A microRNA Cluster-Lefty Pathway is Required for Cellulose Synthesis During Ascidian Larval Metamorphosis
- 1Sars Fang Centre, MoE Key Laboratory of Marine Genetics and Breeding, College of Marine Life Sciences, Ocean University of China, Qingdao, China
- 2Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao, China
Synthesis of cellulose and formation of tunic structure are unique traits in the tunicate animal group. However, the regulatory mechanism of tunic formation remains obscure. Here, we identified a novel microRNA cluster of three microRNAs, including miR4018a, miR4000f, and miR4018b in Ciona savignyi. In situ hybridization and promoter assays showed that miR4018a/4000f/4018b cluster was expressed in the mesenchymal cells in the larval trunk, and the expression levels were downregulated during the later tailbud stage and larval metamorphosis. Importantly, overexpression of miR4018a/4000f/4018b cluster in mesenchymal cells abolished the cellulose synthesis in Ciona larvae and caused the loss of tunic cells in metamorphic larvae, indicating the regulatory roles of miR4018a/4000f/4018b cluster in cellulose synthesis and mesenchymal cell differentiation into tunic cells. To elucidate the molecular mechanism, we further identified the target genes of miR4018a/4000f/4018b cluster using the combination approaches of TargetScan prediction and RNA-seq data. Left–right determination factor (Lefty) was confirmed as one of the target genes after narrow-down screening and an experimental luciferase assay. Furthermore, we showed that Lefty was expressed in the mesenchymal and tunic cells, indicating its potentially regulatory roles in mesenchymal cell differentiation and tunic formation. Notably, the defects in tunic formation and loss of tunic cells caused by overexpression of miR4018a/4000f/4018b cluster could be restored when Lefty was overexpressed in Ciona larvae, suggesting that miR4018a/4000f/4018b regulated the differentiation of mesenchymal cells into tunic cells through the Lefty signaling pathway during ascidian metamorphosis. Our findings, thus, reveal a novel microRNA-Lefty molecular pathway that regulates mesenchymal cells differentiating into tunic cells required for the tunic formation in tunicate species.
Introduction
Development of Urochordate ascidians experiences a swimming larval-to-sessile juvenile metamorphic transition. During this process, part of the larval organs, such as the notochord and sensory organ disappeared, while novel adult organs, such as the heart and digestive apparatus are lately formed (Karaiskou et al., 2015; Sasakura and Hozumi, 2018; Hotta et al., 2020). It has been revealed that certain adult organs are derived from the larval mesenchymal cells (Hirano and Nishida, 1997; Imai et al., 2003; Tokuoka et al., 2005; Fodor et al., 2021).
Mesenchymal stem cells (MSCs) are stromal cells that can self-renew and exhibit multilineage differentiation, with the capability to become an important source of tissue and organ formation and establish the cardiovascular, hematopoietic, bone, and soft tissues during embryonic development (Ding et al., 2011). MSCs in vertebrates, such as humans and mice, are divided into three cell groups: mesenchymal stromal/stem cells, pericytes, and smooth muscle cells. They differentiate into multiple organs, thus, are used wildly in cell engineering and human clinical medicine (Ding et al., 2011; Slukvin and Kumar, 2018; Shin et al., 2020; Storer et al., 2020). In invertebrates, especially in ascidian Ciona, mesenchymal cells are derived from the A7.6, B8.5, and B7.7 cell lineages at the 110-cell stage (Imai et al., 2003; Nakamura et al., 2012). Ascidians Halocynthia sp. and Ciona sp. mesenchymal cells derived from A7.6 cells are named as trunk lateral cells (TLC) (Imai et al., 2003; Nakamura et al., 2012; Satoh, 2014), and they mainly contribute to the formation of adult blood cells, epithelial cells of the first/second branchial cleft, tunic cells, muscle cells of the inlet and outlet, body wall muscle cells, and part of the stomach (Jeffery et al., 2008). Mesenchymal cells derived from B7.5 cells are trunk ventral cells (TVC) (Nakamura et al., 2012). They migrate between the abdominal surface and the endoderm of the trunk, and eventually differentiate into cells in tissues and organs, such as the adult heart, pericardium, and body wall muscles (Hirano and Nishida, 2000; Tokuoka et al., 2005). Mesenchymal cells derived from B7.7 and B8.5 cells differentiate into the tunic and part of the stomach of the adult (Satoh, 2014). In ascidians, mesenchymal cells have conserved behaviors and property as those in vertebrates, such as migration and differentiation (Imai et al., 2003; Tokuoka et al., 2005). Thus, this simple animal is an ideal model at an evolutionary perspective to study the functions of mesenchymal cell and their associations with developmental signaling pathways.
Tunic, composed of cellulose, is a unique structure in a tunicate animal (Dehal et al., 2002). Tunic also has been revealed to be a complex and active tissue with mesenchymal properties (Matos et al., 2020). It had been shown that mesenchymal cells can migrate across epidermal cells into tunic (Cloney and Grimm, 1970; Di Bella et al., 2005). Different types of cells are distributed within tunic (De Leo et al., 1981; Hirose et al., 1994a; Hirose et al., 1994b; Hirose, 2009), playing variable functions, such as accumulation of metabolites and inorganics (Hirose et al., 2006), phagocytosis, pigmentation (Hirose et al., 1999), and defense responses (López-Legentil et al., 2005) in different ascidian species. Cellulose in tunicates is synthesized by a horizontally transferred cellulose synthase gene (CesA) (Nakashima et al., 2004; Matthysse et al., 2005; Sagane et al., 2010). A mutant of CesA in Ciona leads to a variety of phenotypes, including embryonic hatchling defects, notochord alignment, and tail elongation (Sasakura et al., 2005).
MicroRNAs (miRNAs), a class of evolutionally conserved small noncoding RNA molecules with 18–22 nucleotides (Bartel, 2004; Bushati and Cohen, 2007; Leonardo et al., 2012; Bartel, 2018), are commonly identified in eukaryotes and involved in the multiple physiological processes, such as cell differentiation (Foshay and Gallicano, 2009; Sanchez-Vasquez et al., 2019; Yeh et al., 2019), apoptosis (Fujita et al., 2009; Cheng et al., 2018; Sur et al., 2019), and growth and development (Hayashi et al., 2011; Rebustini et al., 2012). In previous studies, we identified 106 known miRNAs and 59 novel miRNAs in Ciona savignyi. Among them, miR4108a, miR4000f, and miR4018b were significantly downregulated during metamorphosis (Zhang et al., 2018). The results of in situ hybridization showed that the three miRNAs formed a gene cluster and were expressed in the larval trunk; however, the roles of these miRNAs in development, their target genes, and the involved molecular signaling pathways remain elusive.
Based on previous results, this study mainly focused on, and studied, the functions of miR4018a/4000f/4018b gene cluster in C. savignyi. We found that this gene cluster was required for cellulose synthesis and tunic formation in larvae and metamorphic juveniles. We further identified its target gene Left–right determination factor (Lefty) and revealed that upregulation of Lefty by the decreased expression miR4018a/4000f/4018b cluster was essential for this process. Thus, we elucidate a novel microRNA-Lefty molecular pathway that regulates mesenchymal cell differentiation required for the synthesis of cellulose in tunicate species.
