Quinazolinones as Competitive Inhibitors of Carbonic Anhydrase-II (Human and Bovine): Synthesis, in-vitro, in-silico, Selectivity, and Kinetics Studies
- 1Natural and Medical Sciences Research Center, University of Nizwa, Nizwa, Oman
- 2International Center for Chemical and Biological Sciences, H. E. J. Research Institute of Chemistry, University of Karachi, Karachi, Pakistan
- 3Department of Chemistry, Government College University, Faisalabad, Pakistan
- 4Institute of Chemical Sciences, Bahauddin Zakariya University, Multan, Pakistan
Carbonic anhydrase-II (CA-II) is associated with glaucoma, malignant brain tumors, and renal, gastric, and pancreatic carcinomas and is mainly involved in the regulation of the bicarbonate concentration in the eyes. CA-II inhibitors can be used to reduce the intraocular pressure usually associated with glaucoma. In search of potent CA-II inhibitors, a series of quinazolinones derivatives (4a-p) were synthesized and characterized by IR and NMR spectroscopy. The inhibitory potential of all the compounds was evaluated against bovine carbonic anhydrase-II (bCA-II) and human carbonic anhydrase-II (hCA-II), and compounds displayed moderate to significant inhibition with IC50 values of 8.9–67.3 and 14.0–59.6 μM, respectively. A preliminary structure-activity relationship suggested that the presence of a nitro group on the phenyl ring at R position contributes significantly to the overall activity. Kinetics studies of the most active inhibitor, 4d, against both bCA-II and hCA-II were performed to investigate the mode of inhibition and to determine the inhibition constants (Ki). According to the kinetics results, 4d is a competitive inhibitor of bCA-II and hCA-II with Ki values of 13.0 ± 0.013 and 14.25 ± 0.017 μM, respectively. However, the selectivity index reflects that the compounds 4g and 4o are more selective for hCA-II. The binding mode of these compounds within the active sites of bCA-II and hCA-II was investigated by structure-based molecular docking. The docking results are in complete agreement with the experimental findings.
Introduction
Carbonic anhydrases (CAs, EC 4.2.1.1) are zinc-containing metallo-enzymes, found in animals, plants, algae, archaea, and eubacteria. CAs are encoded by three gene families, α-CA, β-CA, and γ-CA, that are evolutionarily unrelated (Hewett-Emmett, 2000; Jakubowski et al., 2018). These metallo-enzymes use zinc as a cofactor for the reversible inter-conversion of carbon dioxide and bicarbonate, while α-CAs possess high versatility, being able to catalyze other hydrolytic processes (Hewett-Emmett, 2000). Carbonic anhydrases are a class of hydrolase enzymes (Pocker and Meany, 1967; Arslan, 2001). In humans, more than 16 isoforms of carbonic anhydrase (hCA) are present (Shank et al., 2005; Shaik et al., 2019). CAs are involved in different physiological and pathological processes (Lindskog, 1997; Aggarwal et al., 2013; Ozensoy Guler et al., 2016). Consequently, these enzymes are interesting therapeutic targets for the treatment of pathological disorders (Chegwidden et al., 2000; Krishnamurthy et al., 2008; Supuran, 2008). CA-II is mainly involved in the regulation of the bicarbonate concentration in the eyes. CA-II inhibitors can be used to reduce the intraocular pressure usually associated with glaucoma (Supuran and Scozzafava, 2007; Pastorekova and Supuran, 2014; Ruusuvuori and Kaila, 2014; Zaraei et al., 2019). Moreover, CA-II is also expressed in malignant brain tumors (Parkkila et al., 1995) and renal, gastritis, and pancreatic carcinomas (Frazier et al., 1990; Pastorekova et al., 1997; Parkkila et al., 2000). The inhibitors of CA-II have also been considered as an adjunct in cancer chemotherapy (Zaraei et al., 2019).
These highly abundant proteins are involved in crucial physiological processes related with respiration. These enzymes are mainly involved in pH/CO2 homeostasis, secretion of electrolytes in tissues/organs, and transportation of CO2 and bicarbonate between the lungs and metabolizing tissues. Other than that these enzymes are also involved in many other physiological or pathological processes, such as bone resorption, gluconeogenesis, calcification, lipogenesis and ureagenesis, and tumorgenicity (Hewett-Emmett, 2000). CA-II has also been involved in glaucoma, epilepsy, leukemia, and cystic fibrosis (Achal and Pan, 2011; Sentürk et al., 2011).
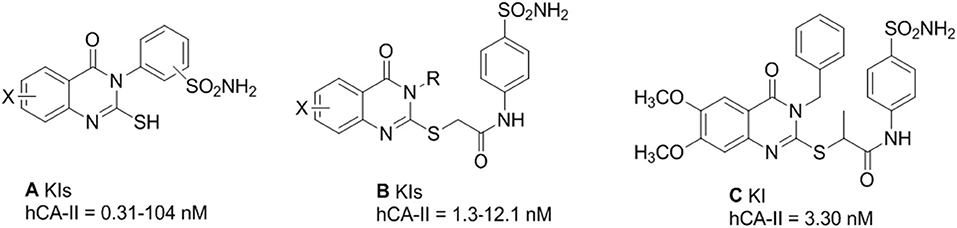
Quinazolinones are N-containing heterocyclic compounds that are widely distributed in nature, including in plants and microorganisms (He et al., 2017). Quinazolinone emerged as a privileged class of heterocyclic compounds with an increasing number of drug candidates and is regularly used in medicinal chemistry (Khan et al., 2014, 2015, 2016). The 2,3-Disubstituted quinazolinones retain anticancer (Al-Suwaidan et al., 2013), anticonvulsant (Gawad et al., 2011), anti-microbial (Al-Amiery et al., 2014), and anti-inflammatory activities (Alaa et al., 2016). Several derivatives of 3-(4-Aminosulfonyl)-phenyl-2-mercapto-substituted-4(3H)-quinazolinones (A) are reported for hCA-I (KIs = 135–282 nM), hCA-II (KIs = 0.25–10.8 nM), hCA-IX (KIs of 3.7–50.4 nM), and CA-XII (KIs of 0.60–52.9 nM) as potent inhibitors (Alafeefy et al., 2016). Several other 2-[(3-substituted-4(3H)-quinazolinon-2-yl)thio]-N-(4-sulfamoylphenyl)acetamides (B) derivatives also showed potent inhibition of α-CA, from Vibrio cholerae (VchCA) and human “hCA-I” and “hCA-II,” at a nanomolar level (Alafeefy et al., 2014) (Figure 1). Recently, 2-[(3-Benzyl-6,7-dimethoxy-4-(3H)-quinazolinon-2-yl)thio]-N-(4-sulfamoylphenyl)propanamide was also reported for hCA-II (KI, 3.30 nM) inhibition (El-Azab et al., 2019).
In this study, we report the synthesis and in-vitro bovine carbonic anhydrase-II (bCA-II) and human carbonic anhydrase-II (hCA-II) inhibitory activities of a series of quinazolinone analogs. Furthermore, the mode of inhibition was further explored by kinetic studies of the active analogs. Additionally, molecular docking studies were carried out to predict the main structural features responsible for the anti-CA-II activities of the compounds to predict the structure-activity relationships.
Experimental
Chemistry
The chemicals used in this study were purchased from Sigma and Aldrich in extra purified form. Melting points for all compounds were recorded by Büchi 434 melting point apparatus. FT-IR spectra (KBr disks) were measured on a Bruker FT-IR IFS48 spectrophotometer. NMR spectra were recorded on Bruker Avance 400 spectrometers in DMSO-d6. The chromatograms were visualized under UV light irradiation. The CHN analysis was performed on a Carlo Erba Strumentazione-Mod-1106.
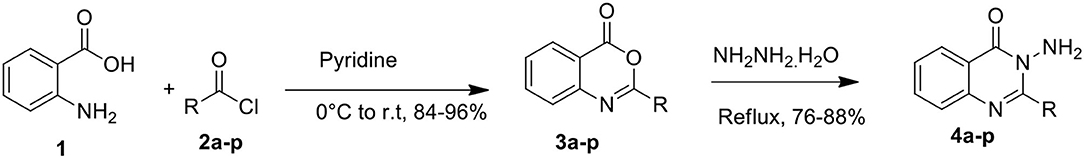
General Procedure for Synthesis of 2-substituted-4H-benzo[d][1,3]oxazin-4-ones (3a-p) (Bari et al., 2020)
To an 100 mL round bottom flask, anthranilic acid 1 (0.01 mol) and dry pyridine (30 mL) were added at room temperature with stirring. The solution was cooled to 0°C, followed by dropwise addition of the corresponding aromatic acid chloride 2a-p (0.02 mol) in 10 mL of dry pyridine with constant stirring. After this addition, the reaction mixture was stirred for a further half an hour at room temperature and set aside for 1 h. The product, obtained as a pasty mass, was diluted with ice cold water (50 mL) and treated with aqueous sodium bicarbonate solution. When the effervescence ceased, the precipitate obtained was filtered and washed with water. The crude benzoxazine obtained was dried and recrystallized from aqueous ethanol.
