Single-Cell Atlas of the Chinese Tongue Sole (Cynoglossus semilaevis) Ovary Reveals Transcriptional Programs of Oogenesis in Fish
- 1College of Fisheries and Life Science, Shanghai Ocean University, Shanghai, China
- 2Key Lab of Sustainable Development of Marine Fisheries, Ministry of Agriculture and Rural Affairs, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, China
- 3BGI-Qingdao, BGI-Shenzhen, Qingdao, China
- 4BGI-Shenzhen, Shenzhen, China
- 5Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 6Qingdao-Europe Advanced Institute for Life Sciences, BGI-Shenzhen, Qingdao, China
- 7College of Marine Technology and Environment, Dalian Ocean University, Dalian, China
- 8School of Marine Sciences, Ningbo University, Ningbo, China
- 9Institut de Ciències Del Mar (ICM), Spanish National Research Council (CSIC), Barcelona, Spain
Oogenesis is a highly orchestrated process that depends on regulation by autocrine/paracrine hormones and growth factors. However, many details of the molecular mechanisms that regulate fish oogenesis remain elusive. Here, we performed a single-cell RNA sequencing (scRNA-seq) analysis of the molecular signatures of distinct ovarian cell categories in adult Chinese tongue sole (Cynoglossus semilaevis). We characterized the successive stepwise development of three germ cell subtypes. Notably, we identified the cellular composition of fish follicle walls, including four granulosa cell types and one theca cell type, and we proposed important transcription factors (TFs) showing high activity in the regulation of cell identity. Moreover, we found that the extensive niche–germline bidirectional communications regulate fish oogenesis, whereas ovulation in fish is accompanied by the coordination of simultaneous and tightly sequential processes across different granulosa cells. Additionally, a systems biology analysis of the homologous genes shared by Chinese tongue sole and macaques revealed remarkably conserved biological processes in germ cells and granulosa cells across vertebrates. Our results provide key insights into the cell-type-specific mechanisms underlying fish oogenesis at a single-cell resolution, which offers important clues for exploring fish breeding mechanisms and the evolution of vertebrate reproductive systems.
Introduction
Oogenesis is a markedly complex, dynamic process, from self-renewal and differentiation of oogonia, development of follicle cells, maturation of oocytes and ovulation (Gershon and Dekel, 2020). Natural selection ensures that the structure and function of each organism provides, under the environment that organism has adapted to, the best conditions for the survival of its germ cells. The transition from oviparity to viviparity is a crucial evolutionary change in the life history of vertebrates. The main reproductive mode of teleosts is oviparity with lecithotrophy, i.e., zebrafish, sea bass and tongue sole, while most mammals show viviparity with matrotrophy, i.e., human, monkey and mouse (Lodé, 2012). However. it is barely known whether vertebrates with different reproductive modes retain the essential transcription characteristics of oogenesis in evolution. At least within mammals, previous research showed that human oogenesis appears to be more complex than oogenesis in other mammals (Sylvestre et al., 2013; Biase, 2017); however, shared gene expression patterns between human and mouse oocytes suggest some evolutionarily conserved mechanisms of mammalian oogenesis (Zhang et al., 2018).
During oogenesis, the development of germ cells is accompanied by the transition from mitosis to meiosis and the expression of stage-specific genes (Zhao et al., 2020b). The dynamic translation of the germ cell state is regulated by stage-specific transcription factors (TFs). In humans, TFs such as GTF2l, CSDE1, SOHLH2, SMARCE1, and TUB are considered to play a crucial role in the transition from the primordial to the primary stage (Zhang et al., 2018). In addition, several TFs, such as Msx1, Msx2, Gata4, Cdx2, and Sox4, have strong effects on the transition from mitosis to meiosis in mouse oogenesis (Zhao et al., 2020b). Nevertheless, the TFs that participate in fish oogenesis have rarely been explored or identified, despite the enormous implications of the proper regulation of fish oogenesis for both fisheries and aquaculture production.
Each follicle is composed of a germ cell surrounded by follicle cells (Sánchez and Smitz, 2012). Follicle cells regulate the maturation and ovulation of oocytes through complex interactions and endocrine processes (Li and Albertini, 2013). In female mammals, follicle cells are anatomically divided into mural granulosa cells and cumulus cells (Wigglesworth et al., 2015). Additionally, some investigators have identified the transcription characteristics of mural granulosa cells and cumulus cells as well as associated signals regulating oogenesis at the single-celllevel (Fan et al., 2019; Zhao et al., 2020a). The BMP15/GDF9 and NOTCH signaling pathways are thought to be two key pathways that regulate oogenesis (Eppig et al., 1997; Zhang et al., 2011). In fish, light and electron microscopy analyses have revealed a follicular complex consisting of a mature oocyte, zona radiata, follicle cells, basement membrane and theca (Viana et al., 2018). However, the subtypes of fish follicle cells and the regulation in oogenesis are not systematically understood.
Another key developmental event in oogenesis is ovulation, which is the process whereby mature oocytes break away from the surrounding somatic cells (granulosa cells and theca cells) under a luteinizing hormone (LH) surge (Curry, 2010). Many investigations of human oogenesis have demonstrated that proteinases such as plasminogen activators, matrix metallopeptidase and a disintegrin and metalloproteinase with thrombospondin type 1 motif (ADAMTS) mediate cumulus oocyte complex (COC) expansion and oocyte release by hydrolysing extracellular matrix (ECM) proteins in the follicular layer (Curry, 2010). Notably, prostaglandins (PGs) are an important signaling molecule involved in ovulation in mammals and teleosts (Takahashi et al., 2018). However, the ovulation mechanism of other vertebrates is still lack of in-depth research, especially in fish.
