Lutzomyia longipalpis TGF-β Has a Role in Leishmania infantum chagasi Survival in the Vector
- 1Laboratório de Biologia Molecular de Parasitas e Vetores, Instituto Oswaldo Cruz, Rio de Janeiro, Brazil
- 2Parasitology Department, Faculty of Science, Charles University, Prague, Czechia
Despite the increasing number of studies concerning insect immunity, Lutzomyia longipalpis immune responses in the presence of Leishmania infantum chagasi infection has not been widely investigated. The few available studies analyzed the role of the Toll and IMD pathways involved in response against Leishmania and microbial infections. Nevertheless, effector molecules responsible for controlling sand fly infections have not been identified. In the present study we investigated the role a signal transduction pathway, the Transforming Growth Factor-beta (TGF-β) pathway, on the interrelation between L. longipalpis and L. i. chagasi. We identified an L. longipalpis homolog belonging to the multifunctional cytokine TGF-β gene family (LlTGF-β), which is closely related to the activin/inhibin subfamily and potentially involved in responses to infections. We investigated this gene expression through the insect development and in adult flies infected with L. i. chagasi. Our results showed that LlTGF-β was expressed in all L. longipalpis developmental stages and was upregulated at the third day post L. i. chagasi infection, when protein levels were also higher as compared to uninfected insects. At this point blood digestion is finished and parasites are in close contact with the insect gut. In addition, we investigated the role of LlTGF-β on L. longipalpis infection by L. i. chagasi using either gene silencing by RNAi or pathway inactivation by addition of the TGF-β receptor inhibitor SB431542. The blockage of the LlTGF-β pathway increased significantly antimicrobial peptides expression and nitric oxide levels in the insect gut, as expected. Both methods led to a decreased L. i. chagasi infection. Our results show that inactivation of the L. longipalpis TGF-β signal transduction pathway reduce L. i. chagasi survival, therefore suggesting that under natural conditions the parasite benefits from the insect LlTGF-β pathway, as already seen in Plamodium infection of mosquitoes.
Introduction
Leishmaniasis is a serious public health concern, affecting millions of people every year. Leishmaniasis is caused by parasites from the genus Leishmania. These parasites are transmitted in the New and Old World by phlebotomine sand flies belonging to the Lutzomyia and Phlebotomus genera, respectively. When a female sand fly feeds on an infected vertebrate host, macrophages containing amastigote parasites are ingested. Inside the insect gut, the aflagellated immotile rounded parasites transform into flagellated motile promastigotes that multiply and then migrate to the anterior midgut (Lainson and Rangel, 2005). To prevent being eliminated these parasites must resist the blood digestion process (Pruzinova et al., 2018) and adhere to the midgut epithelium prior to insect defecation (Wilson et al., 2010). The presence of Leishmania parasites in the insect gut is not harmless to the vector. For instance, parasites secrete a chitinolytic enzyme that attack and damage the sand fly stomodeal valve, leading to the regurgitation of the gut content at the bite site (Rogers et al., 2008). The intricate interactions among Leishmania parasites, gut microbioma and the insect immunity are complex, and evolved to avoid the harmful consequences of an eventual uncontrolled microbial or leishmanial growth inside the insect gut (Sant'Anna et al., 2014; Kelly et al., 2017;Telleria et al., 2018).
Most of the knowledge on insect immune responses was acquired from Drosophila studies. Pathogens in the insect midgut elicit immune responses that have evolved to eliminate or control infections. These immune responses initiate when the insect innate immune system recognizes the invader. This recognition occurs when pathogen-associated molecular patterns (PAMPs) binds to host-derived pattern recognition receptors (PPRs), leading to the activation of signal transduction pathways. These pathways intensify the immune responses, induce the synthesis of antimicrobial molecules and potentiate effector mechanisms. Among these widely conserved receptor-mediated immune mechanisms, Toll, IMD, and Jak-Stat pathways are the most studied in insects. They are regulators and mediators of insect humoral and cellular immune responses, with intracellular negative regulators that keep these pathways under control (reviewed in Lemaitre and Hoffmann, 2007; Kleino and Silverman, 2014; Lindsay and Wasserman, 2014; Myllymäki and Rämet M, 2014). To date few studies have addressed L. longipalpis immunity. The Toll and IMD pathways can be activated in L. longipalpis LL5 embryonic cells in response to microbial and L. i. chagasi infections (Tinoco-Nunes et al., 2016). Bacterial challenges in L. longipalpis larvae leads to a positive modulation of Pirk gene, a suppressor of the IMD pathway at the signal transduction level (Heerman et al., 2015). In adult L. longipalpis the RNAi-mediated gene silencing of the IMD pathway repressor Caspar decreased Leishmania survival (Telleria et al., 2012). Furthermore the expression of a sand fly defensin was upregulated after artificial infection with different bacteria (Boulanger et al., 2004; Telleria et al., 2013). In insects another important strategy for infection control is the gut environment oxidative stress modulation. The oxidative stress is a consequence of aerobic processes that lead to the production of reactive oxygen species (ROS) such as superoxide, hydrogen peroxide, and nitric oxide (NO), among others (reviewed in Murphy et al., 2011, Kodrik et al., 2015). While aerobic processes occur autonomously in many organisms, some stimuli during development and pathogen challenges can induce ROS production as a result of growth factor receptors and activation of cytokines such as mitogen, integrin and wingless (reviewed in Covarrubias et al., 2008).
The Transforming Growth Factor-beta (TGF-β) pathway, a highly conserved signal transduction pathway in animals, has received very little attention in insects. This cytokine superfamily comprises more than 40 members, grouped into subfamilies according to structure or function: TGF-β stritu sensu, bone morphogenic protein (BMP), decapentaplegic (Dpp), Mullerian inhibiting substance, and activin/inhibin (Massague, 1990). These cytokines are involved in several aspects of animal biology including embryonic development, organogenesis, stress responses, and immune modulation (Huminiecki et al., 2009; Massague, 2012; Chen and Ten Dijke, 2016; Morikawa et al., 2016; Mullen and Wrana, 2017). Activins and BMPs are key regulators of immune response having pro- and anti-inflammatory roles (reviewed in Jones et al., 2004; Aleman-Muench and Soldevila, 2012; Lee et al., 2012; Spottiswoode et al., 2017). Activins are secreted in stimulated immune cells and they can act as immune response activator or repressor depending on the which cell type is stimulated (Ogawa and Funaba, 2011).
Although we have some information regarding immunity effectors in sand flies, there is little information on the cytokine-like molecules possibly involved in their regulation. In the present work, we characterized a L. longipalpis TGF-β gene (LlTGF-β). We also investigated the involvement of LlTGF-β in L. i. chagasi infection. The blockage of this signal transduction pathway revealed an effect on insect immune related molecules and NO production with consequences to L. i chagasi infection.
Materials and Methods
Insects
Sand flies were obtained from a laboratory colony of L. longipalpis established from sand flies caught in Jacobina (Bahia, Brazil). Insects were fed on 50–70 % sucrose ad libitum and females were blood fed on anesthetized hamsters or mice once a week. Females 2–3 day old were used on experimental procedures. All procedures involving animals were performed in accordance with the Brazilian Ethics Committee for Animal Use at FIOCRUZ (CEUA L-016/2018).
Artificial Infection With Leishmania
L. i. chagasi (MHOM/BR/1974/PP75) were cultured in M199 medium, pH 7.4, supplemented with 10% fetal bovine serum and collected at exponential growth phase, washed with PBS, and resuspended in inactivated rabbit blood at 107 parasites/mL. Sand flies were infected with promastigotes for practical reasons, since there is evidence that infections initiated with Leishmania promastigotes, axenic amastigotes or macrophage-derived amastigotes yield to equally successful infections of sand flies (Freitas et al., 2012; Sadlova et al., 2017). Insects were fed on infective blood through chick skin using a Hemotek artificial feeder. As control group, insects were fed on blood.
