Recognition of Proteins by Metal Chelation-Based Fluorescent Probes in Cells
- Department of Chemistry, The University of Hong Kong, Hong Kong, China
Fluorescent probes such as thiol-reactive and Ni2+-nitrilotriacetate (NTA) based probes provide a powerful toolbox for real-time visualization of a protein and a proteome in living cells. Herein, we first went through basic principles and applications of thiol-reactive based probes in protein imaging and recognition. We then summarize a family of metal-NTA based fluorescence probes in the visualization of His6-tagged protein and identification of metalloproteins at proteome-wide scale. The pros and cons of the probes, as well as ways to optimize them, are discussed.
Introduction
In the past decades, small molecule-based probes have been extensively used in protein labeling, allowing in situ studies of function, subcellular localization, and dynamics of the proteins of interest (POIs). Because of their relatively small sizes, these probes have less perturbation on the investigated proteins (Giepmans et al., 2006; Sletten and Bertozzi, 2009). Particularly, the small molecule-based tagging system has received growing attention, the POI is genetically fused with a short peptide of specific motifs, which is subsequently recognized by the probes. The recognition of POI by the probe either through chemical reaction or metal chelation usually results in fluorescent responses, which is applicable for biological imaging and quantification. Metal-chelation labeling of a protein is particularly attractive owing to its simplicity and high specificity. Pioneered by Griffin et al. (1998), a family of biarsenical probes have been developed, which specifically recognize a tetracysteine tag. Given that the (histidine)6-tag is more widely used in protein purification, enormous efforts have been made to develop fluorescence probes to selectively recognize the His6-tag (Kapanidis et al., 2001; Hauser and Tsien, 2007). These probes have been further utilized to label endogenous metal-binding proteins, providing a useful tool for metalloproteomics (Zhang et al., 2015; Jiang et al., 2018).
In this mini-review, we focus on metal chelation-based protein labeling, typically on the development of fluorescence probes that specifically recognize tetra-cysteine- and His6-tags in live cells. We highlight the optimization of the probes and their utilities in various systems. We also discuss the optimizations of fluorophores and molecule delivery for probes in tracking intracellular proteins.
The Thiol-Reactive Fluorescent Probes
Protein Recognition Based on Thiol Reaction
Cysteine residues are quite abundant and vital in protein structures to link two cysteine-containing peptides. Among 3,758 identified human proteins, more than 15,000 cysteine residues were reported (Gasser, 2014). The high abundance of cysteine residues has inspired scientists to develop strategies to label proteins via these sites. Although both As(III) and Sb(III) have high affinity to cysteine residues, only As(III) has been selected in the development of sensors. As-S coordination bonds are generated through thiol exchange, enabling As(III) to recognize cysteine residues. Compared with the liner structures created by As(III) and monothiols, the ring structures consisting of As(III) and two closely spaced thiols appear to be much more stable due to the factor of entropy (Stocken and Thompson, 1946). This explains why the formation of dithiol-reactive As(III) complexes is the main trend and the distance between the two cysteine residues was estimated to be 3–4 Å (Adams et al., 1990; Bhattacharjee and Rosen, 1996). When As(III) meets di-vicinal cysteine residues, chelation of As(III) to cysteines is achieved (Figure 1A).
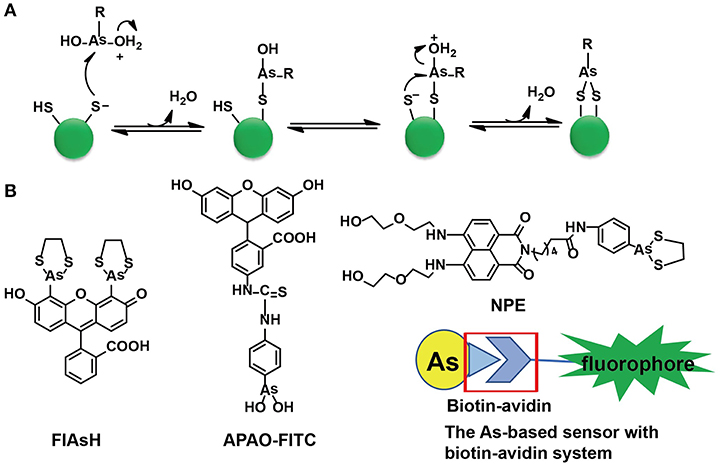
Figure 1. The As(III)-based thiol-reactive fluorescent probes. (A) The recognition mechanism of As(III) binding toward two closely spaced cysteines. The buffer action can make several RAs(OH)2 molecules to be protonated at the hydroxyl oxygen (R stands for any other chemical group). The attack of a thiolate anion on As(III) can release a water molecule. Then a proton transfer between the other thiolate anion and the hydroxyl oxygen is induced by the buffer action whilst the attack of this thiolate anion on As(III) will proceed an intramolecular displacement of another water molecule. (B) The structures of typical As(III)-based probes designed by different strategies. (A) was adapted from Gasser (2014) Copyright 2014 Wiley.
