Apoptosis Triggered by ORF3 Proteins of the Circoviridae Family
- 1Research Center of Avian Disease, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Institute of Preventive Veterinary Medicine, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 3Key Laboratory of Animal Disease and Human Health of Sichuan Province, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
Apoptosis, a form of the programmed cell death, is an indispensable defense mechanism regulating cellular homeostasis and is triggered by multiple stimuli. Because of the regulation of apoptosis in cellular homeostasis, viral proteins with apoptotic activity are particular foci of on antitumor therapy. One representative viral protein is the open reading frame 3 (ORF3) protein, also named as apoptin in the Circoviridae chicken anemia virus (CAV), and has the ability to induce tumor-specific apoptosis. Proteins encoded by ORF3 in other circovirus species, such as porcine circovirus (PCV) and duck circovirus (DuCV), have also been reported to induce apoptosis, with subtle differences in apoptotic activity based on cell types. This article is aimed at reviewing the latest research advancements in understanding ORF3 protein-mediated apoptosis mechanisms of Circoviridae from three perspectives: subcellular localization, interactions with host proteins, and participation in multiple apoptotic signaling pathways, providing a scientific basis for circovirus pathogenesis and a reference on its potential anticancer function.
Introduction
The Circoviridae family, whose members are considered to cause fatal diseases in birds and pigs, is composed of two recognized genera: Circovirus and Cyclovirus (Rosario et al., 2017). Chicken anemia virus (CAV), the only bird virus member of the Cyclovirus family, leads to atrophy of bone marrow hematopoietic tissue and lymphatic tissues in young chickens (Taniguchi et al., 1983). The Circovirus genus contains pathogenic viruses of vertebrates, such as beak and feather disease virus (BFDV), goose circovirus (GoCV), duck circovirus (DuCV) (Hattermann et al., 2003), and porcine circovirus (PCV) (Ha et al., 2020). The presence of Circovirus in invertebrates has also been reported (Wang et al., 2018). Infections with any of these viruses can potentially cause fatal diseases (Todd, 2000; Soike et al., 2004; Todd, 2004; Raidal et al., 2015; Palinski et al., 2016; Dennis et al., 2018; Klaumann et al., 2018; Fatoba and Adeleke, 2019), which are characterized by damage to lymphoid tissues and immunosuppression (Todd, 2000).
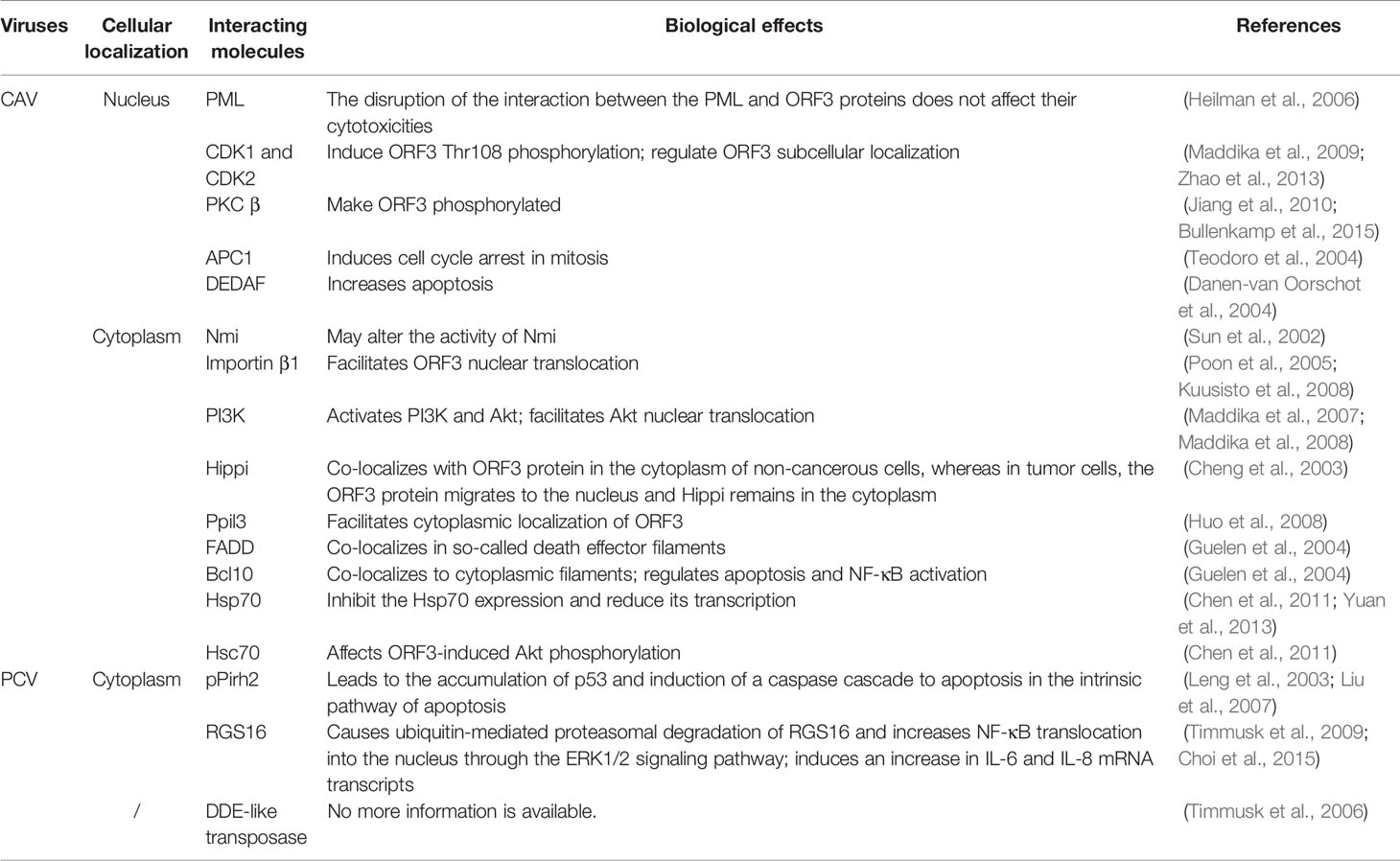
The members of the Circoviridae family are small, nonenveloped viruses with circular single-stranded DNA genomes. The length of genome is approximately 2000 bp, and it contains two major open-reading frames (ORFs)—ORF1 and ORF2, encoding Cap and Rep proteins, respectively. Cap is the sole structural protein of the virus and has a highly conserved basic amino acid sequence, indicating that Cap contains the major antigenic determinant (Nawagitgul et al., 2000; Hattermann et al., 2003). Rep is mainly associated with rolling circle replication (RCR) (Hamel et al., 1998; Luo et al., 2018). Adjacent to two Cap and Rep, the origin of replication with a stem loop structure is located in the intergenic region (Faurez et al., 2009). Especially for PCV, a Rep’ protein can be generated by alternative transcript splicing of ORF1, and the Rep-Rep’ complex is required for promoting virus replication by RCR system (Steinfeldt et al., 2002). In addition to the Rep and Cap, ORF3 of Circovirus encodes a protein that can participate in cell death during viral infection. In PCV, the ORF3 protein plays an important role in the pathogenesis of the virus due to its apoptotic activity in vitro and in vivo, although it is not essential for virus replication (Liu et al., 2005; Liu et al., 2006; Lin et al., 2011). In other kinds of Circoviruses, such as DuCV, the ORF3 protein is thought to induce apoptosis as well (Xiang et al., 2012). In Cyclovirus, reports have implicated that ORF3 encodes a nonstructural protein and participates in the induction of apoptosis and viral cytotoxicity in host cells (Kucharski et al., 2016) (Figure 1A).