For the LlTGF-β receptor inhibition assays, the flies were fed with blood containing L. i. chagasi and 10 μM of the TGF-β receptor inhibitor SB431542 (TOCRYS Biosciences). As control the flies were fed with the same volume of the vehicle dimethyl sulfoxide (DMSO) added to the infective blood meal. For these experiments, pools of 10 fully engorged females were collected at 24, 72, and 144 h post infection.
LlTGF-β Gene Identification
Partial L. longipalpis TGF-β gene (LlTGF-β) sequence was identified through the screening of a L. longipalpis EST library. Sequence identity was determined by similarity using the blastx search tool (Altschul et al., 1990) against the NCBI GenBank. Full cDNA sequence was obtained by PCR using an existing cDNA library (Ramalho-Ortigao et al., 2001) with a LlTGF-β internal primers (LlTGFblibrary1-R or LlTGFblibrary2-R) and cDNA library plasmid primer (T3) (Table 1). Phylogeny analysis of LlTGF-β was performed utilizing Mega 6.06 software, using Maximum Likelihood statistical method with bootstrap method with 500 replicates as phylogeny test. The mutation model used was Jones-Taylor-Thornton with Gamma distribution with invariant sites, and 5 rate categories was chosen according to MEGA 6.06 best mutation model analysis.
LlTGF-β Double Stranded RNA Synthesis
LlTGF-β cDNA template was amplified by PCR using primers containing the T7 promoter at the 5' end (dsTGFb-F and dsTGFb-R—Table 1). The control dsRNA cDNA template was obtained from β-galactosidase gene amplified from plasmid pGEM T-easy vector (PROMEGA) (Tinoco-Nunes et al., 2016). The PCR products were used as templates for transcription reactions with the Megascript RNAi Kit (Ambion) according to the manufacturer's instructions and subsequently concentrated to 40 μg/μL using a vacuum microcentrifuge concentrator.
Sandfly Microinjections
L. longipalpis female dsRNA microinjection procedure was adapted from Sant'Anna et al. (2008). Shortly, 3 day old sandflies were microinjected with 32 nL of 4 μg/μL ds-LlTGF-β or control dsRNA solution using micro-capillary glass needle coupled to a Nanoject II microinjector (Drummond). Injected flies were artificially blood fed or infected with Leishmania as previously described and pools of 5 females were collected at the time-points of 24, 48, and 72 h after feeding.
RNA Extraction and cDNA Synthesis
Total RNA was extracted with TRIzol reagent (Invitrogen-Life Technologies) according to the manufacturer's protocol, from groups of 5 to 10 fully engorged females collected at 24, 48, 72, and 144 h after blood feeding, after artificial L. i. chagasi infection, or after dsRNA microinjection. Total RNA was also extracted from pools of unfed females (0 h), males, eggs, and larvae (L1, L2, L3, and L4 instars). RNA was treated with RQ1 RNase-Free DNase (Promega). First strand cDNA was synthetized with SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen), oligo dT(16) primer, and up to 1 μg of total RNA.
Semi-Quantitative PCR of Larvae and Blood Fed or Infected Sand Flies cDNA
Semi-quantitative PCR was carried out using cDNA samples under the following cycling conditions: 96°C/3 min, followed by 25 cycles at 96°C for 45 s, 60°C for 45 s and 72°C for 45 s, and a final extension of 72°C for 5 min. Primers for LlTGF-β and constitutive expression controls, histone (Telleria et al., 2007) and RP49 (Tinoco-Nunes et al., 2016), are listed in Table 1. PCR products were separated in 2.0% ethidium bromide-stained agarose gel. The intensity of amplified products was determined by densitometry using the ImageJ software 1.48 software (Schneider et al., 2012). LlTGF-β transcription was normalized to histone or RP49 amplification, plotted on GraphPad Prism software (version 6.05—GraphPad Software, Inc). RT-PCR procedures were performed at least 3 times with consistent results. Significance was evaluated by t-test and Mann-Whitney post-test, with p < 0.05.
Quantitative PCR Using cDNA From L. longipalpis Silenced for LlTGF-β or Fed With TGF-β Receptor Inhibitor SB431542
Quantitative PCR was performed using the kit iQTM SYBR Green Supermix (Applied Biosystems) under the following cycling conditions: 96°C/3 min, followed by 42 cycles at 96°C for 30 s, 60°C for 20 s. All experiments were performed using three biological cDNA replicates. The primers used are listed on Table 1. Expression was normalized using the L. longipalpis reference gene RP49, relative levels of RNA expressed were calculated using the ΔΔCT method (Schefe et al., 2006) and plotted on GraphPad Prism software. Q-PCR procedures were performed with 3 replicates with consistent results. Significant differences were evaluated by t-test and Mann-Whitney post-test, with p < 0.05.
LlTGF-β Recombinant Protein (LlTGF-βrec)
A 3′-end fragment (359 bp) of the LlTGF-β gene was amplified by PCR using specific primers (TGF-beta-His-F and TGF-beta-His-R), the PCR product was cloned in pGEM-T easy plasmid (Promega), excised with BamH-I and Hind-III restriction enzymes and then subcloned in pET28a expression plasmid. Protein expression was carried out in E. coli (BL21 DE3 strain) system by IPTG induction followed by purification with Ni2+-NTA Agarose His-tagged protein purification system (QIAGEN). The expressed recombinant protein (~15 kDa) was sequenced for identity confirmation by mass spectrometry at the protein sequencing platform at FIOCRUZ.
LlTGF-β Anti-serum Production
LlTGF-βrec (1 mg) was inoculated into 45 days old New Zealand male rabbit with Freund's complete adjuvant, with two subsequent boosts using incomplete Freund's adjuvant. Animal use was approved by the Instituto Oswaldo Cruz Ethical Commitee on Animal Use (CEUA/IOC-010/2018). Serum titration was carried out by immuno-dot blot (not shown), using the corresponding antigen, and revealed with horseradish peroxydase (HRP)-labeled goat anti-rabbit IgG as the secondary antibody. Western blot assays were also carried to verify LlTGF-β anti-serum specificity in sand fly dissected gut samples (Figure S2).
ELISA
ELISA high binding microplates (NOH® ELISA Plate—LBEP196) were coated overnight in a moist chamber with the protein extract of 12 midguts from L. longipalpis 72 h after feeding on blood or blood containing L. i. chagasi. 5 μg of LlTGF-βrec was also used as a positive control. Wells were blocked overnight at 4°C with 1% BSA in PBS. On the following day, wells were incubated overnight at 4°C with anti-LlTGF-β serum 1:100 diluted in 1% BSA in PBS. Wells were washed three times with PBS and incubated overnight at 4°C with 1:40.000 dilution of goat anti-rabbit IgG peroxidase-conjugated antibody. After washing, plates were revealed by adding to each well 50 μl of 3,3′,5,5′-Tetramethylbenzidine (TMP) substrate solution followed by a 15 min incubation at room temperature. Following this step, 50 μl of stop solution (0.2 M H2SO4) was added to each well and incubated for 30 min. The endpoint absorbance was measured at 450 nm.
Nitrite Detection in Sand Fly Guts
Detection of Nitrite-derived NO was performed with pools of 10 midguts (with Malpighian tubules removed) dissected from LlTGF-β or β-gal (control group) dsRNA injected flies artificially fed on defibrinated rabbit blood, or non-injected insects fed on blood containing 10 μM of the TGF-β receptor inhibitor SB431542 or DMSO (control group). Samples were collected at 24, 48, and 72 h after feeding, homogenized in 0.9% NaCl solution, and centrifuged. The supernatant was split in 2 replicates of 50 μL, loaded in a clear polystyrene medium-binding flat bottom 96-well plate (Corning, SIGMA) for Griess Reagent nitrite detection in 150 μL final volume assay following standard procedures. Nitrite levels were measured under 550 nm wave-length in an Infinite M2000 plate reader (TECAN, Switzerland).