Results
miR4018a/4000f/4018b Cluster was Expressed in the Mesenchymal Cells
Our previous investigation revealed that miR4018a, miR4000f, and miR4018b were consecutively located in scaffold reftig_107 in the genome of the ascidian C. savignyi, forming an miRNA cluster (Zhang et al., 2018). Its expression level was significantly downregulated during the larval stage by RNA-seq data (Zhang et al., 2018), indicating its potential roles in larval metamorphosis. We, thus, further validated the expression levels of miR4018a, miR4000f, and miR4018b by qPCR. The results showed that the expression levels of miR4018a, miR4000f, and miR4018b at 42 h postfertilization (hpf), 2 days postattachment (dpa), and 4 dpa were significantly lower than that at 18 and 21 hpf (Supplementary Figure S1). In addition, our previous data showed that all the three members of the microRNAs cluster expressed in a scattered cell population, which was likely mesenchymal cells at the head and trunk part in tailbud embryos (Zhang et al., 2018). To validate the expression tissues, we made a construct containing 3,025 bp upstream of miR4018a/4000f/4018b fused with TdTomato, miR4018a/4000f/4018b upstream 3k > TdTomato (Figure 1A), and co-electroporated into Ciona eggs with aldoketoreductase (AKR) promoter-GFP construct, AKR-promoter > GFP (Tokuoka et al., 2005; Idris et al., 2013; Satoh, 2014; Cao et al., 2019). AKR is a mesenchymal marker gene, which is expressed in the mesenchymal cells and becomes a mesenchymal marker at the late larval stage and juvenile stages (Idris et al., 2013). Our results showed that TdTomato signals were scattered in the larval trunk colocalized with AKR signaling at different developmental stages (Figure 1B), validating that the miR4018a/4000f/4018b cluster was indeed expressed in the mesenchymal cell population (Figure 1C).
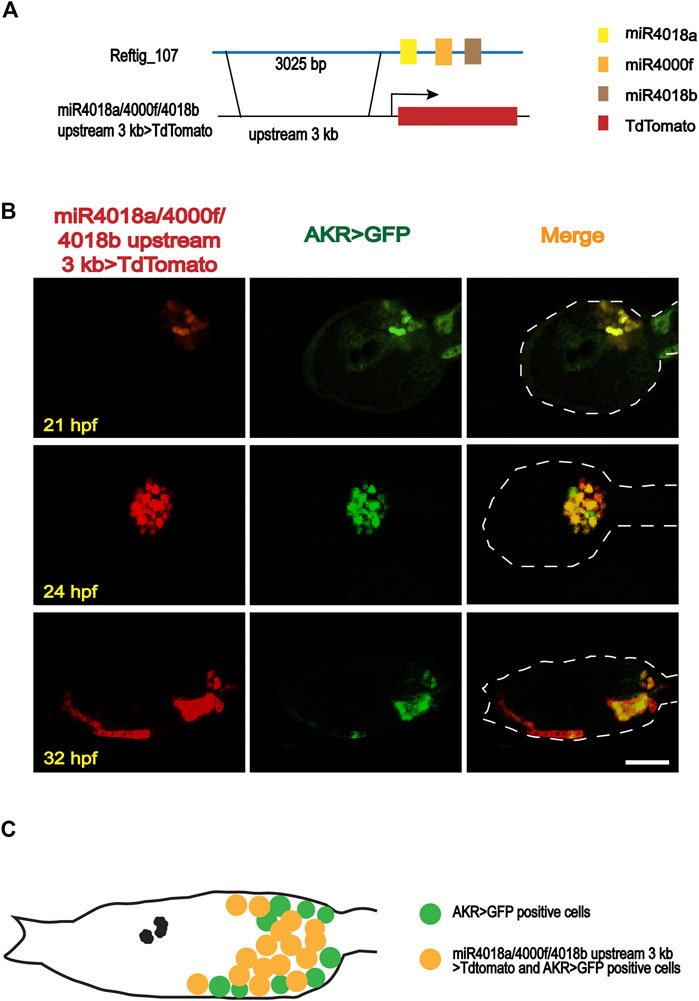
FIGURE 1. Expression patterns of miR4018a/4000f/4018b cluster in Ciona savignyi. (A) Schematic representation of the regulatory fragment of miR4018a/4000f/4018b cluster. A 3,025-bp upstream fragment of the pre-miR4018a/4000f/4018b cluster was fused with fluorescent protein TdTomato to make a construct. (B) MiR4018a/4000f/4018b upstream 3 kb > TdTomato co-expressed with the mesenchymal marker gene AKR promoter fusion construct in the larval trunk at 21 h postfertilization (hpf), 24 hpf, and 32 hpf. (C) The model of the expression pattern of miR4018a/4000f/4018b cluster in the mesenchymal cells. Statistical data showed that miR4018a/4000f/4018b upstream 3 kb > TdTomato-positive cells (n = 18 embryos) were all expressed in the AKR > GFP-positive cells, suggesting that miR4018a/4000f/4018b cluster was expressed in the mesenchymal cell in C. savignyi. Scale bar in (B): 50 μm.
Down-Regulation of miR4018a/4000f/4018b Cluster was Required for Cellulose Synthesis During Ciona Larval Metamorphosis
We wondered whether miR4018a/4000f/4018b cluster is involved in metamorphosis, due to its significant downregulated expression. To validate its roles in mesenchymal cells during larval development, we overexpressed the AKR > TdTomato-miR4018a/4000f/4018b in Ciona larvae using AKR driver, and the TdTomato positive signals were detected in mesenchymal cells in both miR4018a/4000f/4018b cluster-overexpressed and control animals at different stages (Figure 2A). We found that the cellulose was significantly decreased and hardly observed in miR4018a/4000f/4018b cluster-overexpressed individuals in swimming and metamorphic larvae (green arrowheads in Figure 2A). The qPCR results showed that the expression levels of three miRNAs in the miR4018a/4000f/4018b-overexpressed group increased significantly compared with the control animals, indicating that the three miRNAs were all overexpressed in Ciona larvae successfully (Supplementary Figure S2). In addition, we tracked an individual Ciona embryo from the larval stage to juvenile (after metamorphosis) and observed the similar defects of the cellulose synthesis (n = 11/17, Supplementary Figure S3). Cellulose synthesis defects were also observed in the larvae overexpressing miR4018a/4000f/4018b cluster by its own driver (miR4018a/4000f/4018b upstream 3 kb > TdTomato-miR4018a/4000f/4018b) (n = 18/26, Supplementary Figure S4).