General Procedure for the Synthesis of Quinazolinone (4a-p) (Panicker et al., 2010)
To a 50 mL round bottom flask fitted with reflux condenser were added 2-substituted-4H-benzo[d][1,3]oxazin-4-one (0.01 mol) and hydrazine hydrate (0.06 mol) in excess. The reaction mixture was heated under reflux in an oil bath for 10–16 h. The course of the reaction was monitored by thin layer chromatography (TLC). After completion of the reaction as checked by TLC, the reaction mixture was cooled and separated solids were collected by filtration, washed with water/hexane, dried, and purified by column chromatography (methanol/chloroform/water, 10:2:1) to afford the corresponding quinazolinones 4a-p.
Experimental Data
3-Amino-2-Phenylquinazolin-4(3H)-one (4a) (Panicker et al., 2010)
Yield: 82%; mp: 200–202°C; FT-IR (υmax, KBR, cm−1): 3,319, 3,051, 1,660, 1,603, 1,490, 1258; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 5.36 (s, 2H), 7.47–8.18 (m, 9H); Anal. Calcd. for C14H11N3O: C, 70.88; H, 4.64; N, 17.72. Found: C, 70.92; H, 4.67; N, 17.68.
3-Amino-2-(Napthalen-1-yl) Quinazolin-4(3H)-one (4b)
Yield: 80%; m.p: 215–216°C; FT-IR (υmax, KBR, cm−1): 3,316, 3,053, 1,662, 1,600, 1,493, 1,257; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 5.42 (2H, s) 7.55–7.64 (4H, m), 7.7–7.88 (4H, m), 8.20 (1H, d, J = 7.4 Hz),), 8.42 (1H, d, J = 7.2 Hz), 8.38 (1H, d, J = 7.2 Hz); Anal. Calcd. for C14H11N3O: C, 70.88; H, 4.64; N, 17.72. Found: C, 70.92; H, 4.67; N, 17.68.
3-Amino-2-(2-Nitrophenyl) Quinazolin-4(3H)-one (4c) (Pandit and Dodiya, 2013)
Yield: 77%; m.p: 180–182°C; FT-IR (υmax, KBR, cm−1): 3,309, 3,053, 1,673, 1,603, 1,548, 1,489, 1,354, 1,250; 701; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 5.43 (2H, s), 7.51–7.63 (4H, m), 7.91–8.02 (4H, m); Anal. Calcd. for C14H10N4O3: C, 59.57; H, 3.57; N, 19.85; Found: C, 59.56; H, 3.58; N, 19.85.
3-Amino-2-(3-Nitrophenyl) Quinazolin-4(3H)-one (4d) (Pandit and Dodiya, 2013)
Yield: 84%; m.p: 206–208°C; FT-IR (υmax, KBR, cm−1): 3,308, 3,053, 1,675, 1,600; 1,548, 1,490, 1,248, 1,355, 703; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 5.48 (s, 2H), 7.62–7.68 (3H, m), 8.07 (1H, d, J = 7.4 Hz), 8.2 (1H, d, J = 7.2 Hz), 8.27 (1H, d, J = 7.8 Hz), 8.56 (1H, s); Anal. Calcd. for C14H10N4O3: C, 59.57; H, 3.57; N, 19.85. Found: C, 59.56; H, 3.58; N, 19.85.
3-Amino-2-(4-nitrophenyl) quinazolin-4(3H)-one (4e) (Pandit and Dodiya, 2013)
Yield: 76%; m.p: 224–225°C; FT-IR (υmax, KBR, cm−1): 3,307, 3,053, 1,670, 1,603, 1,548, 1,488, 1,353, 1,250; 708; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 5.42 (2H, s), 7.66–7.71 (3H, m), 8.06 (1H, d, J= 7.2 Hz), 8.12 (2H, d, J = 7.8 Hz), 8.26 (2H, d, J = 7.8 Hz); Anal. Calcd. for C14H10N4O3: C, 59.57; H, 3.57; N, 19.85. Found: C, 59.56; H, 3.58; N, 19.85.
3-Amino-2-(2-Bromophenyl) Quinazolin-4(3H)-one (4f)
Yield: 79%; m.p: 204–206°C; FT-IR (υmax, KBR, cm−1): 3,300, 3,053, 1,673, 1,610, 1,010, 690; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 5.25 (2H, s), 7.35–7.39 (2H, t, J = 7.2 Hz), 8.06 (1H, d, J = 8.1 Hz), 7.58–7.66 (5H, m); Anal. Calcd for C14H10BrN3O: C, 53.19; H, 3.19; N, 13.29. Found: C, 53.17; H, 3.20; N, 13.28.
3-Amino-2-(4-Bromophenyl) Quinazolin-4(3H)-one (4g)
Yield: 81%; m.p: 184–185°C; FT-IR (υmax, KBR, cm−1): 3,300, 3,053, 1,676, 1,610, 1,010, 690; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 5.26 (2H, s), 7.52 (2H, d, J = 7.45 Hz), 7.61–7.67 (3H, m), 7.74 (2H, d, J = 7.6 Hz), 8.1 (1H, d, J = 7.55 Hz); Anal. Calcd for C14H10BrN3O: C, 53.19; H, 3.19; N, 13.29. Found: C, 53.17; H, 3.20; N, 13.28.
3-Amino-2-(4-Methylphenyl) Quinazolin-4(3H)-one (4h) (Babu et al., 2014)
Yield: 85%; mp: 294–296°C; FT-IR (υmax, KBR, cm−1): 3,300, 3,053, 2,948, 1,676, 1,610, 1,496; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 2.34 (3H, s), 4.93 (2H, s), 8.08 (1H, d, J = 7.2 Hz), 7.28 (2H,d, J = 7.8 Hz), 7.44 (2H, d, J = 7.8 Hz), 7.55-7.67 (3H, m); Anal. Calcd for C15H13N3O: C, 71.70; H, 5.21; N, 16.72. Found: C, 71.71; H, 5.20; N, 16.73.
3-Amino-2-(2-Fluorophenyl) Quinazolin-4(3H)-one (4i)
Yield: 84%; m.p: 169–170°C; (υmax, KBR, cm−1): 3,308, 3,053, 1,681, 1,610; 1,329, 506 1H NMR (400 MHz, DMSO-d6, δ, ppm): 4.98 (2H, s), 8.20 (1H, d, J = 7.2 Hz), 7.77 (1H, bd, J = 8.4 Hz), 7.59–7.67 (4H, m), 7.29–7.36 (2H, m); Anal. Calcd for C14H10FN3O: C, 65.88; H, 3.95; N, 16.46. Found: C, 65.89; H, 3.94; N, 16.47
3-Amino-2-(3-Fluorophenyl) Quinazolin-4(3H)-one (4j)
Yield: 78%; m.p: 164–165°C; FT-IR (υmax, KBR, cm−1): 3,310, 3,053, 1,679, 1,610; 1,335, 502; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 5.52 (2H, s), 8.01 (1H, d, J = 7.2Hz), 7.78 (1H, d, J = 7.8 Hz), 7.58–7.66 (4H, m), 7.32 (1H, t, J = 7.8 Hz); Anal. Calcd for C14H10FN3O: C, 65.88; H, 3.95; N, 16.46. Found: C, 65.89; H, 3.94; N, 16.47.
3-Amino-2-(4-Fluorophenyl) Quinazolin-4(3H)-one (4k)
Yield: 81%; m.p: 176–178°C; FT-IR (υmax, KBR, cm−1): 3,310, 3,053, 1,680, 1,610; 1,335, 502; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 5.38 (s, 2H), 7.26–7.30 (2H, m), 7.41–7.46 (2H, m), 7.53–7.56 (2H, m), 8.26 (1H,d, J = 7.5 Hz); Anal. Calcd for C14H10FN3O: C, 65.88; H, 3.95; N, 16.46. Found: C, 65.87; H, 3.94; N, 16.47.
3-Amino-2-(2-Chlorophenyl) Quinazolin-4(3H)-one (4l) (Pandit and Dodiya, 2013)
Yield: 85%; m.p: 160–162°C; FT-IR (υmax, KBR, cm−1): 3,310, 3,053, 1,689, 1,610; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 5.08 (2H, s), 7.41–7.45 (2H, t, J = 7.3 Hz), 7.48 (1H, d, J = 7.3 Hz) 7.54 (1H, d, J = 7.3 Hz), 7.58–7.69 (3H, m), 8.07 (1H, d, J = 7.2 Hz); Anal. Calcd for C14H10ClN3O: C, 61.89; H, 3.71; N, 15.47. Found C, 61.87; H, 3.73; N, 15.47.
3-Amino-2-(3-Chlorophenyl) Quinazolin-4(3H)-one (4m) (Pandit and Dodiya, 2013)
Yield: 79%; m.p: 144–145°C; FT-IR (υmax, KBR, cm−1): 3,307, 3,210, 3,051, 2,993, 1,668, 1,591, 1,252, 1,123; 458; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 5.33 (2H, s), 8.14 (1H, d, J = 7.2 Hz), 7.84 (1H, s), 7.63–7.68 (3H, m), 7.46–7.52 (3H, m); Anal. Calcd for C14H10ClN3O: C, 61.89; H, 3.71; N, 15.47. Found C, 61.87; H, 3.73; N, 15.47.