Designs of Arsenic-Based Fluorescent Probes
A family of arsenic(III)-based probes was designed and further modified for in situ visualizations of proteins with genetically fused tetra-cysteine residues. Roger Tsien et al. firstly reported a membrane-permeable and non-fluorescent As(III)-complex, i.e., FLAsH-EDT2 (EDT = ethanedithiol), to site-specifically recognize proteins containing four cysteines at the i, i + 1, i + 4, and i + 5 positions of an α-helix in 1998 (Griffin et al., 1998). A few years later, they found that four cysteines occurring in hairpin conformation were more preferred than the α-helix. Subsequently, a library of FLAsH-EDT2 analogs was developed and ReAsH-EDT2 has been regarded as the most suitable one to target a hairpin structure (Adams et al., 2002).
Both FLAsH-EDT2 and ReAsH-EDT2 are turn-on fluorescent probes as the 1,3,2-dithiarsolane (EDT) results in weak fluorescence before thiol-reaction with cysteines. Such a benefit was inherited in the design of other As-based probes, NPE, and CTNPE. Due to the structural features of vicinal-dithiol-containing proteins (VDPs), only one As(III) was kept for protein recognition, whilst two biocompatible diglycol amine groups were integrated into these probes to improve their water solubility and biological compatibility (Huang et al., 2011). Based on this work, a family of probes with various linkers (6-aminocaproic acid, succinic acid, and piperazine) were designed to detect VDPs in diverse conditions (different pH response, hydrophobicity, etc.; Huang et al., 2013). Later, the same group designed the first generation of radiometric fluorescent probe for VDPs, VTAF. This probe can be used to quantify the VDPs via the readout of the radiometric fluorescence signal with high selectivity (Huang et al., 2014). Besides these EDT-containing molecules, a similar As(III)-based probe, APAO-FITC, was synthesized with only one As-based moiety with the EDT being replaced by two hydroxyl groups (-OH). APAO-FITC has been shown to label native As-binding proteins without strong disturbance on their secondary structures (Femia et al., 2012). To expand the toolbox of As(III)-based probes, the biotin-avidin system was then taken into consideration while the As(III)-binding proteins could be analyzed by the combination of proteome microarray assay with biotin-conjugated As, i.e., Biotin-As. Given the high affinity of biotin toward avidin, the proteins distinguished by Biotin-As via thiol reaction quickly bind to the avidin-fluorophore to turn on the fluorescence (Figure 1B; Zhang et al., 2015).
Targeting Cysteine in Diverse Systems
The first generation of As(III)-based fluorescent probes, FLAsH-EDT2 and ReAsH-EDT2, mainly target proteins with genetically fused tetracysteine tag since this motif seems to be rare among native proteins (Femia et al., 2012). Moreover, two As-based moieties of the probes result in stable protein labeling, and these two probes work successfully in the visualization of intracellular proteins as well as in affinity chromatography to track arsenic-binding proteins (Griffin et al., 1998). Importantly, FLAsH-EDT2 and ReAsH-EDT2 have also been used to study the protein-protein interactions and protein folding by Luedtke et al. (2007). The tetracysteine tag was introduced to several vital biomarkers, e.g., avian pancreatic polypeptide (aPP), to estimate the protein mis-folding in the development of brain diseases by monitoring the fluorescent change of ReAsH-EDT2.
Besides those tetra-cysteine fused proteins, the thiol-groups of native proteins play a principal role in the balance of intracellular redox environment (Ying et al., 2007), whilst those proteins usually have the dithiol-motif such as -CXnC- rather than tetracysteine. Acting as the reductive end of this oxidation-reduction network, VDPs can be labeled by NPE both in vitro and in vivo. Staining of NPE finally lit up the VDPs in live cells without invasiveness (Huang et al., 2011). The analysis of As(III)-proteomes was achieved (Zhang et al., 2015) and the Biotin-As(III) serves as the As(III)-protein sensor following the protein separation by human proteome microarrays. In total 360 As(III)-binding proteins were reported and most of them were involved in the pathway of glycolysis. Subsequently, hexokinase-2 (HK2) was further studied by biochemical and metabolomics analysis. Consequently, they validated the identified As(III)-binding proteins which could be potential drug targets.
Design of Metal-NTA Based Fluorescent Probes
His6-Ni2+-NTA System for Imaging Proteins
The first report on using the His6-Ni2+-NTA system for protein purification was dated back to 1975 (Porath et al., 1975). It became a routine method for purification of His6-tagged proteins (Arnau et al., 2006). The wide usage of the His6-Ni2+-NTA system inspired scientists to visualize proteins by conjugating signaling groups, usually fluorophores. The first fluorescent modification on Ni-NTA was made by Kapanidis et al. (2001) and Katayama in 2001 (Amano et al., 2002). Subsequently, this type of probe was utilized to label membrane His-tagged proteins by Guignet et al. (2004) and Zenmyo et al. (2019).