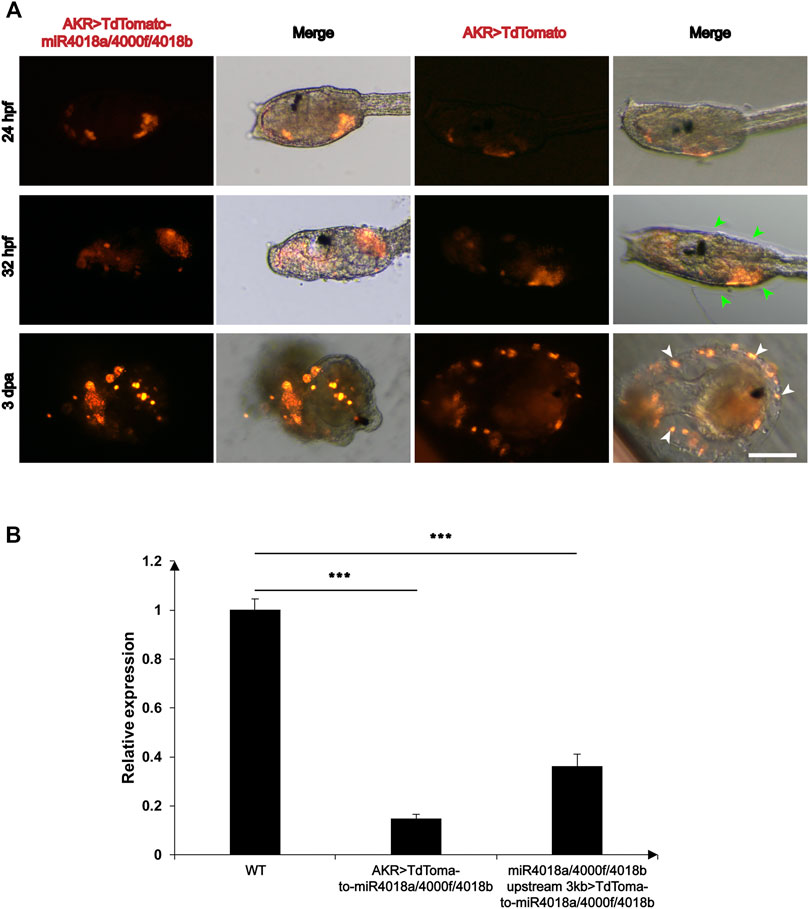
FIGURE 2. Overexpression of miR4018a/4000f/4018b cluster caused failure of cellulose formation in larvae and loss of tunic cells in the metamorphic larvae. (A) AKR > miR4018a/4000f/4018b-TdTomaoto and AKR > TdTomato (control) were electroporated in the fertilized eggs, respectively. At 32 hpf, the cellulose could be observed surrounding the trunk in AKR > TdTomato-expressed larvae (green arrowheads), while in AKR > miR4018a/4000f/4018b-TdTomaoto-expressed larvae, cellulose could not be observed. In 3 dpa control larvae, the cells with TdTomato-positive signal could be observed within the tunic (white arrowheads), while in miR4018a/4000f/4018b cluster-overexpressed larvae, isolated fluorescent-positive cells could be found surrounding the larvae. Scale bar: 50 μm. (B) The expression level of cellulose synthase gene (CesA) in control and miRNA-overexpression groups. The results showed that the expression level of CesA was decreased significantly at the groups of AKR promoter driven and miR4018a/4000f/4018b promoter-driven miRNA cluster overexpression compared with the control group. Asterisks (***) represent statistical significance (p < 0.001).
Our data also showed the defects of tail absorption in miRNAs overexpression groups. There were 25/31 larvae without the whole tail resorption in miRNA overexpression group, while in the control, 13/29 larvae without the whole tail resorption (white-dashed squares in Supplementary Figures S3 and S4). In addition, we found that miRNA-overexpressed larvae had similar phenotypes in tunic construction with “swimming juvenile” mutant, the tail tunic was narrower than that of wild-type larvae, and the normal fin did not develop (Sasakura et al., 2005).
Since cellulose synthesis in tunicates is mediated by CesA (Nakashima et al., 2004; Matthysse et al., 2005; Sagane et al., 2010), and mutant of CesA affects the structure of tunic in ascidian (Sasakura et al., 2005), we then investigated the expression levels of CesA in miR4018a/4000f/4018b cluster-overexpressed larvae. The results showed that the expression level of CesA decreased significantly in miRNA-overexpressed groups compared with that in the control animals (Figure 2B), indicating that miR4018a/4000f/4018b cluster suppresses the expression of CesA.
Overexpression of miR4018a/4000f/4018b Cluster Led to the Loss of Tunic Cells
The ascidian tunic is a complex and active tissue that presents mesenchymal characteristics (Hirose et al., 1994b). With the confocal time-lapse imaging, we observed that the mesenchymal cells that migrated and finally presented within the tunic (Figure 2) likely differentiated into the tunic cells. Surprisingly, we found that the floating mesenchymal marker-positive tunic cells were lost in miR4018a/4000f/4018b cluster-overexpressed metamorphic larvae, but presented normally within the tunic in control animals (Figure 2A white arrowheads). This suggests that miR4018a/4000f/4018b is involved in the differentiation of mesenchymal cells into tunic cells.
Identification of Target Genes of miR4018a/4000f/4018b Cluster
To reveal the underlying molecular mechanisms, we conducted RNA-Seq using miR4018a/4000f/4018b-overexpressed larvae and then sorted 6,902 downregulation expressed genes compared with that in wild-type larvae (Supplementary Figure S5). In addition, we computably predicted 929 target genes based on the seed sequence of miR4018a/4000f/4018b cluster by TargetScan (Supplementary Figure S5). Overlapped genes (563) were selected based on RNA-Seq data and TargetScan prediction. Along with the fact that the expression level of miR4018a/4000f/4018b cluster was lower at 42 hpf, we chose 47 significantly upregulated genes (log2fold <−1) out of 563 candidates. Then we focused on 12 genes based on gene function annotation (Supplementary Table S1).
Lefty was the Target Gene of miR4018a/4000f/4018b Cluster
To validate the target gene of miR4018a/4000f/4018b cluster, the 12 genes that were screened out were further experimentally examined through the luciferase report gene assay. We overexpressed miRNAs in cells transfected with luciferase reporter gene constructs containing different target gene 3′UTRs (Twist is excluded because it does not have a 3′UTR region binding to miRNA). No significant alteration in luciferase activity has been observed in the groups of Pmsa3, Tll1, EeF2k, Pdcd4, Sele, FBXW2, Tab1, MYOT, and TGFR-2 (Supplementary Figure S6). The luciferase activity of miRNA/F11-3′UTR was slightly increased compared with the F11-3′UTR group; however, it was not very significant (p = 0.03, Supplementary Figure S6). Only the Lefty gene (Gene ID: ENSCSAVG00000003020) was finally identified as the target of miR4018a/4000f/4018b cluster with the fact that the luciferase activity of miR4018/Lefty 3′UTR-transfected cells decreased remarkably compared with the miRNA negative control mimic and Lefty mutant [removal of miR4018 binding sites (AUGUUCC)] (Figures 3A, B). Furthermore, we examined the relationship of miR4018a/4000f/4018b cluster and Lefty in HEK-293T cells by Western blotting. The result showed that the level of Lefty protein decreased remarkably in cells cotransfected with CMV > Lefty and miR4018 mimic compared with that in the group of CMV > Lefty only (Figure 3C). These results indicate that Lefty is the target gene of the miR4018a/4000f/4018b cluster.