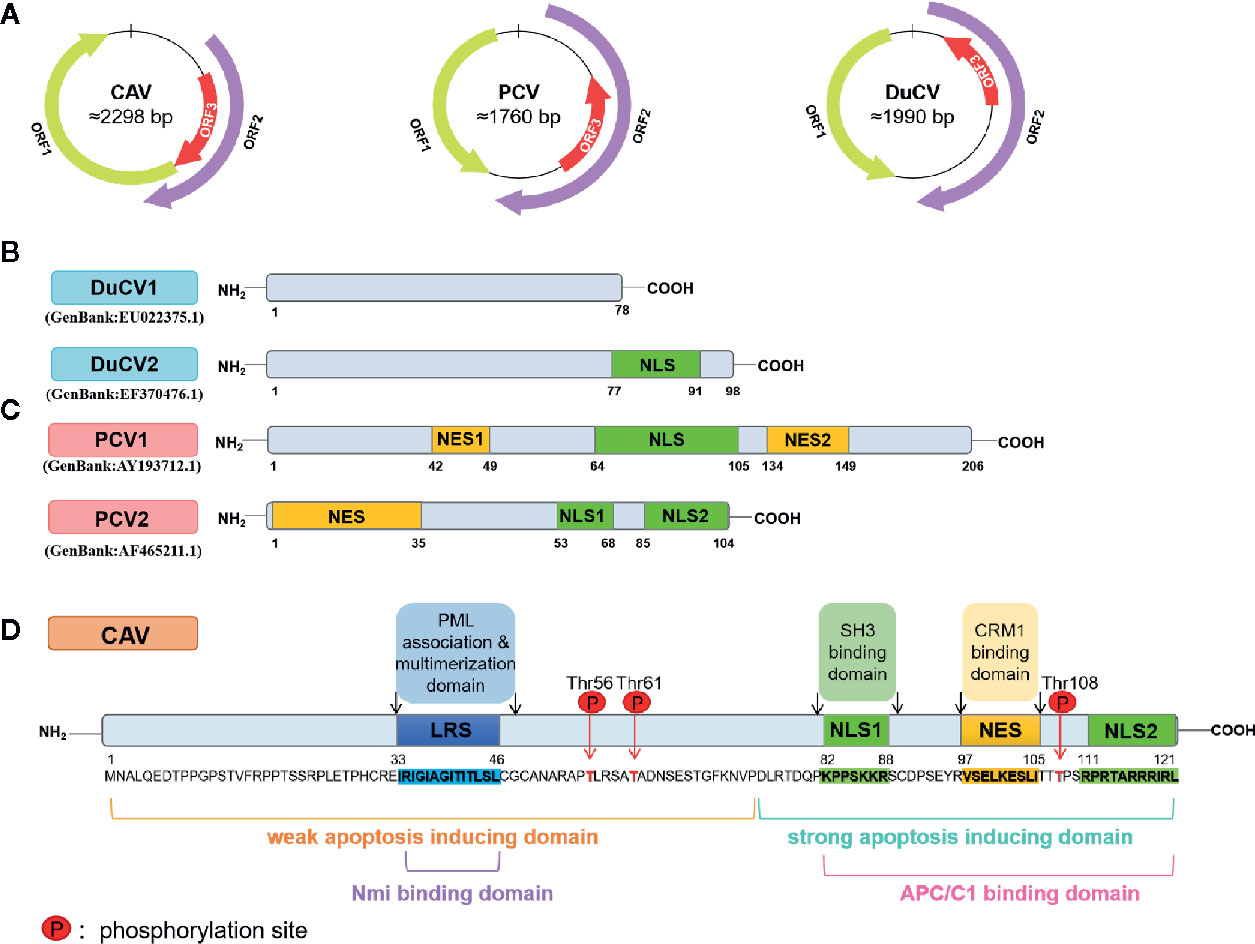
Figure 1 Schematic diagram of the CAV, PCV and DuCV gene structures. (A) The relative localizations of three major ORF proteins in CAV, PCV, and DuCV differ from each other. In particular, the ORF3 proteins of PCV and DuCV are oriented in the direction opposite that of Rep gene, in contrast to the ORF3 protein in CAV. (B) The ORF3 protein structures between DuCV1 and DuCV2 differ. The DuCV2 ORF3 (99 aa) is 20 aa longer than that of DuCV 1, in which an NLS has been identified. (C) The ORF3 structures between PCV1 and PCV2 differ. The length of the ORF3 is 315 bp in PCV2, while in PCV1, it is approximately 612 bp. The NES has been identified in the N-terminus of PCV2 ORF3 protein at residues 1–35, and a bipartite NLS was found at residues 53–68 and 85–104. (D) The primary structures of the CAV ORF3 protein (Los et al., 2009). The following main domains are shown in color: leucine-rich region (blue), phosphorylation site (red), bipartite nuclear localization sequences (green), and nuclear export sequence (yellow).
Apoptosis causes a non-lytic and typically immunologically silent form of cell death (Jorgensen et al., 2017). The extrinsic pathway and intrinsic pathway in host cells, also known as the death receptor pathway and mitochondrial pathway, respectively, are regarded as the two classic apoptotic ways to initiate cell death, as both culminate in the activation of the effector caspases 3 (Elmore, 2007; Jorgensen et al., 2017). Activation of the intrinsic pathway causes mitochondrial outer-membrane permeabilization (MOMP) (Green and Kroemer, 2004; Kroemer et al., 2007) and the release of mitochondrial contents to the cytoplasm (Dewson and Kluck, 2009; Zheng et al., 2016), where cytochrome C forms an apoptosome (Cain et al., 2002; Jiang and Wang, 2004; Taylor et al., 2008; Jorgensen et al., 2017). In the extrinsic pathway, apoptosis is triggered by the binding of a specific ligand and its cognate death receptor, featured with the activation of caspase 8 and caspase 3, which ultimately causes cellular DNA cleavage (Tummers and Green, 2017). Moreover, an endoplasmic reticulum (ER) pathway can elicit cell apoptosis (Kaufman, 2002) by regulating the concentration of Ca2+ and activating the inositol-requiring enzyme 1 (IRE1), protein kinase-like endoplasmic reticulum kinase (PERK), and activating transcription factor (ATF) pathways, which are connected to the mitochondrial pathway (Verma and Datta, 2012). Upon activation of the different intermediate molecules in a signaling cascade, each of these pathways comes across at the same terminal caspase activation step and generally leads to cleavage of various proteins (Ghobrial et al., 2005). The viral ORF3 protein is considered to possess potential apoptotic activity because it interacts with proteins in these apoptosis pathways to induce cell death.
The Structure of the ORF3 Proteins in the Circoviridae Family
In different viruses, such as PCV, DuCV, and CAV, ORF2 is commonly located on the sense strand, and ORF1 and ORF3 are located on the antisense direction. Specifically, for PCV and DuCV, ORF3 is oriented in the opposite direction of ORF2, which is different from CAV (Figure 1A). In PCV2, the ORF3 protein was first described as nonstructural (Liu et al., 2005). Moreover, the length of ORF3 is 315 bp in PCV2 and approximately 612 bp in PCV1, which means that the ORF3 protein in PCV2 is truncated (Figure 1C). In contrast, the CAV ORF3 protein, specifically termed as apoptin, is a compact polypeptide consisting of 121 amino acids (Leliveld et al., 2004). In addition, the amino acid identity of ORF3 in PCV1 and PCV2 is only about 61.5% (Finsterbusch and Mankertz, 2009). The results of sequence comparison analyses revealed that DuCV can be divided into two genotypes: DuCV1 and DuCV2 (Wang et al., 2011; Zhang et al., 2013; Wen et al., 2014) (Figure 1B), and within the same genotype, the ORF3 homology reaches 95.8%–100%. Due to a T/A difference at nucleotide 236 in DuCV1, ORF3 protein (78 aa) is truncated by a premature stop codon. In other words, similar to PCV1 and PCV2, ORF3 protein of DuCV1 is 20 aa shorter at C-terminus than that of DuCV2 (Wu et al., 2018).
Notably, the ORF3 protein of CAV has a short hydrophobic leucine-rich sequence (LRS) at the N-terminus (aa 33-46) mediating self-association and binding of promyelocytic leukemia (PML) protein (Heilman et al., 2006; Janssen et al., 2007) and multiple other cellular partners (Figure 1D). Upon their formation, PML nuclear bodies (PML NBs) recruit the anaphase-promoting complex/cyclosome (APC/C) to these subnuclear structures (Heilman et al., 2006), which takes part in the process of apoptosis. In addition, through the interaction of the proline-rich hydrophobic regions in the N-terminus (aa 1-69), this ORF3 protein forms globular complexes composed of 30-40 monomers (Leliveld et al., 2003a; Leliveld et al., 2003b), which may play important roles in apoptosis regulation.
The Nuclear Localization Signals of ORF3 Proteins
Nuclear localization signals (NLSs) contribute to the understanding of protein subcellular localization. NLSs are classified as either monopartite, characterized by a cluster of basic residues, or bipartite, characterized by two clusters of basic residues separated by several other residues (Canela-Pérez et al., 2019).
A bipartite NLS in CAV ORF3 is composed of NLS1 (aa 82-88) and NLS2 (aa 111-121) (Figure 1D). Due to the recognition of the NLS by members of the importin (IMP) family, the ORF3 protein undergoes active nuclear import. Therefore, in both normal and transformed cells, the ORF3 protein can shuttle in and out of the cell nucleus owing to its NLS and central nuclear export signal (NES) at residues 97-105 (Wang et al., 2004; Poon et al., 2005; Heilman et al., 2006). Moreover, the NLS activity of the CAV ORF3 protein was also found to be regulated by intramolecular masking (Wagstaff and Jans, 2006). For PCV1, an LRS at residues 42-49 and a strong probability of an NES at residues 134-149 in ORF3 were predicted (Hough et al., 2015) (Figure 1C). Interestingly, compared with the sequence of CAV ORF3, the sequence alignment of PCV1 showed an additional NES sequence overlapping a region just upstream of the predicted area for the C-terminal NES (residues 127-136) (Hough et al., 2015). For PCV2, the NLS was confirmed to be in two halves of the C-terminal region at residues 53–68 and 85–104 (Lin et al., 2011). In the N-terminal of the ORF3, an NES motif is located at residues 1–35 (Gu et al., 2016). Furthermore, a variant monopartite type of NLS has been identified at the C-terminal 77–91 residues in the DuCV 2 ORF3, which is essential for its nuclear localization (Wu et al., 2018) (Figure 1B).
The Regulation of ORF3-Induced Apoptosis
In the genome of Circoviridae family members, ORF3s have been recognized as encoding functional proteins connected to apoptosis (Liu et al., 2005; Liu et al., 2006; Karuppannan et al., 2009; Karuppannan and Kwang, 2011; Lin et al., 2011; Xiang et al., 2012; Kucharski et al., 2016). It appears that the subcellular localization of ORF3 proteins and their interactions with specific signaling proteins play crucial roles in selective toxicity prior to the induction of apoptosis.
The Contribution of ORF3 Subcellular Localization to Apoptosis
The subcellular localization of ORF3 proteins is closely related to their NLSs. For instance, in a majority of tumor and transformed cells, CAV ORF3 proteins primarily accumulate in the nucleus (Danen-Van Oorschot et al., 1997; Noteborn, 2009). In addition, the nuclear accumulation of ORF3 proteins in PCV2 and DuCV2 has been confirmed (Lin et al., 2011). Nevertheless, the DuCV1 protein without an NLS is dispersed in the cytoplasm (Wu et al., 2018).