The major drawback of the Ni-NTA system appeared to be the relatively low affinity of NTA toward Ni2+ with a dissociation constant of 1–20 μM (Soh, 2008). Hence, the multivalent tactic by conjugating more than one Ni-NTA moiety was developed (Guignet et al., 2004; Soh, 2008; Jing and Cornish, 2011). Very recently, R. Tampé and his co-workers designed linear, dendritic, and cyclic trisNTA scaffolds and compared their labeling abilities toward His-tagged proteins in fixed cells. Nevertheless, these multi-NTA probes can be hardly used in imaging of intracellular proteins owing to larger molecule size, which limited them to enter cells (Gatterdam et al., 2018). The other disadvantage is the fluorescent quenching triggered by paramagnetic Ni2+ in bio-imaging. By introducing NTA at the 6-position in the fluorescein ring, Lippard et al. developed a new Ni-NTA based probe with slighter fluorescent quenching, due to the molecular spatial conformation upon Ni2+ chelation (Goldsmith et al., 2006). To strengthen the protein binding, photoactive crosslinkers were firstly integrated by Auer et al. into the probes to form a stable covalent bond upon UV-irradiation (Hintersteiner et al., 2008). Hamachi et al. also synthesized a nucleophilic reaction-based Ni-NTA probe to label His-tagged proteins covalently (Uchinomiya et al., 2009). Nevertheless, almost all those probes suffer exclusively poor membrane permeability while a cell penetrating peptide (CPP) carrier was used to address this issue (Uchinomiya et al., 2013; Figure 2A). The usage of Ni-NTA was even combined with Fe3O4 nanoparticles to increase the adsorption efficiency (Guo et al., 2018).
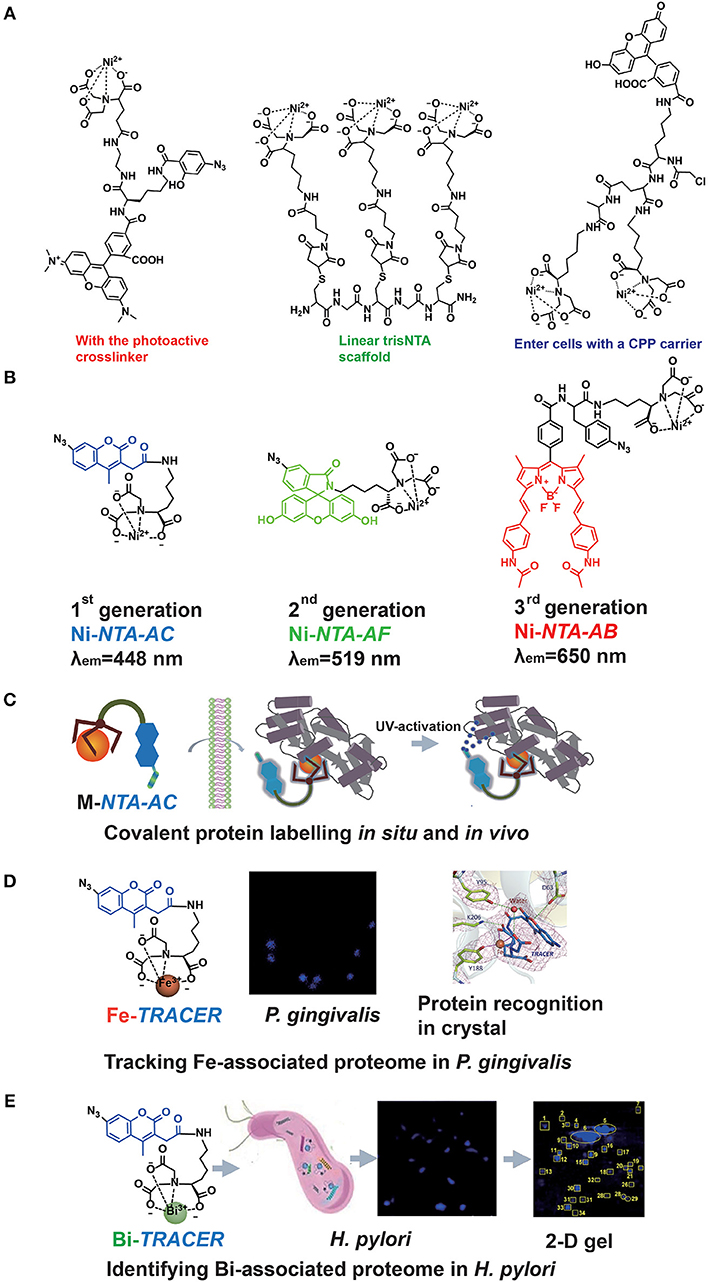
Figure 2. The probe family based upon NTA-metal coordination with their applications in protein tracking. Typical Ni-NTA-based probes were presented in (A) whilst our three generations of homemade probes were listed as (B), including Ni-NTA-AC, Ni-NTA-AF, and Ni-NTA-AB. Then the explorations for metal-associated proteomes in diverse system by using M-TRACER were presented as (C) the covalent labeling ability of M-TRACER toward intracellular proteins with UV-activation, (D) Fe-associated proteome tracked by Fe-TRACER in P. gingivalis and the protein-binding model released by X-ray crystallography, (E) Bi-associated proteome labeled by Bi-TRACER in H. pylori. (C,D) were reproduced from Jiang et al. (2018) with the permission of The Royal Society of Chemistry. (E) was reproduced from Wang et al. (2017) with the permission of The Royal Society of Chemistry.
