrophobins Form a Sheath Preventing Immune Recognition of Airborne Conidia. Virulence 1, 185–187. doi: 10.4161/viru.1.3.11317
Arias, M., Santiago, L., Vidal-García, M., Redrado, S., Lanuza, P., Comas, L., et al. (2018). Preparations for Invasion: Modulation of Host Lung Immunity During Pulmonary Aspergillosis by Gliotoxin and Other Fungal Secondary Metabolites. Front. Immunol. 9, 2549. doi: 10.3389/fimmu.2018.02549
Balenga, N. A., Klichinsky, M., Xie, Z., Chan, E. C., Zhao, M., Jude, J., et al. (2015). A Fungal Protease Allergen Provokes Airway Hyper-Responsiveness in Asthma. Nat. Commun. 6, 6763. doi: 10.1038/ncomms7763
Beauvais, A., Bozza, S., Kniemeyer, O., Formosa, C., Balloy, V., Henry, C., et al. (2013). Deletion of the α-(1,3)-Glucan Synthase Genes Induces a Restructuring of the Conidial Cell Wall Responsible for the Avirulence of Aspergillus Fumigatus. PloS Pathog. 9, e1003716. doi: 10.1371/journal.ppat.1003716
Beauvais, A., Latgé, J. P. (2015). Aspergillus Biofilm In Vitro and In Vivo. Microbiol. Spectr. 3, MB-0017-2015. doi: 10.1128/microbiolspec.mb-0017-2015
Beauvais, A., Maubon, D., Park, S., Morelle, W., Tanguy, M., Huerre, M., et al. (2005). Two α(1-3) Glucan Synthases With Different Functions in Aspergillus Fumigatus. Appl. Environ. Microbiol. 71, 1531–1538. doi: 10.1128/AEM.71.3.1531-1538.2005
Beauvais, A., Schmidt, C., Guadagnini, S., Roux, P., Perret, E., Henry, C., et al. (2007). An Extracellular Matrix Glues Together the Aerial-Grown Hyphae of Aspergillus Fumigatus. Cell. Microbiol. 9, 1588–1600. doi: 10.1111/j.1462-5822.2007.00895.x
Bok, J. W., Chung, D. W., Balajee, S. A., Marr, K. A., Andes, D., Nielsen, K. F., et al. (2006). GliZ, a Transcriptional Regulator of Gliotoxin Biosynthesis, Contributes to Aspergillus Fumigatus Virulence. Infect. Immun. 74, 6761–6768. doi: 10.1128/IAI.00780-06
Bolcome, R. E., Sullivan, S. E., Zeller, R., Barker, A. P., Collier, R. J., Chan, J. (2008). Anthrax Lethal Toxin Induces Cell Death-Independent Permeability in Zebrafish Vasculature. Proc. Natl. Acad. Sci. U. S. A. 105, 2439–2444. doi: 10.1073/pnas.0712195105
Botts, M. R., Giles, S. S., Gates, M. A., Kozel, T. R., Hull, C. M. (2009). Isolation and Characterization of Cryptococcus Neoformans Spores Reveal a Critical Role for Capsule Biosynthesis Genes in Spore Biogenesis. Eukaryot. Cell 8, 595–605. doi: 10.1128/EC.00352-08
Brian, P. W., Dawkins, A. W., Grove, J. F., Hemming, H. G., Lowe, D., Norris, G. L. F. (1961). Phytotoxic Compounds Produced by Fusarium Equiseti. J. Exp. Bot. 12, 1–12. doi: 10.1093/jxb/12.1.1
Briard, B., Fontaine, T., Samir, P., Place, D. E., Muszkieta, L., Malireddi, R. K. S., et al. (2020). Galactosaminogalactan Activates the Inflammasome to Provide Host Protection. Nature 588, 688–692. doi: 10.1038/s41586-020-2996-z
Briard, B., Karki, R., Malireddi, R. K. S., Bhattacharya, A., Place, D. E., Mavuluri, J., et al. (2019). Fungal Ligands Released by Innate Immune Effectors Promote Inflammasome Activation During Aspergillus Fumigatus Infection. Nat. Microbiol. 4, 316–327. doi: 10.1038/s41564-018-0298-0