The CAV NES and NLS, residues 74–121 in the C-terminus constitute tumor cell-specific nuclear targeting signals (Danen-Van Oorschot et al., 2003; Kuusisto et al., 2008). Thus, the ORF3 protein has selective toxicity that induces apoptosis in various transformed cells by gathering in the nucleus, whereas in normal cells, the apoptotic activity of the ORF3 protein disappears because of nuclear accumulation impairment (Danen-Van Oorschot et al., 1997; Poon et al., 2005; Helck et al., 2008; Zhao et al., 2013). There are reports of CAV ORF3 protein toxicity towards SV40-transformed fibroblasts and UV-irradiated cells from individuals with hereditary cancer-prone syndromes as well (Danen-Van Oorschot et al., 1997; Zhang et al., 1999; Guelen et al., 2004). Only one report indicates it’s toxicity towards some non-cancerous cells, such as 1BR3 normal human diploid fibroblasts and the early passage secondary culture of normal human embryonal lung fibroblasts 6689, which has no further research about it’s mechanism. And most studies show that apoptosis induction by ORF3 proteins is confined to a broad panel of human tumor cells (Backendorf et al., 2008). Therefore, this study focuses mainly on the different mechanisms of CAV ORF3 protein in cancer cells and in normal cells. In addition, the nuclear localization of the PCV2 ORF3 is correlated with a triggered apoptotic response in porcine peripheral blood mononuclear cells (PBMCs) (Lin et al., 2011). Although the cytoplasmic localization of the PCV1 ORF3 protein does not differ in primary and transformed cells, the ability of ORF3 protein to induce apoptosis selectively relies on the transformation status of the cell (Hough et al., 2015). Similar to ORF3 proteins of CAV and PCV, DuCV ORF3 protein has also been proven to have apoptotic activity (Xiang et al., 2012), implying that it may play a vital role in the pathogenesis of DuCV (Wu et al., 2018). A study showed that the nuclear localization of the DuCV2 ORF3 protein enhanced its apoptotic activity, compared with that of DuCV1, whose ORF3 protein is dispersed in the cytoplasm (Wu et al., 2018). And the analysis of the apoptotic activities of ORF3 proteins in DuCV1 and DuCV2 suggested that the 20 C-terminal residues in the DuCV2 ORF3 self-inhibit the virus-induced apoptotic activity (Wu et al., 2018); however, the mechanism of this inhibition is unclear. In addition, there is a precise difference in the pathogenesis of these two genotypes.
Exploring the tumor cell-specific nuclear signal of the CAV ORF3 protein, evidence from other studies suggests that the ORF3 protein is phosphorylated robustly in a broad panel of tumor cells but negligibly in normal cells (Rohn et al., 2002). Tumor-specific phosphorylation depends on the phosphorylation site threonine 108 (Thr108), which allows its interaction with other proteins and modification by kinases. Because Thr108 phosphorylation inhibits the activity chromosome region maintenance 1 (CRM1), the ORF3 protein with the NES motif, which depends on a functional CRM1, cannot be driven out of the nucleus in tumor cells (Poon et al., 2005) (Figure 2). Although the adjacent threonine (Thr107) was also found to serve as a compensatory phosphorylation site in the event of Thr108 de-phosphorylation, the activity of the ORF3 protein is diminished (Rohn et al., 2005; Lanz et al., 2012).
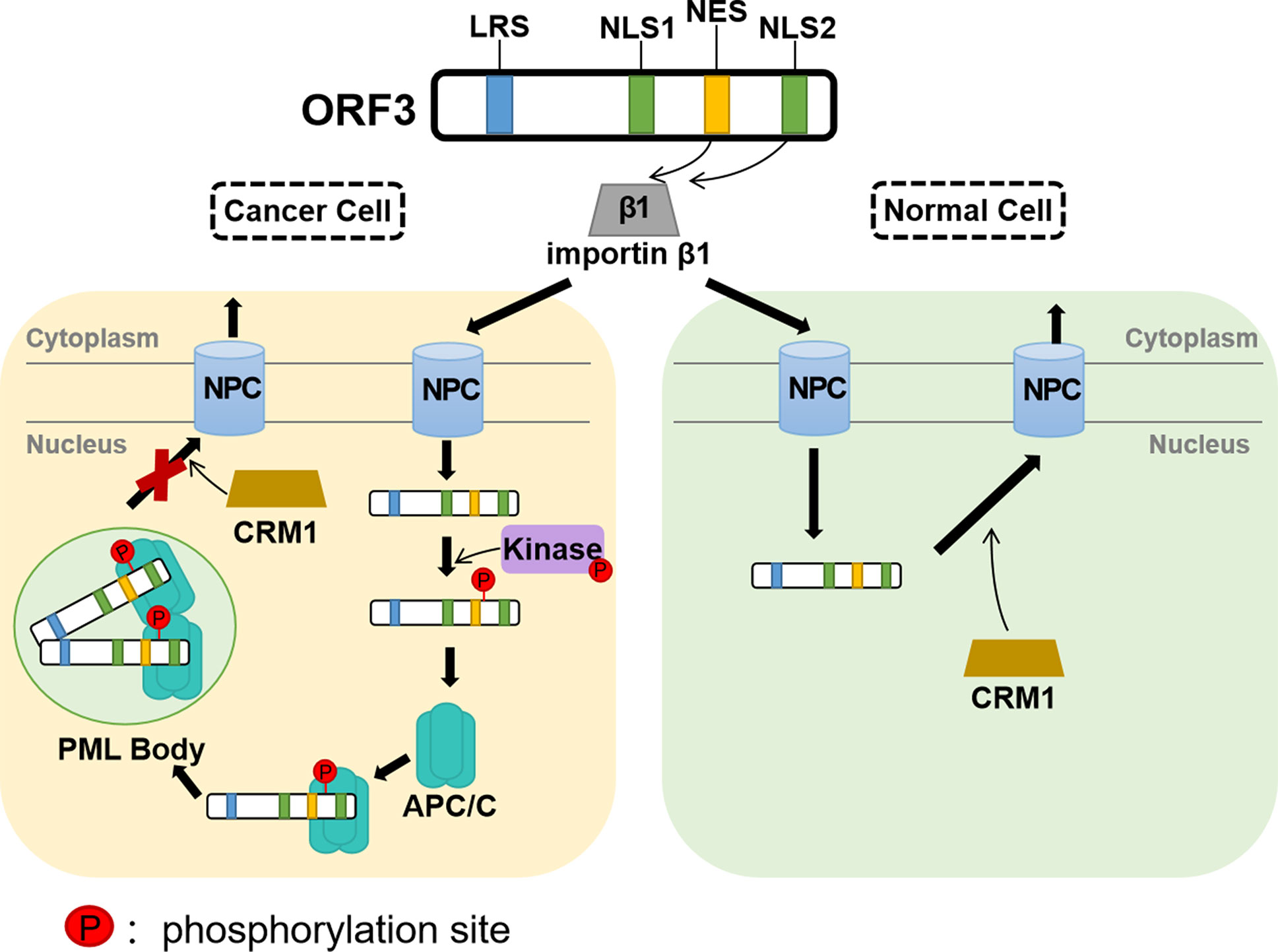
Figure 2 A simulation model of the CAV ORF3 protein associated with various binding partners and its different effects on signal transduction pathways. The ORF3 protein can associate with importin β1, which facilitates its translocation from the cytoplasm to the nucleus. In tumor cells, the ORF3 protein phosphorylated by Kinase, interacts with a subunit of APC/C called APC1 and directly interacts with PML to induce the formation of PML nuclear bodies. The NES of the ORF3 protein is functional in a CRM1-dependent fashion in normal but not in tumor cells, which indicates that in tumor cells, the ORF3 protein is unable to be driven out of the nucleus with the NES out of function when CRM1 inhibited. This hypothetical model may not apply to non-tumor cells in 1BR3 normal human diploid fibroblasts and the early passage secondary culture of normal human embryonal lung fibroblasts 6689 (Guelen et al., 2004).
Further investigation confirmed that the DNA damage response (DDR) regulates the nuclear localization and apoptotic effect of CAV ORF3 through checkpoint kinase1/2 (Chk1/2), which is mediated by the phosphorylation of Thr56 and Thr61 during viral replication (Kucharski et al., 2016; Feng et al., 2020). Upon induction of DNA damage, the ORF3 protein expressed in primary cells is translocated to the nucleus, but in transformed cells, the inhibition of DDR signaling results in the cytoplasmic localization of the protein (Kucharski et al., 2011). To date, the specific molecular mechanisms by which the ORF3 protein moves between subcellular localizations and mediates apoptosis remain to be determined.
Proteins Interacting With ORF3
Since DuCV cannot be isolated and cultured in vitro, researchers have focused more on CAV and PCV; hence the mechanisms of DuCV, particularly the ORF3 protein, remain unclear. The identification of ORF3-associating proteins will guide us to a greater understanding of the mechanism of ORF proteins in apoptosis induction.
CAV
Recently, a number of cellular proteins have been found to interact with the CAV ORF3 protein, including PI3-K (Maddika et al., 2007), PKCβ1 (Jiang et al., 2010; Bullenkamp et al., 2015), CDK1 (Maddika et al., 2009; Zhao et al., 2013), Nmi (Sun et al., 2002), DEDAF (Danen-van Oorschot et al., 2004), APC/C (Teodoro et al., 2004), Hippi (Cheng et al., 2003), Ppil3 (Huo et al., 2008), FADD (Guelen et al., 2004), Bcl10 (Guelen et al., 2004), Hsp70, and Hsc (Chen et al., 2011; Yuan et al., 2013). Among these proteins, PI3-K, Nmi, Hippi, Ppil3, FADD, Bcl10, and Hsp70, were confirmed to interact with the ORF3 protein in the cytoplasm. In the nucleus, the ORF3 protein interactions with DEDAF, PML, and APC were identified (Table 1).