Recently, a new family of Ni-NTA based fluorescent probes, which can rapidly enter cells to label His6-tagged protein covalently, was developed. The first probe, Ni-NTA-AC, consists of a mono-NTA group, a coumarin fluorophore, four-carbon connecting chain, and an arylazide incorporated into the fluorophore (Lai et al., 2015). Integration of photoactive crosslinker, arylazide, is essential not only for strengthening the protein-binding but also for fluorescent enhancement after UV-irradiation. The second and third generations of probes, Ni-NTA-AF and Ni-NTA-AB, were developed by incorporating fluorophores of fluorescein and BODIPY, respectively. It has been demonstrated that these probes could light up His6-tagged proteins in bacterial cells, mammalian cells and even tobacco leaves. Both green (Ni-NTA-AF) and red (Ni-NTA-AB) probes could overcome the cellular intrinsic autofluorescence with better contrast than the blue probe, Ni-NTA-AC. Moreover, these probes can be used to quantify His6-tagged proteins with a wider linear range (25–1,000 ng) compared with Western Blot (Chao et al., 2017; Yang et al., 2019). The same strategy was followed by an independent group in Denmark, and they recently reported imidazole carbamate probes to label His-tagged proteins (Mortensen et al., 2019) (Figure 2B).
Exploring Metalloproteomes
The importance of metals in biology is notable, mostly because metals can play critical roles in life processes, especially acting as the catalytic or structural cofactors (Hoppert, 2011). Metals are also frequently incorporated into pharmaceuticals for diagnostic and therapeutic purposes. Given the complexity of metal-protein interactions, tracking, and mining metalloproteins in vivo are great challenges. As a metal chelator, NTA can chelate a wide range of transition metal ions (Nancharaiah et al., 2006). Thus, the application of Ni-NTA-AC could be further extended. By replacing Ni2+ with other metal ions, such as Fe3+, Cu2+, and Bi3+, we designed a metal tunable fluorescence probe, M-TRACER (Tunable Reagent of Arylazide-Conjugated fluorescER), where M stands for metals (Lai et al., 2017). Incubating cells with the M-TRACER, the probe enters live cells and binds metal-associated proteins. After UV activation, those bound proteins are anchored to the probe via covalent bonds, enabling downstream analysis. Cell lysates from these stained cells were subjected to the combination of 2-D gel for protein separation and matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS) for protein identification. The identified proteins were then subjected to Gene Ontology (GO) enrichment analysis and mapped into protein networks. This approach implements the exploration of endogenous metal-associated proteins in various biological systems, including those proteins that bind metal ions weakly or transiently (Figure 2C).
The specific recognition of metal-associated proteins by the probe was demonstrated by using Fe-TRACER and human serum transferrin as a showcase. The x-ray structure of the protein adduct shows that Fe-TRACER binds to the specific ferric iron binding site in the C-lobe of transferrin, with Fe3+ coordinating to tyrosine 188 (Y188), while asparagic acid 63 (D63), tyrosine 95 (Y95), and lysine 206 (K206) participate in the formation of H-bonding to stabilize the structure. Interestingly, the probe folds a sandwich-like structure and Fe3+ coordinates to only two carboxylates and one nitrogen of the probe and one oxygen from Y188, leaving enough vacant site for interacting with proteins (Figure 2D).
Using Ni-TRACER, Ni-associated proteome in Helicobacter pylori (H. pylori) was firstly mined and 44 Ni2+-associated proteins were identified (Lai et al., 2017). Similarly, 63 Bi3+-binding proteins were identified by Bi-TRACER, in combination with quantitative proteomics. Then totally 119 Bi-regulated proteins were identified (Wang et al., 2017), providing rich resources to understand the mechanism of bismuth drugs, though these drugs have been applied to treat H. pylori infection clinically for decades (Gerrits et al., 2006) (Figure 2E). To deeply understand the metal homeostasis in pathogens, e.g., iron in Porphyromonas gingivalis (P. gingivalis), a keystone species to trigger periodontitis (Hajishengallis et al., 2012), Fe-TRACER was employed to unveil Fe-associated proteome in P. gingivalis, for this kind of bacteria is highly iron-dependent with a full arsenal of heme-binding proteins (Ciuraszkiewicz et al., 2014; Hajishengallis, 2015). Finally, 17 Fe-associated proteins were identified and they were mostly enriched in 8 biological processes and 2 molecular functions (Jiang et al., 2018). Similarly, the endogenous metalloproteome in live eukaryote cells can be tracked and mined by this approach. In total 54 Cu-associated proteins were identified in HeLa cells by Cu-TRACER, with one-third being confirmed previously by IMAC (Lai et al., 2017).