3-Amino-2-(4-Chlorophenyl) Quinazolin-4(3H)-one (4n) (Castillo et al., 2012)
Yield: 86%; m.p: 165–166°C; FT-IR (υmax, KBR, cm−1): 3,310, 3,213, 3,053, 2,990, 1,670, 1,591, 1,252, 1,123; 462 1H NMR (400 MHz, DMSO-d6, δ, ppm): 4.86 (s, 2H) 7.34 (2H, d, J = 7.4 Hz), 7.56 (2H, d, J = 7.4 Hz), 7.68–7.75 (3H, m), 8.04 (1H, d, J = 7.2 Hz); Anal. Calcd for C14H10ClN3O: C, 61.89; H, 3.71; N, 15.47. Found C, 61.87; H, 3.73; N, 15.47.
3-Amino-2-(4-Methoxyphenyl) Quinazolin-4(3H)-one (4o) (Pandit and Dodiya, 2013)
Yield: 88%; m.p: 221–223°C; FT-IR (υmax, KBR, cm−1): 3,310, 3,053, 2,940, 1,689, 1,610, 1,490; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 4.92 (2H,s), 3.76 (3H, s), 6.96 (2H, d, J =7.5 Hz), 7.34 (2H, d, J = 7.5 Hz),7.60–7.68 (3H, m), 8.09 (1H, d, J = 7.3 Hz); Anal. Calcd. for C15H13N3O2: C, 67.40; H, 4.90; N, 15.72; Found C, 67.42; H, 4.88; N, 15.71.
3-Amino-2-(3,4,5-Trimethoxyphenyl) Quinazolin-4(3H)-one (4p)
Yield: 78%; m.p: 221–222°C; FT-IR (υmax, KBR, cm−1): 3,310, 3,053, 2,950, 1,689, 1,610, 1,495; 1H NMR (400 MHz, DMSO-d6, δ, ppm): 3.79 (6H, s), 3.82 (3H, s), 4.88 (2H, s), 6.62 (2H, s), 7.48–7.52 (3H, m), 8.01 (1H, d, J = 7.3 Hz); Anal. Calcd. for C17H17N3O4: C, 62.38; H, 5.23; N, 12.84; Found C, 62.38; H, 5.24; N, 12.83.
In-vitro Assay Protocol
In-vitro bCA-II and hCA-II activities were measured by following the spectrophotometric method described by Pocker and Meany with slight modifications (Pocker and Meany, 1967; Ur Rehman et al., 2020). The spectrophotometric assay was conducted in HEPES-Tris buffer of pH 7.4 (20 mM) at 25°C. Each inhibitory well-consisted of 140 μL of HEPES-Tris buffer solution, 20 μL of bCA-II enzyme solution (0.1 mg/mL HEPES-Tris buffer), and 20 μL of test compound in HPLC grade DMSO (maintain 10% of the final concentration). The mixture solution was pre-incubated for 15 min at 25°C. Substrate p-nitrophenyl acetate (p-NPA) (0.7 mM) was prepared in HPLC grade methanol and the reaction was started by adding 20 μL to a well in a 96-well-plate. The amount of product formed was measured for 30 min continuously at 1 min intervals at 400 nm in a 96-well-plate using xMARK microplate spectrophotometer, Bio-Rad (USA). The activity of the controlled compound was taken as 100%. All experiments were carried out in triplicates of each used concentration, and results are represented as a mean of the triplicate.
Molecular Docking Protocol
The docking studies were performed on the crystal structures of bCA-II (1V9E, resolution = 1.95 Å) (Saito et al., 2004) and hCA-II (1BN1, resolution = 2.1 Å) (Boriack-Sjodin et al., 1998). The selected target receptors were retrieved from Protein Data Bank (www.rcsb.org) in PDB format. Chain A of each protein (1V9E and 1BN1) was chosen for docking. For the molecular docking simulation, Molecular Operating Environment (MOE) docking suite was used (MOEv2014.09). The crystal structures were prepared using Protein Preparation Wizard implemented in the MOE with the default settings. Hydrogen atoms were added in the MOE using protonate 3D protocol and the protein structures were minimized with an MMFF94x force field until an RMSD gradient of 0.1 kcal·mol−1Å−1 was achieved. The 3D-structures of ligands were prepared by Chemdraw and minimized by default option of MOE (Force field = MMFF94x, RMS gradient = 0.1 kcal·mol−1Å−1). For docking, Triangle Matcher placement method and London dG scoring function were applied (Edelsbrunner, 1992; Naïm et al., 2007). After docking, thirty docked poses of each compound were saved and the best-scoring docked pose of each ligand was visualized.
Results and Discussion
Chemistry
The target compounds were synthesized according to the route depicted in Scheme 1. Anthranilic acid 1 was treated with corresponding acid chlorides (2a-p) in the presence of pyridine to form precursor heterocycle (3a-p) via literature method (Scheme 1) (Bari et al., 2020). The corresponding benzoxazinones were refluxed in excess of hydrazine hydrate to furnish the 3-Amino-2-aryl quinazolin-4(3H)-ones (4a-p) in moderate to excellent yields of 76–88% (Table 1) (Panicker et al., 2010).
The structures of compounds (4a-p) were established using microanalysis (CHN) and spectral data, i.e., IR and 1H NMR. The C=N band in FTIR appeared in the range of 1,489–1,521 cm−1. Moreover, the carbonyl group of compounds (4a-p) appeared at 1,660–1,689 cm−1. The amino moiety (-NH2) of compounds (4a-p) was verified by the characteristic peak at 3,300–3,319 cm−1 in FT-IR spectra. The amino-moiety of compounds (4a-p) was verified by 1H NMR spectra which appeared as a singlet for amino protons in a range from δ 4.92–5.52 ppm. The spectral data of other aromatic and aliphatic protons was also in accordance with the structures of anticipated compounds. CHN analysis also supported the anticipated structures (4a-p) and the observed values were in good agreement with the values found.
Biology
In-vitro bCA-II and hCA-II inhibitions
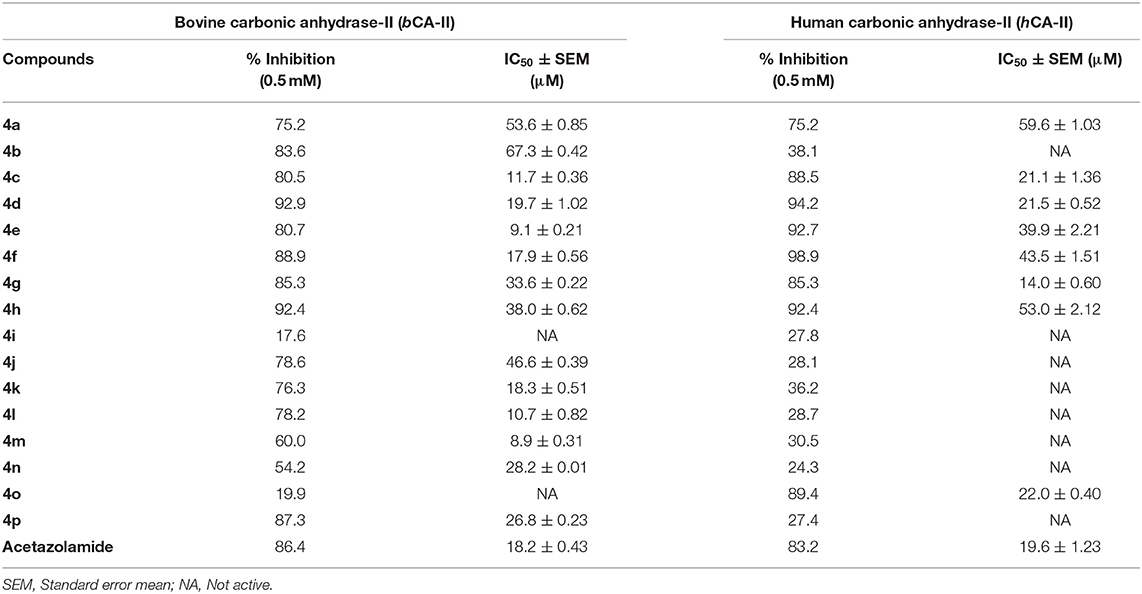
All quinazolinone analogs (4a-p) were evaluated against bovine carbonic anhydrase-II (bCA-II) and human carbonic anhydrase-II (hCA-II) for their ability to act as an inhibitor of CA-II. All the assays were carried out at a micromolar level using acetazolamide as a standard inhibitor. After initial screening, all the analogs demonstrated significant inhibitory activity against bCA-II with IC50 values in the range of 8.9 ± 0.31 −67.3 ± 0.42 μM, except compounds 4i and 4o, which were found to be inactive (Table 2). Moreover, compounds 4c, 4e, 4f, 4l, and 4m showed more superior activity than the standard drug ‘acetazolamide' (IC50 = 18.2 ± 0.43 μM). Compound 4m was the most active inhibitor (IC50 = 8.9 ± 0.31 μM), followed by 4e (IC50 = 9.1 ± 0.21 μM), 4l (IC50 = 10.7 ± 0.82 μM), 4c (IC50 = 11.7 ± 0.36 μM), and 4f (IC50 = 17.9 ± 0.56 μM). However, compounds 4d, 4k, 4n, and 4p depicted biological activity comparable to the standard acetazolamide with IC50 values in range of 18.3 ± 0.51–28.2 ± 0.01 μM, while compounds 4a, 4b, 4g, 4h, and 4j were found to be the least active hits of the series. For hCA-II, compounds 4a, 4c-4h, and 4o were found to be active with IC50 values in the range of 14.0 ± 0.60–59.6 ± 1.03 μM, as compared to acetazolamide (IC50 = 19.6 ± 1.23 μM) (Table 2). Among all the compounds, 4g (IC50 = 14.0 ± 0.60 μM) exhibited better activity than the standard drug, while compounds 4c (IC50 = 21.1 ± 1.36 μM), 4d (IC50 = 21.5 ± 0.52 μM), and 4o (IC50 = 22.0 ± 0.40 μM) demonstrated activities comparable to acetazolamide. Compounds 4e (IC50 = 39.9 ± 2.21 μM), 4f (IC50 = 43.5 ± 1.51 μM), 4h (IC50 = 53.0 ± 2.12 μM), and 4a (IC50 = 59.6 ± 1.03 μM) were shown to be the least active hits in this series. The in-vitro results indicated that compounds 4g and 4o are more selective inhibitors for hCA-II. The biological activities of all the compounds are tabulated in Table 2.
