Commentary: The icmF3 Locus is Involved in Multiple Adaptation- and Virulence-related Characteristics in Pseudomonas aeruginosa PAO1
- Ecosystem and Public Health, University of Calgary, Calgary, AB, Canada
A commentary on
The icmF3 locus is involved in multiple adaptation- and virulence-related characteristics in Pseudomonas aeruginosa PAO1
by Lin, J., Cheng, J., Chen, K., Guo, C., Zhang, W., Yang, X., et al. (2015). Front. Cell. Infect. Microbiol. 5:70. doi: 10.3389/fcimb.2015.00070
Pseudomonas aeruginosa is an important pathogen commonly isolated from patients with burns, wounds and cystic fibrosis (Lyczak et al., 2000; Gellatly and Hancock, 2013). The P. aeruginosa strain PAO1 was originally reported as a wound isolate from a patient in Australia in 1955 (Holloway, 1955), and has since been studied in many laboratories as a reference strain (Stover et al., 2000). However, a number of genetic variants of PAO1 in different laboratories have been reported including a large 2.2 Mb inversion and a number of single nucleotide variants and insertion-deletion mutations (Stover et al., 2000; Heurlier et al., 2005; Klockgether et al., 2010).
The type 6 secretion system (T6SS) functions as a molecular weapon that delivers toxic effectors to prokaryotic and eukaryotic target cells (Ho et al., 2014). The T6SS was first functionally characterized in Vibrio cholerae and P. aeruginosa PAO1 by the Mekalanos group in 2006 (Mougous et al., 2006; Pukatzki et al., 2006), and thereafter the PAO1 strain has been used as an important model to study the T6SS functions (Ho et al., 2014; Russell et al., 2014). PAO1 possesses three distinct T6SS clusters (H1, H2, and H3) of which the H1-T6SS delivers six known antimicrobial substrates (Hood et al., 2010; Whitney et al., 2014). The H2- and the H3-T6SS are implicated in both antimicrobial and anti-eukaryotic activities and can secrete PldA and PldB phospholipases, respectively (Lesic et al., 2009; Sana et al., 2012; Russell et al., 2013; Jiang et al., 2014).
The T6SS main structure consists of an outer sheath, an inner tube, and a membrane-bound anchor complex (Basler, 2015). Contraction of the outer sheath ejects the inner tube and its associated effector proteins to the extracellular environment (Ho et al., 2014; Basler, 2015). IcmF, a key T6SS protein, carries an ATPase domain (Ma et al., 2012) and interacts with TssL and TssJ to form a membrane-spanning complex with a hollow space that allows the inner tube and effectors to travel through (Basler, 2015; Durand et al., 2015). The recently published paper by Lin et al. (Lin et al., 2015) reported multiple interesting yet moderate phenotypes associated with the IcmF3 of the H3-T6SS, including iron acquisition, bacterial killing, motility, antibiotic resistance, and virulence. However, the molecular mechanism of IcmF3 involvement in such diverse cellular processes is not clear. Because IcmF is required for T6SS assembly, those reported phenotypes suggest a broad versatile role of the H3-T6SS in PAO1 (Lin et al., 2015). Interestingly, the T6SS in Yersinia pseudotuberculosis is involved in Zn2+ transportation (Wang et al., 2015). An alternative but not mutually exclusive explanation is that IcmF3 itself regulates other cellular processes in addition to its primary role as a key T6SS component.
Lin et al. tested Escherichia coli survival after co-incubation with PAO1 or the icmF3 mutant for 36 h and reported that the CFU of E. coli remained at a high level (108 CFU/ml, see Table S3 in Lin et al., 2015). We found this surprising because the PAO1 strain in our lab (L-PAO1), obtained from J. Mekalanos (Mougous et al., 2006; Basler et al., 2013) and originally from S. Lory, can efficiently kill E. coli cells after 24 h co-incubation. Considering the reported genome divergence of PAO1 in different laboratories (Klockgether et al., 2010), we hypothesized that different PAO1 sublines may have variable killing abilities against E. coli. To test this hypothesis, we carried out E. coli killing assays using several PAO1 strains from different sources (Figure 1A), of which the L-PAO1 has been used extensively in T6SS research (Mougous et al., 2006; Hood et al., 2010; Basler et al., 2013; Ho et al., 2013). The H-PAO1 and the M-PAO1 are isolates from R. Hancock and C. Manoil, respectively, and are the host strains for two defined PAO1 transposon mutant libraries (Jacobs et al., 2003; Lewenza et al., 2005). The P-PAO1 strain and the V-PAO1 strain are from M. Parsek (Colvin et al., 2011) and E. Banin (Cohen et al., 2015), respectively. We followed the reported protocol by Lin et al. (2015) with minor modifications, primarily that the killing was done on LB medium directly instead of a filter membrane. Overnight cultures of PAO1 and E. coli MG1655 carrying a pPSV37 plasmid vector (gentamycin resistance) (Lee et al., 2010) were washed with fresh LB and then mixed at a 10 to 1 ratio, followed by co-incubation on a LB agar plate at 37°C for 36 h. Survival of E. coli was enumerated by serial dilutions on LB-gentamycin plates. Our results demonstrate that L-PAO1, V-PAO1, and H-PAO1 eliminated E. coli MG1655 after co-incubation, whereas P-PAO1 and M-PAO1 had little killing activity against E. coli (Figures 1B,C). To test if the T6SS is required for the observed killing, we constructed the tssB1 deletion mutant (ΔH1-T6SS) and the tssB1-3 triple deletion mutant (ΔT6SS) in the L-PAO1 strain. Our results show that both mutants killed E. coli efficiently, suggesting L-PAO1 possesses other antimicrobial mechanisms independent of the T6SS clusters.