Molecular Modifications and Delivery Majorization
Development of Fluorophores
The development of delocalized electronic structure, i.e., the π system, just leads to the evaluation of fluorophores from blue to red fluorescence for the energy matching with π/π* energy gap finally resulted in fluorescent emission. Among numerous blue fluorophores, the coumarins appeared to be the oldest ones (Kumar et al., 2015). Their easy synthesis, photostability, and large Stokes shift have made our minds to select a coumarin derivative in the design of NTA-AC. Fluoresciens are the most recognized fluorophores with green emissions, which means a collection of available synthetic routes (Fu and Finney, 2018). NTA-AF incorporated a fluorescein fluorophore with the arylazide group for a significant turn-on effect after UV-activation. Rhodamines and BODIPYs act as the red fluorophores. The latter ones are comparatively new and the rigidity of BODIPYs limits their vibrational state density (Fu and Finney, 2018). Such a limitation endows BODIPYSs with sharp absorption and emission bands, matching with the design of NTA-AB. The use of near infra-red (NIR) fluorophores became a fast-growing and exciting field because of low absorption through biological matrixes (Sevick-Muraca, 2012). A series of NIR fluorescent nanoparticles were designed based upon benzo[1, 2-b: 4, 5-b']dithiophene 1, 1, 5, 5-tetraoxide (BDTO), and they showed good photostability and biocompatibility in cells and animals (Zhen et al., 2018). Imaging at the second near-infrared window (NIR-II; 1,000–1,700 nm) has also been achieved, and antiquenching cyanine fluorophores were reported by providing the optical penetration as deep as 8 mm with high contrast and super photostability (Wang et al., 2019). Herein, the development of NIR probes is urgently required to visualize metalloproteins.
Delivery of Probes Into Cells
To track intracellular POI, the delivery of fluorescent probes appeared to be a critical aspect. To break the limitations of lipophilic cell membrane toward large, charged molecules, about five kinds of approaches have been reported for living cell imaging (Lymperopoulos et al., 2010). Among them, electroporation enabled a high efficiency of fluorophores and proteins with notable size (up to 60 kDa) into living E. coli cells. Even though high voltages certainly decrease the cell viability, researchers can select higher ionic strengths with lower electroporation voltages and use glycerol to slightly increase the threshold of suitable ionic strength (Sustarsic et al., 2014). Addition of detergents, polyamidoamine and peptide dendrimers can be available for both prokaryotic and eukaryotic cells whilst bacteria usually required a larger amount. Tween 80 (0.25%) assisted Ni-NTA-AB to label intracellular His6-tagged protein whilst we found no significant effect of Tween 80 on cell viability (Chao et al., 2017). Besides these, arginine-rich CPPs have been demonstrated to induce multilamellarity of the cell membrane and subsequently enter cells via the formation of a fusion pore (Allolio et al., 2018). The design of a CPP carrier successfully improved the membrane permeability of Ni-based probes in mammalian cells (Uchinomiya et al., 2013). The last two approaches were microinjection and use of pore-forming agents such as streptolysin-O (SLO). In particular, SLO, a bacterial toxin, can form temporary pores in the membrane for a wide range of probes including small ligands. The efficiency achieved as high as 85% while around 95% of cells were reported to contain intact membrane after recovery of SLO treatments (Teng et al., 2016).
Concluding Remarks
Two families of representative probes based on metal-oriented protein recognition, i.e., As(III)-based probes and metal-NTA-based probes, were introduced and summarized from their binding mechanisms, molecule structures, to biological applications. These probes initially targeted fused proteins with either tetracysteine or His6-tags. For imaging of extracellular and intracellular proteins, almost all As(III)-based probes entered cells successfully whilst an array of Ni-NTA-based probes lack cellular membrane permeability. Despite diverse structure optimizations on these Ni-NTA-based probes, no clear clue of cellular membrane permeability was presented. Consequently, the different permeability between these two classes of probes resulted in their different applications in biology besides labeling relevant peptide-fused proteins. As(III)-based probes light up the native As(III)-binding proteins (Huang et al., 2011) while the proteomic analysis is achieved by the combination of the microarray with Biotin-As (Zhang et al., 2015). For the Ni-NTA-based probes, the Ni2+-binding proteins are detected in cell lysates and membrane parts (Kapanidis et al., 2001; Guignet et al., 2004; Arnau et al., 2006; Soh, 2008; Jing and Cornish, 2011).
Our probes were built upon metal-NTA coordination chemistry, and Ni2+ was selected as a showcase to inherit the success of Ni-NTA-based probes. In total, three probes were synthesized, i.e., Ni-NTA-AC, Ni-NTA-AF, and Ni-NTA-AB, with fluorescent emission ranging from blue to red. Importantly, they can track intracellular His-tagged proteins in live cells (Lai et al., 2015; Chao et al., 2017). The multiple-metal-binding ability of NTA expands the toolbox for these probes. Chelating with a variety of transition metals, the probes (M-TRACER) can label the relevant metal-associated proteins through metal ion-oriented recognition. As a result, the networks of Ni2+, Bi3+, Cu2+, and Fe2+/3+-associated proteins (and their proteomes) have been lit up in situ and in vivo (Lai et al., 2017; Wang et al., 2017; Jiang et al., 2018; Li et al., 2018). Subsequent protein identification and function analysis can be achieved by proteomics and other omics techniques. More sensitive and selective probes will be generated from further modifications of fluorophores, as well as optimizations of molecule delivery. They may provide a new horizon to understand bacterial and human metalloproteomes, and their relationship with human health such as infectious disease and drug development.