Results
LlTGF-β Sequence and Phylogenetic Tree
The full LlTGF-β sequence (GenBank: MF074142) contains 1,158 nucleotides that encode a 386 amino acid sequence (Figure S1). The amino acid sequence contains eight conserved cysteine residues involved in disulfide bonds and a RXXR motif that is a putative catalytic domain. The TGF-β superfamily domain was identified from amino acids 280 to 385.
TGF-β family aminoacid sequences available on NCBI data base were aligned in order to create a TGF-β family tree and assess subgroup similarities. Sequences obtained from organisms of different taxa but belonging to the same TGF-β subfamily were grouped such as activin/inhibin, TGF-β sensu stricto, and glass bottom boat (GBB) member of the BMP subfamily (Figure 1). LlTGF-β grouped with G. morsitans and P. papatasi activin/inhibin sequences with high bootstrap support.
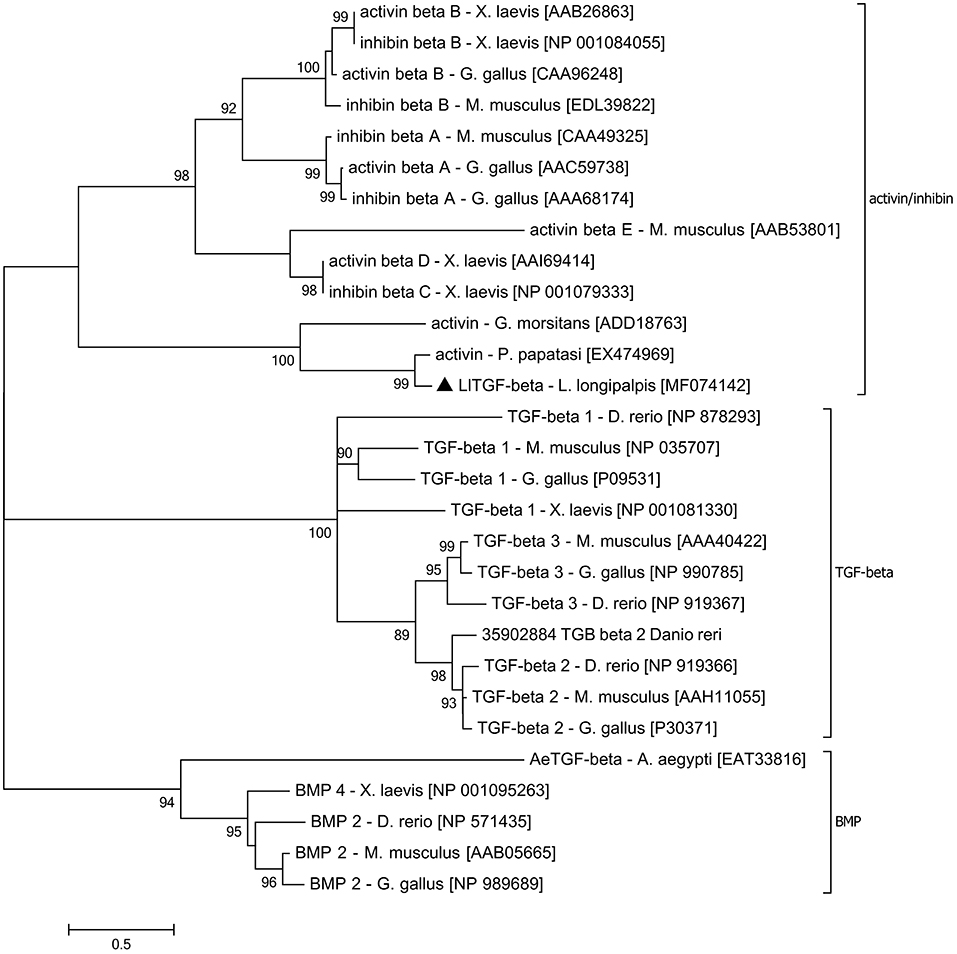
Figure 1. LlTGF-β phylogenetic tree. The LlTGF-β deduced amino acid sequence was aligned with other TGF-β sequences from the insect vectors Aedes aegypti, Glossina morsitans, Phlebotomus papatasi, and the vertebrates Danio rerio, Gallus gallus, Mus musculus, Xenopus laevis. The evolutionary history was inferred using MEGA 6.06 software with Maximum Likelihood method based on the JTT matrix-based model. Evolutionary rate differences among sites [5 categories (+G, parameter = 3.2942)] were modeled by discrete Gamma distribution. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 2.1884% sites). Positions with gaps and missing data were eliminated. The TGF-β subfamily groups are indicated on the right side of the phylogram. Each sequence is labeled with species name followed by GenBank accession number. Branch length represents numbers of substitutions per site. Numbers on the tree nodes indicate bootstrap values higher than 85% (500 replicates).
LlTGF-β Gene Expression in Immature and Adult Insects, and in Infected Females
Due to the activin/inhibin subfamily association to growth and development regulation we investigated its expression in sand fly larval and adult stages. The LlTGF-β gene was ubiquitously expressed in all L. longipalpis developmental stages (Figure 2A), with a slight non-significant increase in adult male and female insects. Comparison among sugar fed, blood fed and Leishmania infected sand flies females revealed that blood ingestion induced LlTGF-β expression when compared to sugar fed females (Figure 2B). Additionally, LlTGF-β gene was significantly upregulated in Leishmania infected females when compared to blood fed insects at 72 h post feeding (Figure 2B). This increased LlTGF-β transcription occurred in both carcass and dissected gut (Figure 2C). Our attempts to use Western blot to verify the presence of LlTGF-β in L. longipalpis guts were hindered by the recognition of mammal TGF-β in blood fed insects (Figure S2). Nevertheless, these bands disappeared rapidly and were totally absent at 24, 48, and 72 h post-blood-feeding. Using the more sensitive ELISA detection we also observed an increased production of LlTGF-β at the protein level in Leishmania infected compared to blood fed females at 72 h after feeding (Figure 2D).
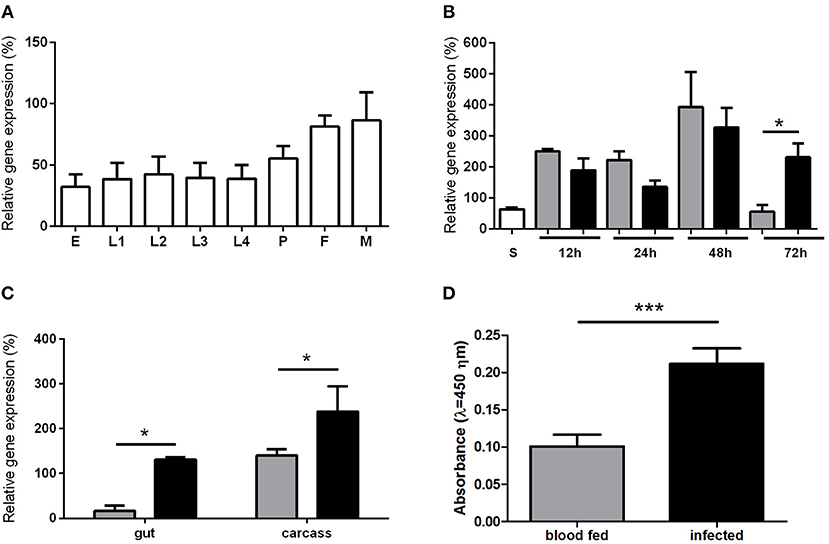
Figure 2. LlTGF-β gene expression in sand flies. (A) L. longipalpis development stages: egg (E), larval stages (L1, L2, L3, L4), pupae (P), adult female (F), and male (M). (B) L. longipalpis fed on sucrose (S) are represented by white bar. Flies collected at 12, 24, 48, and 72 h post blood meal are represented by gray bars or after feeding on infective blood represented by black bars. (C) LlTGF-β expression in gut and carcass 72 h after ingestion of blood without (gray bars) or with (black bars) Leishmania. Gene expression by semi-quantitative PCR was calculated relative to histone or RP49 L. longipalpis reference genes. (D) ELISA for detection of LlTGF-β protein 72 h after blood or infective meal. Bars represent mean with standard error of 3 replicates. Significant differences were evaluated by t-test and Mann-Whitney post-test (*p < 0.05; ***p < 0.001).