Chinese tongue sole (Cynoglossus semilaevis) is a flatfish with asynchronous oocyte development and batch spawning since oocytes of diverse developmental stages are found in the ovary in the same growth phase (Chen et al., 2010; Wen et al., 2015), which lends itself to the investigation of fish oogenesis. The whole-genome sequence of Chinese tongue sole was completed in 2014 (Chen et al., 2014), which provides a foundation for the transcriptomic researches. In the present study, we performed single-cell RNA sequencing (scRNA-seq) to analyse the key events throughout oogenesis in Chinese tongue sole. We described the progression of germ cells and transcriptional characteristics of somatic niche cells. Importantly, we identified diverse granulosa cell subtypes and inferred the key highly active TFs involved in follicle cell fate decisions. Moreover, we found that the niche–germline bidirectional communications regulate fish oogenesis, whereas the interaction between different granulosa cells contributes to the ovulation of fish. Furthermore, by cross-species comparison of ovarian cells from Chinese tongue sole and macaques, we revealed the conserved gene expression features in oogenesis across mammals and fish. Our results provide a solid reference dataset for research on fish propagation-related biology and sheds light on the adaptability of reproductive strategies in lower vertebrates.
Materials and Methods
Hematoxylin-Eosin Staining of Chinese Tongue Sole Ovary
To make paraffin sections, the ovary tissue was fixed in the fixing agent for at least 24 h. For dehydration and paraffin embedding, samples were placed in 75% ethanol for 4 h, 85% ethanol for 2 h, 90% alcohol for 2 h, 95% ethanol for 1 h, 100% ethanol I for 30 min, 100% ethanol II for 30 min, ethanol benzene for 5–10 min, xylene II for 5–10 min, 65°C melting paraffin I for 1 h, 65°C melting paraffin II for 1 h, and 65°C melting paraffin III for 1 h. The wax-soaked tissue was embedded in an embedding machine. Samples were cooled at −20°C on a freezing table. After the wax solidified, the wax block was removed from the embedding frame and repaired. The trimmed wax block was sectioned on a microtome paraffin slicer with a thickness of 4 μm. For Hematoxylin-eosin staining, dewaxing was performed by rinsing in xylene I for 20 min, xylene II for 20 min, 100% ethanol I for 5 min, 100% ethanol II for 5 min, and 75% ethanol for 5 min. Sections were then rinsed with tap water, stained with hematoxylin solution for 3–5 min, and rinsed with tap water. Then, the sections were treated with Hematoxylin Differentiation solution and rinsed with tap water. The sections were treated with Hematoxylin Scott Tap Bluing and rinsed with tap water. They were then treated with 85% ethanol for 5 min and 95% ethanol for 5 min. Finally, sections were stained with Eosin dye for 5 min. For dehydration, sections were treated with 100% ethanol I for 5 min, 100% ethanol II for 5 min, 100% ethanol III for 5 min, xylene I for 5 min, and xylene II for 5 min, followed by sealing with neutral gum. Sections were observed by microscopy and images were obtained for analysis.
Dissociation of Ovarian Cells in Female Chinese Tongue Sole
To obtain a high-quality cell suspension, we strictly controlled the preparation time. First, six adult female Chinese tongue sole were dissected and their ovaries were removed. The ovaries of Chinese tongue sole were dissected on ice and washed twice with DMEM-NaCl solution, and other tissues that adhered to the ovaries were removed with tweezers. Then, the combined enzyme digestion solution was prepared. Add 1 ml of trypsin with a concentration of 0.25% and 200 μL of collagenase with a concentration of 1 mg/ml to 3.8 ml of DMEM-NaCl solution. The ovarian tissue was cut with a blade to produce a tissue homogenate, which was then transferred to a combined enzyme digestion solution. After heating in a 30°C water bath for 7 min, digestion was rapidly terminated with 10 ml DMEM-NaCl solution. The digested cell suspension was filtered with a 40 μm cell strainer to collect approximately 7 ml of the filtered cell suspension in a 15 ml tube. Centrifugation was performed with a horizontal rotor in a cryogenic centrifuge at 300 × g for 3 min. After removing the supernatant, 5 ml of PBS-NaCl solution was added, centrifugation was conducted at 300 × g for 3 min, and washing was performed twice. Finally, 0.05% BSA in PBS was added to resuspend the cells. Finally, 10 μL single-cell suspensions was mixed with 1 μL 0.4% dye trypan blue solution to measure the total cell concentration (1,000–2000 cells per 1 μL) as well as the ratio of live cells (greater than 80 percent) using a hemocytometer.
scRNA-Seq Library Preparation and Sequencing
We used the DNBelab C Series Single Cell RNA Library Preparation Kit (MGI Tech Co., Ltd.) based on droplet microfluidics technology for library construction. In detail, we pooled 110,000 cells per library with an average concentration of 1,000 cells/μL and activity greater than 80%. Cells were prepared as droplets, in which cell lysis and mRNA capture were performed using the DNBelab C4 portable single-cell system. Single-cell microdroplets were recovered by using the emulsion breaking recovery system, after which magnetic bead-captured mRNA was transcribed into cDNA and subjected to reverse transcription (42°C for 90 min; 14 cycles of 50°C for 2 min and 42°C for 2 min; holding at 4°C) for cDNA enrichment. Finally, the cDNA products were used to prepare single-stranded DNA libraries via steps including shearing, end repair, ligation, PCR (95°C for 3 min; 11 cycles of 98°C for 20 s, 58°C for 30 s and 72°C for 30 s; 72°C for 10 min; holding at 4°C), denaturation, circularization, and digestion. Then, 10 ng of the digested products was collected for sequencing on the MGISEQ 2000 platform.
scRNA-Seq Data Processing
The raw data obtained by scRNA-seq on the MGISEQ2000 platform were filtered and demultiplexed using PISA (version 1.10.2) (https://github.com/shiquan/PISA). Reads were aligned to the reference genome from NCBI using STAR (version 2.7.9a) (https://github.com/alexdobin/STAR) and were sorted using Sambamba (version 0.7.0) (https://github.com/biod/sambamba/releases/tag/v0.7.0). The cell versus gene UMI count matrix was generated using PISA.