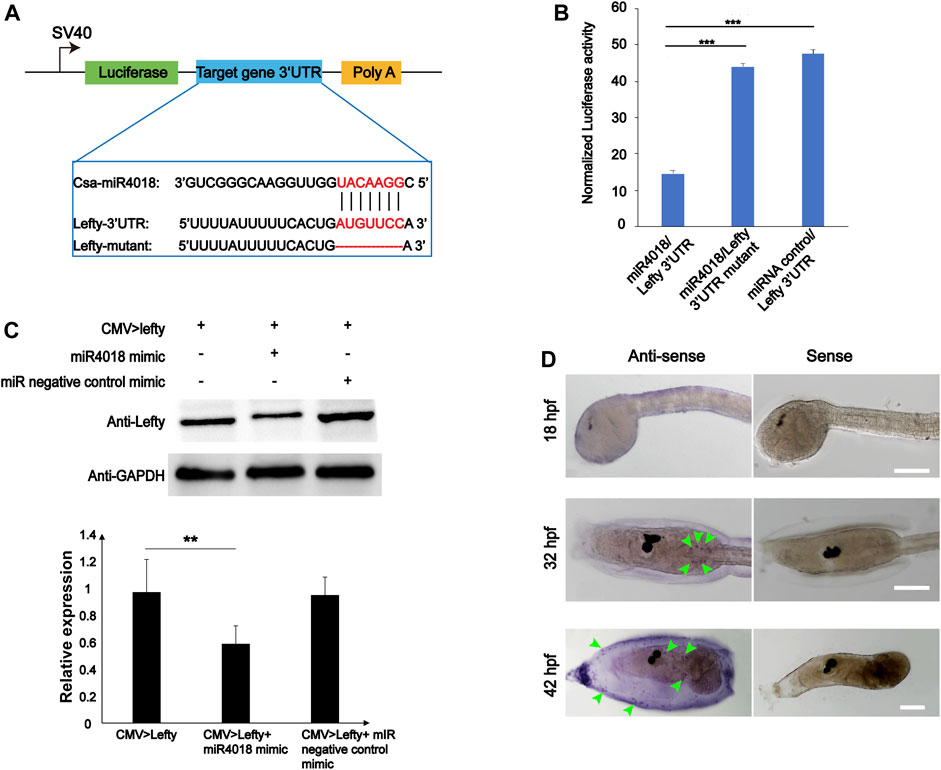
FIGURE 3. Lefty is the target gene of miR4018. (A) Schematic representation of luciferase reporter constructs carrying the 3′-UTR regions of target genes. Lefty-3′ UTR, Lefty-3′ UTR mutants were cloned into the downstream luciferase open reading frame of the pmirGLO dual-luciferase vector. Sequence alignments of csa-miR-4018 and its predicted target gene and Lefty-3′UTR and mutants are displayed in the blue line box. (B) Relative luciferase activity in HEK-293T cells cotransfected with pmirGLO-Lefty 3′ UTR and miR4018 mimics, pmirGLO-Lefty 3′ UTR mutants and miR4018 mimics, Lefty-3′ UTR mutants and miRNA negative control mimics, respectively. Firefly luciferase values were normalized Renilla luciferase activity. Student’s t-test was used to evaluate the significance of luciferase data. Asterisks (***) represent statistical significance (p < 0.001). (C) Lefty expression at the protein level in human embryonic kidney 293T cells (HEK293T). GAPDH was used as an internal control in the Western blotting experiments. CMV > Lefty, CMV > Lefty and miR4018 mimics, CMV > Lefty and miRNA negative control mimics were cotransfected into HEK-293T cells, respectively. Expression level of Lefty in CMV > Lefty and miR4018 mimic group was significantly decreased compared with that in the additional two groups. Asterisks (**) represent statistical significance (p < 0.01). (D) Expression patterns of Lefty detected by whole-mount in situ hybridization. Hybridized signals were specifically detected in a scattered cell population in the trunk in the embryos and larvae at 31 and 42 hpf (green arrowheads). No visible signal was examined in the embryos at 18 hpf. Scale bar: 100 μm.
miR4018a/4000f/4018b Cluster Regulated Cellulose Synthesis via Lefty Signaling
Single-cell RNA sequencing data showed that Lefty is normally expressed in the notochord, B-line mesenchyme, and TVC in C. savignyi embryos (Zhang et al., 2020). We detected the expression pattern of Lefty in C. savignyi larvae at different stages by whole-mount in situ hybridization. The results showed that it was specifically detected in a scattered cell population at 31 and 42 hpf, similar to the expression pattern of miR4018a/4000f/4018b cluster (Figure 3D), indicating that Lefty was likely also expressed in the mesenchymal cells. The similar expression patterns of Lefty with miR4018a/4000f/4018b cluster implies that Lefty may act as a downstream factor of miR4018a/4000f/4018b cluster to regulate cellulose synthesis. To test if Lefty is involved in tunic formation as the target gene of the miR4018a/4000f/4018b cluster, we co-overexpressed miR4018a/4000f/4018b and Lefty in Ciona larvae, in which we found that both tunic and tunic cells were presented (n = 22/35, Figure 4). In miR4018a/4000f/4018b-only overexpression group, 25/35 larvae were without the tunic (Figure 4). This result implies that Lefty could largely rescue the phenotypes in miR4018a/4000f/4018b-overexpressed larvae. This experiment demonstrates that miR4018a/4000f/4018b regulated cellulose synthesis and tunic formation via Lefty signaling.
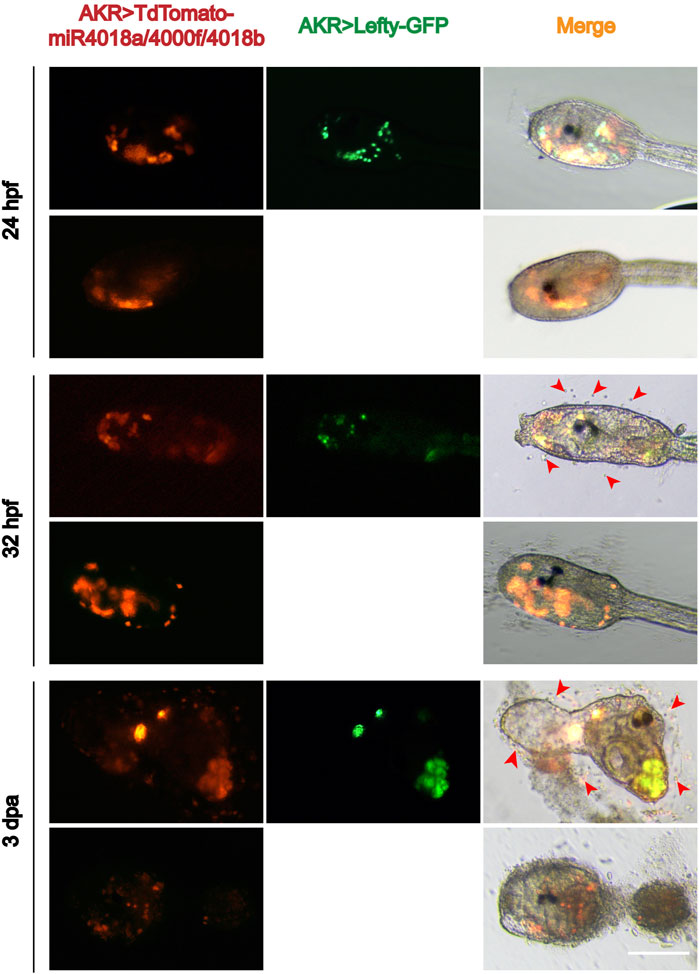
FIGURE 4. Overexpression of Lefty rescue the phenotype in the miR4018a/4000f/4018b-overexpressed larvae. Cellulose and the sporadic tunic cells could be observed in AKR > TdTomato-miR4018a/4000f/4018b and AKR > Lefty-GFP coexpressed swimming and metamorphotic larvae at 32 hpf and 3 dpa (red arrowhead) (n = 22/35). In miR4018a/4000f/4018b-only overexpression group, 25/35 larvae without the tunic. Scale: bar 50 μm.
Discussion
The tunic is a multifunctional tissue that provides support and mechanical rigidity. It is mainly composed of cellulose. Tunicates is the only animal group that can synthesize cellulose in the epidermis by a horizontally transferred cellulose synthase gene (CesA) (Nakashima et al., 2004; Matthysse et al., 2005; Sagane et al., 2010). In ascidians, the tunic was first observed in the late tailbud stage, and gradually formed a tunic covering the trunk and tail, respectively, during swimming stage. With the development of the metamorphosis of larvae, the tunic of the tail was lost, and a complete tunic was formed in juveniles (Cloney and Cavey, 1982; Stato et al., 1997). During this process, there are free tunic cells between the tunic and epidermis. Studies have shown that tunic cells are derived from mesenchymal cells of the B8.5 and B7.7 cell lineages (Satoh, 2014), and can be divided into different cell types that perform different biological functions, such as accumulation of metabolites and inorganic products (Hirose et al., 2006), pigmentation (Hirose et al., 1999), defense responses (López-Legentil et al., 2005), and phagocytosis (Li et al., 2020). In addition, tunic formation requires the presence of tunic cells. Some free tunic cells contain and deposit molecules, which modify the larval tunic, so they may be involved in the formation of tunic in the ascidian embryos (Bishop et al., 2010). Tunic cells can migrate during the whole embryonic development process, form a large number of aggregates, and migrate to the outer surface of the embryo to perform their functions (Bishop et al., 2010).