Author Contributions
NJ drafted this manuscript. HL revised this whilst. HS supervised this.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adams, E., Jeter, D., Cordes, A. W., and Kolis, J. W. (1990). Chemistry of organometalloid complexes with potential antidotes: structure of an organoarsenic (III) dithiolate ring. Inorg. Chem. 29, 1500–1503. doi: 10.1021/ic00333a012
Adams, S. R., Campbell, R. E., Gross, L. A., Martin, B. R., Walkup, G. K., Yao, Y., et al. (2002). New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 124, 6063–6076. doi: 10.1021/ja017687n
Allolio, C., Magarkar, A., Jurkiewicz, P., Baxová, K., Javanainen, M., Mason, P. E., et al. (2018). Arginine-rich cell-penetrating peptides induce membrane multilamellarity and subsequently enter via formation of a fusion pore. Proc. Natl. Acad. Sci. U.S.A. 115, 11923–11928. doi: 10.1073/pnas.1811520115
Amano, H., Ohuchi, Y., Katayama, Y., and Maeda, M. (2002). “A new fluorescent reagent for the detection of proteins having histidine-tag (his-tag),” in Analytical Sciences/Supplements Proceedings of IUPAC International Congress on Analytical Sciences 2001 (ICAS 2001) The Japan Society for Analytical Chemistry, Tokyo, 2002, i1469–i1471.
Arnau, J., Lauritzen, C., Petersen, G. E., and Pedersen, J. (2006). Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein Expres. Purif. 48, 1–13. doi: 10.1016/j.pep.2005.12.002
Bhattacharjee, H., and Rosen, B. P. (1996). Spatial proximity of Cys113, Cys172, and Cys422 in the metalloactivation domain of the ArsA ATPase. J. Bio. Chem. 271, 24465–24470. doi: 10.1074/jbc.271.40.24465
Chao, A., Jiang, N., Yang, Y., Li, H., and Sun, H. (2017). A Ni-NTA-based red fluorescence probe for protein labelling in live cells. J. Mater. Chem. B 5, 1166–1173. doi: 10.1039/C6TB02848A
Ciuraszkiewicz, J., Smiga, M., Mackiewicz, P., Gmiterek, A., Bielecki, M., Olczak, M., et al. (2014). Fur homolog regulates Porphyromonas gingivalis virulence under low-iron/heme conditions through a complex regulatory network. Mol. Oral Microbol. 29, 333–353. doi: 10.1111/omi.12077
Femia, A. L., Temprana, C. F., Santos, J., Carbajal, M. L., Amor, M. S., Grasselli, M., et al. (2012). An arsenic fluorescent compound as a novel probe to study arsenic-binding proteins. Protein J. 31, 656–666. doi: 10.1007/s10930-012-9441-6
Fu, Y., and Finney, N. S. (2018). Small-molecule fluorescent probes and their design. RSC Adv. 8, 29051–29061. doi: 10.1039/C8RA02297F
Gasser, G. (ed.). (2014). Inorganic Chemical Biology: Principles, Techniques and Applications. Chichester: John Wiley & Sons, Ltd. Available online at: https://www.wiley.com/en-us/Inorganic+Chemical+Biology%3A+Principles%2C+Techniques+and+Applications-p-9781118510025
Gatterdam, K., Joest, E. F., Gatterdam, V., and Tampé, R. (2018). Rücktitelbild: the scaffold design of trivalent chelator heads dictates affinity and stability for labeling his-tagged proteins in vitro and in cells. Angew. Chem. 130, 12766–12766. doi: 10.1002/ange.201808284
Gerrits, M. M., Van Vliet, A. H. M., Kuipers, E. J., and Kusters, J. G. (2006). Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect. Dis. 6, 699–709. doi: 10.1016/S1473-3099(06)70627-2
Giepmans, B. N. G., Adams, S. R., Ellisman, M. H., and Tsien, R. Y. (2006). The fluorescent toolbox for assessing protein location and function. Science 312, 217–224. doi: 10.1126/science.1124618
Goldsmith, C. R., Jaworski, J., Sheng, M., and Lippard, S. J. (2006). Selective labeling of extracellular proteins containing polyhistidine sequences by a fluorescein–nitrilotriacetic acid conjugate. J. Am. Chem. Soc. 128, 418–419. doi: 10.1021/ja0559754
Griffin, B. A., Adams, S. R., and Tsien, R. Y. (1998). Specific covalent labeling of recombinant protein molecules inside live cells. Science 281, 269–272. doi: 10.1126/science.281.5374.269
Guignet, E. G., Hovius, R., and Vogel, H. (2004). Reversible site-selective labeling of membrane proteins in live cells. Nat. Biotechnol. 22, 440–444. doi: 10.1038/nbt954
Guo, H., Li, M., Tu, S., and Sun, H. (2018). Selective binding and magnetic separation of his-tagged proteins using Fe3O4/PAM/NTA-Ni2+ magnetic nanoparticles. IOP Conf. Ser. 322, 022017. doi: 10.1088/1757-899X/322/2/022017