Cell Clustering and Identification of Cell Types
For each library, we used the count matrix and followed the Seurat vignette (https://satijalab.org/seurat/pbmc3k_tutorial.html) to create the Seurat object. In brief, cells were first prefiltered according to the expression of at least 200 genes in each cell and the expression of each gene expressed in at least 3 cells. Thereafter, cells with less than 5% mitochondrial genes and UniQ gene counts between 200 and 3500 were retained. For the remaining cells, the gene expression count data for each sample were normalized, 3,000 highly variable genes were selected, and scaling was conducted with the “SCTransform” function. After normalization, principal component analysis (PCA) was used to reduce the number of dimensions of the gene expression matrix. The number of components used was determined with the “ElbowPlot” function. The first 30 PCs were selected as the input for the “FindNeighbors” and “RunUMAP” functions (default parameters), and a parameter of resolution of 0.5 was chosen for the “FindClusters” function. Subsequently, “DoubletFinder” was employed to identify doublets using the same PCs of the above PCA, assuming a 4% or 10% doublet formation rate for the loaded cells of each sample in a droplet channel. The optimal pK values were determined for each organ based on the mean-variance normalized bimodality coefficient. After doublet removal of all library, we merge the six matrices and use the “selectintegrationfeatures” function to select the top 3,000 genes with the greatest variation for integration, after which we used the “prepsctintegration” function to aggregate SCT objects, and then the “findintegrationanchors” function with the normalization. method parameter as “SCT”, the top 3,000 genes as the anchor. features and k. filter parameter as 200 to identify the sources of common variation between conditions via canonical correlation analysis (CCA). This procedure was conducted find the anchor or nearest neighbour (MNN) for cross-dataset identification, after which the anchor points were filtered, incorrect anchor points were removed, and the data were finally integrated by using the “integration data” function with the normalization. method parameter as “SCT”. After integrating and removing batch effects, we ran the PCA (30PCs) and UMAP (default parameters) analyses again, and we finally used the “findallmarker” function (with only. pos equal to TRUE, min. pct and logfc. threshold equal to 0.25 and test. use equal to “wilcox”)to calculate the DEGs between different clusters.
Gene Function Enrichment Analysis
Gene function enrichment analysis was performed using Metascape (https://metascape.org/gp/index.html). The Chinese tongue sole genes used for the enrichment analysis were homologous to zebrafish genes.
Cell Trajectory Analysis
Single-cell pseudotime trajectories were constructed with the Monocle 2 package (version 2.14.0) (http://cole-trapnell-lab.github.io/monocle-release/docs/). In describing the process of oogenesis, we transformed the Seurat dataset into the cell_data_set of Monocle2. Then, we chose the ordered genes to define cell progress, we used DDRTree to reduce the space to two dimensions, and all cells were ordered with the orderCells function with the default parameters.
Identification of DEGs Between Two Different Clusters
The DESeq2 (v1.30.1) R package was used to find additional DEGs. The package uses non-normalized counts as an input. The data from each sample were aggregated, and the DESeqDataSetFromMatrix function was used to construct the count matrix. DESeq and the results function were used for the differential expression analysis and to generate results tables.
Transcriptional Regulation Analysis of Follicle Cell Subtypes
The SCENIC analysis was run as described on the follicle cell subclusters, using the 10-thousand motifs database for RcisTarget and GENIE3. The input matrix was the normalized expressional matrix, oriented from the one-to-one homologous genes of human and Chinese tongue sole.
Cytokine/Receptor Interaction Analysis
ITALK was used to analyse the cell-cell communication between three germ cell subtypes and five granulosa cells subtypes. Firstly, we transformed the matrix of tongue sole into the matrix of human. The one-to-one homologous genes of human and Chinese tongue sole were obtained by blast. Then we set the matrix and cell types as the input data, and the ligand-receptor pairs were detected from top 50% highly expressed genes. We set growth factors, cytokines, checkpoint and other as the communication types. Finally, we using LRPlot function to visualize the network plot.
Conservation of Marker Gene Expression
Macaque UMI matrices were obtained from the NCBI Gene Expression Omnibus (GEO) database (GSE149629). Germ and somatic cells were extracted from the matrix according to the barcode of the cell type reported previously (Zhao et al., 2020a).
Results
The Global Transcriptome Profile Identifies Eight Cell Types in Chinese Tongue Sole Ovaries
To capture all the cellular information in the ovary of Chinese tongue sole, we collected the ovaries of six sexually mature females in October. At this time, the ovary was in stage IV and contained oocytes in all stages of development, and the follicles exhibited a double-layer membrane structure. Using the iDrop system, six libraries were constructed and sequenced (Figure 1A). After quality control, we obtained the transcriptional spectrum of 7,185 cells. Using mutual nearest neighbour methodology, we identified 15 clusters, which were visualized via Uniform Manifold Approximation and Projection (UMAP) (Supplementary Figure S1A). Our data showed no apparent batch effect, and each sample contained all clusters (Supplementary Figures S1B, S1C).