It was identified that CAV ORF3 protein interacts with the p85 Src homology 3 (SH3) domain of phosphatidylinositol 3-kinase (PI3-K) in tumor cells (Maddika et al., 2007). The initiation of the PI3-K/Akt pathway triggers the nuclear translocation and activation of Akt, which results in the induction of cyclin-dependent kinase 2 (CDK2), which in turn leads to the phosphorylation of the ORF3 protein (Maddika et al., 2008; Maddika et al., 2009) (Figure 3). PKCβ in tumor cells has also been proven to play a crucial role in the phosphorylation and nuclear migration of the CAV ORF3 protein, which induces the activation of multiple signaling events involving caspase 9, caspase 3 activation, and cleavage of PKCδ (Jiang et al., 2010). APC1 is a subunit of the anaphase-promoting complex/cyclosome (APC/C) (Teodoro et al., 2004), which is a major regulator of cell cycle function. Upon ORF3 protein shuttling into the nucleus, APC1 is inhibited, resulting in APC/C disruption, and then, the cell undergoes apoptosis following arrest in the G2/M phase of the cell cycle (Teodoro et al., 2004).
Figure 3 Assumption of the CAV ORF3 protein-induced apoptosis of tumor cells (Los et al., 2009). Through association with PI3-K, the ORF3 protein can lead to constitutive activation of PI3-K and the subsequent phosphorylation of Akt. The nuclear translocation of Akt activates CDK2 by direct and indirect phosphorylation, which subsequently phosphorylates the ORF3 protein at Thr108. Then, the ORF3 protein is forced to accumulate in the nucleus. In the cytoplasm, the ORF3 protein can interact with various proteins, including Nmi, Hippi, Ppil3, FADD, Bcl10, and Hsp70. In the nucleus, the ORF3 protein associates with other types of interaction partners, such as DEDAF, PML and APC. In addition, the ORF3 protein can trigger Nurr77 phosphorylation and then make its nuclear export. Nurr77 in the cytoplasm is known to regulate the Bcl-2 family such that apoptosis is induced via the mitochondrial pathway.
A report provides evidence that the association of ORF3 protein with Ppil3 or Hippi may lead to ORF3 protein sequestration in the cytoplasm and prevent the apoptosis of normal cells (Los et al., 2009). Hippi is a protein that interacts with huntingtin-interacting protein 1 (Hip1) (Cheng et al., 2003), and the Hip-1-Hippi complex has been shown to induce apoptosis through the recruitment and activation of the cysteine protease caspase 8 (Gervais et al., 2002). It has also been shown that both in vitro and in human cells, Hippi can bind with the self-multimerization domain of the ORF3 protein, and the ORF3 protein binds to the C-terminal half of Hippi, including its death effector domain-like motif (Cheng et al., 2003). Moreover, in tumor cells, the ORF3 protein and Hippi are primarily located separately in the nucleus and cytoplasm, respectively, whereas in normal cells, they can co-localize in the cytoplasm (Cheng et al., 2003). These features support the theory that, in normal cells, the co-expression of the ORF3 protein and Hippi may suppress the apoptotic activity of the ORF3 protein (Majumder et al., 2006).
PCV
In a study on the modulation of cellular functions by the PCV2, the ORF3 protein was found to interact directly with the E3 ubiquitin ligase pPirh2 (also known as RCHY1) (Liu et al., 2007), which targets p53, a tumor suppressor and a transcription factor (Hong et al., 2014) (Table 1). The interaction suppresses pPirh2 stabilization and causes a decrease in degradation of p53, leading to increased accumulation of p53, which eventually induces a caspase signaling cascade that leads to apoptosis via the intrinsic pathway (Leng et al., 2003) (Figure 4). It was proven that the amino acid residues 20–65 in the ORF3 protein play a crucial role in the interaction of the ORF3 protein with pPirh2, which is competitive over p53 (Karuppannan et al., 2010).
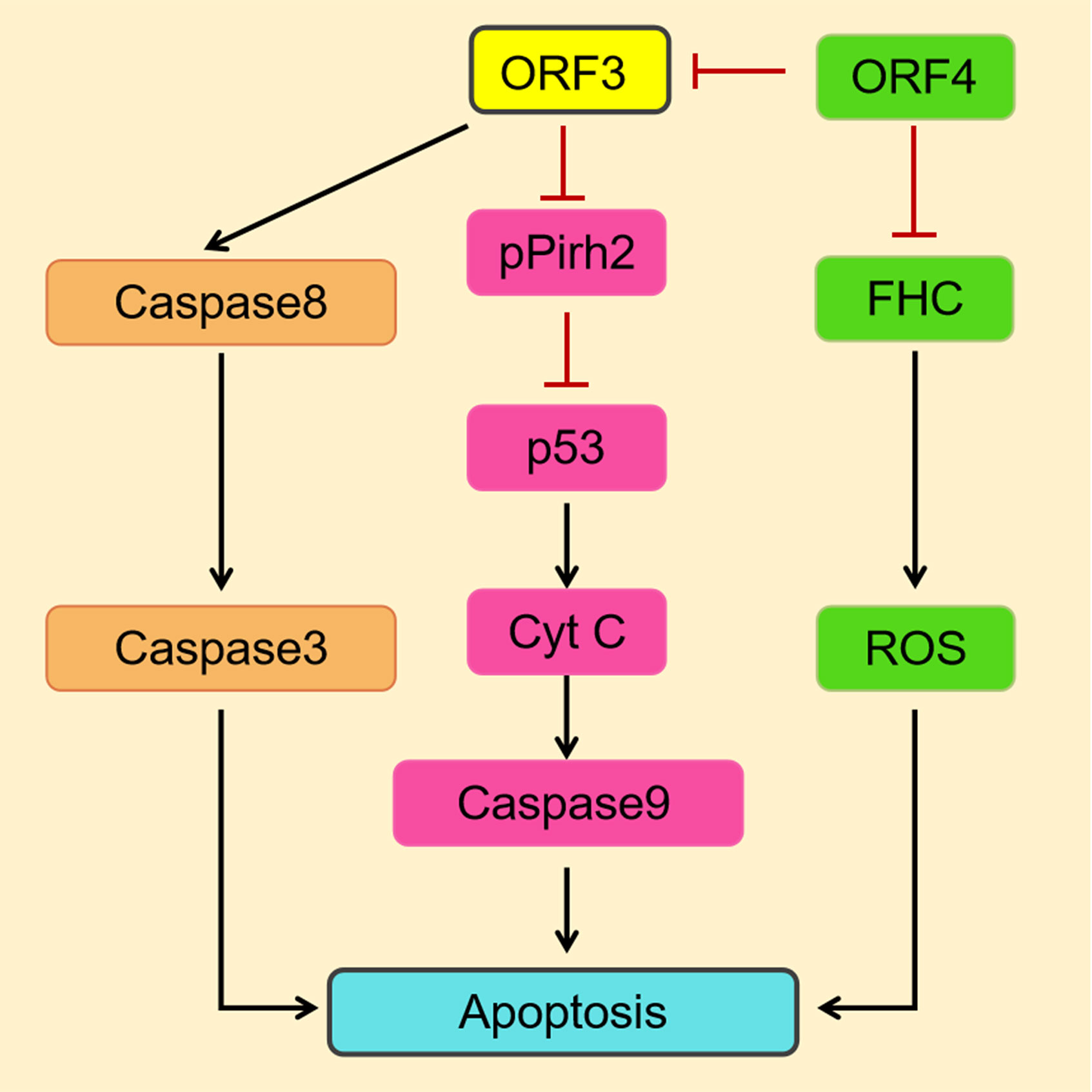
Figure 4 A simulation model describes the mechanisms of PCV2 ORF3-induced apoptosis (Pan et al., 2018). On the one hand, the PCV2 ORF3 protein induces apoptosis through the death receptor pathway via the activation of caspase 8 and caspase 3. On the other hand, the interaction of the ORF3 protein and pPirh2 causes the accumulation of p53 and then upregulates factors downstream of p53 to induce apoptosis. The ORF4 protein inhibits apoptosis by suppressing the activation of ORF3. And by interacting with FHC, which has been suggested to prevent cells from undergoing apoptosis induced by a variety of stimuli, it can ultimately suppress apoptosis by inhibiting the production of ROS.
Previously, specific interactions of PCV2 ORF3 with regulator of G protein signaling 16 (RGS16) and DDE-like transposase, the sequence of which is very similar to that of Tn10 transposase sequence, have been reported (Timmusk et al., 2006; Timmusk et al., 2009) (Table 1). RGSs, a protein family that modifies signaling via G protein-coupled receptors (GPCRs), have a conserved domain of approximately 120 amino acids, which binds to the activated Gα subunit of G-proteins and thereby terminates G-protein signaling (Jean-Baptiste et al., 2006; Xie and Palmer, 2007). More evidence suggests that the direct interaction between PCV2 ORF3 and RGS16 within the cytoplasm leads to the ubiquitin-mediated proteasomal degradation of RGS16, further enhancing NF-κB translocation into the nucleus through the ERK1/2 signaling pathway and increasing the number of IL-6 and IL-8 mRNA transcripts (Choi et al., 2015). However, there is no additional information about the interaction of the DDE-like transposase ORF3 protein.
Currently, studies have indicated that the number of proteins associated with pPirh2 and p53 is greater than 20 and 320, respectively (Jung et al., 2012), which means that the process of ORF3-induced apoptosis may be much more complicated (Pan et al., 2018). Accordingly, future studies may consider whether other factors are involved in regulating apoptosis through their interactions with ORF3 and should determine exactly the roles that the proteins play.
Pathways of ORF3-Mediated Apoptosis
The precise mechanisms of cell death mediated by the ORF3 protein from the Circoviridae family are not clear, although there is a consensus regarding some of the specific molecular events (Table 1).