Kinetics Studies
To investigate the mechanism of action of these compounds, kinetics studies were performed. The kinetics studies were used to discover the type of inhibition and dissociation constant (Ki). Kinetics studies of the most active compound, 4d, against both bCA-II and hCA-II were performed, using different substrate concentrations on one side with different concentrations of 4d on the other side. Compound 4d inhibited both the bCA-II and hCA-II enzymes in a concentration-dependent manner with Ki values of 13.0 ± 0.013 and 14.25 ± 0.017 μM, respectively. From the kinetics studies, it was deduced that the compound 4d is a competitive inhibitor for both bCA-II and hCA-II. The Lineweaver-Burk plots were used for determination of the type of inhibition, in which the reciprocal of substrate concentrations was plotted against the reciprocal of the rate of the reaction to monitor the effect of the inhibitor on both Km and Vmax. The Lineweaver-Burk plots of 4d against both bCA-II and hCA-II clearly showed that the mode of inhibition of 4d is competitive (Figures 2A, 3A). In competitive inhibition, the Km of enzyme increased, while Vmax are not affected. The Lineweaver-Burk plots of compounds 4d (Figures 2A, 3A) showed that in the presence of compounds 4d, the Km of both enzymes bCA-II and hCA-II increased significantly on applying compounds 4d, while the Vmax remain unchanged, which described the competitive behavior of compounds 4d and its interaction in the active site of the enzyme.
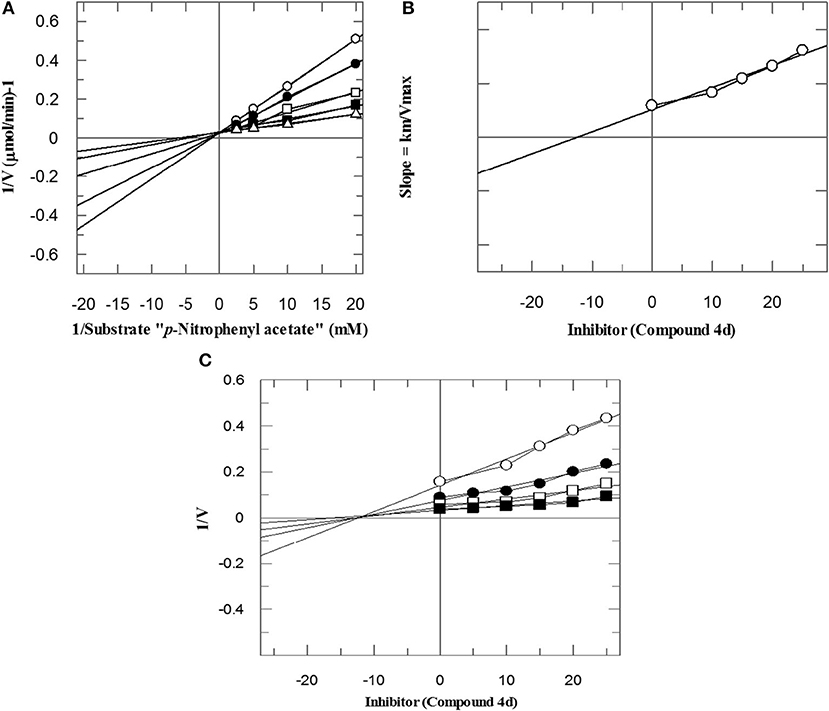
Figure 2. The mode of inhibition of bCA-II by compound 4d. (A) Lineweaver–Burk plot of reciprocal rate of reaction (velocities) vs. reciprocal of substrate (p-nitrophenyl acetate) in the absence (Δ), and in the presence of 14 μM (■), 17 μM (□), 20 μM (•), and 23 μM (∘) of compound 4d. (B) Secondary re-plot of Lineweaver–Burk plot between the slopes of each line on Lineweaver–Burk plot vs. different concentrations of compound 4d. (C) Dixon plot of reciprocal rate of reaction (velocities) vs. different concentrations of compound 4d.
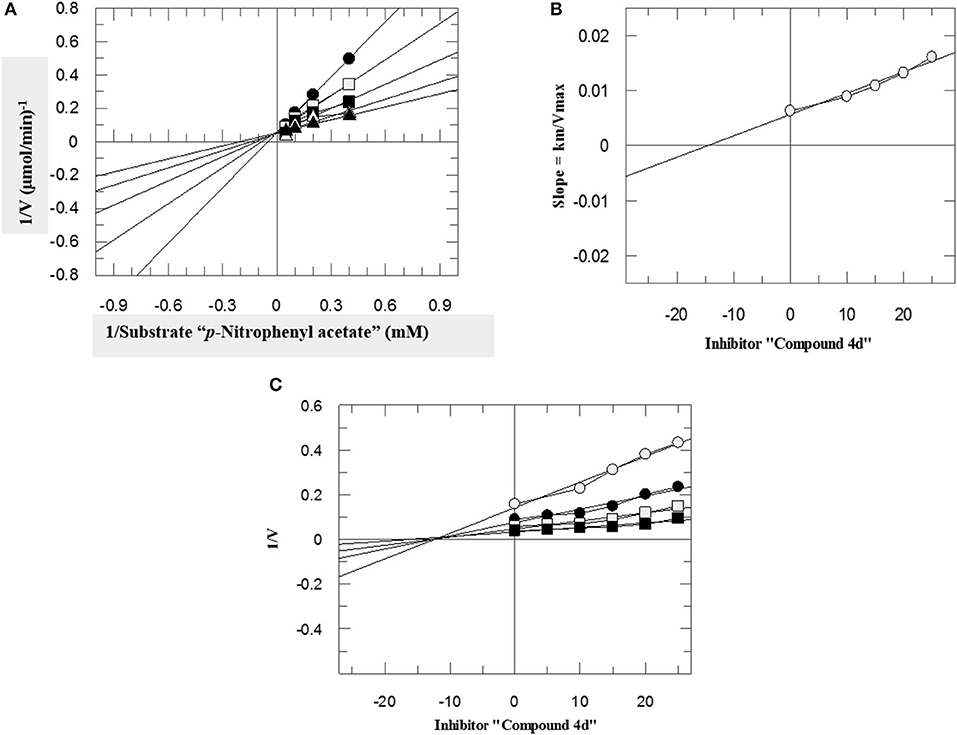
Figure 3. The mode of inhibition of hCA-II by compound 4d. (A) Lineweaver–Burk plot of reciprocal rate of reaction (velocities) vs. reciprocal of substrate (p-nitrophenyl acetate) in the absence (▴), and in the presence of 10 μM (Δ), 15 μM (■), 20 μM (□), and 25 μM (•) of compound 4d. (B) Secondary re-plot of Lineweaver–Burk plot between the slopes of each line on Lineweaver–Burk plot vs. different concentrations of compound 4d. (C) Dixon plot of reciprocal rate of reaction (velocities) vs. different concentrations of compound 4d.
The Ki values of compounds 4d for both enzymes bCA-II and hCA-II were deduced by secondary replots of Lineweaver-Burk plots by plotting the slope of each line in the Lineweaver-Burk plots against different concentrations of compound 4d (Figures 2B, 3B). The Ki values were confirmed by Dixon plot after plotting the reciprocal of the rate of reaction against different concentrations of compound 4d (Figures 2C, 3C).