Leishmania Infection of Sandflies Silenced for LlTGF-β or Fed With TGF-β Receptor Inhibitor SB431542
To investigate the involvement of the TGF-β signal transduction pathway on the evolution of Leishmania infection in L. longipalpis this pathway was abrogated using RNAi to silence LlTGF-β (Figure S3A) or employing a TGF-β receptor inhibitor. After each treatment L. longipalpis were infected with L. i. chagasi. Both approaches had as consequence a significant decrease in Leishmania infection at 72 and 144 h post infection (Figures 3A,B).
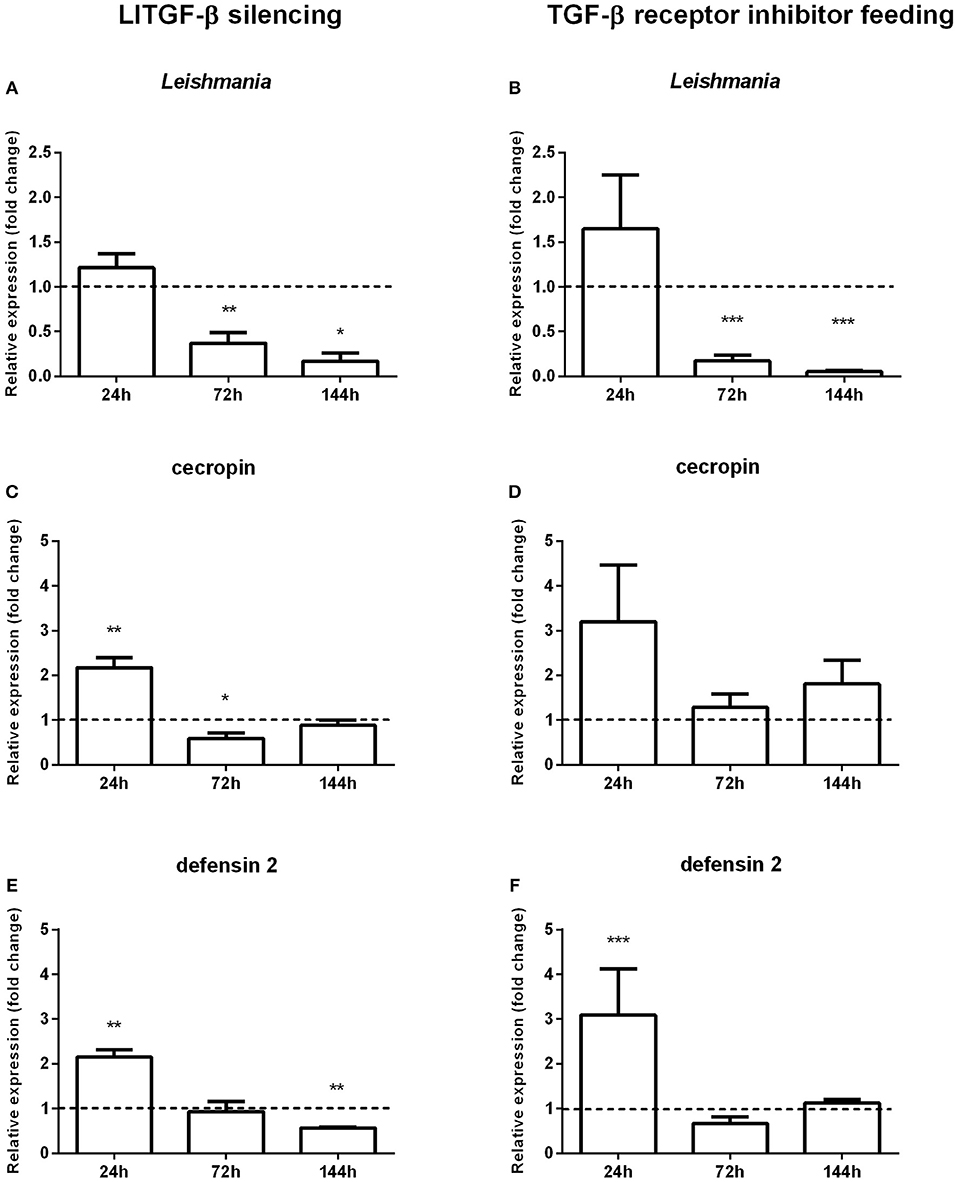
Figure 3. Gene expression in sand flies infected by L. i. chagasi after abrogation of the LlTGF-β signaling pathway. (A,C,E) Gene expression of LlTGF-β silenced insects. (B,D,F) Gene expression of insects fed with TGF-β receptor inhibitor. (A,B) Quantification of L. i. chagasi in sand flies. (C,D) Relative expression of cecropin. (E,F) Relative expression of defensin 2. Samples were collected at 24, 72, and 144 h post infection. Dotted lines indicate gene expression of β-galactosidase dsRNA injected (A,C,E) or DMSO fed flies (B,D,F) control groups, or. Both test and control groups were infected by L. i. chagasi. Comparisons were done between silenced vs. non-silenced, or inhibitor treated vs. non-treated sand fly groups. Bars represent mean with standard error of fold change in gene expression relative to control groups of 3 independent experiments. Significant differences were evaluated by t-test and Mann–Whitney post-test (*p < 0.05; **p < 0.01; ***p < 0.001).
Gene Expression of Immune Effectors After LlTGF-β Knock-Down or Feeding With TGF-β Receptor Inhibitor SB431542 Followed by Leishmania Infection
The decreased L. longipalpis infection by Leishmania upon the TGF-β signaling pathway inhibition led us to investigate the expression of some effector molecules. We assessed the effect of LlTGF-β silencing or TGF-β receptor inhibitor treatment on Toll regulated AMPs, such as defensin 2 and cecropin. In sand flies with TGF-beta silenced and infected with L. i. chagasi a statistically significant increase of both cecropin (Figure 3C) and defensin 2 (Figures 3E,F) expression was observed only at 24 h post infection, with a significant decrease at 72 and 144, respectively. In sand flies fed with the TGF-β receptor inhibitor and infected with Leishmania, cecropin expression levels increased at 24 h, although not significantly (Figure 3D).
Regulation of NO Production by LlTGF-β
To test the possible role of LlTGF-β transduction signaling pathway on NO production, we assessed iNOS expression by qPCR. iNOS levels were increased in LlTGF-β silenced insects at 24 h post infection (Figure 4A) and presented a slight but not significant increase on TGF-β receptor inhibitor SB431542 fed insects at 24 h post infection (Figure 4B). We tested the nitrite production levels in LlTGF-β silenced insects. In non-fed silenced insects nitrite levels were very low and no detectable modulation occurred (Figure S3B). Since the insect acquire Leishmania parasites through blood feeding we also measured the nitrite levels after LlTGF-β gene silencing or TGF-β receptor inhibitor treatements on blood fed L. longipalpis. LlTGF-β gene silencing was followed by an increase of nitrite levels in sand fly guts at 48 h post blood feeding when compared to control groups (Figure 4C). A similar result was obtained in TGF-β receptor inhibitor treated females, with an increase of nitrite at 48 h post blood feeding (Figure 4D).