In contrast to PCV and DuCV, the CAV ORF3 protein has the unique ability to induce tumor-specific cell apoptosis independent of p53 (Zhuang et al., 1995). It is now well established that ORF3 protein expression in CAV results in the activation of caspases (Danen-van Oorschot et al., 2000). Moreover, the mitochondrial pathway is activated by the ORF3 protein through a Nurr77-dependent pathway (Maddika et al., 2005) (Figure 3). What’s particularly interesting is that Nur77, which is shuttled from the nucleus to the cytoplasm upon the transient expression of ORF3 protein, is able to transmit apoptotic signals from the nucleus to mitochondria (Maddika et al., 2005). Upon transfer to the cytoplasm, Nur77 may directly or indirectly cause cytochrome C and AIF release and activate the mitochondrial apoptosis pathway (Los et al., 2009). Moreover, the data indicates that Apaf-1 is required for ORF3-mediated apoptosis (Burek et al., 2006). Taken together, these studies suggest that the ORF3 protein ultimately induces apoptosis modulated by Bcl2 family members through the loss of mitochondrial membrane potential and the cleavage of caspase 3 and caspase 7 (Maddika et al., 2005; Chaabane et al., 2014).
As opposed to the G2/M arrest observed with the CAV ORF3 protein, PCV1 ORF3 induces dramatic G1 cell cycle arrest (Hough et al., 2015). In contrast to that of PCV2, the ORF3 protein of PCV1 appears to be more cytotoxic by activating a caspase-dependent apoptotic pathway, and potentially initiates a caspase-independent poly ADP-ribose polymerase (PARP) cleavage pathway (Chaiyakul et al., 2010). But the precise apoptotic signaling networks have yet to be discovered. Reports have indicated that the PCV2 ORF3 protein induces apoptosis via the death receptor pathway, ultimately by activating both caspase 3 and caspase 8 (Liu et al., 2005) (Figure 4), and the apoptotic response is correlated with its nuclear localization (Kiupel et al., 2005). However, in melanoma cells and mouse primary splenocytes, it showed to induce apoptosis in a completely different manner—a p53-dependent pathway, which is independent of caspase 3 and caspase 8 (Teras et al., 2018).
To date, studies have successfully expressed the ORF3 protein of DuCV in DF-1, CHO, and Sf9 cells (Xiang et al., 2012; Wu et al., 2018), which suggests a cellular tendency of ORF3 by performing as a different expression in cell lines. The expression levels of caspase 3 and caspase 8 mRNA are up-regulated after the transfection of ORF3 protein, indicating that the DuCV ORF3 protein may induce apoptosis through the death receptor pathway, similar to PCV2 ORF3. However, the specific mechanism of the signaling pathway remains to be further determined.
Apoptotic Effects of the Interactions Between ORF3 and Other Viral Proteins
The co-localization and association of the CAV ORF3 and ORF2 protens have been observed. It was revealed that the ORF2 protein directly interacts with the ORF3 protein in the nucleus to downregulate apoptosis by altering the phosphorylation status of the latter, but not completely abolish it (Lai et al., 2017). In the meantime, the de-phosphorylation of ORF3 protein at Thr108 via this interaction seems to participate in modulating the CAV infection process (Lai et al., 2017). There is doubt about whether other phosphorylation sites on the ORF3 protein are similarly regulated by ORF2 or if there are more undiscovered sites.
For PCV2, a newly identified putative protein ORF4 was shown to inhibit apoptosis by suppressing the activation of ORF3, as indicated by significant decreases in caspase 3 and caspase 8 (Gao et al., 2014) (Figure 4). It was confirmed that the ORF4 protein interacts with ferritin heavy chain(FHC), the only one subunit of ferritin that has ferroxidase activity, resulting in a reduction in FHC content, and ultimately suppresses apoptosis by inhibiting the production of ROS (Lv et al., 2015). On the other hand, the ORF4 protein can induce apoptosis via the mitochondrial pathway by interacting with adenine nucleotide translocase 3 (ANT3) (Lin et al., 2018). As stated above, the ORF4 protein has a dual function in the induction of apoptosis, one of which is its association with ORF3 to suppress apoptosis.
Conclusions and Perspectives
The main histological changes associated with infections by Circoviridae family viruses are lymphoid tissues, which show with lymphocyte depletion and necrosis (Woods and Latimer, 2000; Palinski et al., 2016; Dennis et al., 2018; Klaumann et al., 2018) that may be connected to the apoptotic activity of ORF3 proteins. The more studies on ORF3 proteins that are executed, the more evidence confirming its significant role in the pathogenicity of viruses is revealed. Regarding molecular structures, the findings of NLSs in CAV (Rohn et al., 2002), PCV (Lin et al., 2011; Hough et al., 2015), and DuCV (Wu et al., 2018) ORF3 proteins suggest the possibility of these proteins being located in the nucleus. The characterization of the NLSs and NESs of the ORF3 proteins (Wang et al., 2004; Poon et al., 2005; Hough et al., 2015; Gu et al., 2016) has begun to shed light on the subcellular localization of ORF3 proteins, which contributes to their different apoptotic activities. Particularly for the CAV ORF3 protein, phosphorylation sites (Rohn et al., 2002; Rohn et al., 2005; Lanz et al., 2012; Kucharski et al., 2016; Feng et al., 2020), such as Thr108, Thr107, Thr56, and Thr61, are considered to be of vital importance to its nuclear localization and apoptotic effect, whereas it has not yet been discovered in the ORF3 proteins of PCV and DuCV. On the other hand, the CAV ORF3 protein, in particular, has a tumor cell-specific nuclear targeting signal (Danen-Van Oorschot et al., 2003; Kuusisto et al., 2008), which indicates tumor-selective toxicity induced upon its accumulation in the nuclei of transformed cells (Danen-Van Oorschot et al., 1997; Poon et al., 2005; Helck et al., 2008; Zhao et al., 2013). However, whether the ORF3 protein of PCV or DuCV has tumor-selective cytotoxicity remains to be discovered. Hence, these findings suggest that more attention is needed on the aspects highlighted herein to elucidate the mechanisms of the ORF3 proteins.
For the aspect of inducing apoptosis pathways, the PCV2 ORF3 protein activates both caspase 3 and caspase 8 through the death receptor pathway (Liu et al., 2005), which may also be triggered by the DuCV ORF3. The p53-dependent pathway, which is independent of caspase 3 and caspase 8, is considered to be another different manner activated by the PCV2 ORF3 protein. The CAV ORF3 protein is distinct in its ability to induce the selective killing of transformed cells independent of p53 (Zhuang et al., 1995; Noteborn, 2004). More than one-half of human cancers are not responsive to many chemotherapeutics owing to mutations in p53 (Leroy et al., 2014). Therefore, the study on the CAV ORF3 protein provides a unique system for identifying apoptosis pathways (Maddika et al., 2006) that can kill cancer cells selectively independent of p53. Currently, many studies on the CAV ORF3 protein aim to deliver the protein as a potentially safe cancer chemotherapy drug (Ruiz-Martínez et al., 2017; Castro et al., 2018; Wyatt et al., 2019). Clearly, continued studies of pathways related to cell death have obvious therapeutic value (Hough et al., 2015), although the induction activities of the PCV and DuCV ORF3 proteins are weaker than the activity of the CAV ORF3 protein.
In conclusion, although the specific mechanisms of these ORF3 proteins differ in CAV, PCV, and DuCV of the Circoviridae family, they are certainly accompanied by the induction of apoptosis (Guelen et al., 2004; Liu et al., 2005; Xiang et al., 2012), which may provide a theoretical foundation to explain their pathogenesis. Based on a wide array of reported works on the CAV, PCV and DuCV ORF3 proteins, this review offers the first glimpse into the ORF3-induced apoptosis by Circoviridae viruses, which involves specific structures, subcellular localizations and apoptosis pathways of the ORF3 protein, and suggests that further investigations into these apoptosis mechanisms be researched. As stated above, further study will help us obtain a deeper understanding of the molecular mechanisms of ORF3-induced apoptosis and provide a new perspective on the pathogenesis of viruses in the Circoviridae family.
Author Contributions
YZ conceived and wrote the paper. XZ conceived and modified the paper. RJ, ZY, and JH contributed to English proofreading. MW and AC were responsible for revising the manuscript critically for expert content. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (CARS-SVDIP), and China Agricultural Research System (CARS-42-17).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the colleagues who shared their experience regarding infections in avian disease.