In vertebrates, bone morphogenetic protein (Chen et al., 2019), fibroblast-binding factors (FGFs) (Sun and Stathopoulos, 2018), transforming growth factor beta (Hao et al., 2019), Wingless-type/β-catenin (Jarvinen et al., 2018; Chen et al., 2019), and other signaling pathways are involved in the differentiation of mesenchymal stem cells into diverse cell types, such as chondrocytes (Yang et al., 2018), odontogenic cells (Cui et al., 2018), and muscle cells (De Bari et al., 2003; Kumar et al., 2017). Also, miR-145 mediates the differentiation of mesenchymal cells into smooth muscle cells (Yeh et al., 2019), and mesenchymal stem cell-derived exosome miR-223 regulates neuronal cell apoptosis (Wei et al., 2020). In ascidian, it has been reported that FGF, pmRACK1, Tbx6, and twist-like signaling play a center role in the fate determination from mesenchymal cell to other organs, such as the muscle, somatic, gonad, nerve cord, tunic, and heat (Tokuoka et al., 2005; Kim et al., 2007; Tatzuke et al., 2012; Kumano et al., 2014). In this study, we showed that a novel ascidian miRNA cluster (miR4018a/4000f/4018b) was expressed in the mesenchymal cells of larvae and is downregulated during larval metamorphosis. Overexpression of miR4018a/4000f/4018b cluster caused the failure of cellulose synthesis in swimming larvae and loss of tunic in metamorphic juveniles.
We speculated that miR4018a/4000f/4018b cluster might be responsible for tunic cell development. Overexpression of this miRNA cluster affected its downstream gene expression and inhibited the differentiation of mesenchymal cells into tunic cells. Defective tunic cell specification may have an impact on epidermal cells and eventually disrupted the expression of the essential genes (e.g., CesA) related to cellulose synthesis. In order to explore the molecular mechanism, we conducted a screening based on RNA-Seq and TargetScan prediction to find the target genes of miR4018a/4000f/4018b. Lefty was finally identified as the target after screening and luciferase assay. Lefty is a member of the TGF-β subfamily, and its encoded protein can regulate stem cell differentiation by competitively binding TGF-β receptors (Tabibzadeh and Hemmati-Brivanlou, 2006). Lefty is known to regulate the balance of cell pluripotency and cell differentiation through the Lefty/Nodal signaling pathway (Dai et al., 2016; Li et al., 2018; Zhang et al., 2019). In addition, Lefty can also cooperate with Smad (mothers against decapentaplegic homolog), Wnt, and the transcriptional factor octamer-binding factor 3/4 to support cell stemness (Tabibzadeh and Hemmati-Brivanlou, 2006). We further showed that Lefty overexpression could rescue the defects of tunic formation and loss of tunic cells caused by miR4018a/400f/4018b overexpression, confirming that miRNA cluster-Lefty pathway is required for the differentiation of Ciona mesenchymal cells into tunic cells and cellulose synthesis.
Our study also found that miRNA overexpression caused a more severe tunic-deficiency phenotype than that in CesA mutant (Sasakura et al., 2005). It is possible that overexpression of miRNA cluster regulates additional targets that affect tunic synthesis process, such as extracellular matrix components.
In conclusion, we report a novel miR4018a/4000f/4018b cluster-Lefty pathway that plays an important role in the differentiation of mesenchymal cells into tunic cells and synthesis of cellulose, providing insights into the understanding of the mesenchymal cell development and differentiation, and formation of tunic structure in tunicates.
Methods
Adult Animals, Fertilization, and Electroporation
The ascidian C. savignyi adults were collected from the eastern coast of Qingdao (Shandong, China) and maintained in a seawater tank for 3 days under constant light to accumulate gametes in the laboratory. The eggs and sperm were obtained from different individuals and then fertilized in the seawater for 5 min. After fertilization, eggs were dechorionated, electroporated, and then cultured at 16°C for development. The electroporation was performed as the previously described protocol (Lu et al., 2020).
Promoter Cloning and Plasmid Construction
The PCR of a promoter was performed with the Phanta Max Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China). The PCR product was purified by GeneJET Gel Extraction Kit (Thermo Scientific, Waltham, MA, USA) and then ligated into the vector containing the reporter gene GFP or TdTomato. All constructs were generated using the one ClonExpress MultiS One Step Cloning Kit (Vazyme, Nanjing, China) and confirmed by sequencing. The sequences of primers used for plasmid construction are shown in Supplementary Table S2.
Quantitative PCR
Two types of samples were used for quantitative PCR (qPCR). One was the embryos/larvae at the different stages (18, 21, 25, 42 hpf and 2, 4 dpa), another was the wild-type and miR4018a/4000f/4018b-overexpressed larvae collected at 31 hpf. The total RNA was extracted with RNAiso Reagent (Takara). All cDNAs were synthesized using the miRNA First Strand cDNA Synthesis Kit (by stem-loop) (Vazyme, Nanjing, China). qPCR amplification was performed using the miRNA Universal SYBR qPCR Master Mix Kit (Vazyme, Nanjing, China) on the Roche Applied Science LightCyclerTM 480 Real-Time PCR System. According to the supplier’s protocol, the reaction was carried out with the following conditions: 95°C for 5 min, 40 cycles of 95°C for 10 s and 60°C for 30 s, 95°C for 15 s, 60°C for 60 s, and 95°C for 15 s. Data were calculated using the 2−ΔΔCt method, and statistical analyses were performed using paired Student’s t-tests. A value of p < 0.05 was considered statistically significant.
Whole-Mount in situ Hybridization
The ascidian larvae were fixed in 4% paraformaldehyde for 2 h at RT. In situ hybridization was performed as previously described (Zhang et al., 2018). Labeled antisense RNA probes were transcribed from linearized DIG RNA Labeling Mixture (Roche America) according to the manufacturer’s instructions. The Lefty probe size is 780 bp, and the sequences of primers are as follows: forward primer, 5′ TTATCGTCCTGTTCCTCGCA 3′ and reverse primer, 5′ GCTGGTTCCTTCACGTTTGT 3′.
RNA-Seq and the Target Gene Prediction
The wild-type and miR4018a/4000f/4018b cluster-overexpressed larvae were collected at 32 hpf, and the total RNA was extracted and sent for RNA-seq. The gene libraries were constructed according to the NEB common library construction method, and the original data was formed by Illumina sequencing. Clean reads for subsequent analysis were obtained after filtration of original data, sequencing error rate check, and GC content distribution check. Clean reads after quality control were compared with the genome for gene annotation, and the sequencing of the reference transcription construction library was completed. Differential gene enrichment analysis was performed using NovoMagic (https://magic.novogene.com/).
The target genes of miR4018a, 4000f, and 4018b were predicted using TargetScan. 3′UTRs of C. savignyi mRNAs obtained from the ENSEMBL database were used as potential target sequences. The predicted targets with downregulated expression after miR4018a/4000f/4018b overexpression were collected for further analysis (Zhang et al., 2018).