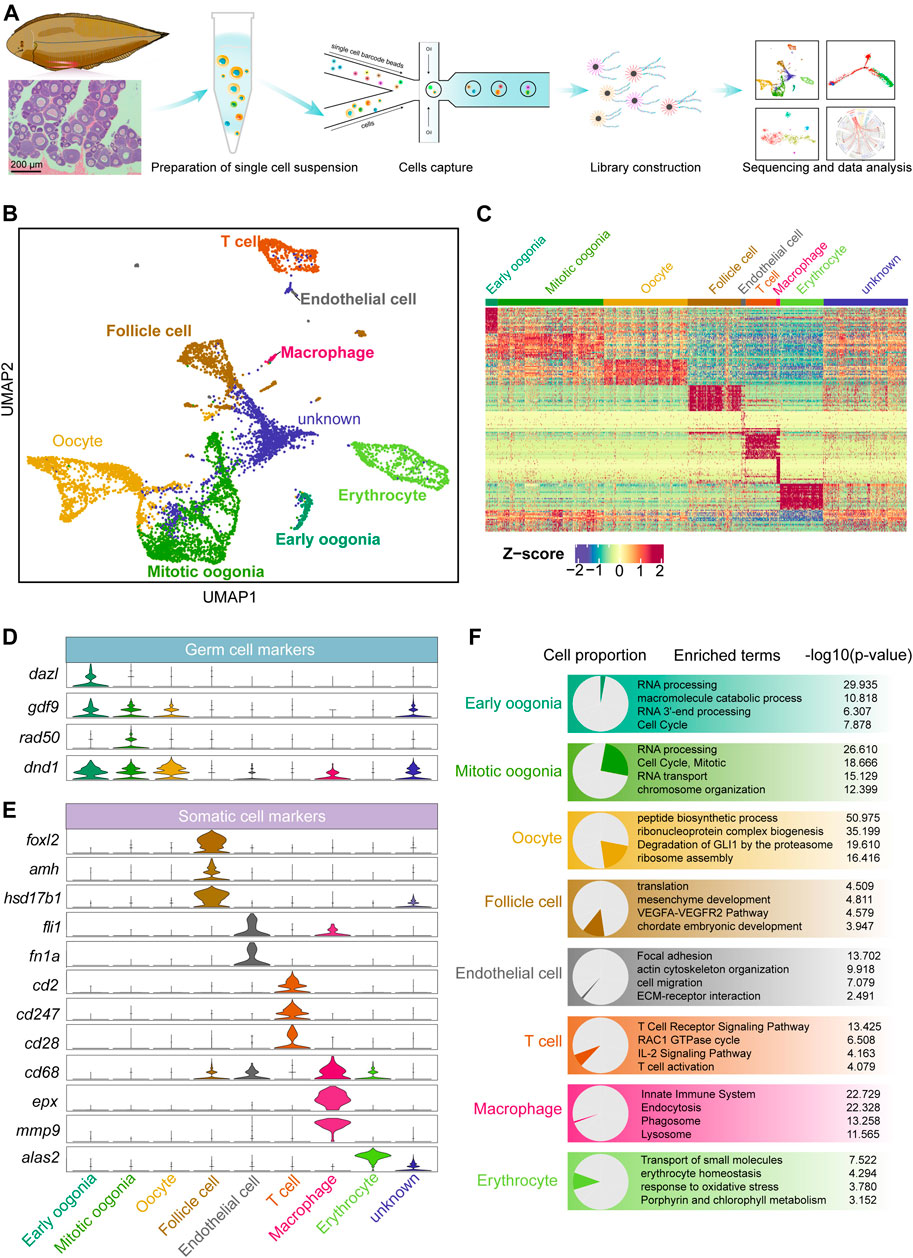
FIGURE 1. Overview of germ and somatic ovarian cell types determined by scRNA sequencing in Chinese tongue sole. (A) Schematic representation of the scRNA-seq analysis of Chinese tongue sole ovarian cells. (B) Visualization of major ovarian cell types among 7,185 cells in UMAP (unknown is undefined). Different cell types are shown in distinct colours. (C) Heatmap showing the expression of the top 20 DEGs of the main cell types. The colours from blue to red indicate low to high gene expression levels, respectively. Z-scores were calculated by subtracting the average value for the set of data from the value for each cell and dividing by the standard deviation. (D) Violin plots of the normalized expression of marker genes in the three germ cell types. (E) Violin plots of the normalized expression of marker genes in the five somatic cell types. (F) Enriched terms of DEGs are shown for ovarian cell types (p-values are shown). The proportion of each cell type is shown on the left.
Three germ cell populations and five somatic cell populations were identified by gene expression pattern analysis (Figures 1B,C). Typical marker genes of oogenesis, such as gdf9 and dnd1 (Liu and Ge, 2007; Yang et al., 2012), were widely expressed in the three identified germ cell types. Dazl (Bachvarova et al., 2004; Li et al., 2016; Bertho et al., 2021) was highly expressed in early oogonia, while the mitotic marker rad50 (Kostyrko et al., 2015; Völkening et al., 2020; Liu et al., 2021) was mainly expressed in mitotic oogonia (Figure 1D). In addition, we identified somatic cells based on the following markers: follicle cells (foxl2, amh and hsd17b1) (Pisarska et al., 2011; Luo et al., 2020; Moolhuijsen and Visser, 2020; Li et al., 2021), endothelial cells (fli1 and fn1a) (Nagai et al., 2018; Wu et al., 2021), T cells (cd2, cd247 and cd28) (Hurley et al., 1994; Lundholm et al., 2010), macrophages (cd68, epx and mmp9) (Hunyady et al., 1996; Sendler et al., 2018) and erythrocytes (alas2) (Munday et al., 2000; Tsang et al., 2017) (Figure 1E). The top 20 differentially expressed genes (DEGs) in each cell were shown in Supplementary Figure S1D. Furthermore, an enrichment analysis of the DEGs of each cell type was performed based on the Metascape resource. The results showed that “RNA processing” and “cell cycle, mitotic” were enriched in early oogonia and mitotic oogonia, respectively, while “peptide biosynthetic process” was enriched in oocytes. In contrast, follicle cells were enriched in the terms “mesenchyme development”, and endothelial cells were enriched in “focal adhesion” and “ECM-receptor interaction”. In addition, “T cell receptor signaling pathway”, “innate immune system” and “erythrocyte homeostasis” were enriched terms in T cells, macrophages and erythrocytes, respectively (Figure 1F).