Butler, G. (2010). Fungal Sex and Pathogenesis. Clin. Microbiol. Rev. 23, 140–159. doi: 10.1128/CMR.00053-09
Calvo, A. M., Bok, J., Brooks, W., Keller, N. P. (2004). veA Is Required for Toxin and Sclerotial Production in Aspergillus Parasiticus. Appl. Environ. Microbiol. 70, 4733–4739. doi: 10.1128/AEM.70.8.4733-4739.2004
Choi, Y. K., Kang, E. H., Park, H. M. (2014). Role of LAMMER Kinase in Cell Wall Biogenesis During Vegetative Growth of Aspergillus Nidulans. Mycobiology 42, 422–426. doi: 10.5941/MYCO.2014.42.4.422
Dagenais, T. R. T. T., Keller, N. P. (2009). Pathogenesis of Aspergillus Fumigatus in Invasive Aspergillosis. Clin. Microbiol. Rev. 22, 447–465. doi: 10.1128/CMR.00055-08
de Sena-Tomás, C., Sutherland, J. H., Milisavljevic, M., Nikolic, D. B., Pérez-Martín, J., Kojic, M., et al. (2015). LAMMER Kinase Contributes to Genome Stability in Ustilago Maydis. DNA Repair (Amst.) 33, 70–77. doi: 10.1016/j.dnarep.2015.05.011
de Vries, R. P., Visser, J. (2001). Aspergillus Enzymes Involved in Degradation of Plant Cell Wall Polysaccharides. Microbiol. Mol. Biol. Rev. 65, 497–522. doi: 10.1128/mmbr.65.4.497-522.2001
Dubey, L. K., Moeller, J. B., Schlosser, A., Sorensen, G. L., Holmskov, U. (2014). Induction of Innate Immunity by Aspergillus Fumigatus Cell Wall Polysaccharides Is Enhanced by the Composite Presentation of Chitin and Beta-Glucan. Immunobiology 219, 179–188. doi: 10.1016/j.imbio.2013.10.003
Dubourdeau, M., Athman, R., Balloy, V., Huerre, M., Chignard, M., Philpott, D. J., et al. (2006). Aspergillus Fumigatus Induces Innate Immune Responses in Alveolar Macrophages Through the MAPK Pathway Independently of TLR2 and TLR4. J. Immunol. 177, 3994–4001. doi: 10.4049/jimmunol.177.6.3994
Du, C., McGuffin, M. E., Dauwalder, B., Rabinow, L., Mattox, W. (1998). Protein Phosphorylation Plays an Essential Role in the Regulation of Alternative Splicing and Sex Determination in Drosophila. Mol. Cell 2, 741–750. doi: 10.1016/s1097-2765(00)80289-0
Duran, R. M., Cary, J. W., Calvo, A. M. (2006). Production of Cyclopiazonic Acid, Aflatrem, and Aflatoxin by Aspergillus Flavus Is Regulated by Vea, a Gene Necessary for Sclerotial Formation. Appl. Microbiol. Biotechnol. 73, 1158–1168. doi: 10.1007/s00253-006-0581-5
Etxebeste, O., Garzia, A., Espeso, E. A., Ugalde, U. (2010). Aspergillus Nidulans Asexual Development: Making the Most of Cellular Modules. Trends Microbiol. 18, 569–576. doi: 10.1016/j.tim.2010.09.007
Fallon, J. P., Reeves, E. P., Kavanagh, K. (2010). Inhibition of Neutrophil Function Following Exposure to the Aspergillus Fumigatus Toxin Fumagillin. J. Med. Microbiol. 59, 625–633. doi: 10.1099/jmm.0.018192-0
Faro-Trindade, I., Willment, J. A., Kerrigan, A. M., Redelinghuys, P., Hadebe, S., Reid, D. M., et al. (2012). Characterisation of Innate Fungal Recognition in the Lung. PloS One 7, e35675. doi: 10.1371/journal.pone.0035675