Computational Studies
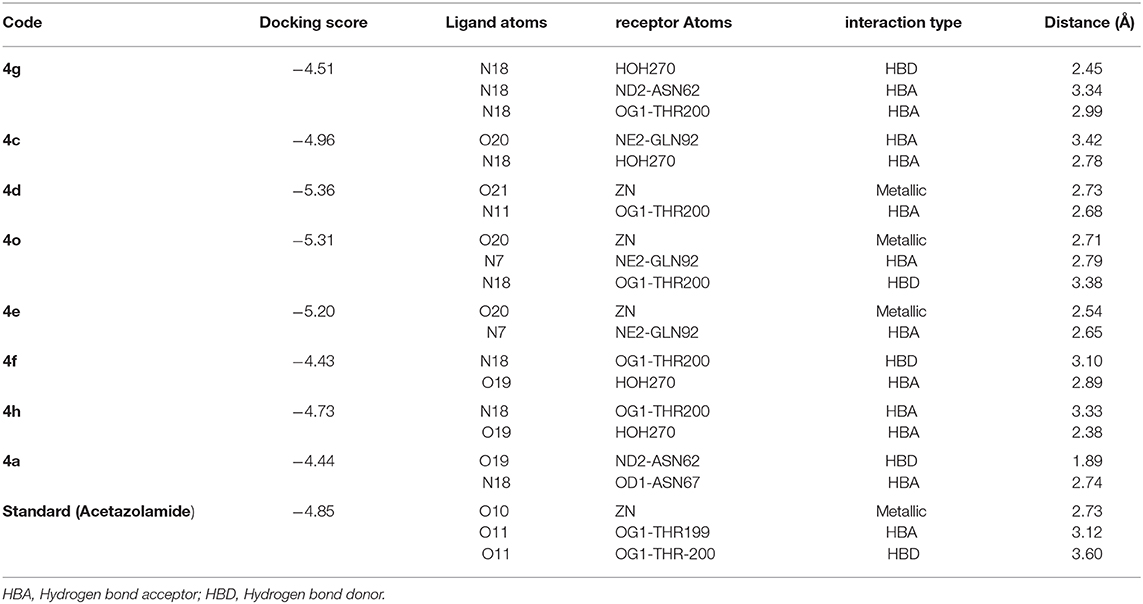
Computational medicinal chemistry has expedited the pace of drug design and discovery over the last four decades (Lin et al., 2020). Docking is a computational method which is widely used to study and understand the interaction between two macro-molecules (for e.g., protein-protein or protein-DNA) or between a macromolecule and ligand (e.g., for a drug-receptor, drug-DNA, or drug-RNA) and effectively predict the inhibitory mechanism of drugs. Therefore, we predicted the mode of interactions of all the active compounds though molecular docking. The reference inhibitor (acetazolamide) and quinazolinones were docked into the catalytic active pocket of hCA-II and bCA-II. The binding modes and receptor-ligand interactions in the binding site of bCA-II and hCA-II in the three-dimensional and two-dimensional form were carefully examined by visual analysis through MOE. The results are presented in Tables 3, 4. Molecular docking revealed that all the active quinazolinones derivatives were exactly fitted into the active catalytic pocket of both enzymes (hCA-II and bCA-II). The docked orientation of compounds revealed direct interactions of ligands with the zinc ion present in the active site. Moreover, interaction of compounds with the active site residues and water molecule stabilize the compounds in the active site.
The docking process was first validated by re-docking the standard inhibitor acetazolamide in the active site of enzyme and is presented in Figures 4D, 6D.
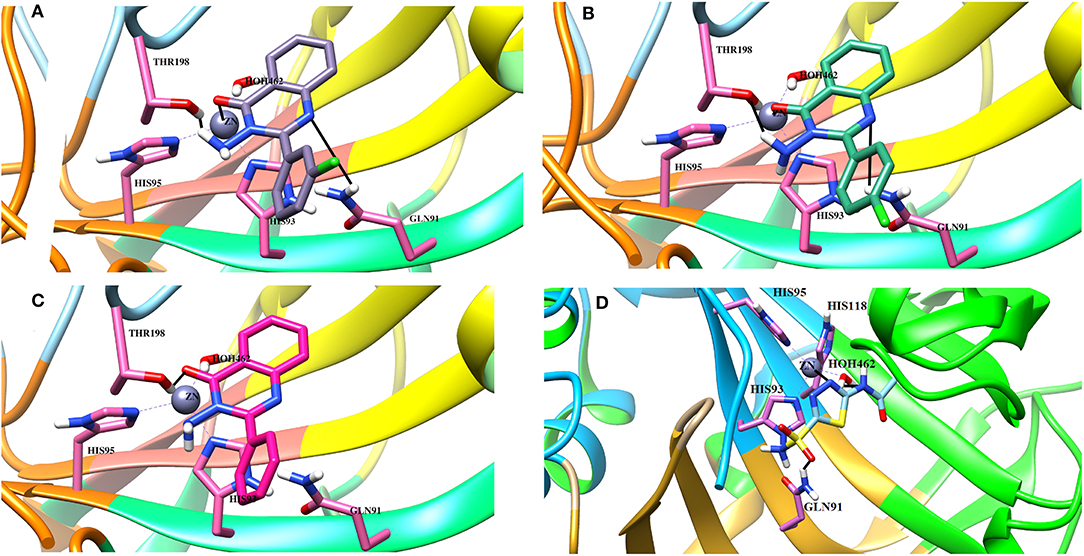
Figure 4. The 3D interaction of the (A) most active compound (4 m, shown in gray stick model), (B) moderate active compound (4n, green stick model), (C) least active compound (4a, pink stick model), and (D) standard drug (acetazolamide, shown in cyan sticks) in the active site of bCA-II. The active site residues are depicted in pink stick model. The hydrogen bonds are presented in black lines.
Binding Interactions of Active Compounds and Their Predictive SAR Against bCA-II
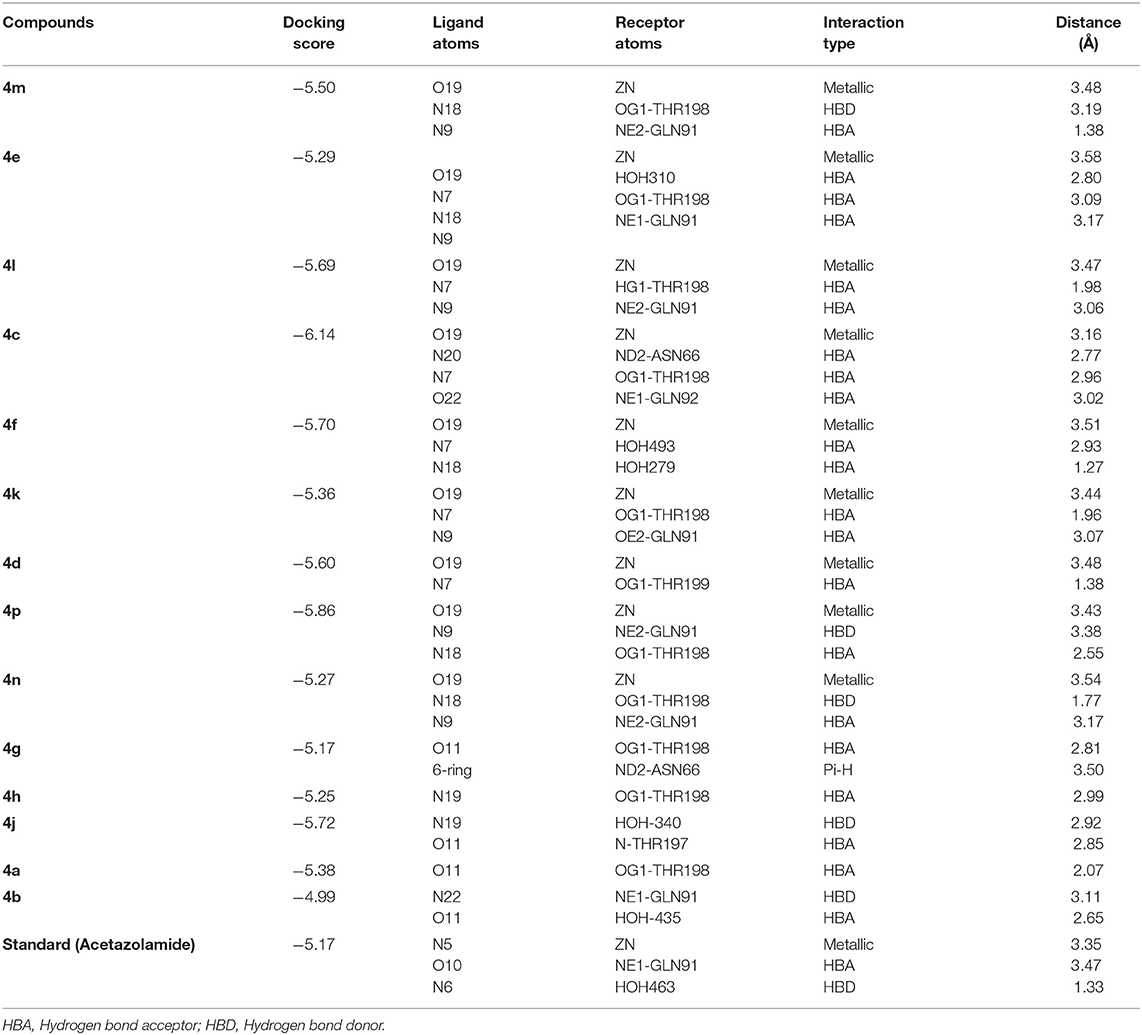
From the docked poses of the all active compounds, it was confirmed that the compounds directly interact with Zn2+ ion through their quinazolinone-carbonyl moiety. Compounds 4c, 4d, 4e, 4f, 4k, 4l, and 4m were found to be potent inhibitors of bCA-II with IC50 values in the range of 8.9–19.7 μM. Compound 4m was the most potent inhibitor (IC50 = 8.9 ± 0.31μM), followed by 4e > 4l > 4c > 4f > 4k > 4d, while compounds 4p > 4n displayed moderate active, and 4g > 4h > 4j > 4a > 4b were retrieved as the least active, as compared to standard acetazolamide. The docked pose of 4m indicates that the quinazolinone-carbonyl moiety of 4m formed a metallic bond (3.44 Å) with the Zn2+ ion, while the quinazolinone-nitrogen formed hydrogen bonds with the side chain of Thr198 and Gln91 at a distance of 2.28 and 3.41 Å, respectively. Similarly, carbonyl oxygen and the nitro groups of 4e mediated a metallic bond with Zn2+ ion (3.58 Å), and H-bonds with the side chains of Thr198 (2.80 Å) and Gln91 (3.09 Å). Additionally, the quinazolinone substituted amino group of 4e mediated a H-bond with a water molecule in the active site (3.17 Å). The third most active compounds, 4l and 4c, also followed a similar type of interaction, however, 4c was also stabilized by the side chains of Asn66 and Gln92 through hydrogen bonding. The carbonyl oxygen of 4f and 4k interacted with the Zn2+ ion, however 4f lost the interactions with Thr198 and Gln91, instead mediating H-bonding with two water molecules (WAT493 and WAT279), however 4k retained H-bonds with Thr198 and Gln91. Compound 4d showed a metallic and a H-bond with the Zn2+ ion and Thr199, respectively, however it lost the interactions with Gln91 and water molecules.