Dynamics of Gene Expression Across Germ Cells Revealed by a Pseudotime Trajectory
To explore the development of female germ cells in fish, Monocle2 was employed for cell trajectory analysis of the three identified germ cell types to recapitulate the temporal progression of oogenesis. Notably, in the unsupervised pseudotime analysis, early oogonia, mitotic oogonia and oocytes followed a continuous trajectory. Early oogonia were observed at the beginning of the pseudotime, followed by mitotic oogonia and, finally, oocytes at the end point (Figure 2A). Furthermore, hierarchical clustering analysis of the ordered genes showed three groups with different expression patterns. Many markers of oogonia, such as dazl, dnmt1, sox2, sox4, sall4 and lin28a, were enriched in the first group. Moreover, the second group contained many mitosis-related genes, such as cdk2, cdk4, hsp90b1, bub1b, bub3, smc1 and smc5. The third group showed striking meiotic features, such as meiob, tex30 and rad51 expression (Figure 2B). This suggests that the cells arranged in pseudotime order undergo a transition from non-division to mitosis and then meiosis. Additionally, we analysed the differential expression of these genes in three germ cell types. As expected, two markers of germ stem cells, dazl and sox4, were significantly overexpressed in early oogonia. Subsequently, the expression levels of the mitotic markers mcm3 and cenpn significantly decreased during the transition from mitotic oogonia to oocytes, while the expression of meiotic markers, such as meiob and tex30, increased strikingly (Figure 2C). This was consistent with observations in mice (Zhao et al., 2020b). Taken together, these findings suggest that fish oogenesis is a biological process similar to mammalian oogenesis, in which proliferation via mitosis is followed by meiosis.
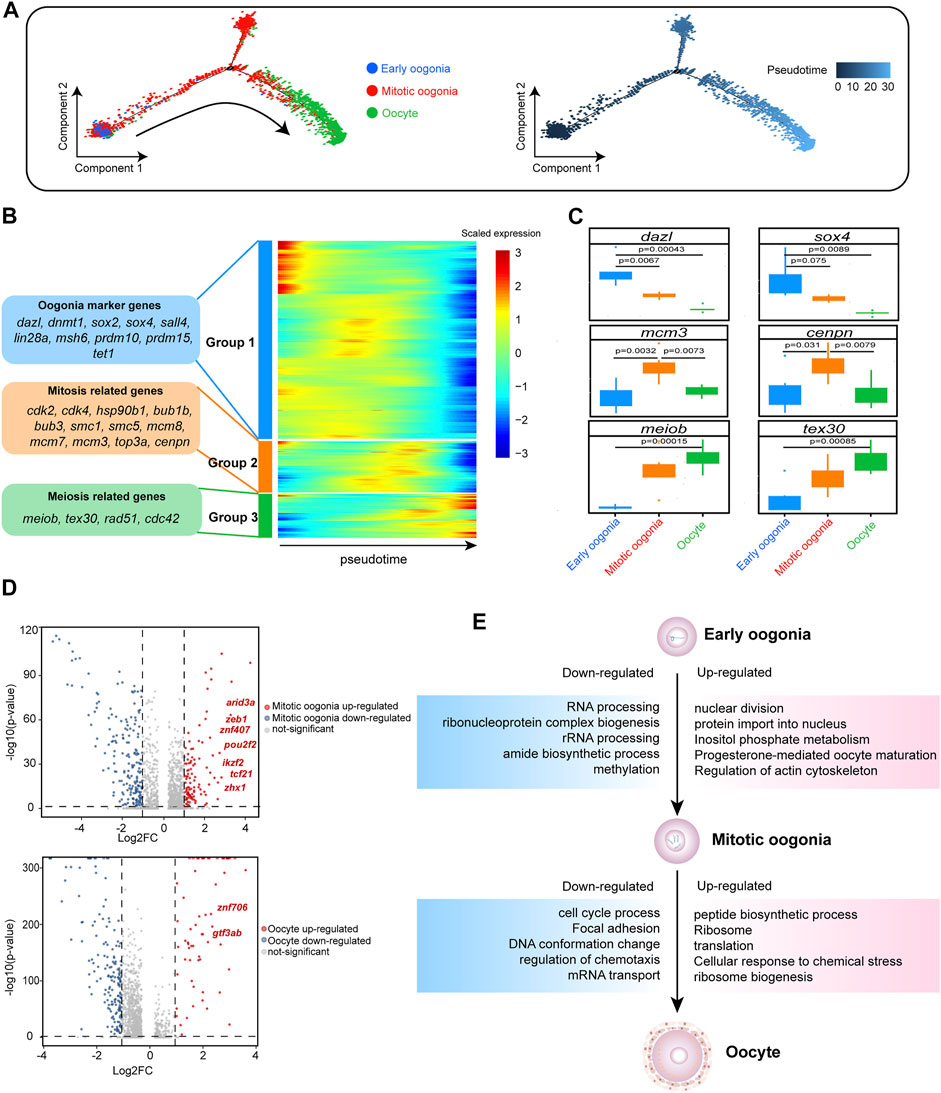
FIGURE 2. Gene expression dynamics and transcriptional characteristics of ovarian germ cells. (A) Developmental pseudotime trajectory of germ cells. Different colours in the figure on the left side represent different cell types. The figure on the right represents the degree of differentiation of the cell types in the pseudotime trajectory (where darker colours indicate a lower degree of differentiation). (B) Gene expression heatmap over pseudotime. After clustering analysis, genes were divided into three groups (Groups 1–3) (representative genes are shown on the left). Colours from blue to red indicate low to high gene expression levels, respectively. (C) Boxplot showing the mean expression levels of oogenesis-associated genes in each germ cell subtype. Two-tailed t test p-values are indicated. (D) Volcano plot showing the DEGs of germ cells, setting a fold change >2 as the threshold (dotted line). In the upper figure, upregulated genes in mitotic oogonia relative to early oogonia are indicated in red, and downregulated genes are indicated in blue. In the lower figure, upregulated genes in oocytes relative to mitotic oogonia are indicated in red, and downregulated genes are indicated in blue. Upregulated transcription factors (TFs) are labelled. (E) DEGs and associated enriched terms (identified using Metascape) characteristic of germ cell developmental transitions, based on Figure 2D. Upregulated terms are annotated in pink boxes, and downregulated terms are annotated in blue boxes.