Hajishengallis, G. (2015). Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15, 30–44. doi: 10.1038/nri3785
Hajishengallis, G., Darveau, R. P., and Curtis, M. A. (2012). The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10, 717–725. doi: 10.1038/nrmicro2873
Hauser, C. T., and Tsien, R. Y. (2007). A hexahistidine-Zn2+-dye label reveals STIM1 surface exposure. Proc. Natl. Acad. Sci. U.S.A. 104, 3693–3697. doi: 10.1073/pnas.0611713104
Hintersteiner, M., Weidemann, T., Kimmerlin, T., Filiz, N., Buehler, C., and Auer, M. (2008). Covalent fluorescence labeling of his-tagged proteins on the surface of living cells. ChemBioChem 9, 1391–1395. doi: 10.1002/cbic.200800089
Hoppert, M. (2011). Metalloenzymes. Encyclopedia of Geobiology. Dordrecht: Springer. doi: 10.1007/978-1-4020-9212-1_134
Huang, C., Jia, T., Tang, M., Yin, Q., Zhu, W., Zhang, C., et al. (2014). Selective and ratiometric fluorescent trapping and quantification of protein vicinal dithiols and in situ dynamic tracing in living cells. J. Am. Chem. Soc. 136, 14237–14244. doi: 10.1021/ja5079656
Huang, C., Yin, Q., Meng, J., Zhu, W., Yang, Y., Qian, X., et al. (2013). Versatile probes for the selective detection of vicinal-dithiol-containing proteins: design, synthesis, and application in living cells. Chem. Eur. J. 19, 7739–7747. doi: 10.1002/chem.201300567
Huang, C., Yin, Q., Zhu, W., Yang, Y., Wang, X., Qian, X., et al. (2011). Highly selective fluorescent probe for vicinal-dithiol-containing proteins and in situ imaging in living cells. Angew. Chem. Int. Edit. 50, 7551–7556. doi: 10.1002/anie.201101317
Jiang, N., Cheng, T., Wang, M., Chan, G. C., Jin, L., Li, H., et al. (2018). Tracking iron-associated proteomes in pathogens by a fluorescence approach. Metallomics 10, 77–82. doi: 10.1039/C7MT00275K
Jing, C., and Cornish, V. W. (2011). Chemical tags for labeling proteins inside living cells. Accounts Chem. Res. 44, 784–792. doi: 10.1021/ar200099f
Kapanidis, A. N., Ebright, Y. W., and Ebright, R. H. (2001). Site-specific incorporation of fluorescent probes into protein: hexahistidine-tag-mediated fluorescent labeling with (Ni2+: nitrilotriacetic acid) n–fluorochrome conjugates. J. Am. Chem. Soc. 123, 12123–12125. doi: 10.1021/ja017074a
Kumar, K. A., Renuka, N., Pavithra, G., and Kumar, G. V. (2015). Comprehensive review on coumarins: molecules of potential chemical and pharmacological interest. J. Chem. Pharm. Res. 7, 67–81. Available online at: https://www.researchgate.net/profile/K_Kumar25/publication/282566993_Comprehensive_review_on_coumarins_Molecules_of_potential_chemical_and_pharmacological_interest/links/56120dc808aec422d1172c1a.pdf
Lai, Y. T., Chang, Y. Y., Hu, L., Yang, Y., Chao, A., Du, Z. Y., et al. (2015). Rapid labeling of intracellular his-tagged proteins in living cells. Proc. Natl. Acad. Sci. U.S.A. 112, 2948–2953. doi: 10.1073/pnas.1419598112
Lai, Y. T., Yang, Y., Hu, L., Cheng, T., Chang, Y. Y., Koohi-Moghadam, M., et al. (2017). Integration of fluorescence imaging with proteomics enables visualization and identification of metallo-proteomes in living cells. Metallomics 9, 38–47. doi: 10.1039/C6MT00169F
Li, H., Wang, R., and Sun, H. (2018). Systems approaches for unveiling the mechanism of action of bismuth drugs: new medicinal applications beyond Helicobacter pylori infection. Accounts Chem. Res. 52, 216–227. doi: 10.1021/acs.accounts.8b00439
Luedtke, N. W., Dexter, R. J., Fried, D. B., and Schepartz, A. (2007). Surveying polypeptide and protein domain conformation and association with FlAsH and ReAsH. Nat. Chem. Biol. 3:779. doi: 10.1038/nchembio.2007.49
Lymperopoulos, K., Kiel, A., Seefeld, A., Stöhr, K., and Herten, D. P. (2010). Fluorescent probes and delivery methods for single-molecule experiments. ChemPhysChem 11, 43–53. doi: 10.1002/cphc.200900359
Mortensen, M. R., Nielsen, N. L., Palmfeldt, J., and Gothelf, K. V. (2019). Imidazole carbamate probes for affinity guided azide-transfer to metal-binding proteins. Org. Biomol. Chem. 17, 1379–1383. doi: 10.1039/C8OB03017K
Nancharaiah, Y. V., Schwarzenbeck, N., Mohan, T. V. K., Narasimhan, S. V., Wilderer, P. A., and Venugopalan, V. P. (2006). Biodegradation of nitrilotriacetic acid (NTA) and ferric–NTA complex by aerobic microbial granules. Water Res. 40, 1539–1546. doi: 10.1016/j.watres.2006.02.006