The quinazolinone moiety of the moderate active compounds (4p and 4n) interacted with the Zn2+ ion, while the quinazolinone moiety of both the compounds mediated hydrogen bonding with the side chains of Gln91 and Thr198.
The docked view of the least active compounds (4g, 4h, 4j, 4a, and 4b) showed that quinazolinone carbonyl of 4g interacted with the side chain of Thr198 via H-bond (2.81 Å) and the side chain of Asn66 offered a hydrophobic interaction to the compound (3.50 Å). Whereas, quinazolinone-amide group of 4h mediated a H- bond with the side chain of Thr198 (2.99 Å), and the amide group of 4j interacted with a water molecule (WAT340) present in the active site. Similarly, the carbonyl oxygen and amide nitrogen of compounds 4a and 4b mediated H-bonds with the side chain of Thr197 and Gln91, respectively. Additionally, 4b formed a bond with a water molecule (WAT435). The binding interactions and docking scores of each docked compound are tabulated in Table 3. Figure 4 shows the binding interactions of the most active (4m), moderate (4n), and the least active (4a) compounds, however, a docked view of the most potent compound (4d) and all the compounds are presented in Figure 5.
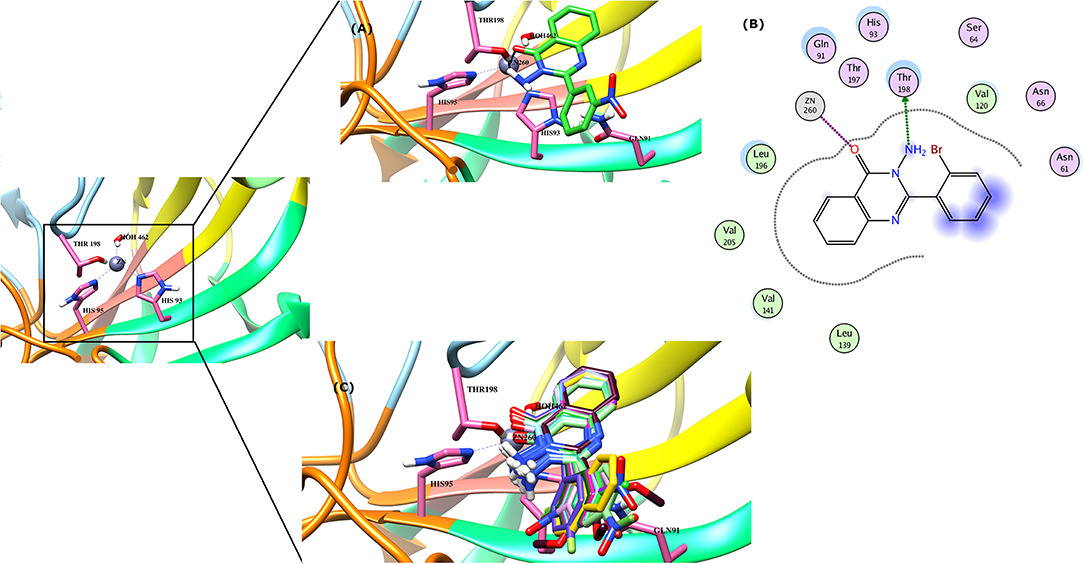
Figure 5. (A) The docked view of compound 4d is shown in the active site of bCA-II in 3D-form. (B) The binding interactions of 4d in two-dimensional form is demonstrated. (C) The binding modes of all the active compounds are shown. The hydrogen bonds are shown in black lines.
Binding Interactions of Active Compounds and Their Predictive SAR Against hCA-II
Docking analysis deduced that the quinazolinone moieties of these compounds are responsible for the formation of metallic interactions with the Zn2+ atom in the catalytic cavity. Compound 4g (IC50 = 14.0 ± 0.60 μM) was the most potent and selective inhibitor of hCA-II, as compared to standard acetazolamide (IC50 = 19.6 ± 1.23 μM), followed by compounds 4c > 4d > 4o and least active 4e > 4f > 4h > 4a. The quinazolinone moiety of 4g interacted with the side chain of Asn62, Thr200, and Wat270 with bond lengths of 3.34, 2.99, and 2.45 Å, respectively. Similarly, 4c interacted with the side chain of Gln92 and Wat270, whereas 4d was linked with Zn2+ atom through its nitro-oxygen group. The nitro-oxygen and aminoquinoline moiety of 4o interacted with Zn2+ atom and the side chains of Gln92 and Thr200 via hydrogen bonds. However, the quinazolinone moiety of 4e interacted with the Zn2+ ion and Gln92. On the other hand, the least active compounds, 4a, 4f, and 4h, interacted with the side chains of Asn62, Asn67, Thr200, and water molecules within the active site. The detailed binding interactions of active compounds and their docking scores are tabulated in Table 4. The binding interactions of most active (4g), moderate (4c), and least active (4a) compounds in the active site of hCA-II are presented in Figure 6. The docked orientation of all active compounds and the binding interaction of compound 4d are shown in Figure 7.
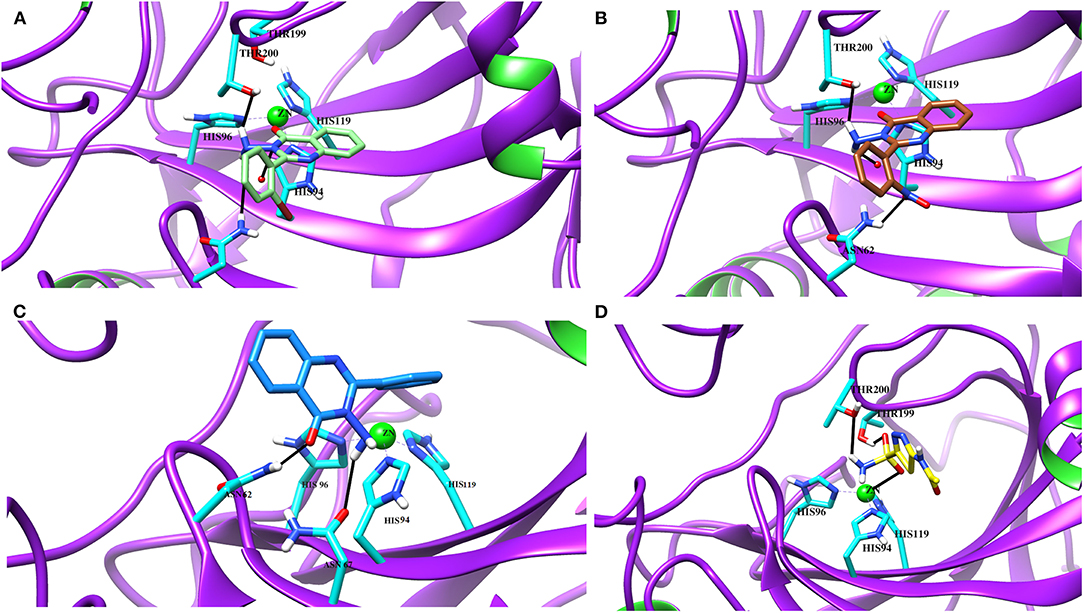
Figure 6. The 3D docked view of the (A) most active compound (4g, showed as lime green stick model), (B) moderate active compound (4c, shown as green stick model) (C) least active compound (4a, shown as sky blue stick model) and (D) standard (acetazolamide shown as yellow stick model) against hCA-II. The active site residues are depicted in cyan stick model. The hydrogen bonds are presented in black lines.