The molecular mechanism promoting the state transition of fish germ cells is unclear. Therefore, we conducted a differential analysis of the transcripts of different germ cells. Compared with early oogonia, there were 135 significantly upregulated genes in mitotic oogonia (| log2FC | > 1), including seven TFs (arid3a, znf407, zeb1, pou2f2, ikzf2, tcf21 and zhx1). In addition, there were 62 significantly upregulated genes in oocytes (| log2FC | > 1) compared with mitotic oogonia, including two TFs (gtf3ab and znf706) (Figure 2D). We hypothesized that these TFs might play a critical role in the transition of germ cells. Simultaneously, we found that “RNA processing” in early oogonia was replaced by “nuclear division” in mitotic oogonia, and “cell cycle process” in mitotic oogonia was replaced by “peptide biological process” in oocytes according to the enrichment analysis of DEGs (Figure 2E). These results indicated that the dynamic transcriptome regulates fish oogenesis in a stage-specific manner.
Identification of Follicle Cell Subtypes in Fish Ovaries
Ovarian follicle cells play an important role in oocyte maturation and ovulation. Mature fish oocytes are surrounded by two major cell layers, an outer thecal cell layer and an inner granulosa cell layer (Nagahama and Yamashita, 2008; Nagahama et al., 1995; Viana et al., 2018). To explore whether fish follicle cells have different subtypes, similar to those of mammals, we performed a re-clustering analysis of the follicle cells and revealed the existence of four distinct granulosa cell subclusters (GC1, GC2, GC3 and GC4) and one theca cell type (Tc) (Figure 3A). To systematically investigate the similarities and differences between subclusters, we performed a correlation analysis together with the hierarchical clustering of subclusters. This approach highlighted that GC1, GC2, GC3, and GC4 were more closely grouped together, while Tc was different from the other four clusters (Supplementary Figure S2A). To examine the features of the subclusters, we detected the DEGs in each cluster, and the genes with highly variable expression were identified. The results showed that there were significant differences in gene expression among different subclusters (Figure 3B). We found that marker genes of mammalian cumulus cells, such as foxl2, inha and inhbb, were highly expressed in GC1, while marker genes of mammalian mural granulosa cells, such as krt8 and krt18, exhibited peak expression specifically in GC3. Other granulosa cell marker genes such as adamts1, fgfr4, sox9, and tnfaip6 were specific expressed in GC4. Additionally, we observed that ribosomal protein coding genes were highly expressed in GC2 (Figure 3B). Notably, cell-cycle phase prediction showed that GC2 were more proliferative than other clusters, suggesting that GC2 was a proliferating granulosa cell type (Supplementary Figure S2B). Androgen synthesis related genes such as nr5a1 and hsd3b1 were highly expressed in Tc (Figure 3B). It is well established that the theca cells of fish are the source of androgens, which are transferred to granulosa cells where they are converted to oestrogens by the activity of aromatase (Nagahama and Yamashita, 2008).
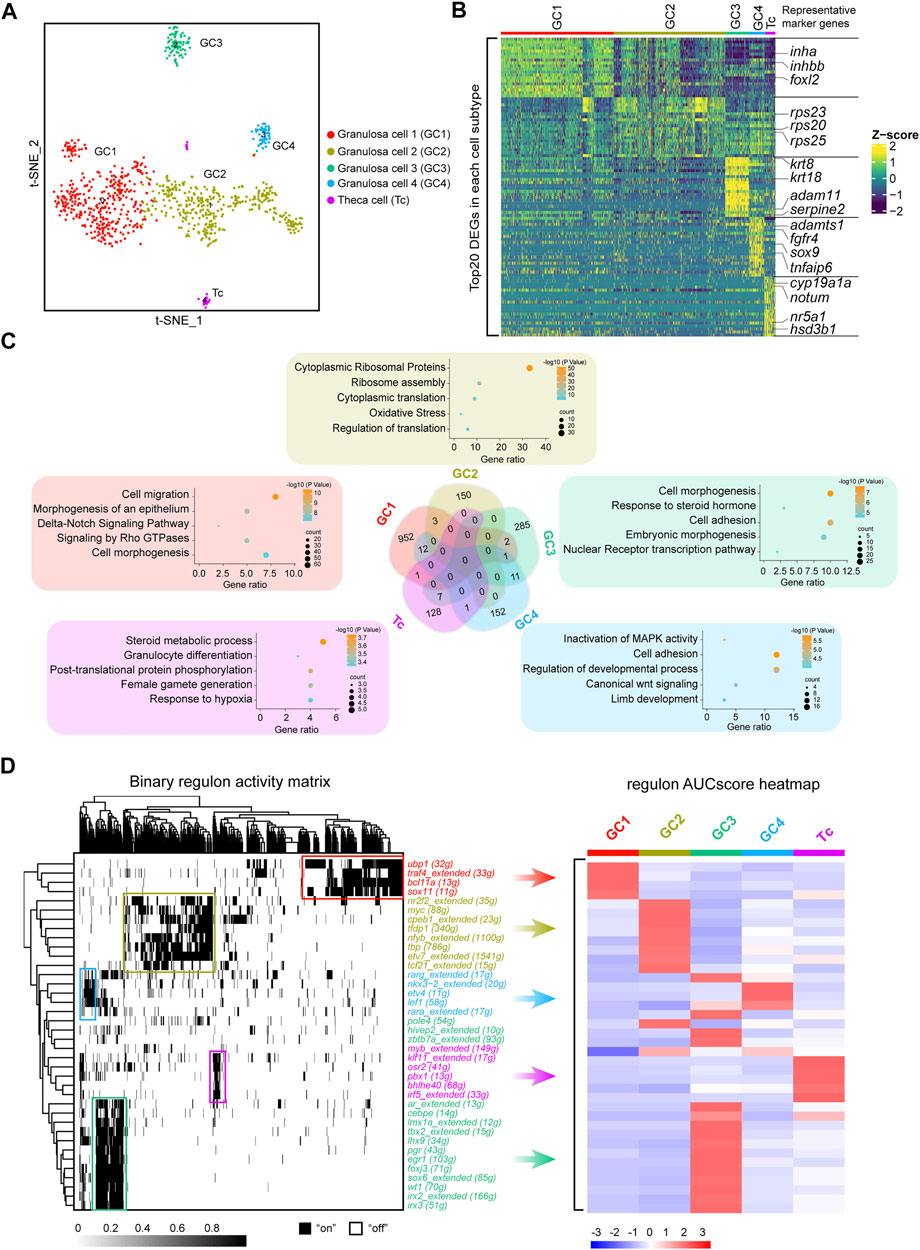