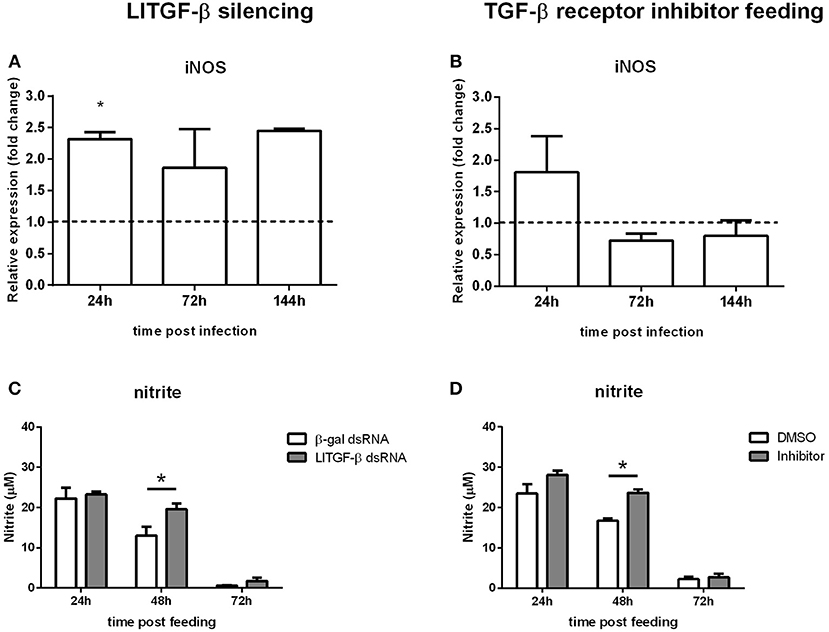
Figure 4. Nitric oxide production in sand flies abrogated for the LlTGF-β signaling pathway. (A) iNOS gene expression of LlTGF-β silenced insects, (B) fed with blood containing TGF-β receptor inhibitor, both (A,B) infected by L. i. chagasi. Samples were collected at 24, 72, and 144 h post infection. Dotted line indicates gene expression of control group, β-gal dsRNA injected or blood containing DMSO fed flies. Bars represent mean with standard error of fold change in gene expression relative to control groups of three independent experiments. Significant differences were evaluated by t-test and Mann-Whitney post-test. (C) NO measurement in LlTGF-β silenced sand flies fed on blood. (D) Nitrite measurement in flies fed on blood containing TGF-β receptor inhibitor. Bars represent mean with standard error of nitrite measurements in test and control groups of 3 independent experiments. Significant differences were evaluated by t-test and Mann-Whitney post-test (*p < 0.05).
Discussion
TGF-β belongs to a family of multifunctional cytokines found in organisms that go from arthropods to mammals (Massague, 1990). TGF-β molecules belonging to different subfamilies has been identified in many insect species including Bombyx mori (Hu et al., 2016), Drosophila (reviewed in Hinck et al., 2016), and the mosquitoes Culex pipiens (Hickner et al., 2015) and Anopheles stephensi (Crampton and Luckhart, 2001). In the malaria vector A. stephensi, a TGF-β homolog named As60A was implicated in the insect immune response to Plasmodium (Crampton and Luckhart, 2001). It was also determined that the mammalian TGF-β 1 present in the ingested blood regulates NO production, modulating the A. stephensi immune response against the parasite (Luckhart et al., 2003).
In sand flies, this is the first report on the characterization of a TGF-β family gene. The identified LlTGF-β sequence contains the catalytic and conserved signature domains of the TGF-β superfamily. Phylogenetic analysis showed LlTGF-β to be highly similar to the activin/inhibin subfamily.
In insects, as much as in other organisms, TGF-β family molecules are involved in ontogeny (O'Connor et al., 2006). The expression profile of the L. longipalpis TGF-β gene shows that it is expressed in all developmental stages and may thus be involved in sand fly ontogeny as well. Additionaly, members of the TGF-β family have a role in nutrient detection and therefore may regulate nutrient acquisition (Chng et al., 2014). In the case of L. longipalpis LlTGF-β expression in blood fed females was increased after blood intake, indicating that the presence of nutrients stimulates activin production. Regarding the effect of L. longipalpis infection with Leishmania, LlTGF-β expression was increased at 3 days post infection, when parasites are in close contact with the insect midgut epithelium, shown both by RT-PCR as well as by ELISA. This is in line with the fact that the activin/inhibin subfamily may be involved in immune response.
TGF-β is considered an anti-inflammatory and immunosuppressive cytokine in mammals. When mice chronically infected with L. major were treated with an antibody anti-TGF-β a pro-inflammatory environment was created with consequent decrease of parasite numbers at the lesion site (Li et al., 2018). In addition, in vitro assays showed that the incubation of recombinant TGF-β with L. braziliensis infected macrophages increased parasite loads (Barral et al., 1995). In an inflammatory situation, Toll-like receptors (TLRs) mediate the activation of activins that consequently suppresses the immune response (negative feedback; Sideras et al., 2013).
To investigate the putative participation of the insect LlTGF-β signaling pathway in response to parasitic infection, we blocked this signaling pathway. Both blocking gene translation or protein interaction with a receptor generated a strong decrease on parasite numbers in late time points, revealing that the suppression of this cytokine-like has a deleterious effect on the parasite. This indicates that this molecule most probably has an anti-inflammatory role in L. longipalpis, as already seen in mammals.
We assessed the effects of suppressing the TGF-β signaling on sand fly immune effector molecules. The silencing of LlTGF-β or inhibition of a TGF-β receptor resulted in an early increased expression of the AMPs cecropin and defensin suggesting that the L. longipalpis activin-like molecule can have a suppressing effect over AMPs expression. AMPs production is one of the main responses against infection in insects and is regulated mainly by the Toll and IMD pathways (Leulier et al., 2003; Kaneko et al., 2004). In Drosophila, AMPs are crucial for the maintenance of gut homeostasis, especially in what concerns microbiota regulation (Bosco-Drayon et al., 2012), and the dpp and dawdle, members of the TGF-β family, have a suppressing effect on AMPs expression (Coggins et al., 2012).
Besides the repressor effect on the insect immunity, activins were previously shown to be involved in immune response regulation throught other pathways. Studies with parasitic infections in vertebrates showed that TGF-β family members are responsible not only for regulating initial pro-inflammatory and late anti-inflammatory responses (Vodovotz et al., 2004), but are also involved in repressing inducible nitric oxide synthase (iNOS) expression, stability and activity in different cell types through mechanisms that were not fully understood (Berg et al., 2007; Aleman-Muench and Soldevila, 2012).
In A. stephensi, the TGF-β gene As60A expression was induced by infection with Plasmodium berghei (Crampton and Luckhart, 2001). Interestingly, human blood derived TGF-β was able to activate the mosquito TGF-β pathway leading to the activation of the MAPK/ERK cascade and downregulation of NO production (Surachetpong et al., 2009). Both A. stephensi and Drosophila are able to modulate ROS levels to combat pathogen infections (Luckhart et al., 1998; Ha et al., 2005; Shrinet et al., 2014). On the other hand, NO mediated an early Drosophila innate immune responses through diptericin and drosomycin expression (Foley and O'Farrell, 2003) indicating that it is an important signaling molecule.