Cell Culture and Luciferase Assay
Human embryonic kidney 293T cells (HEK293T) were cultured in the following conditions: Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, USA) supplemented with 10% fetal bovine serum (Gibco, USA), penicillin (100 U/ml), and streptomycin (100 μg/ml) grown in a humidified atmosphere of 5% CO2 at 37°C.
The wild-type 3′UTR fragments of 12 candidate target genes containing predicted cas-miR4018 or cas-miR4000f seed sites were amplified by PCR using cDNA as templates. The PCR products were cloned into the firefly luciferase coding region of the pmirGLO plasmid to construct reporters for luciferase assays. For the luciferase assay, 6 h before cell transfections, HEK293T cells were passaged in 96-well plates with 3 × 104 cells per well. Then the cells were cotransfected with 0.1 μg of luciferase reporter plasmid and 100 nM miRNA mimics. Twelve hours after transfection, a fresh medium was placed into each well. Forty-eight hours after transfection, firefly and Renilla luciferase activities were measured using the dual-luciferase reporter assay system (Beyotime, China), and relative reporter activity was normalized to Renilla luciferase activity. The data statistical analyses were performed using paired Student’s t-tests. Experiments were performed in triplicate.
Western Blot
The total protein (20 μg) was used for 10% SDS-polyacrylamide gel electrophoresis at 150 V for 1 h and then transferred onto polyvinylidene fluoride (PVDF) membranes in Western transfer buffer at 60 mA for 3 h in an ice bath. Nonspecific binding sites were blocked with 5% fat-free powdered milk in Tris-buffered saline plus Tween 20 [0.2% (vol/vol)] (TBST) (blocking solution) at room temperature (RT) for 2 h. After washing three times with TBST (each for 15 min), the membranes were incubated overnight at 4°C with primary Lefty antibody (Lefty antibody: TBST = 1:1,000) (Abcam Chain). After the incubation, the PVDF membranes were washed three times and then incubated at RT for 2 h with the horseradish peroxidases (HRP)-conjugated secondary antibody. After washing three times with TBST (each for 15 min), proteins were detected using enhanced chemiluminescence (Advansta, Menlo Park, CA, USA). Quantification was performed using the Quantity One program (Bio-Rad). The experiments were performed three times in triplicate. Gray scale analysis was performed using ImageJ, and data statistical analyses were performed using paired Student’s t-tests.
Data Availability Statement
The data presented in the study are deposited in the SRA repository, accession number PRJNA790731.
Author Contributions
BD and XZ conceived the project. Most experiments were carried out by XS with the help of XZ. Data were analyzed and discussed by XS and LY. The initial draft was drafted by XS. The final version of the manuscript was prepared by BD.
Funding
This work was supported by the National Key Research and Development Program of China (2019YFE0190900, 2018YFD0900705), the National Natural Science Foundation of China (Grant No. 31771649), and the Taishan Scholar Program of Shandong Province, China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Wei Wang and Zhiyi LV for comments and suggestions on the revision of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.835906/full#supplementary-material
References
Bartel, D. P. (2004). MicroRNAs: Genomics,Biogenesis, Mechanism, and Function'. Cell 116, 281–297. doi:10.1016/s0092-8674(04)00045-5
Bishop, C. D., Hall, B. K., and Bates, W. R. (2010). HSP90 Expression in Two Migratory Cell Types during Ascidian Development: Test Cells deposit HSP90 on the Larval Tunic. Int. J. Dev. Biol. 54 (8-9), 1337–1346. doi:10.1387/ijdb.082730cb
Bushati, N., and Cohen, S. M. (2007). microRNA Functions. Annu. Rev. Cel Dev. Biol. 23, 175–205. doi:10.1146/annurev.cellbio.23.090506.123406
Cao, C., Lemaire, L. A., Wang, W., Yoon, P. H., Choi, Y. A., Parsons, L. R., et al. (2019). Comprehensive Single-Cell Transcriptome Lineages of a Proto-Vertebrate. Nature 571 (7765), 349–354. doi:10.1038/s41586-019-1385-y
Chen, X., Liu, J., Li, N., Wang, Y., Zhou, N., Zhu, L., et al. (2019). Mesenchymal Wnt/β-Catenin Signaling Induces Wnt and BMP Antagonists in Dental Epithelium. Organogenesis 15 (2), 55–67. doi:10.1080/15476278.2019.1633871
Cheng, X., Zhang, G., Zhang, L., Hu, Y., Zhang, K., Sun, X., et al. (2018). Mesenchymal Stem Cells Deliver Exogenous miR-21viaexosomes to Inhibit Nucleus Pulposus Cell Apoptosis and Reduce Intervertebral Disc Degeneration. J. Cel. Mol. Med. 22 (1), 261–276. doi:10.1111/jcmm.13316
Cloney, R. A., and Cavey, M. J. (1982). Ascidian Larval Tunic: Extraembryonic Structures Influence Morphogenesis. Cell Tissue Res 222, 547–562. doi:10.1007/BF00213854
Cloney, R. A., and Grimm, L. (1970). Transcellular Emigration of Blood Cells during Ascidian Metamorphosis. Z. Zellforsch. 107, 157–173. doi:10.1007/bf00335222
Cui, D., Li, H., Wan, M., Peng, Y., Xu, X., Zhou, X., et al. (2018). The Origin and Identification of Mesenchymal Stem Cells in Teeth: from Odontogenic to Non-odontogenic. Curr. Stem Cel Res Ther 13 (1), 39–45. doi:10.2174/1574888X12666170913150403
Dai, H.-Q., Wang, B.-A., Yang, L., Chen, J.-J., Zhu, G.-C., Sun, M.-L., et al. (2016). TET-mediated DNA Demethylation Controls Gastrulation by Regulating Lefty-Nodal Signalling. Nature 538 (7626), 528–532. doi:10.1038/nature20095
De Bari, C., Dell'Accio, F., Vandenabeele, F., Vermeesch, J. R., Raymackers, J.-M., and Luyten, F. P. (2003). Skeletal Muscle Repair by Adult Human Mesenchymal Stem Cells from Synovial Membrane. J. Cel Biol 160 (6), 909–918. doi:10.1083/jcb.200212064
De Leo, G., Patricolo, E., and Frittitta, G. (1981). Fine Structure of the Tunic ofCiona intestinalisL. II. Tunic Morphology, Cell Distribution and Their Functional Importance. Acta Zoologica (Stockh) 62, 259–271. doi:10.1111/j.1463-6395.1981.tb00634.x
Dehal, P., Satou, Y., Campbell, R. K., Chapman, J., Degnan, B., De Tomaso, A., et al. (2002). The Draft Genome of Ciona intestinalis : Insights into Chordate and Vertebrate Origins. Science 298 (5601), 2157–2167. doi:10.1126/science.1080049