Fogh, J., Fogh, J. M., Orfeo, T. (1977). One Hundred and Twenty Seven Cultured Human Tumor Cell Lines Producing Tumors in Nude Mice. J. Natl. Cancer Inst. 59, 221–226. doi: 10.1093/jnci/59.1.221
Fontaine, T., Beauvais, A., Loussert, C., Thevenard, B., Fulgsang, C. C., Ohno, N., et al. (2010). Cell Wall α1-3glucans Induce the Aggregation of Germinating Conidia of Aspergillus Fumigatus. Fungal Genet. Biol. 47, 707–712. doi: 10.1016/j.fgb.2010.04.006
Fontaine, T., Delangle, A., Simenel, C., Coddeville, B., van Vliet, S. J., van Kooyk, Y., et al. (2011). Galactosaminogalactan, a New Immunosuppressive Polysaccharide of Aspergillus Fumigatus. PloS Pathog. 7, e1002372. doi: 10.1371/journal.ppat.1002372
Gastebois, A., Aimanianda, V., Bachellier-Bassi, S., Nesseir, A., Firon, A., Beauvais, A., et al. (2013). SUN Proteins Belong to a Novel Family of β-(1,3)-Glucanmodifying Enzymes Involved in Fungal Morphogenesis. J. Biol. Chem. 288, 13387–13396. doi: 10.1074/jbc.M112.440172
Gibbons, J. G., Beauvais, A., Beau, R., McGary, K. L., Latgé, J. P., Rokas, A. (2012). Global Transcriptome Changes Underlying Colony Growth in the Opportunistic Human Pathogen Aspergillus Fumigatus. Eukaryot. Cell 11, 68–78. doi: 10.1128/EC.05102-11
Gravelat, F. N., Beauvais, A., Liu, H., Lee, M. J., Snarr, B. D., Chen, D., et al. (2013). Aspergillus Galactosaminogalactan Mediates Adherence to Host Constituents and Conceals Hyphal β-Glucan From the Immune System. PloS Pathog. 9, e1003575. doi: 10.1371/journal.ppat.1003575
Hatinguais, R., Willment, J. A., Brown, G. D. (2020). PAMPs of the Fungal Cell Wall and Mammalian PRRs. Curr. Top. Microbiol. Immunol. 425, 187–223. doi: 10.1007/82_2020_201
Hohl, T. M., Van Epps, H. L., Rivera, A., Morgan, L. A., Chen, P. L., Feldmesser, M., et al. (2005). Aspergillus Fumigatus Triggers Inflammatory Responses by Stage-Specific β-Glucan Display. PloS Pathog. 1, 0232–0240. doi: 10.1371/journal.ppat.0010030
Kang, E.-H., Kim, J., Oh, H.-W., Park, H.-M. (2013). LAMMER Kinase LkhA Plays Multiple Roles in the Vegetative Growth and Asexual and Sexual Development of Aspergillus Nidulans. PloS One 8, e58762. doi: 10.1371/journal.pone.0058762
Kauffman, H. F., Christomee, J. F., Van De Riet, M. A., Timmerman, A. J. B., Borger, P. (2000). Protease-Dependent Activation of Epithelial Cells by Fungal Allergens Leads to Morphologic Changes and Cytokine Production. J. Allergy Clin. Immunol. 105, 1185–1193. doi: 10.1067/mai.2000.106210
Kim, H. S., Han, K. Y., Kim, K. J., Han, D. M., Jahng, K. Y., Chae, K. S. (2002). The veA Gene Activates Sexual Development in Aspergillus Nidulans. Fungal Genet. Biol. 37, 72–80. doi: 10.1016/S1087-1845(02)00029-4
Knox, B. P., Deng, Q., Rood, M., Eickhoff, J. C., Keller, N. P., Huttenlocher, A. (2014). Distinct Innate Immune Phagocyte Responses to Aspergillus Fumigatus Conidia and Hyphae in Zebrafish Larvae. Eukaryot. Cell 13, 1266–1277. doi: 10.1128/EC.00080-14
Kong, Q., Wang, L., Liu, Z., Kwon, N., Kim, S. C., Yu, J.-H. (2013). Gβ-Like CpcB Plays a Crucial Role for Growth and Development of Aspergillus Nidulans and Aspergillus Fumigatus. PloS One 8, e70355. doi: 10.1371/journal.pone.0070355