ROS production in L. longipalpis immune response against two different pathogens, Leishmania mexicana and Serratia marcescens, elicited different immune responses. S. marcescens infection increased ROS levels in the insect gut whereas L. mexicana did not (Diaz-Albiter et al., 2012). The RNAi gene silencing of L. longipalpis catalase, a ROS-detoxifying gene, led to decreased L. mexicana infection. These results suggest that ROS are harmful to Leishmania and S. marcescens but Leishmania is able to modulate the L. longipalpis oxidative stress response while S. marcescens is not (Diaz-Albiter et al., 2012).
Our results showed that blocking the TGF-β pathway increased iNOS expression in early times post Leishmania infection. In the following days infection levels were reduced indicating that, in the absence of an intact TGF-β pathway, increased iNOS levels can be deleterious to Leishmania. We would expect that under normal LlTGF-β conditions it would control iNOS expression and consequently nitrite levels. To test this hypothesis we measured nitrite levels in blood fed flies under the effect of both TGF-β pathway blockage methods. We observed that there was a significant increase of nitrite levels 2 days post blood feeding. In hematophagous insects blood digestion requires an efficient control of oxidative stress (Souza et al., 1997) and in L. longipalpis the nitrite levels in blood fed gut reached nearly 100 times higher than sugar fed insects. In order to control the harmful effects of these free radicals insects have a plethora of mechanisms to keep the ROS balance in the gut. In L. longipalpis the TGF-β blockade significantly hindered the nitrite balance in the gut on the second day of blood feeding, time when the subproducts of blood digestion are released in high levels (Graça-Souza et al., 2006). High doses of NO are implicated in parasite killing, but in the case of Leishmania infection the parasites proliferate in the insect gut apparently benefitting from LlTGF-β controlling NO levels.
Our results show the conservation of the TGF-β-mediated signaling in the presence of Leishmania infection in vector and vertebrate hosts, and a role for this citokine-like molecule in the regulation of AMPs expression and nitrite levels.
Data Availability
The datasets generated for this study can be found in GenBank, MF074142.
Author Contributions
TD-B, ET, AT, YT-C contributed conception and design of the study; TD-B, ET, CM, RMC, MS-N, MJ performed experiments; TD-B, ET, AT, YT-C participated in manuscript writing; PV contributed with reagents. All authors contributed to manuscript revision, read and approved the submitted version.
Funding
This work was supported by Instituto Oswaldo Cruz-Fiocruz, PAPES VI Fiocruz, and CNPq. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001. ET was also supported by the International Mobility of Researchers at Charles University number CZ.02.2.69/0.0/0.0/16_027/0008495.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Adriana Oliveira de Araujo, Daiana Zucoloto, and Rute Maria Julio for rearing insects and Nikola Polanska for technical support on nitrite detection assays. We are also grateful to the Genomic Platform—DNA Sequencing—RPT01A, and Mass Spectometry—RPT02A (Rede de Plataformas Tecnológicas FIOCRUZ) and Dr. Mariana Waghabi, IOC-Fiocruz, for the generous gift of the TGF-β receptor inhibitor.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00071/full#supplementary-material
Figure S1. LlTGF-β cDNA and deduced amino acid sequences. Numbers on the left side indicate nucleotides. Numbers on right indicate amino acid residues. Cleavage signal peptide site is indicated by a black triangle (between residues 24 and 25). Conserved cysteine residues are shown in bold characters (residues 282, 283, 311, 315, 350, 351, 383, and 385). Putative catalytic domain motif is indicted by a rectangle (residues 268 to 271). The TGF-β superfamily domain is indicated by underlined characters (residues 280 to 385).
Figure S2. LlTGF-β antiserum assay detecting a single polypeptide from L. longipalpis gut samples. Samples corresponding to 2 guts dissected at 0 (non-fed), 2, 6, 12, 24, 48, and 72 h after blood feeding, and recombinant LlTGF-β as positive control (750 ng) were separated by 12% SDS-PAGE under constant 100 V, and transferred to nitrocellulose membrane during 1 h at 4°C. Membranes were blocked with 5% low-fat dried milk in Tris buffered saline (TBS) supplemented with 0.05% Tween-20. Membranes were washed three times with TBS and incubated for 1 h with anti-LlTGF-β serum at 1:500 dilution. HRP-conjugated goat anti-rabbit IgG at a 1:40,000 dilution was used as secondary antibody. The relative molecular mass of the reactive polypeptides was calculated by comparison with the mobility of molecular mass standards, using ImageJ 1.42q software (NIH, USA).
Figure S3. LlTGF-β silencing: Non-blood fed flies were injected with LlTGF-β dsRNA or β-gal dsRNA (control group) and kept with sugar meal ad libitum. (A) LlTGF-β gene expression in non-blood fed females after LlTGF-β dsRNA microinjection is expressed relative to control group indicated by dotted line as described previously in manuscript qPCR methods; (B) Guts were dissected at 24, 48, and 72 h pos dsRNA injection for nitrite detection assays. Gray bars represent nitrite levels in LlTGF-β dsRNA injected insects. White bars represent nitrite levels in β-gal dsRNA injected insects. Graphics represent mean with standard error of 3 independent experiments. Significant differences were evaluated by t-test and Mann-Whitney post-test (*p < 0.05; **p < 0.01).
References
Aleman-Muench, G. R., and Soldevila, G. (2012). When versatility matters: activins/inhibins as key regulators of immunity. Immunol. Cell. Biol. 90, 137–148. doi: 10.1038/icb.2011.32
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Barral, A., Teixeira, M., Reis, P., Vinhas, V., Costa, J., Lessa, H., et al. (1995). Transforming growth factor-beta in human cutaneous leishmaniasis. Am. J. Pathol. 147, 947–954.