Di Bella, M. A., Carbone, M. C., and De Leo, G. (2005). Aspects of Cell Production in Mantle Tissue of Ciona intestinalis L. (Tunicata, Ascidiacea). Micron 36 (5), 477–481. doi:10.1016/j.micron.2005.01.007
Ding, D.-C., Shyu, W.-C., and Lin, S.-Z. (2011). Mesenchymal Stem Cells. Cel Transpl. 20 (1), 5–14. doi:10.3727/096368910x
Fodor, A., Liu, J., Turner, L., and Swalla, B. J. (2021). Transitional Chordates and Vertebrate Origins: Tunicates. Evol. Develop. Biol. 141, 149. doi:10.1016/bs.ctdb.2020.10.001
Foshay, K. M., and Gallicano, G. I. (2009). miR-17 Family miRNAs Are Expressed during Early Mammalian Development and Regulate Stem Cell Differentiation. Develop. Biol. 326 (2), 431–443. doi:10.1016/j.ydbio.2008.11.016
Fujita, K., Mondal, A. M., Horikawa, I., Nguyen, G. H., Kumamoto, K., Sohn, J. J., et al. (2009). p53 Isoforms Δ133p53 and P53β Are Endogenous Regulators of Replicative Cellular Senescence. Nat. Cel Biol 11 (9), 1135–1142. doi:10.1038/ncb1928
Hao, Y., Baker, D., and Ten Dijke, P. (2019). TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int. J. Mol. Sci. 20 (11), 2767. doi:10.3390/ijms20112767
Hayashi, T., Koyama, N., Azuma, Y., and Kashimata, M. (2011). Mesenchymal miR-21 Regulates Branching Morphogenesis in Murine Submandibular Gland In Vitro. Develop. Biol. 352 (2), 299–307. doi:10.1016/j.ydbio.2011.01.030
Hirano, T., and Nishida, H. (1997). Developmental Fates of Larval Tissues after Metamorphosis in AscidianHalocynthia Roretzi. Develop. Biol. 192, 199–210. doi:10.1006/dbio.1997.8772
Hirano, T., and Nishida, H. (2000). Developmental Fates of Larval Tissues after Metamorphosis in the Ascidian, Halocynthia Roretzi. Develop. Genes Evol. 210, 55–63. doi:10.1007/s004270050011
Hirose, E. (2009). Ascidian Tunic Cells: Morphology and Functional Diversity of Free Cells outside the Epidermis. Invertebrate Biol. 128 (1), 83–96. doi:10.1111/j.1744-7410.2008.00153.x
Hirose, E., Hirose, M., and Neilan, B. A. (2006). Localization of Symbiotic Cyanobacteria in the Colonial Ascidian Trididemnum Miniatum (Didemnidae, Ascidiacea). Zoolog. Sci. 23 (5), 435–442. doi:10.2108/zsj.23.435
Hirose, E., Ishii, T., Saito, Y., and Taneda, Y. (1994a). Phagocytic Activity of Tunic Cells in the Colonial Ascidian Aplidium Yamazii (Polyclinidae, Aplousobranchia). Zoolog. Sci. 11, 203–208.
Hirose, E., Ishii, T., Saito, Y., and Taneda, Y. (1994b). Seven Types of Tunic Cells in the Colonial Ascidian Aplidium Yamazii (Polyclinidae, Aploosobranchia): Morphology, Classification, and Possible Functions. Zoolog. Sci. 11, 737–743.
Hirose, E., Kimura, S., Itoh, T., and Nishikawa, J. (1999). Tunic Morphology and Cellulosic Components of Pyrosomas, Doliolids, and Salps (Thaliacea, Urochordata). Biol. Bull. 196 (1), 113–120. doi:10.2307/1543173
Hotta, K., Dauga, D., and Manni, L. (2020). The Ontology of the Anatomy and Development of the Solitary Ascidian Ciona: the Swimming Larva and its Metamorphosis. Sci. Rep. 10, 17916. doi:10.1038/s41598-020-73544-9
Idris, M. M., Thorndyke, M. C., and Brown, E. R. (2013). Evidence for Dynamic and Multiple Roles for Huntingtin in Ciona intestinalis. Invert Neurosci. 13 (2), 151–165. doi:10.1007/s10158-013-0158-9
Imai, K. S., Satoh, N., and Satou, Y. (2003). A Twist-like bHLH Gene Is a Downstream Factor of an Endogenous FGF and Determines Mesenchymal Fate in the Ascidian Embryos. Development 130 (18), 4461–4472. doi:10.1242/dev.00652
Järvinen, E., Shimomura-Kuroki, J., Balic, A., Jussila, M., and Thesleff, I. (2018). Mesenchymal Wnt/β-Catenin Signaling Limits Tooth Number. Development 145 (4), 158048. doi:10.1242/dev.158048
Jeffery, W. R., Chiba, T., Krajka, F. R., Deyts, C., Satoh, N., and Joly, J.-S. (2008). Trunk Lateral Cells Are Neural Crest-like Cells in the Ascidian Ciona intestinalis: Insights into the Ancestry and Evolution of the Neural Crest. Develop. Biol. 324 (1), 152–160. doi:10.1016/j.ydbio.2008.08.022
Karaiskou, A., Swalla, B. J., Sasakura, Y., and Chambon, J.-P. (2015). Metamorphosis in Solitary Ascidians. Genesis 53 (1), 34–47. doi:10.1002/dvg.22824
Kim, G. J., Kumano, G., and Nishida, H. (2007). Cell Fate Polarization in Ascidian Mesenchyme/muscle Precursors by Directed FGF Signaling and Role for an Additional Ectodermal FGF Antagonizing Signal in Notochord/nerve Cord Precursors. Development 134 (8), 1509–1518. doi:10.1242/dev.02825
Kumano, G., Negoro, N., and Nishida, H. (2014). Transcription Factor Tbx6 Plays a central Role in Fate Determination between Mesenchyme and Muscle in Embryos of the ascidian,Halocynthia Roretzi. Develop. Growth Differ. 56 (4), 310–322. doi:10.1111/dgd.12133
Kumar, A., D’Souza, S. S., Moskvin, O. V., Toh, H., Wang, B., Zhang, J., et al. (2017). Specification and Diversification of Pericytes and Smooth Muscle Cells from Mesenchymoangioblasts. Cel Rep. 19 (9), 1902–1916. doi:10.1016/j.celrep.2017.05.019
Leonardo, T. R., Schultheisz, H. L., Loring, J. F., and Laurent, L. C. (2012). The Functions of microRNAs in Pluripotency and Reprogramming. Nat. Cel Biol 14 (11), 1114–1121. doi:10.1038/ncb2613
Li, H., Cao, R., Bai, L., Qiao, X.-m., and Zhao, Y.-q. (2018). Lefty Promotes the Proliferation and Invasion of Trophoblast Cells by Inhibiting Nodal Expression. Cell Biol Int 42 (9), 1259–1264. doi:10.1002/cbin.10976
Li, K. L., Nakashima, K., Inoue, J., and Satoh, N. (2020). Phylogenetic Analyses of Glycosyl Hydrolase Family 6 Genes in Tunicates: Possible Horizontal Transfer. Genes (Basel) 11 (8), 937. doi:10.3390/genes11080937
López-Legentil, S., Dieckmann, R., Bontemps-Subielos, N., Turon, X., and Banaigs, B. (2005). Qualitative Variation of Alkaloids in Color Morphs of Cystodytes (Ascidiacea). Biochem. Syst. Ecol. 33 (11), 1107–1119. doi:10.1016/j.bse.2005.03.011
Lu, Q., Gao, Y., Fu, Y., Peng, H., Shi, W., Li, B., et al. (2020). Ciona Embryonic Tail Bending Is Driven by Asymmetrical Notochord Contractility and Coordinated by Epithelial Proliferation. Development 147 (24), 185868. doi:10.1242/dev.185868