FIGURE 3. Identification of five discrete transcriptional states of follicle cells. (A) UMAP of follicle cell re-clustering. (B) Heatmap of the top 20 DEGs in different subclusters of follicle cells. The colours from dark blue to yellow indicate low to high gene expression levels, respectively. Representative marker genes are shown in right. (C) Venn diagram showing the intersection of DEGs of the five subclusters. Representative enriched terms for DEGs are shown. (D) Left: SCENIC binary regulon activity heatmap depicting follicle cell-enriched regulons. “On” indicates active, while “Off” indicates inactive; Right: Heatmap of the AUC scores of follicle cell subtype-specific regulons. The regulons correspond to the graph on the left.
Subsequently, we analysed the overlap of subclusters of DEGs. Venn diagrams showed that only a small number of DEGs overlapped between each subcluster. To further ascertain the features of the subclusters, we performed gene enrichment analysis of each cluster of specific DEGs. According to the gene enrichment scoring results, the genes expressed in GC1 were enriched in “cell migration” and the “Delta-Notch signaling pathway,” and GC2 was enriched in “cytoplasmic ribosomal proteins”. In addition, “response to steroid hormone” and “cell adhesion” were observed in GC3, while “inactivation of MAPK activity” and “cell adhesion” were enriched in GC4 (Figure 3C). Notably, theca cells were enriched in the terms “steroid metabolic process” and “female gamete generation” (Figure 3C). Collectively, these results indicated that subpopulations of follicle cells play distinct roles in fish oogenesis.
After the recapitulation of the follicle cell subtype transcriptome landscape, we used single-cell regulatory network inference and clustering (SCENIC) to deduce the key highly active TFs involved in follicle cell fate decisions. Distinct follicle cell subtypes were enriched in diverse regulons. For example, GC1 was enriched in regulons such as ubp1, traf4, bcl11a, and sox11; GC2 was enriched in regulons such as nr2f2, myc, cpeb1, and tfdp1; GC3 was enriched in regulons such as ar, lhx9, wt1, and irx3; GC4 was enriched in regulons such as nkx3-2, etv4 and lef1; and theca cells were enriched in regulons such as klf11, osr2 and pbx1 (Figure 3D). These TFs may be the driving genes for the differentiation of different follicle cell subtypes.
The Multilineage Interactome Network in Fish Oogenesis
To investigate the complex interactions involved in fish oogenesis, we performed an unbiased ligand–receptor interaction analysis between the germ and somatic cell subclusters by using iTALK (Wang et al., 2019). We found that bidirectional communication occurred between germ cells and granulosa cells, while germ cells and theca cells only showed unidirectional regulation from theca cells to germ cells. Additionally, we noted that extensive interactions occurred in distinct somatic cells but no interactions were found between germ cells (Figure 4A). These findings indicate the niche–germline bidirectional communication is mandatory for the development of germ cells and granulosa cells in fish ovaries. The development of theca cells is more dependent on granulosa cells than on germ cells.