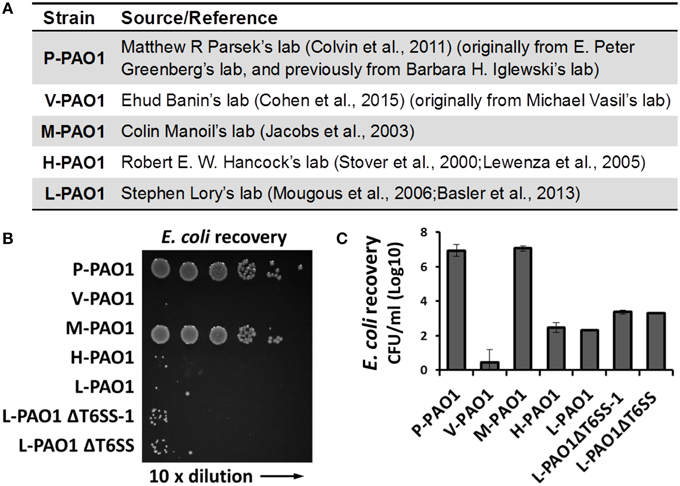
Figure 1. Survival of E. coli MG1655 cells after incubation with different Pseudomonas aeruginosa PAO1 strains. (A) The source of strains used in this study. (B) Survival of E. coli MG1655 pPSV37 enumerated by serial dilutions. L-PAO1ΔT6SS-1 is the tssB1 mutant defective in the H1-T6SS, and L-PAO1ΔT6SS is the tssB1-3 triple mutant defective in all three T6SS systems. (C) Summary of the killing assays. Experiments were performed three times. The mean values and the standard errors are shown.
The genome divergence of different PAO1 strains is known to cause phenotypic variations in virulence (Preston et al., 1995; Klockgether et al., 2010). Here we show different PAO1 strains also differ in their capability of killing neighboring cells. In complex multispecies environments such as the cystic fibrosis patient's lung, it is conceivable that competition between different species may select for PAO1 mutants with enhanced killing abilities. However, how PAO1 strains during lab passage diverge to gain/lose antimicrobial properties is not intuitively apparent. Nonetheless, because PAO1 is widely used as a model strain, researchers should be aware of the strain variations and should provide detailed description of the strain source to allow the P. aeruginosa community to better interpret the results.
Author Contributions
LT, XL, RM performed the experiments. TD conceived the study and designed the experiment. LT and TD wrote the paper.
Funding Statement
This work was supported by a start-up grant of the University of Calgary and a Canadian Institutes of Health Research (CIHR) operating grant to TD. TD is also supported by a Government of Canada Research Chair award, a Canadian Foundation for Innovation grant (CFI-JELF), and an Alberta Innovation and Advanced Education grant.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to John Mekalanos for providing L-PAO1, Shawn Lewenza for H-PAO1 and Joe J. Harrison for P-PAO1, M-PAO1, and V-PAO1 strains. We thank the Advancing Canadian Wastewater Assets (ACWA) for providing infrastructure support.