Berg, D. T., Gupta, A., Richardson, M. A., O'Brien, L. A., Calnek, D., and Grinnell, B. W. (2007). Negative regulation of inducible nitric-oxide synthase expression mediated through transforming growth factor-beta-dependent modulation of transcription factor TCF11. J. Biol. Chem. 282, 36837–36844. doi: 10.1074/jbc.M706909200
Bosco-Drayon, V., Poidevin, M., Boneca, I. G., Narbonne-Reveau, K., Royet, J., and Charroux, B. (2012). Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe 12, 153–165. doi: 10.1016/j.chom.2012.06.002
Boulanger, N., Lowenberger, C., Volf, P., Ursic, R., Sigutova, L., Sabatier, L., et al. (2004). Characterization of a defensin from the sand fly Phlebotomus duboscqi induced by challenge with bacteria or the protozoan parasite Leishmania major. Infect. Immun. 72, 7140–7146. doi: 10.1128/IAI.72.12.7140-7146.2004
Chen, W., and Ten Dijke, P. (2016). Immunoregulation by members of the TGFbeta superfamily. Nat. Rev. Immunol. 16, 723–740. doi: 10.1038/nri.2016.112
Chng, W. A., Sleiman, M. S. B., Schüpfer, F., and Lemaitre, B. (2014). Transforming growth factor beta/activin signaling functions as a sugar-sensing feedback loop to regulate digestive enzyme expression. Cell Rep. 9, 336–348. doi: 10.1016/j.celrep.2014.08.064
Coggins, S. A., Estévez-Lao, T. Y., and Hillyer, J. F. (2012). Increased survivorship following bacterial infection by the mosquito Aedes aegypti as compared to Anopheles gambiae correlates with increased transcriptional induction of antimicrobial peptides. Dev. Comp. Immunol. 37, 390–401. doi: 10.1016/j.dci.2012.01.005
Covarrubias, L., Hernandez-Garcia, D., Schnabel, D., Salas-Vidal, E., and Castro-Obregon, S. (2008). Function of reactive oxygen species during animal development: passive or active? Dev. Biol. 320, 1–11. doi: 10.1016/j.ydbio.2008.04.041
Crampton, A., and Luckhart, S. (2001). The role of As60A, a TGF-beta homolog, in Anopheles stephensi innate immunity and defense against Plasmodium infection. Infect. Genet. Evol. 1, 131–141. doi: 10.1016/S1567-1348(01)00017-X
Diaz-Albiter, H., Sant' Anna, M. R., Genta, F. A., and Dillon, R. J. (2012). Reactive oxygen species-mediated immunity against Leishmania mexicana and Serratia marcescens in the phlebotomine sand fly Lutzomyia longipalpis. J. Biol. Chem. 287, 23995–24003. doi: 10.1074/jbc.M112.376095
Foley, E., and O'Farrell, P. H. (2003). Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev. 17, 115–125. doi: 10.1101/gad.1018503
Freitas, V. C., Parreiras, K. P., Duarte, A. P., Secundino, N. F., and Pimenta, P. F. (2012). Development of Leishmania (Leishmania) infantum chagasi in its natural sandfly vector Lutzomyia longipalpis. Am. J. Trop. Med. Hyg. 86, 606–612. doi: 10.4269/ajtmh.2012.11-0386
Graça-Souza, A. V., Maya-Monteiro, C., Paiva-Silva, G. O., Braz, G. R., Paes, M. C., Sorgine, M. H., et al. (2006). Adaptations against heme toxicity in blood-feeding arthropods. Insect Biochem. Mol. Biol. 36, 322–335. doi: 10.1016/j.ibmb.2006.01.009
Ha, E. M., Oh, C. T., Ryu, J. H., Bae, Y. S., Kang, S. W., Jang, I. H., et al. (2005). An antioxidant system required for host protection against gut infection in Drosophila. Dev. Cell. 8, 125–132. doi: 10.1016/j.devcel.2004.11.007
Heerman, M., Weng, J. L., Hurwitz, I., Durvasula, R., and Ramalho-Ortigao, M. (2015). Bacterial infection and immune responses in lutzomyia longipalpis sand fly larvae midgut. PLoS Negl. Trop. Dis. 9:e0003923. doi: 10.1371/journal.pntd.0003923
Hickner, P. V., Mori, A., Zeng, E., Tan, J. C., and Severson, D. W. (2015). Whole transcriptome responses among females of the filariasis and arbovirus vector mosquito Culex pipiens implicate TGF-beta signaling and chromatin modification as key drivers of diapause induction. Funct. Integr. Genomics 15, 439–447. doi: 10.1007/s10142-015-0432-5
Hinck, A. P., Mueller, T. D., and Springer, T. A. (2016). Structural biology and evolution of the TGF-beta family. Cold Spring Harb Perspect. Biol. 8:12. doi: 10.1101/cshperspect.a022103
Hu, X., Jiang, Y., Gong, Y., Zhu, M., Zhu, L., Chen, F., et al. (2016). Important roles played by TGF-beta member of Bmdpp and Bmdaw in BmNPV infection. Mol. Immunol. 73, 122–129. doi: 10.1016/j.molimm.2016.04.004
Huminiecki, L., Goldovsky, L., Freilich, S., Moustakas, A., Ouzounis, C., Heldin, C., et al. (2009). Emergence, development and diversification of the TGF-beta signalling pathway within the animal kingdom. BMC Evol. Biol. 9:28. doi: 10.1186/1471-2148-9-28
Jones, K. L., de Kretser, D. M., Patella, S., and Phillips, D. J. (2004). Activin A and follistatin in systemic inflammation. Mol. Cell. Endocrinol. 225, 119–125. doi: 10.1016/j.mce.2004.07.010
Kaneko, T., Goldman, W. E., Mellroth, P., Steiner, H., Fukase, K., Kusumoto, S., et al. (2004). Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20, 637–649. doi: 10.1016/S1074-7613(04)00104-9
Kelly, P. H., Bahr, S. M., Serafim, T. D., Ajami, N. J., Petrosino, J. F., Meneses, C., et al. (2017). The gut microbiome of the vector lutzomyia longipalpis is essential for survival of leishmania infantum. MBio 8:1. doi: 10.1128/mBio.01121-16
Kleino, A., and Silverman, N. (2014). The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 42, 25–35. doi: 10.1016/j.dci.2013.05.014
Kodrík, D., Bednárová, A., Zemanová, M., and Krishnan, N. (2015). Hormonal regulation of response to oxidative stress in insects-an update. Int. J. Mol. Sci. 16, 25788–25816. doi: 10.3390/ijms161025788
Lainson, R., and Rangel, E. F. (2005). Lutzomyia longipalpis and the eco-epidemiology of American visceral leishmaniasis, with particular reference to Brazil: a review. Mem. Inst. Oswaldo Cruz 100, 811–827. doi: 10.1590/S0074-02762005000800001
Lee, K. B., Taghavi, C. E., Murray, S. S., Song, K. J., Keorochana, G., and Wang, J. C. (2012). BMP induced inflammation: a comparison of rhBMP-7 and rhBMP-2. J. Orthop. Res. 30, 1985–1994. doi: 10.1002/jor.22160
Lemaitre, B., and Hoffmann, J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743. doi: 10.1146/annurev.immunol.25.022106.141615
Leulier, F., Parquet, C., Pili-Floury, S., Ryu, J. H., Caroff, M., Lee, W. J., et al. (2003). The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 4, 478–484. doi: 10.1038/ni922
Li, H. Y., Lin, X. W., Geng, S. L., and Xu, W. H. (2018). TGF-beta and BMP signals regulate insect diapause through Smad1-POU-TFAM pathway. Biochim. Biophys. Acta Mol. Cell. Res. 1865, 1239–1249. doi: 10.1016/j.bbamcr.2018.06.002
Lindsay, S. A., and Wasserman, S. A. (2014). Conventional and non-conventional Drosophila Toll signaling. Dev. Comp. Immunol. 42, 16–24. doi: 10.1016/j.dci.2013.04.011
Luckhart, S., Crampton, A. L., Zamora, R., Lieber, M. J., Dos Santos, P. C., Peterson, T. M., et al. (2003). Mammalian transforming growth factor beta1 activated after ingestion by Anopheles stephensi modulates mosquito immunity. Infect. Immun. 71, 3000–3009. doi: 10.1128/IAI.71.6.3000-3009.2003
Luckhart, S., Vodovotz, Y., Cui, L., and Rosenberg, R. (1998). The mosquito Anopheles stephensi limits malaria parasite development with inducible synthesis of nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 95, 5700–5705. doi: 10.1073/pnas.95.10.5700
Massague, J. (1990). The transforming growth factor-beta family. Annu. Rev. Cell. Biol. 6, 597–641. doi: 10.1146/annurev.cb.06.110190.003121