References
Backendorf C., Visser A. E., de Boer A. G., Zimmerman R., Visser M., Voskamp P., et al. (2008). Apoptin: therapeutic potential of an early sensor of carcinogenic transformation. Annu. Rev. Pharmacol. Toxicol. 48, 143–169. doi: 10.1146/annurev.pharmtox.48.121806.154910
Bullenkamp J., Gaken J., Festy F., Chong E.Z., Ng T., Tavassoli M.. (2015). Apoptin interacts with and regulates the activity of protein kinase C beta in cancer cells. Apoptosis 20, 831–842. doi: 10.1007/s10495-015-1120-6
Burek M., Maddika S., Burek C. J., Daniel P. T., Schulze-Osthoff K., Los M.. (2006). Apoptin-induced cell death is modulated by Bcl-2 family members and is Apaf-1 dependent. Oncogene 25, 2213–2222. doi: 10.1038/sj.onc.1209258
Cain K., Bratton S. B., Cohen G. M. (2002). The Apaf-1 apoptosome: a large caspase-activating complex. Biochimie 84, 203–214. doi: 10.1016/s0300-9084(02)01376-7
Canela-Pérez I., López-Villaseñor I., Mendoza L., Cevallos A. M., Hernández R. (2019). Nuclear localization signals in trypanosomal proteins. Mol. Biochem. Parasitol. 229, 15–23. doi: 10.1016/j.molbiopara.2019.02.003
Castro J., Ribo M., Benito A., Vilanova M. (2018). Apoptin, A Versatile Protein with Selective Antitumor Activity. Curr. Med. Chem. 25, 3540–3559. doi: 10.2174/0929867325666180309112023
Chaabane W., Cieslar-Pobuda A., El-Gazzah M., Jain M. V., Rzeszowska-Wolny J., Rafat M., et al. (2014). Human-gyrovirus-Apoptin triggers mitochondrial death pathway–Nur77 is required for apoptosis triggering. Neoplasia 16, 679–693. doi: 10.1016/j.neo.2014.08.001
Chaiyakul M., Hsu K., Dardari R., Marshall F., Czub M. (2010). Cytotoxicity of ORF3 proteins from a nonpathogenic and a pathogenic porcine circovirus. J. Virol. 84, 11440–11447s. doi: 10.1128/JVI.01030-10
Chen K., Luo Z., Tang J., Zheng S. J. (2011). A critical role of heat shock cognate protein 70 in Apoptin-induced phosphorylation of Akt. Biochem. Biophys. Res. Commun. 409, 200–204. doi: 10.1016/j.bbrc.2011.04.119
Cheng C. M., Huang S. P., Chang Y. F., Chung W. Y., Yuo C. Y. (2003). The viral death protein Apoptin interacts with Hippi, the protein interactor of Huntingtin-interacting protein 1. Biochem. Biophys. Res. Commun. 305, 359–364. doi: 10.1016/s0006-291x(03)00764-2
Choi C. Y., Rho S. B., Kim H. S., Han J., Bae J., Lee S. J., et al. (2015). The ORF3 protein of porcine circovirus type 2 promotes secretion of IL-6 and IL-8 in porcine epithelial cells by facilitating proteasomal degradation of regulator of G protein signalling 16 through physical interaction. J. Gen. Virol. 96, 1098–1108. doi: 10.1099/vir.0.000046
Danen-van Oorschot A. A., van Der Eb A. J., Noteborn M. H. (2000). The chicken anemia virus-derived protein apoptin requires activation of caspases for induction of apoptosis in human tumor cells. J. Virol. 74, 7072–7078. doi: 10.1128/jvi.74.15.7072-7078.2000
Danen-Van Oorschot A. A., Zhang Y. H., Leliveld S. R., Rohn J. L., Seelen M. C., Bolk M. W., et al. (2003). Importance of nuclear localization of apoptin for tumor-specific induction of apoptosis. J. Biol. Chem. 278, 27729–27736. doi: 10.1074/jbc.M303114200
Danen-van Oorschot A. A. A. M., Voskamp P., Seelen M. C. M. J., van Miltenburg M. H. A. M., Bolk M. W., Tait S. W., et al. (2004). Human death effector domain-associated factor interacts with the viral apoptosis agonist Apoptin and exerts tumor-preferential cell killing. Cell Death Differ. 11, 564–573. doi: 10.1038/sj.cdd.4401391
Dennis T. P. W., Flynn P. J., de Souza W. M., Singer J. B., Moreau C. S., Wilson S. J., et al. (2018). Insights into Circovirus Host Range from the Genomic Fossil Record. J. Virol. 92, e00145–e00118. doi: 10.1128/JVI.00145-18
Dewson G., Kluck R. M. (2009). Mechanisms by which Bak and Bax permeabilise mitochondria during apoptosis. J. Cell Sci. 122, 2801–2808. doi: 10.1242/jcs
Elmore S. (2007). Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 35, 495–516. doi: 10.1080/01926230701320337
Fatoba A. J., Adeleke M. A. (2019). Chicken anemia virus: A deadly pathogen of poultry. Acta Virol. 63, 19–25. doi: 10.4149/av_2019_110
Faurez F., Dory D., Grasland B., Jestin A. (2009). Replication of porcine circoviruses. Virol. J. 6, 60. doi: 10.1186/1743-422X-6-60
Feng C., Liang Y., Teodoro J. G. (2020). The Role of Apoptin in Chicken Anemia Virus Replication. Pathogens 9, 294–307. doi: 10.3390/pathogens9040294
Finsterbusch T., Mankertz A. (2009). Porcine circoviruses–Small but powerful. Virus Res. 143, 177–183. doi: 10.1016/j.virusres.2009.02.009
Gao Z., Dong Q., Jiang Y., Opriessnig T., Wang J., Quan Y., et al. (2014). ORF4-protein deficient PCV2 mutants enhance virus-induced apoptosis and show differential expression of mRNAs in vitro. Virus Res. 183, 56–62. doi: 10.1016/j.virusres.2014.01.024
Gervais F. G., Singaraja R., Xanthoudakis S., Gutekunst C. A., Leavitt B. R., Metzler M., et al. (2002). Recruitment and activation of caspase-8 by the Huntingtin-interacting protein Hip-1 and a novel partner Hippi. Nat. Cell Bio 4, 95–105. doi: 10.1038/ncb735
Ghobrial I. M., Witzig T. E., Adjei A. A. (2005). Targeting apoptosis pathways in cancer therapy. CA Cancer J. Clin. 55, 178–194. doi: 10.3322/canjclin.55.3.178
Green D. R., Kroemer G. (2004). The pathophysiology of mitochondrial cell death. Science 305, 626–629. doi: 10.1126/science.1099320
Gu J., Wang L., Jin Y., Lin C., Wang H., Zhou N., et al. (2016). Characterization of specific antigenic epitopes and the nuclear export signal of the Porcine circovirus 2 ORF3 protein. Vet. Microbiol. 184, 40–50. doi: 10.1016/j.vetmic.2016.01.006
Guelen L., Paterson H., Gaken J., Meyers M., Farzaneh F., Tavassoli M.. (2004). TAT-apoptin is efficiently delivered and induces apoptosis in cancer cells. Oncogene 23, 1153–1165. doi: 10.1038/sj.onc.1207224
Ha Z., Li J. F., Xie C. Z., Li C. H., Zhou H. N., Zhang Y., et al. (2020). First detection and genomic characterization of porcine circovirus 3 in mosquitoes from pig farms in China. Vet. Microbiol. 240:108522. doi: 10.1016/j.vetmic.2019.108522
Hamel A. L., Lin L. L., Nayar G. P. (1998). Nucleotide Sequence of Porcine Circovirus Associated with Postweaning Multisystemic Wasting Syndrome in Pigs. J. Virol. 72, 5262–5267. doi: 10.1128/JVI.72.6.5262-5267.1998
Hattermann K., Schmitt C., Soike D., Mankertz A. (2003). Cloning and sequencing of Duck circovirus (DuCV). Arch. Virol. 148, 2471–2480. doi: 10.1007/s00705-003-0181-y
Heilman D. W., Teodoro J. G., Green M. R. (2006). Apoptin Nucleocytoplasmic Shuttling Is Required for Cell Type-Specific Localization, Apoptosis, and Recruitment of the Anaphase-Promoting Complex/Cyclosome to PML Bodies. J. Virol. 80(15), 7535–7545. doi: 10.1128/JVI.02741-05
Helck S., Regenbogen M., Sturzu A., Gharabaghi A., Feil G., Beck A., et al. (2008). Value of apoptin’s 40-amino-acid C-terminal fragment for the differentiation between human tumor and non-tumor cells. Apoptosis 13, 495–508. doi: 10.1007/s10495-007-0174-5
Hong B., van den Heuvel A. P., Prabhu V. V., Zhang S., El-Deiry W. S. (2014). Targeting tumor suppressor p53 for cancer therapy: strategies, challenges and opportunities. Curr. Drug Targets 15, 80–89. doi: 10.2174/1389450114666140106101412
Hough K. P., Rogers A. M., Zelic M., Paris M., Heilman D. W. (2015). Transformed cell-specific induction of apoptosis by porcine circovirus type 1 viral protein 3. J. Gen. Virol. 96, 351–359. doi: 10.1099/vir.0.070284-0
Huo D. H., Yi L. N., Yang J. (2008). Interaction with Ppil3 leads to the cytoplasmic localization of Apoptin in tumor cells. Biochem. Biophys. Res. Commun. 372, 14–18. doi: 10.1016/j.bbrc.2008.04.178
Janssen K., Hofmann T. G., Jans D. A., Hay R. T., Schulze-Osthoff K., Fischer U., et al. (2007). Apoptin is modified by SUMO conjugation and targeted to promyelocytic leukemia protein nuclear bodies. Oncogene 26, 1557–1566. doi: 10.1038/sj.onc.1209923
Jean-Baptiste G., Yang Z., Greenwood M. T. (2006). Regulatory mechanisms involved in modulating RGS function. Cell Mol. Life Sci. 63, 1969–1985. doi: 10.1007/s00018-006-6066-y
Jiang X., Wang X. (2004). Cytochrome C-mediated apoptosis. Annu. Rev. Biochem. 73, 87–106. doi: 10.1146/annurev.biochem.73.011303.073706
Jiang J., Cole D., Westwood N., Macpherson L., Farzaneh F., Mufti G., et al. (2010). Crucial roles for protein kinase C isoforms in tumor-specific killing by apoptin. Cancer Res. 70, 7242–7252. doi: 10.1158/0008-5472.CAN-10-1204
Jorgensen I., Rayamajhi M., Miao E. A. (2017). Programmed cell death as a defence against infection. Nat. Rev. Immunol. 17, 151–164. doi: 10.1038/nri.2016.147
Jung Y.-S., Qian Y., Chen X. (2012). Pirh2 RING-finger E3 ubiquitin ligase: Its role in tumorigenesis and cancer therapy. FEBS Lett. 586, 1397–1402. doi: 10.1016/j.febslet.2012.03.052
Karuppannan A. K., Kwang J. (2011). ORF3 of porcine circovirus 2 enhances the in vitro and in vivo spread of the of the virus. Virology 410, 248–256. doi: 10.1016/j.virol.2010.11.009
Karuppannan A. K., Jong M. H., Lee S. H., Zhu Y., Selvaraj M., Lau J., et al. (2009). Attenuation of porcine circovirus 2 in SPF piglets by abrogation of ORF3 function. Virology 383 (2), 338–347. doi: 10.1016/j.virol.2008.10.024
Karuppannan A. K., Liu S., Jia Q., Selvaraj M., Kwang J. (2010). Porcine circovirus type 2 ORF3 protein competes with p53 in binding to Pirh2 and mediates the deregulation of p53 homeostasis. Virology 398, 1–11. doi: 10.1016/j.virol.2009.11.028