Kubodera, T., Yamashita, N., Nishimura, A. (2000). Pyrithiamine Resistance Gene (Ptra) of Aspergillus Oryzae : Cloning, Characterization and Application as a Dominant Selectable Marker for Transformation. Biosci. Biotechnol. Biochem. 64, 1416–1421. doi: 10.1271/bbb.64.1416
Kupfahl, C., Geginat, G., Hof, H. (2006). Gliotoxin-Mediated Suppression of Innate and Adaptive Immune Functions Directed Against Listeria Monocytogenes. Med. Mycol. 44, 591–599. doi: 10.1080/13693780600815411
Kupfahl, C., Michalka, A., Lass-Flörl, C., Fischer, G., Haase, G., Ruppert, T., et al. (2008). Gliotoxin Production by Clinical and Environmental Aspergillus Fumigatus Strains. Int. J. Med. Microbiol. 298, 319–327. doi: 10.1016/j.ijmm.2007.04.006
Lee, M. J., Liu, H., Barker, B. M., Snarr, B. D., Gravelat, F. N., Al Abdallah, Q., et al. (2015). The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps. PloS Pathog. 11, e1005187. doi: 10.1371/journal.ppat.1005187
Lewis, R. E., Wiederhold, N. P., Lionakis, M. S., Prince, R. A., Kontoyiannis, D. P. (2005). Frequency and Species Distribution of Gliotoxin-Producing Aspergillus Isolates Recovered From Patients at a Tertiary-Care Cancer Center. J. Clin. Microbiol. 43, 6120–6122. doi: 10.1128/JCM.43.12.6120-6122.2005
Lim, J. Y., Kang, E. H., Park, Y. H., Kook, J. H., Park, H. M. (2020a). Survival Factor SvfA Plays Multiple Roles in Differentiation and Is Essential for Completion of Sexual Development in Aspergillus Nidulans. Sci. Rep. 10, 5586. doi: 10.1038/s41598-020-62455-4
Lim, J. Y., Park, H. M. (2019a). Expression of Sexual Genes in Aspergillus Fumigatus Homogeneous Culture Produced by Vegetative Mass Mating. J. Microbiol. 57, 1–6. doi: 10.1007/s12275-019-9094-7
Lim, J. Y., Park, H. M. (2019b). The Dual-Specificity LAMMER Kinase Affects Stress-Response and Morphological Plasticity in Fungi. Front. Cell. Infect. Microbiol. 9, 213. doi: 10.3389/fcimb.2019.00213
Lim, J. Y., Park, Y. H., Pyon, Y. H., Yang, J. M., Yoon, J. Y., Park, S. J., et al. (2020b). The LAMMER Kinase Is Involved in Morphogenesis and Response to Cell Wall- and DNA-Damaging Stresses in Candida Albicans. Med. Mycol. 58, 240–247. doi: 10.1093/mmy/myz049
Liu, H., Sang, S., Wang, H., Ren, X., Tan, Y., Chen, W., et al. (2018a). Comparative Proteomic Analysis Reveals the Regulatory Network of the veA Gene During Asexual and Sexual Spore Development of Aspergillus Cristatus. Biosci. Rep. 0, 1–19. doi: 10.1042/BSR20180067
Liu, H., Xu, W., Solis, N. V., Woolford, C., Mitchell, A. P., Filler, S. G. (2018b). Functional Convergence of Glip and Aspf1 in Aspergillus Fumigatus Pathogenicity. Virulence 9, 1062–1073. doi: 10.1080/21505594.2018.1482182
Lohmar, J. M., Puel, O., Cary, J. W., Calvo, A. M. (2019). The Aspergillus Flavus rtfA Gene Regulates Plant and Animal Pathogenesis and Secondary Metabolism. Appl. Environ. Microbiol. 85, e02446–e02418. doi: 10.1128/AEM.02446-18