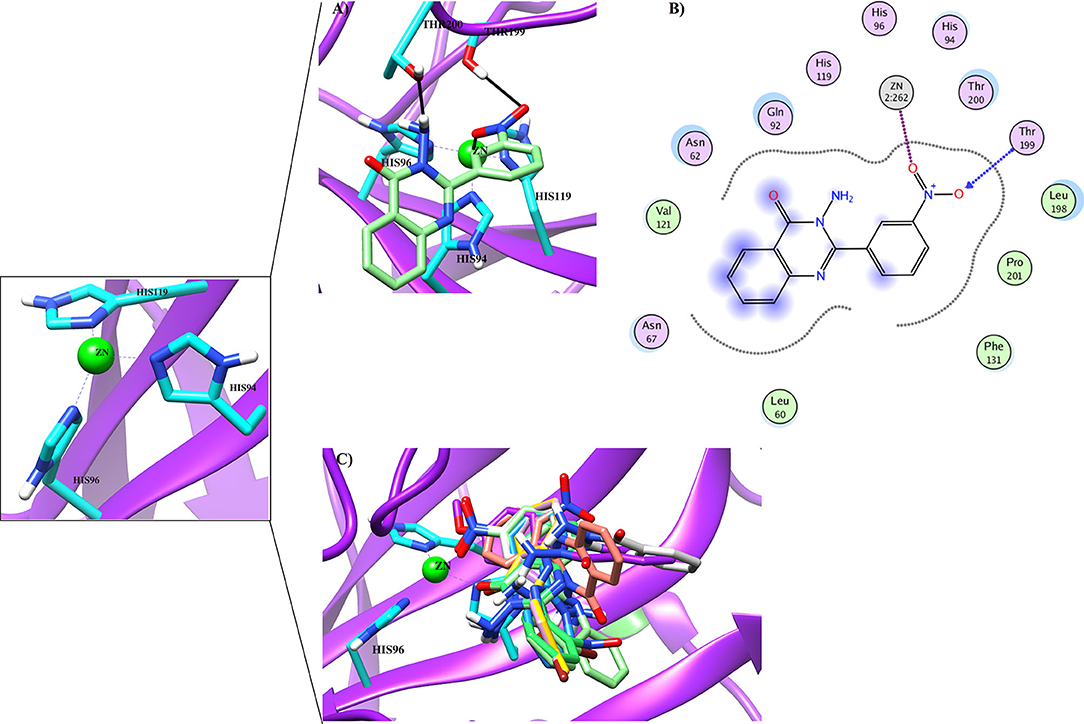
Figure 7. (A) The docked view of compound 4d (lime green stick mode) in the active site of hCA-II. (B) The 2D-binding interactions of 4d are presented (C) The docked orientation of all the active compound in the active site of hCA-II are shown.
Conclusion
Quinazolinone derivatives (4a-p) were synthesized in search of new therapeutic agents for glaucoma and other diseases associated with hyper activity of CA-II. The in-vitro results showed that these skeletons displayed moderate to significant inhibition of both the enzymes (bCA-II and hCA-II). Compounds 4c, 4e, 4f, 4l, and 4m showed superior activity against bCA-II, while compounds 4g, 4c, 4d, and 4o were found to be significantly active against hCA-II. The structure-activity relationship reflected that the nitro group on phenyl ring at R position plays an important role in the overall inhibitory activities of compounds. Among the tested compounds, 4g and 4o are more selective for hCA-II. Moreover, kinetics studies showed the competitive behavior of inhibition of this series. Additionally, molecular docking predicted that the compounds bind efficiently with Zinc ion and several residues within the active site, therefore, through appropriate fitting and binding, these compounds effectively inhibit both bCA-II and hCA-II enzymes.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
AA-H and ZS conceived and designed the study. ZK synthesized all compounds. AK and MK performed in-vitro assay. MK and SH performed the computational studies and analyzed the data. AK wrote the manuscript with input and comments from MK, SH, ZK, ZS, and AA-H. All authors have read and approved the final version of the manuscript.
Funding
The generous support of this project through project number (BFP/RGP/HSS/19/198).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a past co-authorship with one of the authors with the authors AK, SH, and AA-H.
Acknowledgments
The authors would like to thank the University of Nizwa for the generous support of this project. We thank technical staff for assistance.
References
Achal, V., and Pan, X. (2011). Characterization of urease and carbonic anhydrase producing bacteria and their role in calcite precipitation. Curr. Microbiol. 62, 894–902. doi: 10.1007/s00284-010-9801-4
Aggarwal, M., Boone, C. D., Kondeti, B., and McKenna, R. (2013). Structural annotation of human carbonic anhydrases. J. Enzyme Inhib. Med. Chem. 28, 267–277. doi: 10.3109/14756366.2012.737323
Alaa, A.-M., Abou-Zeid, L. A., ElTahir, K. E. H., Mohamed, M. A., El-Enin, M. A. A., and El-Azab, A. S. (2016). Design, synthesis of 2, 3-disubstitued 4 (3H)-quinazolinone derivatives as anti-inflammatory and analgesic agents: COX-1/2 inhibitory activities and molecular docking studies. Bioorg. Med. Chem. 24, 3818–3828. doi: 10.1016/j.bmc.2016.06.026
Alafeefy, A. M., Carta, F., Ceruso, M., Al-Tamimi, A.-M. S., Al-Kahtani, A. A., and Supuran, C. T. (2016). Development of 3-(4-aminosulphonyl)-phenyl-2-mercapto-3H-quinazolin-4-ones as inhibitors of carbonic anhydrase isoforms involved in tumorigenesis and glaucoma. Bioorg. Med. Chem. 24, 1402–1407. doi: 10.1016/j.bmc.2016.02.011
Alafeefy, A. M., Ceruso, M., Al-Tamimi, A.-M. S., Del Prete, S., Capasso, C., and Supuran, C. T. (2014). Quinazoline–sulfonamides with potent inhibitory activity against the α-carbonic anhydrase from Vibrio cholerae. Bioorg. Med. Chem. 22, 5133–5140. doi: 10.1016/j.bmc.2014.08.015
Al-Amiery, A. A., Kadhum, A. A. H., Shamel, M., Satar, M., Khalid, Y., and Mohamad, A. B. (2014). Antioxidant and antimicrobial activities of novel quinazolinones. Med. Chem. Res. 23, 236–242. doi: 10.1007/s00044-013-0625-1
Al-Suwaidan, I. A., Alanazi, A. M., Alaa, A.-M., Mohamed, M. A., and El-Azab, A. S. (2013). Design, synthesis and biological evaluation of 2-mercapto-3-phenethylquinazoline bearing anilide fragments as potential antitumor agents: molecular docking study. Bioorg. Med. Chem. Lett. 23, 3935–3941. doi: 10.1016/j.bmcl.2013.04.056
Arslan, O. (2001). Inhibition of bovine carbonic anhydrase by new sulfonamide compounds. Biochemistry 66, 982–983. doi: 10.1023/A:1012365424900
Babu, Y. R., Bhagavanraju, M., Reddy, G. D., Peters, G. J., and Prasad, V. V. R. (2014). Design and synthesis of quinazolinone tagged acridones as cytotoxic agents and their effects on EGFR tyrosine kinase. Arch. Pharm. 347, 624–634. doi: 10.1002/ardp.201400065
Bari, A., Khan, Z. A., Shahzad, S. A., Naqvi, S. A. R., Khan, S. A., Amjad, H., et al. (2020). Design and syntheses of 7-nitro-2-aryl-4H-benzo [d][1, 3] oxazin-4-ones as potent anticancer and antioxidant agents. J. Mol. Struct. 1214:128252. doi: 10.1016/j.molstruc.2020.128252
Boriack-Sjodin, P. A., Zeitlin, S., Christianson, D. W., Chen, H. H., Crenshaw, L., Gross, S., et al. (1998). Structural analysis of inhibitor binding to human carbonic anhydrase II. Protein Sci. 7, 2483–2489. doi: 10.1002/pro.5560071201
Castillo, M. V., Romano, E., Raschi, A. B., Yurquina, A., and Brandán, S. A. (2012). Structural study and vibrational spectra of 3-amino-2-(4-chlorophenyl) quinazolin-4 (3H)-one. Comput. Theor. Chem. 995, 43–48. doi: 10.1016/j.comptc.2012.06.029
Chegwidden, W. R., Dodgson, S. J., and Spencer, I. M. (2000). “The roles of carbonic anhydrase in metabolism, cell growth and cancer in animals,” in The Carbonic Anhydrases, Vol. 90, eds. W. R. Chegwidden, N. D. Carter, and Y. H. Edwards (Basel: Birkhäuser). doi: 10.1007/978-3-0348-8446-4_16
Edelsbrunner, H. (1992). Weighted Alpha Shapes. University of Illinois at Urbana-Champaign (Champaign, IL).