References
Basler, M. (2015). Type VI secretion system: secretion by a contractile nanomachine. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 370:20150021. doi: 10.1098/rstb.2015.0021
Basler, M., Ho, B. T., and Mekalanos, J. J. (2013). Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894. doi: 10.1016/j.cell.2013.01.042
Cohen, D., Mechold, U., Nevenzal, H., Yarmiyhu, Y., Randall, T. E., Bay, D. C., et al. (2015). Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 112, 11359–11364. doi: 10.1073/pnas.1421450112
Colvin, K. M., Gordon, V. D., Murakami, K., Borlee, B. R., Wozniak, D. J., Wong, G. C. L., et al. (2011). The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7:e1001264. doi: 10.1371/journal.ppat.1001264
Durand, E., Nguyen, V. S., Zoued, A., Logger, L., Péhau-Arnaudet, G., Aschtgen, M.-S., et al. (2015). Biogenesis and structure of a type VI secretion membrane core complex. Nature 523, 555–560. doi: 10.1038/nature14667
Gellatly, S. L., and Hancock, R. E. W. (2013). Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis. 67, 159–173. doi: 10.1111/2049-632X.12033
Heurlier, K., Dénervaud, V., Haenni, M., Guy, L., Krishnapillai, V., and Haas, D. (2005). Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J. Bacteriol. 187, 4875–4883. doi: 10.1128/JB.187.14.4875-4883.2005
Ho, B. T., Basler, M., and Mekalanos, J. J. (2013). Type 6 secretion system-mediated immunity to type 4 secretion system-mediated gene transfer. Science 342, 250–253. doi: 10.1126/science.1243745
Ho, B. T., Dong, T. G., and Mekalanos, J. J. (2014). A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15, 9–21. doi: 10.1016/j.chom.2013.11.008
Holloway, B. W. (1955). Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13, 572–581. doi: 10.1099/00221287-13-3-572
Hood, R. D., Singh, P., Hsu, F., Güvener, T., Carl, M. A., Trinidad, R. R. S., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. doi: 10.1016/j.chom.2009.12.007
Jacobs, M. A., Alwood, A., Thaipisuttikul, I., Spencer, D., Haugen, E., Ernst, S., et al. (2003). Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 100, 14339–14344. doi: 10.1073/pnas.2036282100
Jiang, F., Waterfield, N. R., Yang, J., Yang, G., and Jin, Q. (2014). A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe 15, 600–610. doi: 10.1016/j.chom.2014.04.010
Klockgether, J., Munder, A., Neugebauer, J., Davenport, C. F., Stanke, F., Larbig, K. D., et al. (2010). Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J. Bacteriol. 192, 1113–1121. doi: 10.1128/JB.01515-09
Lee, P.-C., Stopford, C. M., Svenson, A. G., and Rietsch, A. (2010). Control of effector export by the Pseudomonas aeruginosa type III secretion proteins PcrG and PcrV. Mol. Microbiol. 75, 924–941. doi: 10.1111/j.1365-2958.2009.07027.x
Lesic, B., Starkey, M., He, J., Hazan, R., and Rahme, L. G. (2009). Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiol. Read. Engl. 155, 2845–2855. doi: 10.1099/mic.0.029082-0
Lewenza, S., Falsafi, R. K., Winsor, G., Gooderham, W. J., McPhee, J. B., Brinkman, F. S. L., et al. (2005). Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 15, 583–589. doi: 10.1101/gr.3513905
Lin, J., Cheng, J., Chen, K., Guo, C., Zhang, W., Yang, X., et al. (2015). The icmF3 locus is involved in multiple adaptation- and virulence-related characteristics in Pseudomonas aeruginosa PAO1. Front. Cell. Infect. Microbiol. 5:70. doi: 10.3389/fcimb.2015.00070
Lyczak, J. B., Cannon, C. L., and Pier, G. B. (2000). Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. Inst. Pasteur 2, 1051–1060. doi: 10.1016/S1286-4579(00)01259-4
Ma, L.-S., Narberhaus, F., and Lai, E.-M. (2012). IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J. Biol. Chem. 287, 15610–15621. doi: 10.1074/jbc.M111.301630
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Preston, M. J., Fleiszig, S. M., Zaidi, T. S., Goldberg, J. B., Shortridge, V. D., Vasil, M. L., et al. (1995). Rapid and sensitive method for evaluating Pseudomonas aeruginosa virulence factors during corneal infections in mice. Infect. Immun. 63, 3497–3501.
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Russell, A. B., LeRoux, M., Hathazi, K., Agnello, D. M., Ishikawa, T., Wiggins, P. A., et al. (2013). Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512. doi: 10.1038/nature12074
Russell, A. B., Peterson, S. B., and Mougous, J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. doi: 10.1038/nrmicro3185
Sana, T. G., Hachani, A., Bucior, I., Soscia, C., Garvis, S., Termine, E., et al. (2012). The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J. Biol. Chem. 287, 27095–27105. doi: 10.1074/jbc.M112.376368
Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. doi: 10.1038/35023079
Wang, T., Si, M., Song, Y., Zhu, W., Gao, F., Wang, Y., et al. (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. 11:e1005020. doi: 10.1371/journal.ppat.1005020
Keywords: Pseudomonas aeruginosa, PAO1, antimicrobial activity, T6SS, IcmF, strain divergence
Citation: Tang L, Liang X, Moore R and Dong TG (2015) Commentary: The icmF3 Locus is Involved in Multiple Adaptation- and Virulence-related Characteristics in Pseudomonas aeruginosa PAO1. Front. Cell. Infect. Microbiol. 5:83. doi: 10.3389/fcimb.2015.00083
Received: 20 October 2015; Accepted: 03 November 2015;
Published: 18 November 2015.
Edited by:
Matthew S. Francis, Umeå University, SwedenReviewed by:
Michael L. Vasil, University of Colorado School of Medicine, USAAnnabelle Merieau, University of Rouen, France
Copyright © 2015 Tang, Liang, Moore and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao G. Dong, tdong@ucalgary.ca
 Le Tang
Le Tang Xiaoye Liang
Xiaoye Liang Richard Moore
Richard Moore  Tao G. Dong
Tao G. Dong