Lv, P., Ma, D., Gao, S., Zhang, Y., Bae, Y. K., Liang, G., et al. (2020). Generation of Foxn1/Casper Mutant Zebrafish for Allograft and Xenograft of Normal and Malignant Cells. Stem Cell Rep. 15, 749–760. doi: 10.1016/j.stemcr.2020.07.020
Marcos, C. M., de Oliveira, H. C., de Melo, W., de, C. M. A., da Silva, J. de F., Assato, P. A., et al. (2016). Anti-Immune Strategies of Pathogenic Fungi. Front. Cell. Infect. Microbiol. 6, 142. doi: 10.3389/fcimb.2016.00142
Matsunaga, K., Klein, T. W., Friedman, H., Yamamoto, Y. (2001). Alveolar Macrophage Cell Line MH-S Is Valuable as an In Vitro Model for Legionella Pneumophila Infection. Am. J. Respir. Cell Mol. Biol. 24, 326–331. doi: 10.1165/ajrcmb.24.3.4359
Mellado, E., Aufauvre-Brown, A., Gow, N. A. R., Holden, D. W. (1996). The Aspergillus Fumigatus chsC and chsG Genes Encode Class III Chitin Synthases With Different Functions. Mol. Microbiol. 20, 667–679. doi: 10.1046/j.1365-2958.1996.5571084.x
Myung, K., Zitomer, N. C., Duvall, M., Glenn, A. E., Riley, R. T., Calvo, A. M. (2012). The Conserved Global Regulator VeA Is Necessary for Symptom Production and Mycotoxin Synthesis in Maize Seedlings by Fusarium Verticillioides. Plant Pathol. 61, 152–160. doi: 10.1111/j.1365-3059.2011.02504.x
Ni, M., Yu, J. H. (2007). A Novel Regulator Couples Sporogenesis and Trehalose Biogenesis in Aspergillus Nidulans. PloS One 2, e970. doi: 10.1371/journal.pone.0000970
Osherov, N. (2012). Interaction of the Pathogenic Mold Aspergillus Fumigatus With Lung Epithelial Cells. Front. Microbiol. 3:346. doi: 10.3389/fmicb.2012.00346
Park, H. S., Bayram, Ö., Braus, G. H., Kim, S. C., Yu, J. H. (2012). Characterization of the Velvet Regulators in Aspergillus Fumigatus. Mol. Microbiol. 86, 937–953. doi: 10.1111/mmi.12032
Park, S. J., Mehrad, B. (2009). Innate Immunity to Aspergillus Species. Clin. Microbiol. Rev. 22, 535–551. doi: 10.1128/CMR.00014-09
Park, D. S., Yu, Y. M., Kim, Y. J., Maeng, P. J. (2015). Negative Regulation of the Vacuole-Mediated Resistance to K+ Stress by a Novel C2H2 Zinc Finger Transcription Factor Encoded by aslA in Aspergillus Nidulans. J. Microbiol. 53, 100–110. doi: 10.1007/s12275-015-4701-8
Perez-Cuesta, U., Aparicio-Fernandez, L., Guruceaga, X., Martin-Souto, L., Abad-Diaz-de-Cerio, A., Antoran, A., et al. (2020). Melanin and Pyomelanin in Aspergillus Fumigatus: From Its Genetics to Host Interaction. Int. Microbiol. 23, 55–63. doi: 10.1007/s10123-019-00078-0
Rao, K. M. (2001). MAP Kinase Activation in Macrophages. J. Leukoc. Biol. 69, 3–10. doi: 10.1189/jlb.69.1.3
Richie, D. L., Hartl, L., Aimanianda, V., Winters, M. S., Fuller, K. K., Miley, M. D., et al. (2009). A Role for the Unfolded Protein Response (UPR) in Virulence and Antifungal Susceptibility in Aspergillus Fumigatus. PloS Pathog. 5, e1000258. doi: 10.1371/journal.ppat.1000258
Rocha, M. C., De Godoy, K. F., De Castro, P. A., Hori, J. I., Pedro Bom, V. L., Brown, N. A., et al. (2015). The Aspergillus Fumigatus Pkcag579r Mutant Is Defective in the Activation of the Cell Wall Integrity Pathway But Is Dispensable for Virulence in a Neutropenic Mouse Infection Model. PloS One 10, e0135195. doi: 10.1371/journal.pone.0135195