El-Azab, A. S., Alaa, A.-M., Bua, S., Nocentini, A., El-Gendy, M. A., Mohamed, M. A., et al. (2019). Synthesis of benzensulfonamides linked to quinazoline scaffolds as novel carbonic anhydrase inhibitors. Bioorg. Chem. 87, 78–90. doi: 10.1016/j.bioorg.2019.03.007
Frazier, M. L., Lilly, B. J., Wu, E. F., Ota, T., and Hewett-Emmett, D. (1990). Carbonic anhydrase II gene expression in cell lines from human pancreatic adenocarcinoma. Pancreas 5, 507–514. doi: 10.1097/00006676-199009000-00002
Gawad, N. M. A., Georgey, H. H., Youssef, R. M., and El Sayed, N. A. (2011). Design, synthesis, and anticonvulsant activity of novel quinazolinone analogues. Med. Chem. Res. 20, 1280–1286. doi: 10.1007/s00044-010-9465-4
He, D., Wang, M., Zhao, S., Shu, Y., Zeng, H., Xiao, C., et al. (2017). Pharmaceutical prospects of naturally occurring quinazolinone and its derivatives. Fitoterapia 119, 136–149. doi: 10.1016/j.fitote.2017.05.001
Hewett-Emmett, D. (2000). “Evolution and distribution of the carbonic anhydrase gene families,” in The Carbonic Anhydrases, Vol. 90, eds W. R. Chegwidden, N. D. Carter, and Y. H. Edwards (Basel: Birkher). doi: 10.1007/978-3-0348-8446-4_3
Jakubowski, M., Szahidewicz-Krupska, E., and Doroszko, A. (2018). The human carbonic anhydrase II in platelets: an underestimated field of its activity. Biomed Res. Int. 2018:4548353. doi: 10.1155/2018/4548353
Khan, I., Ibrar, A., Abbas, N., and Saeed, A. (2014). Recent advances in the structural library of functionalized quinazoline and quinazolinone scaffolds: synthetic approaches and multifarious applications. Eur. J. Med. Chem. 76, 193–244. doi: 10.1016/j.ejmech.2014.02.005
Khan, I., Ibrar, A., Ahmed, W., and Saeed, A. (2015). Synthetic approaches, functionalization and therapeutic potential of quinazoline and quinazolinone skeletons: the advances continue. Eur. J. Med. Chem. 90, 124–169. doi: 10.1016/j.ejmech.2014.10.084
Khan, I., Zaib, S., Batool, S., Abbas, N., Ashraf, Z., Iqbal, J., et al. (2016). Quinazolines and quinazolinones as ubiquitous structural fragments in medicinal chemistry: an update on the development of synthetic methods and pharmacological diversification. Biorg. Med. Chem. 24, 2361–2381. doi: 10.1016/j.bmc.2016.03.031
Krishnamurthy, V. M., Kaufman, G. K., Urbach, A. R., Gitlin, I., Gudiksen, K. L., Weibel, D. B., et al. (2008). Carbonic anhydrase as a model for biophysical and physical-organic studies of proteins and protein– ligand binding. Chem. Rev. 108, 946–1051. doi: 10.1021/cr050262p
Lin, X., Li, X., and Lin, X. (2020). A review on applications of computational methods in drug screening and design. Molecules 25:1375. doi: 10.3390/molecules25061375
Lindskog, S. (1997). Structure and mechanism of carbonic anhydrase. Pharmacol. Ther. 74, 1–20. doi: 10.1016/S0163-7258(96)00198-2
Naïm, M., Bhat, S., Rankin, K. N., Dennis, S., Chowdhury, S. F., Siddiqi, I., et al. (2007). Solvated interaction energy (SIE) for scoring protein– ligand binding affinities. 1. exploring the parameter space. J. Chem. Inf. Model 47, 122–133. doi: 10.1021/ci600406v
Ozensoy Guler, O., Capasso, C., and Supuran, C. T. (2016). A magnificent enzyme superfamily: Carbonic anhydrases, their purification and characterization. J. Enzyme Inhib. Med. Chem. 31, 689–694. doi: 10.3109/14756366.2015.1059333
Pandit, U., and Dodiya, A. (2013). Synthesis and antitubercular activity of novel pyrazole–quinazolinone hybrid analogs. Med. Chem. Res. 22, 3364–3371. doi: 10.1007/s00044-012-0351-0
Panicker, C. Y., Varghese, H. T., Ambujakshan, K., Mathew, S., Ganguli, S., Nanda, A. K., et al. (2010). Vibrational spectra and computational study of 3-amino-2-phenyl quinazolin-4 (3H)-one. J. Mol. Struct. 963, 137–144. doi: 10.1016/j.molstruc.2009.10.026
Parkkila, A. K., Herva, R., Parkkila, S., and Rajaniemi, H. (1995). Immunohistochemical demonstration of human carbonic anhydrase isoenzyme II in brain tumours. Histochem. J. 27, 974–982. doi: 10.1007/BF02389687
Parkkila, S., Rajaniemi, H., Parkkila, A.-K., Kivelä, J, Waheed, A., Pastoreková, S., et al. (2000). Carbonic anhydrase inhibitor suppresses invasion of renal cancer cells in vitro. Proc. Natl. Acad. Sci. U.S.A. 97, 2220–2224. doi: 10.1073/pnas.040554897
Pastorekova, S., Parkkila, S., Parkkila, A. K., Opavsky, R., Zelnik, V., Saarnio, J., et al. (1997). Carbonic anhydrase IX, MN/CA IX: analysis of stomach complementary DNA sequence and expression in human and rat alimentary tracts. Gastroenterology 112, 398–408. doi: 10.1053/gast.1997.v112.pm9024293
Pastorekova, S., and Supuran, C. T. (2014). “Carbonic anhydrase IX: from biology to therapy,” in Hypoxia and Cancer (Springer), 21–153. doi: 10.1007/978-1-4614-9167-5_6
Pocker, Y., and Meany, J. (1967). The catalytic versatility of erythrocyte carbonic anhydrase. II. Kinetic studies of the enzyme-catalyzed hydration of pyridine aldehydes. Biochemistry 6, 239–246. doi: 10.1021/bi00853a037
Ruusuvuori, E., and Kaila, K. (2014). “Carbonic anhydrases and brain pH in the control of neuronal excitability,” in Carbonic Anhydrase: Mechanism, Regulation, Links to Disease, and Industrial Applications: Subcellular Biochemistry, Vol. 75 (Dordrecht: Springer). doi: 10.1007/978-94-007-7359-2_14
Saito, R., Sato, T., Ikai, A., and Tanaka, N. (2004). Structure of bovine carbonic anhydrase II at 1.95 Å resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 792–795. doi: 10.1107/S0907444904003166
Sentürk, M., Gülçin, I., Beydemir, S., Küfrevioglu, Ö. I., and Supuran, C. T. (2011). In vitro inhibition of human carbonic anhydrase I and II isozymes with natural phenolic compounds. Chem. Biol. Drug Des. 77, 494–499. doi: 10.1111/j.1747-0285.2011.01104.x
Shaik, N. A., Bokhari, H. A., Masoodi, T. A., Shetty, P. J., Ajabnoor, G. M., Elango, R., et al. (2019). Molecular modelling and dynamics of CA2 missense mutations causative to carbonic anhydrase 2 deficiency syndrome. J. Biomol. Struct. Dyn. 38, 4067–4080. doi: 10.1080/07391102.2019.1671899
Shank, R. P., Doose, D. R., Streeter, A. J., and Bialer, M. (2005). Plasma and whole blood pharmacokinetics of topiramate: the role of carbonic anhydrase. Epilepsy Res. 63, 103–112. doi: 10.1016/j.eplepsyres.2005.01.001
Supuran, C. T. (2008). Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 7, 168–181. doi: 10.1038/nrd2467
Supuran, C. T., and Scozzafava, A. (2007). Carbonic anhydrases as targets for medicinal chemistry. Bioorg. Med. Chem. 15, 4336–4350. doi: 10.1016/j.bmc.2007.04.020
Ur Rehman, N., Halim, S. A., Khan, M., Hussain, H., Yar Khan, H., Khan, A., et al. (2020). Antiproliferative and carbonic Anhydrase II inhibitory potential of chemical constituents from Lycium shawii and Aloe vera: evidence from in silico target fishing and in vitro testing. Pharmaceuticals 13:94. doi: 10.3390/ph13050094
Keywords: quinazolinones, bovine carbonic anhydrase-II, human carbonic anhydrase-II, structure-activity relationship, kinetics, molecular docking
Citation: Khan A, Khan M, Halim SA, Khan ZA, Shafiq Z and Al-Harrasi A (2020) Quinazolinones as Competitive Inhibitors of Carbonic Anhydrase-II (Human and Bovine): Synthesis, in-vitro, in-silico, Selectivity, and Kinetics Studies. Front. Chem. 8:598095. doi: 10.3389/fchem.2020.598095
Received: 23 August 2020; Accepted: 28 October 2020;
Published: 01 December 2020.
Edited by:
Imtiaz Khan, The University of Manchester, United KingdomReviewed by:
Mohammed El-Gamal, University of Sharjah, United Arab EmiratesDr. Suresh Reddy Chidipudi, Spiro Organics Private Limited, India
Copyright © 2020 Khan, Khan, Halim, Khan, Shafiq and Al-Harrasi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zahid Shafiq, zahidshafiq@bzu.edu.pk; Ahmed Al-Harrasi, aharrasi@unizwa.edu.om
 Ajmal Khan
Ajmal Khan Majid Khan
Majid Khan Sobia Ahsan Halim
Sobia Ahsan Halim Zulfiqar Ali Khan
Zulfiqar Ali Khan Zahid Shafiq
Zahid Shafiq Ahmed Al-Harrasi
Ahmed Al-Harrasi