Kaufman R. J. (2002). Orchestrating the unfolded protein response in health and disease. J. Clin. Invest. 110, 1389–1398. doi: 10.1172/jci0216886
Kiupel M., Stevenson G. W., Galbreath E. J., North A., HogenEsch H., Mittal S. K.. (2005). Porcine circovirus type 2 (PCV2) causes apoptosis in experimentally inoculated BALB/c mice. BMC Vet. Res. 1, 7. doi: 10.1186/1746-6148-1-7
Klaumann F., Correa-Fiz F., Franzo G., Sibila M., Nunez J. I., Segales J., et al. (2018). Current Knowledge on Porcine circovirus 3 (PCV-3): A Novel Virus With a Yet Unknown Impact on the Swine Industry. Front. Vet. Sci. 5, 315. doi: 10.3389/fvets.2018.00315
Kroemer G., Galluzzi L., Brenner C. (2007). Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 87, 99–163. doi: 10.1152/physrev.00013.2006
Kucharski T. J., Gamache I., Gjoerup O., Teodoro J. G. (2011). DNA damage response signaling triggers nuclear localization of the chicken anemia virus protein Apoptin. J. Virol. 85, 12638–12649. doi: 10.1128/JVI.05009-11
Kucharski T. J., Ng T. F., Sharon D. M., Navid-Azarbaijani P., Tavassoli M., Teodoro J. G., et al. (2016). Activation of the Chicken Anemia Virus Apoptin Protein by Chk1/2 Phosphorylation Is Required for Apoptotic Activity and Efficient Viral Replication. J. Virol. 90, 9433–9445. doi: 10.1128/JVI.00936-16
Kuusisto H. V., Wagstaff K. M., Alvisi G., Jans D. A. (2008). The C-terminus of apoptin represents a unique tumor cell-enhanced nuclear targeting module. Int. J. Cancer 123, 2965–2969. doi: 10.1002/ijc.23884
Lai G. H., et al. (2017). VP2 of Chicken Anaemia Virus Interacts with Apoptin for Down-regulation of Apoptosis through De-phosphorylated Threonine 108 on Apoptin. Sci. Rep. 7, 14799. doi: 10.1038/s41598-017-14558-8
Lanz H. L., Florea B. II, Noteborn M. H., Backendorf C. (2012). Development and application of an in vitro apoptin kinase assay. Anal. Biochem. 421, 68–74. doi: 10.1016/j.ab.2011.10.030
Leliveld S. R., Dame R. T., Mommaas M. A., Koerten H. K., Wyman C., Danen-van Oorschot A. A., et al. (2003a). Apoptin protein multimers form distinct higher-order nucleoprotein complexes with DNA. Nucleic Acids Res. 31, 4805–4813. doi: 10.1093/nar/gkg661
Leliveld S. R., Zhang Y. H., Rohn J. L., Noteborn M. H. M., Abrahams J. P. (2003b). Apoptin Induces Tumor-specific Apoptosis as a Globular Multimer. J. Biol. Chem. 278, 9042–9051. doi: 10.1074/jbc.M210803200
Leliveld S. R., Dame R. T., Rohn J. L., Noteborn M. H. M., Abrahams J. P. (2004). Apoptin’s functional N- and C-termini independently bind DNA. FEBS Lett. 557, 155–158. doi: 10.1016/s0014-5793(03)01465-0
Leng R. P., Lin Y., Ma W., Wu H., Lemmers B., Chung S., et al. (2003). Pirh2, a p53-Induced Ubiquitin-Protein Ligase, Promotes p53 Degradation. Cell 112, 779–791. doi: 10.1016/s0092-8674(03)00193-4
Leroy B., Anderson M., Soussi T. (2014). TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum. Mutat. 35, 672–688. doi: 10.1002/humu.22552
Lin W. L., Chien M. S., Wu P. C., Lai C. L., Huang C. (2011). The Porcine Circovirus Type 2 Nonstructural Protein ORF3 Induces Apoptosis in Porcine Peripheral Blood Mononuclear Cells. Open Virol. J. 5, 148–153. doi: 10.2174/1874357901105010148
Lin C., Gu J., Wang H., Zhou J., Li J., Wang S., et al. (2018). Caspase-Dependent Apoptosis Induction via Viral Protein ORF4 of Porcine Circovirus 2 Binding to Mitochondrial Adenine Nucleotide Translocase 3. J. Virol. 92, e00238–e00218. doi: 10.1128/JVI.00238-18
Liu J., Chen I., Kwang J. (2005). Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J. Virol. 79, 8262–8274. doi: 10.1128/JVI.79.13.8262-8274.2005
Liu J., Chen I., Du Q., Chua H., Kwang J. (2006). The ORF3 protein of porcine circovirus type 2 is involved in viral pathogenesis in vivo. J. Virol. 80, 5065–5073. doi: 10.1128/JVI.80.10.5065-5073.2006
Liu J., Zhu Y., Chen I., Lau J., He F., Lau A., et al. (2007). The ORF3 Protein of Porcine Circovirus Type 2 Interacts with Porcine Ubiquitin E3 Ligase Pirh2 and Facilitates p53 Expression in Viral Infection. J. Virol. 81, 9560–9567. doi: 10.1128/jvi.00681-07
Los M., Panigrahi S., Rashedi I., Mandal S., Stetefeld J., Essmann F., et al. (2009). Apoptin, a tumor-selective killer. Biochim. Biophys. Acta 1793, 1335–1342. doi: 10.1016/j.bbamcr.2009.04.002
Luo G., Zhu X., Lv Y., Lv B., Fang J., Cao S., et al. (2018). Crystal Structure of the Dimerized N Terminus of Porcine Circovirus Type 2 Replicase Protein Reveals a Novel Antiviral Interface. J. Virol. 92, e00724–e00718. doi: 10.1128/jvi.00724-18
Lv Q., Guo K., Wang T., Zhang C., Zhang Y. (2015). Porcine circovirus type 2 ORF4 protein binds heavy chain ferritin. J. Biosci. 40, 477–485. doi: 10.1007/s12038-015-9551-3
Maddika S., Booy E. P., Johar D., Gibson S. B., Ghavami S., Los M. (2005). Cancer-specific toxicity of apoptin is independent of death receptors but involves the loss of mitochondrial membrane potential and the release of mitochondrial cell-death mediators by a Nur77-dependent pathway. J. Cell Sci. 118, 4485–4493. doi: 10.1242/jcs.02580
Maddika S., Mendoza F. J., Hauff K., Zamzow C. R., Paranjothy T., Los M. (2006). Cancer-selective therapy of the future: Apoptin and its mechanism of action. Cancer Biol. Ther. 5, 10–19. doi: 10.4161/cbt.5.1.2400
Maddika S., Bay G. H., Kroczak T. J., Ande S. R., Maddika S., Wiechec E., et al. (2007). Akt is transferred to the nucleus of cells treated with apoptin, and it participates in apoptin-induced cell death. Cell Prolif. 40, 835–848. doi: 10.1111/j.1365-2184.2007.00475.x
Maddika S., Wiechec E., Ande S. R., Poon I. K., Fischer U., Wesselborg S., et al. (2008). Interaction with PI3-kinase contributes to the cytotoxic activity of Apoptin. Oncogene 27, 3060–3065. doi: 10.1038/sj.onc.1210958
Maddika S., Panigrahi S., Wiechec E., Wesselborg S., Fischer U., Schulze-Osthoff K., et al. (2009). Unscheduled Akt-triggered activation of cyclin-dependent kinase 2 as a key effector mechanism of apoptin’s anticancer toxicity. Mol. Cell Biol. 29, 1235–1248. doi: 10.1128/MCB.00668-08
Majumder P., Chattopadhyay B., Mazumder A., Das P., Bhattacharyya N. P. (2006). Induction of apoptosis in cells expressing exogenous Hippi, a molecular partner of huntingtin-interacting protein Hip1. Neurobiol. Dis. 22, 242–256. doi: 10.1016/j.nbd.2005.11.003
Nawagitgul P., Morozov I., Bolin S. R., Harms P. A., Sorden S. D., Paul P. S. (2000). Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 81, 2281–2287. doi: 10.1099/0022-1317-81-9-2281
Noteborn M. H. (2004). Chicken anemia virus induced apoptosis: underlying molecular mechanisms. Vet. Microbiol. 98, 89–94. doi: 10.1016/j.vetmic.2003.10.003
Noteborn M. H. M. (2009). Proteins selectively killing tumor cells. Eur. J. Pharmacol. 625, 165–173. doi: 10.1016/j.ejphar.2009.06.068
Danen-Van Oorschot A. A., Fischer D., Grimbergen J. M., Klein B., Zhuang S., Falkenburg J. H., et al. (1997). Apoptin induces apoptosis in human transformed and malignant cells but not in normal cells. Proc. Natl. Acad. Sci. 94, 5843–5847. doi: 10.1073/pnas.94.11.5843
Palinski R., Piñeyro P., Shang P., Yuan F., Guo R., Fang Y., et al. (2016). A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 91, e01879–e01816. doi: 10.1128/JVI.01879-16
Pan Y., Li P., Jia R., Wang M., Yin Z., Cheng A. (2018). Regulation of Apoptosis During Porcine Circovirus Type 2 Infection. Front. Microbiol. 9, 2086. doi: 10.3389/fmicb.2018.02086
Poon I. K., Oro C., Dias M. M., Zhang J., Jans D. A. (2005). Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res. 65, 7059–7064. doi: 10.1158/0008-5472.CAN-05-1370
Raidal S. R., Sarker S., Peters A. (2015). Review of psittacine beak and feather disease and its effect on Australian endangered species. Aust. Vet. J. 93, 466–470. doi: 10.1111/avj.12388
Rohn J. L., Zhang Y. H., Aalbers R. I., Otto N., Den Hertog J., Henriquez N. V., et al. (2002). A Tumor-specific Kinase Activity Regulates the Viral Death Protein Apoptin. J. Biol. Chem. 277, 50820–50827. doi: 10.1074/jbc.M208557200
Rohn J. L., Zhang Y. H., Leliveld S. R., Danen-van Oorschot A. A., Henriquez N. V., Abrahams J. P., et al. (2005). Relevance of apoptin’s integrity for its functional behavior. J. Virol. 79, 1337–1338. doi: 10.1128/JVI.79.2.1337-1338.2005
Rosario K., Breitbart M., Harrach B., Segales J., Delwart E., Biagini P., et al. (2017). Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch. Virol. 162, 1447–1463. doi: 10.1007/s00705-017-3247-y
Ruiz-Martínez S., Castro J., Vilanova M., Bruix M., Laurents D. V., Ribó M., et al. (2017). A truncated apoptin protein variant selectively kills cancer cells. Investigat. New Drugs 35, 260–268. doi: 10.1007/s10637-017-0431-6
Soike D., Albrecht K., Hattermann K., Schmitt C., Mankertz A. (2004). Novel circovirus in mulard ducks with developmental and feathering disorders. Vet. Rec. 154, 792–793. doi: 10.1136/vr.154.25.792
Steinfeldt T., Finsterbusch T., Mankertz A. (2002). Rep and Rep’ Protein of Porcine circovirus Type 1 Bind to the Origin of Replication in Vitro. Virology 291, 152–160. doi: 10.1006/viro.2001.1203
Sun G. J., Tong X., Dong Y., Mei Z. Z., Sun Z. X. (2002). Identification of a Protein Interacting with Apoptin from Human Leucocyte cDNA Library by Using Yeast Two-hybrid Screening. Acta Biochim. Et Biophys. Sin. 34, 369–372. doi: 10.1007/BF02943277
Taniguchi T., Yuasa N., Maeda M., Horiuchi T. (1983). Chronological observations on hemapathological changes in chicks inoculated with chicken anemia agent. Natl. Inst. Anim. Health Q. (Tokyo) 23, 1–12.