Massague, J. (2012). TGFbeta signalling in context. Nat. Rev. Mol. Cell. Biol. 13, 616–630. doi: 10.1038/nrm3434
Morikawa, M., Derynck, R., and Miyazono, K. (2016). TGF-beta and the TGF-beta family: context-dependent roles in cell and tissue physiology. Cold Spring Harb. Perspect. Biol. 8:5. doi: 10.1101/cshperspect.a021873
Mullen, A. C., and Wrana, J. L. (2017). TGF-beta family signaling in embryonic and somatic stem-cell renewal and differentiation. Cold Spring Harb. Perspect. Biol. 9:7. doi: 10.1101/cshperspect.a022186
Murphy, M. P., Holmgren, A., Larsson, N. G., Halliwell, B., Chang, C. J., Kalyanaraman, B., et al. (2011). Unraveling the biological roles of reactive oxygen species. Cell. Metab. 13, 361–366. doi: 10.1016/j.cmet.2011.03.010
Myllymäki, H., and Rämet M, M. (2014). JAK/STAT pathway in Drosophila immunity. Scand. J. Immunol. 79, 377–385. doi: 10.1111/sji.12170
O'Connor, M. B., Umulis, D., Othmer, H. G., and Blair, S. S. (2006). Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development 133, 183–193. doi: 10.1242/dev.02214
Ogawa, K., and Funaba, M. (2011). Activin in humoral immune responses. Vitam. Horm. 85, 235–253. doi: 10.1016/B978-0-12-385961-7.00012-3
Pruzinova, K., Sadlova, J., Myskova, J., Lestinova, T., Janda, J., and Volf, P. (2018). Leishmania mortality in sand fly blood meal is not species-specific and does not result from direct effect of proteinases. Parasit. Vectors 11:37. doi: 10.1186/s13071-018-2613-2
Ramalho-Ortigao, J. M., Temporal, P., de Oliveira, S. M. P., Barbosa, A. F., Vilela, M. L., Rangel, E., et al. (2001). Characterization of constitutive and putative differentially expressed mRNAs by means of expressed sequence tags, differential display reverse transcriptase-PCR and randomly amplified polymorphic DNA-PCR from the sand fly vector Lutzomyia longipalpis. Mem. Inst. Oswaldo Cruz. 96, 105–111. doi: 10.1590/S0074-02762001000100012
Rogers, M. E., Hajmová, M., Joshi, M. B., Sadlova, J., Dwyer, D. M., Volf, P., et al. (2008). Leishmania chitinase facilitates colonization of sand fly vectors and enhances transmission to mice. Cell Microbiol. 10, 1363–1372. doi: 10.1111/j.1462-5822.2008.01132.x
Sadlova, J., Myskova, J., Lestinova, T., Votypka, J., Yeo, M., and Volf, P. (2017). Leishmania donovani development in Phlebotomus argentipes: comparison of promastigote- and amastigote-initiated infections. Parasitology 144, 403–410. doi: 10.1017/S0031182016002067
Sant'Anna, M. R., Alexander, B., Bates, P. A., and Dillon, R. J. (2008). Gene silencing in phlebotomine sand flies: xanthine dehydrogenase knock down by dsRNA microinjections. Insect Biochem. Mol. Biol. 38, 652–660. doi: 10.1016/j.ibmb.2008.03.012
Sant'Anna, M. R., Diaz-Albiter, H., Aguiar-Martins, K., Al Salem, W. S., Cavalcante, R. R., Dillon, V., et al. (2014). Colonisation resistance in the sand fly gut: Leishmania protects Lutzomyia longipalpis from bacterial infection. Parasit. Vectors 7:329. doi: 10.1186/1756-3305-7-329
Schefe, J. H., Lehmann, K. E., Buschmann, I. R., Unger, T., and Funke-Kaiser, H. (2006). Quantitative real-time RT-PCR data analysis: current concepts and the novel gene expression's CT difference formula. J. Mol. Med. 84, 901–910. doi: 10.1007/s00109-006-0097-6
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675.
Shrinet, J., Nandal, U. K., Adak, T., Bhatnagar, R. K., and Sunil, S. (2014). Inference of the oxidative stress network in Anopheles stephensi upon Plasmodium infection. PLoS ONE 9:e114461. doi: 10.1371/journal.pone.0114461
Sideras, P., Apostolou, E., Stavropoulos, A., Sountoulidis, A., Gavriil, A., Apostolidou, A., et al. (2013). Activin, neutrophils, and inflammation: just coincidence? Semin. Immunopathol. 35, 481–499. doi: 10.1007/s00281-013-0365-9
Souza, A. V., Petretski, J. H., Demasi, M., Bechara, E. J., and Oliveira, P. L. (1997). Urate protects a blood-sucking insect against hemin-induced oxidative stress. Free Radic. Biol. Med. 22, 209–214. doi: 10.1016/S0891-5849(96)00293-6
Spottiswoode, N., Armitage, A. E., Williams, A. R., Fyfe, A. J., Biswas, S., Hodgson, S. H., et al. (2017). Role of activins in hepcidin regulation during malaria. Infect. Immun. 85:12. doi: 10.1128/IAI.00191-17
Surachetpong, W., Singh, N., Cheung, K. W., and Luckhart, S. (2009). MAPK ERK signaling regulates the TGF-beta1-dependent mosquito response to Plasmodium falciparum. PLoS Pathog. 5:e1000366. doi: 10.1371/journal.ppat.1000366
Telleria, E. L., Martins-da-Silva, A., Tempone, A. J., and Traub-Cseko, Y. M. (2018). Leishmania, microbiota and sand fly immunity. Parasitology 2018, 1–18. doi: 10.1017/S0031182018001014
Telleria, E. L., Pitaluga, A., O Ortigão-Farias, N. J., de Araújo, R. A. P., Ramalho-Ortigão, J. M., and Traub-Cseko, Y. M. (2007). Constitutive and blood meal-induced trypsin genes in Lutzomyia longipalpis. Arch. Insect. Biochem. Physiol. 66, 53–63. doi: 10.1002/arch.20198
Telleria, E. L., Sant'Anna, M. R., Alkurbi, M. O., Pitaluga, A. N., Dillon, R. J., and Traub-Cseko, Y. M. (2013). Bacterial feeding, Leishmania infection and distinct infection routes induce differential defensin expression in Lutzomyia longipalpis. Parasit. Vectors 6:12. doi: 10.1186/1756-3305-6-12
Telleria, E. L., Sant'Anna, M. R. V., Ortigao-Farias, J. R., Pitaluga, A. N., Dillon, V. M., Bates, P. A., et al. (2012). Caspar-like gene depletion reduces leishmania infection in sand fly host lutzomyia longipalpis. J. Biol. Chem. 287, 12985–12993. doi: 10.1074/jbc.M111.331561
Tinoco-Nunes, B., Telleria, E. L., Silva-Neves, M., Marques, C., Azevedo-Brito, D. A., Pitaluga, A. N. (2016). The sandfly Lutzomyia longipalpis LL5 embryonic cell line has active Toll and Imd pathways and shows immune responses to bacteria, yeast and Leishmania. Parasit Vectors 9:4. doi: 10.1186/s13071-016-1507-4
Vodovotz, Y., Zamora, R., Lieber, M. J., and Luckhart, S. (2004). Cross-talk between nitric oxide and transforming growth factor-beta1 in malaria. Curr. Mol. Med. 4, 787–797. doi: 10.2174/1566524043359999
Keywords: Lutzomyia longipalpis, Leishmania, vector-parasite interaction, innate immunity, TGF-β, activin
Citation: Di-Blasi T, Telleria EL, Marques C, Couto RM, Silva-Neves M, Jancarova M, Volf P, Tempone AJ and Traub-Csekö YM (2019) Lutzomyia longipalpis TGF-β Has a Role in Leishmania infantum chagasi Survival in the Vector. Front. Cell. Infect. Microbiol. 9:71. doi: 10.3389/fcimb.2019.00071
Received: 12 December 2018; Accepted: 04 March 2019;
Published: 27 March 2019.
Edited by:
Claudia Ida Brodskyn, Gonçalo Moniz Institute (IGM), BrazilReviewed by:
Diego Luis Costa, National Institute of Allergy and Infectious Diseases (NIAID), United StatesIsabel Mauricio, Instituto de Higiene e Medicina Tropical, Universidade NOVA de Lisboa, Portugal
Copyright © 2019 Di-Blasi, Telleria, Marques, Couto, Silva-Neves, Jancarova, Volf, Tempone and Traub-Csekö. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yara Maria Traub-Csekö, ytraub@ioc.fiocruz.br
 Tatiana Di-Blasi1
Tatiana Di-Blasi1  Erich Loza Telleria
Erich Loza Telleria Magdalena Jancarova
Magdalena Jancarova Petr Volf
Petr Volf Antonio Jorge Tempone
Antonio Jorge Tempone Yara Maria Traub-Csekö
Yara Maria Traub-Csekö