Matos, A., Domínguez-Pérez, D., Almeida, D., Agüero-Chapin, G., Campos, A., Osório, H., et al. (2020). Shotgun Proteomics of Ascidians Tunic Gives New Insights on Host-Microbe Interactions by Revealing Diverse Antimicrobial Peptides. Mar. Drugs 18 (7), 362. doi:10.3390/md18070362
Matthysse, A. G., Marry, M., Krall, L., Kaye, M., Ramey, B. E., White, A. R., et al. (2005). The Effect of Cellulose Overproduction on Binding and Biofilm Formation on Roots by Agrobacterium Tumefaciens. MPMI 18, 1002–1010. doi:10.1094/mpmi-18-1002
Nakamura, M. J., Terai, J., Okubo, R., Hotta, K., and Oka, K. (2012). Three-dimensional Anatomy of the Ciona intestinalis Tailbud Embryo at Single-Cell Resolution. Develop. Biol. 372 (2), 274–284. doi:10.1016/j.ydbio.2012.09.007
Nakashima, K., Yamada, L., Satou, Y., Azuma, J.-i., and Satoh, N. (2004). The Evolutionary Origin of Animal Cellulose Synthase. Develop. Genes Evol. 214 (2), 81–88. doi:10.1007/s00427-003-0379-8
Rebustini, I. T., Hayashi, T., Reynolds, A. D., Dillard, M. L., Carpenter, E. M., and Hoffman, M. P. (2012). miR-200c Regulates FGFR-dependent Epithelial Proliferation via Vldlr during Submandibular Gland Branching Morphogenesis. Development 139 (1), 191–202. doi:10.1242/dev.070151
Sagane, Y., Zech, K., Bouquet, J.-M., Schmid, M., Bal, U., and Thompson, E. M. (2010). Functional Specialization of Cellulose Synthase Genes of Prokaryotic Origin in Chordate Larvaceans. Development 137 (9), 1483–1492. doi:10.1242/dev.044503
Sánchez-Vásquez, E., Bronner, M. E., and Strobl-Mazzulla, P. H. (2019). Epigenetic Inactivation of miR-203 as a Key Step in Neural Crest Epithelial-To-Mesenchymal Transition. Development 146 (7), 1017. doi:10.1242/dev.171017
Sasakura, Y., and Hozumi, A. (2018). Formation of Adult Organs through Metamorphosis in Ascidians. Wiley Interdiscip. Rev. Dev. Biol. 7 (2), 304. doi:10.1002/wdev.304
Sasakura, Y., Nakashima, K., Awazu, S., Matsuoka, T., Nakayama, A., Azuma, J.-i., et al. (2005). Transposon-mediated Insertional Mutagenesis Revealed the Functions of Animal Cellulose Synthase in the Ascidian Ciona intestinalis. Proc. Natl. Acad. Sci. 102, 15134–15139. doi:10.1073/pnas.0503640102
Shin, W., Rosin, N. L., Sparks, H., Sinha, S., Rahmani, W., Sharma, N., et al. (2020). Dysfunction of Hair Follicle Mesenchymal Progenitors Contributes to Age-Associated Hair Loss. Develop. Cel 53 (2), 185–198. doi:10.1016/j.devcel.2020.03.019
Slukvin, I. I., and Kumar, A. (2018). The Mesenchymoangioblast, Mesodermal Precursor for Mesenchymal and Endothelial Cells. Cell. Mol. Life Sci. 75 (19), 3507–3520. doi:10.1007/s00018-018-2871-3
Stato, Y., Terakado, K., and Morisawa, M. (1997). Test Cell Migration and Tunic Formation during Pos-Hatching Development of the Ascidian, Ciona intestinalis. Develop. Growth Differ. 39, 117–126.
Storer, M. A., Mahmud, N., Karamboulas, K., Borrett, M. J., Yuzwa, S. A., Gont, A., et al. (2020). Acquisition of a Unique Mesenchymal Precursor-like Blastema State Underlies Successful Adult Mammalian Digit Tip Regeneration. Develop. Cel 52 (4), 509–524. doi:10.1016/j.devcel.2019.12.004
Sun, J., and Stathopoulos, A. (2018). FGF Controls Epithelial-Mesenchymal Transitions during Gastrulation by Regulating Cell Division and Apicobasal Polarity. Development 145 (19), 1927. doi:10.1242/dev.161927
Sur, S., Steele, R., Shi, X., and Ray, R. B. (2019). miRNA-29b Inhibits Prostate Tumor Growth and Induces Apoptosis by Increasing Bim Expression. Cells 8 (11), 1455. doi:10.3390/cells8111455
Tabibzadeh, S., and Hemmati‐Brivanlou, A. (2006). Lefty at the Crossroads of "stemness" and Differentiative Events. Stem Cells 24 (9), 1998–2006. doi:10.1634/stemcells.2006-0075
Tatzuke, Y., Sunanaga, T., Fujiwara, S., and Kawamura, K. (2012). RACK1 Regulates Mesenchymal Cell Recruitment during Sexual and Asexual Reproduction of Budding Tunicates. Develop. Biol. 368 (2), 393–403. doi:10.1016/j.ydbio.2012.06.006
Tokuoka, M., Satoh, N., and Satou, Y. (2005). A bHLH Transcription Factor Gene, Twist-Like1, Is Essential for the Formation of Mesodermal Tissues of Ciona Juveniles. Develop. Biol. 288 (2), 387–396. doi:10.1016/j.ydbio.2005.09.018
Wei, H., Xu, Y., Chen, Q., Chen, H., Zhu, X., and Li, Y. (2020). Mesenchymal Stem Cell-Derived Exosomal miR-223 Regulates Neuronal Cell Apoptosis. Cell Death Dis 11 (4), 290. doi:10.1038/s41419-020-2490-4
Yang, Y., Lin, H., Shen, H., Wang, B., Lei, G., and Tuan, R. S. (2018). Mesenchymal Stem Cell-Derived Extracellular Matrix Enhances Chondrogenic Phenotype of and Cartilage Formation by Encapsulated Chondrocytes In Vitro and In Vivo. Acta Biomater. 69, 71–82. doi:10.1016/j.actbio.2017.12.043
Yeh, Y.-T., Wei, J., Thorossian, S., Nguyen, K., Hoffman, C., del Álamo, J. C., et al. (2019). MiR-145 Mediates Cell Morphology-Regulated Mesenchymal Stem Cell Differentiation to Smooth Muscle Cells. Biomaterials 204, 59–69. doi:10.1016/j.biomaterials.2019.03.003
Zhang, H., Chen, S., Shang, C., Wu, X., Wang, Y., and Li, G. (2019). Interplay between Lefty and Nodal Signaling Is Essential for the Organizer and Axial Formation in Amphioxus Embryos. Develop. Biol. 456 (1), 63–73. doi:10.1016/j.ydbio.2019.08.006
Zhang, T., Xu, Y., Imai, K., Fei, T., Wang, G., Dong, B., et al. (2020). A Single-Cell Analysis of the Molecular Lineage of Chordate Embryogenesis. Sci. Adv. 6 (6), eabc4773. doi:10.1126/sciadv.abc4773
Keywords: microRNA cluster, mesenchymal cell, cellulose synthesis, lefty, Ciona, tunicates
Citation: Sun X, Zhang X, Yang L and Dong B (2022) A microRNA Cluster-Lefty Pathway is Required for Cellulose Synthesis During Ascidian Larval Metamorphosis. Front. Cell Dev. Biol. 10:835906. doi: 10.3389/fcell.2022.835906
Received: 15 December 2021; Accepted: 31 January 2022;
Published: 15 March 2022.
Edited by:
Alberto Stolfi, Georgia Institute of Technology, United StatesCopyright © 2022 Sun, Zhang, Yang and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Dong, bodong@ouc.edu.cn
 Xueping Sun1
Xueping Sun1  Bo Dong
Bo Dong