Rosowski, E. E., Knox, B. P., Archambault, L. S., Huttenlocher, A., Keller, N. P., Wheeler, R. T., et al. (2018a). The Zebrafish as a Model Host for Invasive Fungal Infections. J. Fungi (Basel Switzerland) 4:136. doi: 10.3390/jof4040136
Rosowski, E. E., Raffa, N., Knox, B. P., Golenberg, N., Keller, N. P., Huttenlocher, A. (2018b). Macrophages Inhibit Aspergillus Fumigatus Germination and Neutrophil-Mediated Fungal Killing. PloS Pathog. 14, 1–28. doi: 10.1371/journal.ppat.1007229
Savaldi-Goldstein, S. (2003). Alternative Splicing Modulation by a LAMMER Kinase Impinges on Developmental and Transcriptome Expression. Plant Cell Online 15, 926–938. doi: 10.1105/tpc.011056
Schlam, D., Canton, J., Carreño, M., Kopinski, H., Freeman, S. A., Grinstein, S., et al. (2016). Gliotoxin Suppresses Macrophage Immune Function by Subverting Phosphatidylinositol 3,4,5-Trisphosphate Homeostasis. MBio 7, e02242. doi: 10.1128/mBio.02242-15
Seo, H., Kang, S., Park, Y. S., Yun, C. W. (2019). The Role of Zinc in Gliotoxin Biosynthesis of Aspergillus Fumigatus. Int. J. Mol. Sci. 20, 6192. doi: 10.3390/ijms20246192
Serrano-Gómez, D., Domínguez-Soto, A., Ancochea, J., Jimenez-Heffernan, J. A., Leal, J. A., Corbí, A. L. (2004). Dendritic Cell-Specific Intercellular Adhesion Molecule 3-Grabbing Nonintegrin Mediates Binding and Internalization of Aspergillus Fumigatus Conidia by Dendritic Cells and Macrophages. J. Immunol. 173, 5635–5643. doi: 10.4049/jimmunol.173.9.5635
Shemesh, E., Hanf, B., Hagag, S., Attias, S., Shadkchan, Y., Fichtman, B., et al. (2017). Phenotypic and Proteomic Analysis of the Aspergillus Fumigatus Δprtt, Δxprg and Δxprg/Δprtt Protease-Deficient Mutants. Front. Microbiol. 8:2490. doi: 10.3389/fmicb.2017.02490
Sriranganadane, D., Waridel, P., Salamin, K., Reichard, U., Grouzmann, E., Neuhaus, J. M., et al. (2010). Aspergillus Protein Degradation Pathways With Different Secreted Protease Sets at Neutral and Acidic pH. J. Proteome Res. 9, 3511–3519. doi: 10.1021/pr901202z
Sugui, J. A., Losada, L., Wang, W. (2011). Identification and Characterization of an Aspergillus Fumigatus Supermater Pair. MBio 2, 1–9. doi: 10.1128/mBio.00234-11.Editor
Sugui, J. A., Pardo, J., Chang, Y. C., Zarember, K. A., Nardone, G., Galvez, E. M., et al. (2007). Gliotoxin Is a Virulence Factor of Aspergillus Fumigatus: GliP Deletion Attenuates Virulence in Mice Immunosuppressed With Hydrocortisone. Eukaryot. Cell 6, 1562–1569. doi: 10.1128/EC.00141-07
Szewczyk, E., Nayak, T., Oakley, C. E., Edgerton, H., Xiong, Y., Taheri-Talesh, N., et al. (2006). Fusion PCR and Gene Targeting in Aspergillus Nidulans. Nat. Protoc. 1, 3111–3120. doi: 10.1038/nprot.2006.405
Vallim, M. A., Miller, K. Y., Miller, B. L. (2000). Aspergillus SteA (Sterile12-Like) Is a Homeodomain-C2/H2-Zn+2 Finger Transcription Factor Required for Sexual Reproduction. Mol. Microbiol. 36, 290–301. doi: 10.1046/j.1365-2958.2000.01874.x