Taylor R. C., Cullen S. P., Martin S. J. (2008). Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 9, 231–241. doi: 10.1038/nrm2312
Teodoro J. G., Heilman D. W., Parker A. E., Green M. R. (2004). The viral protein Apoptin associates with the anaphase-promoting complex to induce G2/M arrest and apoptosis in the absence of p53. Genes Dev. 18, 1952–1957. doi: 10.1101/gad.1198404
Teras M., Viisileht E., Pahtma-Hall M., Rump A., Paalme V., Pata P., et al. (2018). Porcine circovirus type 2 ORF3 protein induces apoptosis in melanoma cells. BMC Cancer 18, 1237. doi: 10.1186/s12885-018-5090-2
Timmusk S., Fossum C., Berg M. (2006). Porcine circovirus type 2 replicase binds the capsid protein and an intermediate filament-like protein. J. Gen. Virol. 87, 3215–3223. doi: 10.1099/vir.0.81785-0
Timmusk S., Merlot E., Lövgren T., Järvekülg L., Berg M., Fossum C.. (2009). Regulator of G protein signalling 16 is a target for a porcine circovirus type 2 protein. J. Gen. Virol. 90, 2425–2436. doi: 10.1099/vir.0.008896-0
Todd D. (2000). Circoviruses: immunosuppressive threats to avian species: a review. Avian Pathol. 29, 373–394. doi: 10.1080/030794500750047126
Todd D. (2004). Avian circovirus diseases: lessons for the study of PMWS. Vet. Microbiol. 98, 169–174. doi: 10.1016/j.vetmic.2003.10.010
Tummers B., Green D. R. (2017). Caspase-8: regulating life and death. Immunol. Rev. 277, 76–89. doi: 10.1111/imr.12541
Verma G., Datta M. (2012). The critical role of JNK in the ER-mitochondrial crosstalk during apoptotic cell death. J. Cell Physiol. 227, 1791–1795. doi: 10.1002/jcp.22903
Wagstaff K. M., Jans D. A. (2006). Intramolecular masking of nuclear localization signals: Analysis of importin binding using a novel AlphaScreen-based method. Anal. Biochem. 348, 49–56. doi: 10.1016/j.ab.2005.10.029
Wang Q. M., Fan G. C., Chen J. Z., Chen H. P. (2004). A putative NES mediates cytoplasmic localization of Apoptin in normal cells. Acta Biochim. Biophys. Sin. 36, 817–823. doi: 10.1093/abbs/36.12.817
Wang D., Xie X., Zhang D., Ma G., Wang X., Zhang D.. (2011). Detection of duck circovirus in China: A proposal on genotype classification. Vet. Microbiol. 147, 410–415. doi: 10.1016/j.vetmic.2010.07.014
Wang B., Sun L. D., Liu H. H., Wang Z. D., Zhao Y. K., Wang W., et al. (2018). Molecular detection of novel circoviruses in ticks in northeastern China. Ticks Tick Borne Dis. 9, 836–839. doi: 10.1016/j.ttbdis.2018.03.017
Wen H., Wu Y., Yang C., Zhang X., Lian C., Chen H., et al. (2014). Comments on duck circovirus (DuCV) genotype definition. Gene 538, 207–208. doi: 10.1016/j.gene.2014.01.004
Woods L. W., Latimer K. S. (2000). Circovirus Infection of Nonpsittacine Birds. J. Avian Med. Surg. 14, 154–163. doi: 10.1647/1082-6742(2000)014[0154:CIONB]2.0.CO;2
Wu Z. C., Zhang R. H., Li Y. M., Shao D. H., Chen H., Jiang S. J., et al. (2018). C-terminal 20 residues of ORF3 protein of duck circovirus genotype 2 regulates the nuclear localization and inhibits apoptotic activity of ORF3 protein. Vet. Microbiol. 214, 21–27. doi: 10.1016/j.vetmic.2017.12.002
Wyatt J., Müller M. M., Tavassoli M. (2019). Cancer Treatment Goes Viral: Using Viral Proteins to Induce Tumour-Specific Cell Death. Cancers 11, 1975. doi: 10.3390/cancers11121975
Xiang Q. W., Wang X., Xie Z. J., Sun Y. N., Zhu Y. L., Wang S. J., et al. (2012). ORF3 of duck circovirus: a novel protein with apoptotic activity. Vet. Microbiol. 159, 251–256. doi: 10.1016/j.vetmic.2012.03.045
Xie G. X., Palmer P. P. (2007). How Regulators of G Protein Signaling Achieve Selective Regulation. J. Mol. Bio 366, 349–365. doi: 10.1016/j.jmb.2006.11.045
Yuan L., Zhang L., Dong X., Zhao H., Li S., Han D., et al. (2013). Apoptin selectively induces the apoptosis of tumor cells by suppressing the transcription of HSP70. Tumour Biol. 34, 577–585. doi: 10.1007/s13277-012-0585-y
Zhang Y. H., Abrahams P. J., van der Eb A. J., Noteborn M. H. (1999). The viral protein Apoptin induces apoptosis in UV-C-irradiated cells from individuals with various hereditary cancer-prone syndromes. Cancer Res. 59, 3010–3015.
Zhang Z., Jia R., Lu Y., Wang M., Zhu D., Chen S., et al. (2013). Identification, genotyping, and molecular evolution analysis of duck circovirus. Gene 529, 288–295. doi: 10.1016/j.gene.2013.07.028
Zhao J., Han S. X., Ma J. L., Ying X., Liu P., Li J., et al. (2013). The role of CDK1 in apoptin-induced apoptosis in hepatocellular carcinoma cells. Oncol. Rep. 30, 253–259. doi: 10.3892/or.2013.2426
Zheng J. H., Viacava Follis A., Kriwacki R. W., Moldoveanu T. (2016). Discoveries and controversies in BCL-2 protein-mediated apoptosis. FEBS J. 283, 2690–2700. doi: 10.1111/febs.13527
Keywords: Circoviridae, open reading frame 3 protein, apoptosis, signaling pathways, interaction proteins
Citation: Zhang Y, Zhang X, Cheng A, Wang M, Yin Z, Huang J and Jia R (2021) Apoptosis Triggered by ORF3 Proteins of the Circoviridae Family. Front. Cell. Infect. Microbiol. 10:609071. doi: 10.3389/fcimb.2020.609071
Received: 22 September 2020; Accepted: 21 December 2020;
Published: 02 February 2021.
Edited by:
Pablo Pineyro, Iowa State University, United StatesReviewed by:
Ana Lorena Gutiérrez-Escolano, Instituto Politécnico Nacional de México, MexicoAlan G. Goodman, Washington State University, United States
Copyright © 2021 Zhang, Zhang, Cheng, Wang, Yin, Huang and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renyong Jia, jiary@sicau.edu.cn; Anchun Cheng, chenganchun@vip.163.com
†These authors have contributed equally to this work
 Yanting Zhang
Yanting Zhang Xingcui Zhang1,2,3†
Xingcui Zhang1,2,3†  Anchun Cheng
Anchun Cheng Mingshu Wang
Mingshu Wang Zhongqiong Yin
Zhongqiong Yin Renyong Jia
Renyong Jia