Porath, J., Carlsson, J. A. N., Olsson, I., and Belfrage, G. (1975). Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 258, 598. doi: 10.1038/258598a0
Sevick-Muraca, E. M. (2012). Translation of near-infrared fluorescence imaging technologies: emerging clinical applications. Annu. Rev. Med. 63, 217–231. doi: 10.1146/annurev-med-070910-083323
Sletten, E. M., and Bertozzi, C. R. (2009). Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Edit. 48, 6974–6998. doi: 10.1002/anie.200900942
Soh, N. (2008). Selective chemical labeling of proteins with small fluorescent molecules based on metal-chelation methodology. Sensors 8, 1004–1024. doi: 10.3390/s8021004
Stocken, L. A., and Thompson, R. H. S. (1946). British anti-lewisite: 3. Arsenic and thiol excretion in animals after treatment of lewisite burns. Biochem. J. 40:548. doi: 10.1042/bj0400548
Sustarsic, M., Plochowietz, A., Aigrain, L., Yuzenkova, Y., Zenkin, N., and Kapanidis, A. (2014). Optimized delivery of fluorescently labeled proteins in live bacteria using electroporation. Histochem. Cell. Bio. 142, 113–124. doi: 10.1007/s00418-014-1213-2
Teng, K. W., Ishitsuka, Y., Ren, P., Youn, Y., Deng, X., Ge, P., et al. (2016). Labeling proteins inside living cells using external fluorophores for microscopy. eLife 5:e20378. doi: 10.7554/eLife.20378
Uchinomiya, S., Nonaka, H., Fujishima, S., Tsukiji, S., Ojida, A., and Hamachi, I. (2009). Site-specific covalent labeling of His-tag fused proteins with a reactive Ni (II)–NTA probe. Chem. Commun. 39, 5880–5882. doi: 10.1039/b912025d
Uchinomiya, S., Nonaka, H., Wakayama, S., Ojida, A., and Hamachi, I. (2013). In-cell covalent labeling of reactive His-tag fused proteins. Chem. Commun. 49, 5022–5024. doi: 10.1039/c3cc41979g
Wang, S., Fan, Y., Li, D., Sun, C., Lei, Z., Lu, L., et al. (2019). Anti-quenching NIR-II molecular fluorophores for in vivo high-contrast imaging and pH sensing. Nat. Commun. 10:1058. doi: 10.1038/s41467-019-09043-x
Wang, Y., Hu, L., Xu, F., Quan, Q., Lai, Y. T., Xia, W., et al. (2017). Integrative approach for the analysis of the proteome-wide response to bismuth drugs in Helicobacter pylori. Chem. Sci. 8, 4626–4633. doi: 10.1039/C7SC00766C
Yang, Y., Jiang, N., Lai, Y. T., Chang, Y. Y., Yang, X., Sun, H., et al. (2019). Green fluorescent probe for imaging His6-tagged proteins inside living cells. ACS Sensors 4, 1190–1196. doi: 10.1021/acssensors.8b01128
Ying, J., Clavreul, N., Sethuraman, M., Adachi, T., and Cohen, R. A. (2007). Thiol oxidation in signaling and response to stress: detection and quantification of physiological and pathophysiological thiol modifications. Free Radical Bio. Med. 43, 1099–1108. doi: 10.1016/j.freeradbiomed.2007.07.014
Zenmyo, N., Tokumaru, H., Uchinomiya, S., Fuchida, H., Tabata, S., Hamachi, I., et al. (2019). Optimized reaction pair of the cyshis tag and Ni (II)-NTA probe for highly selective chemical labeling of membrane proteins. Bull. Chem. Soc. Jpn. 92, 995–1000. doi: 10.1246/bcsj.20190034
Zhang, H., Yang, L., Ling, J., Czajkowsky, D. M., Wang, J. F., Zhang, X. W., et al. (2015). Systematic identification of arsenic-binding proteins reveals that hexokinase-2 is inhibited by arsenic. Proc. Natl. Acad. Sci. U.S.A. 112, 15084–15089. doi: 10.1073/pnas.1521316112
Zhen, S., Wang, S., Li, S., Luo, W., Gao, M., Ng, L. G., et al. (2018). Efficient red/near-infrared fluorophores based on benzo [1, 2-b: 4, 5-b′] dithiophene 1, 1, 5, 5-tetraoxide for targeted photodynamic therapy and in vivo two-photon fluorescence bioimaging. Adv. Funct. Mater. 28:1706945. doi: 10.1002/adfm.201706945
Keywords: His-tag, metallomics, metalloproteomics, molecular imaging, thiol-reaction
Citation: Jiang N, Li H and Sun H (2019) Recognition of Proteins by Metal Chelation-Based Fluorescent Probes in Cells. Front. Chem. 7:560. doi: 10.3389/fchem.2019.00560
Received: 09 May 2019; Accepted: 23 July 2019;
Published: 09 August 2019.
Edited by:
Huib Ovaa, Leiden University Medical Center, NetherlandsReviewed by:
Giovanni Signore, Scuola Normale Superiore di Pisa, ItalyAlexander Shekhtman, University at Albany, United States
Copyright © 2019 Jiang, Li and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongzhe Sun, hsun@hku.hk
 Nan Jiang
Nan Jiang Hongyan Li
Hongyan Li