Valsecchi, I., Dupres, V., Stephen-Victor, E., Guijarro, J. I., Gibbons, J., Beau, R., et al. (2018). Role of Hydrophobins in Aspergillus Fumigatus. J. Fungi 4, 2. doi: 10.3390/jof4010002
Van De Veerdonk, F. L., Gresnigt, M. S., Romani, L., Netea, M. G., Latgé, J. P. (2017). Aspergillus Fumigatus Morphology and Dynamic Host Interactions. Nat. Rev. Microbiol. 15, 661–674. doi: 10.1038/nrmicro.2017.90
Wang, D. N., Toyotome, T., Muraosa, Y., Watanabe, A., Wuren, T., Bunsupa, S., et al. (2014). GliA in Aspergillus Fumigatus Is Required for its Tolerance to Gliotoxin and Affects the Amount of Extracellular and Intracellular Gliotoxin. Med. Mycol. 52, 504–516. doi: 10.1093/mmy/myu007
Watson, D. S., Feng, X., Askew, D. S., Jambunathan, K., Kodukula, K., Galande, A. K. (2011). Substrate Specifity Profiling of the Aspergillus Fumigatus Proteolytic Secretome Reveals Consensus Motifs With Predominance of Ile/Leu and Phe/Tyr. PloS One 6, e21001. doi: 10.1371/JOURNAL.PONE.0021001
Werner, J. L., Metz, A. E., Horn, D., Schoeb, T. R., Hewitt, M. M., Schwiebert, L. M., et al. (2009). Requisite Role for the Dectin-1 β-Glucan Receptor in Pulmonary Defense Against Aspergillus Fumigatus. J. Immunol. 182, 4938–4946. doi: 10.4049/jimmunol.0804250
Yu, J. H. (2010). Regulation of Development in Aspergillus Nidulans and Aspergillus Fumigatus. Mycobiology 38:229. doi: 10.4489/MYCO.2010.38.4.229
Yu, E. Y., Lee, J. H., Kang, W. H., Park, Y. H., Kim, L., Park, H. M. (2013). Fission Yeast LAMMER Kinase Lkh1 Regulates the Cell Cycle by Phosphorylating the CDK-Inhibitor Rum1. Biochem. Biophys. Res. Commun. 432, 80–85. doi: 10.1016/j.bbrc.2013.01.082
Yun, B., Farkas, R., Lee, K., Rabinow, L. (1994). The Doa Locus Encodes a Member of a New Protein Kinase Family and Is Essential for Eye and Embryonic Development in Drosophila Melanogaster. Genes Dev. 8, 1160–1173. doi: 10.1101/GAD.8.10.1160
Keywords: Aspergillus fumigatus, gene regulation, fungal development, interactions with host cells, molecular mechanisms of fungal pathogenesis, pathogen associated molecular patterns (PAMPs)
Citation: Lim J-Y, Kim YJ, Woo SA, Jeong JW, Lee Y-R, Kim C-H and Park H-M (2021) The LAMMER Kinase, LkhA, Affects Aspergillus fumigatus Pathogenicity by Modulating Reproduction and Biosynthesis of Cell Wall PAMPs. Front. Cell. Infect. Microbiol. 11:756206. doi: 10.3389/fcimb.2021.756206
Received: 10 August 2021; Accepted: 27 September 2021;
Published: 13 October 2021.
Edited by:
Robert T. Wheeler, University of Maine, United StatesReviewed by:
Jarrod R. Fortwendel, University of Tennessee Health Science Center, United StatesBanu Metin, Istanbul Sabahattin Zaim University, Turkey
Copyright © 2021 Lim, Kim, Woo, Jeong, Lee, Kim and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hee-Moon Park, hmpark@cnu.ac.kr
 Joo-Yeon Lim
Joo-Yeon Lim Yeon Ju Kim1
Yeon Ju Kim1  Jae Wan Jeong
Jae Wan Jeong Cheol-Hee Kim
Cheol-Hee Kim Hee-Moon Park
Hee-Moon Park