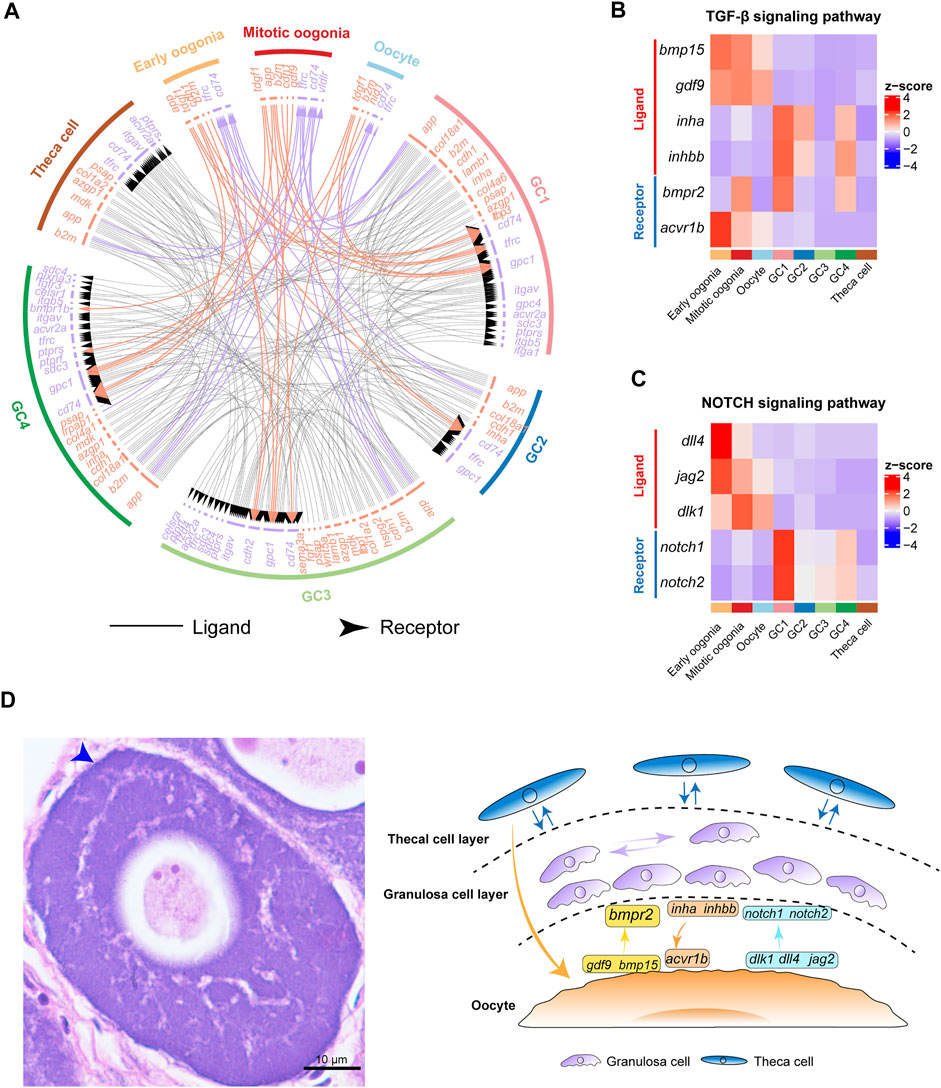
FIGURE 4. Cell–cell communication networks and signaling pathways involved in fish oogenesis. (A) The multilineage interactome network among different cell clusters. Associated interaction pairs are shown. (B) Heatmap showing the expression pattern of ligands and receptors of the TGF-β signaling pathway. (C) Heatmap shows the expression pattern of ligands and receptors of the NOTCH signaling pathway. (D) Left: H&E staining of ovary sections from female Chinese tongue sole. Scale bars represent 10 μm. The blue arrow shows double cell layers of follicle. Right: Schematic summary of interactions involved in fish oogenesis.
Furthermore, we analysed the expression of ligands and receptors of key cell signaling pathways, including the TGF-β and NOTCH signaling pathways. Interestingly, the results showed that the expression of gdf9 and bmp15 exhibited peak expression in germ cells (Figure 4B), whereas their receptor bmpr2 was expressed in both germ cells and granulosa cells. This is consistent with the results observed in human ovaries (Zhang et al., 2018). In contrast, the inha and inhbb ligands were highly expressed in GC1, whereas their receptor, acvr1b, exhibited peak expression specifically in oogonia, highlighting the role of TGF-β signaling in the control of granulosa cell-mediated oogonial proliferation (Figure 4B). These findings suggest that fish oogenesis is regulated by TGF-β signaling, which could be initiated by either germ cells or somatic cells.
Subsequently, we analysed the expression of key components of the NOTCH signaling pathway in germ cells and somatic cells. We found that the dlk1, dll4, and jag2 ligands of the NOTCH signaling pathway were predominantly expressed in germ cells. Their receptors, notch1 and notch2, were highly expressed in GC1 (Figure 4C). These results indicate that NOTCH signaling is an important signal that mediates the regulation of granulosa cell proliferation and differentiation by germ cells during fish oogenesis (Figure 4D).
Ovulation in Fish Is Accompanied by the Synergistic Actions of Different Follicle Cells
To investigate the functions of different follicle cell subtypes in fish ovulation, we analysed the expression of pivotal genes in ovulation. Our data showed that lhcgr, the LH receptor, was specifically expressed in theca cells, indicating that theca cells receive and transmit LH stimulation during ovulation (Figure 5A). This is consistent with previous studies in mammals and fish (Curry, 2010; Takahashi et al., 2018). Additionally, pgr and ptger4b, the receptors of progesterone and prostaglandin E2, were highly expressed in GC3, suggesting that progesterone and prostaglandin mainly affect fish ovulation through GC3 (Figure 5A). Adamts1 and adamts9 have been reported to be associated with follicle rupture in mammals, especially adamts1, which is a protein lyase proven to play an important role in ovulation in a gene knockout experiment (Shozu et al., 2005; Brown et al., 2006). We found that adamts1 was predominantly expressed in GC4, whereas adamts9 was expressed in only a few cells (Figure 5A). This suggests that adamts1 secreted by GC4 might promote follicular rupture during fish ovulation. Notably, ecm1 was expressed in all subtypes, indicating that adamts1 acts on the extracellular matrix proteins of distinct granulosa cells to induce different biological changes in the follicle wall (Figure 5A). Taken together, our results suggest that fish ovulation is based on the synergistic action of diverse granulosa cells and theca cells (Figure 5B).
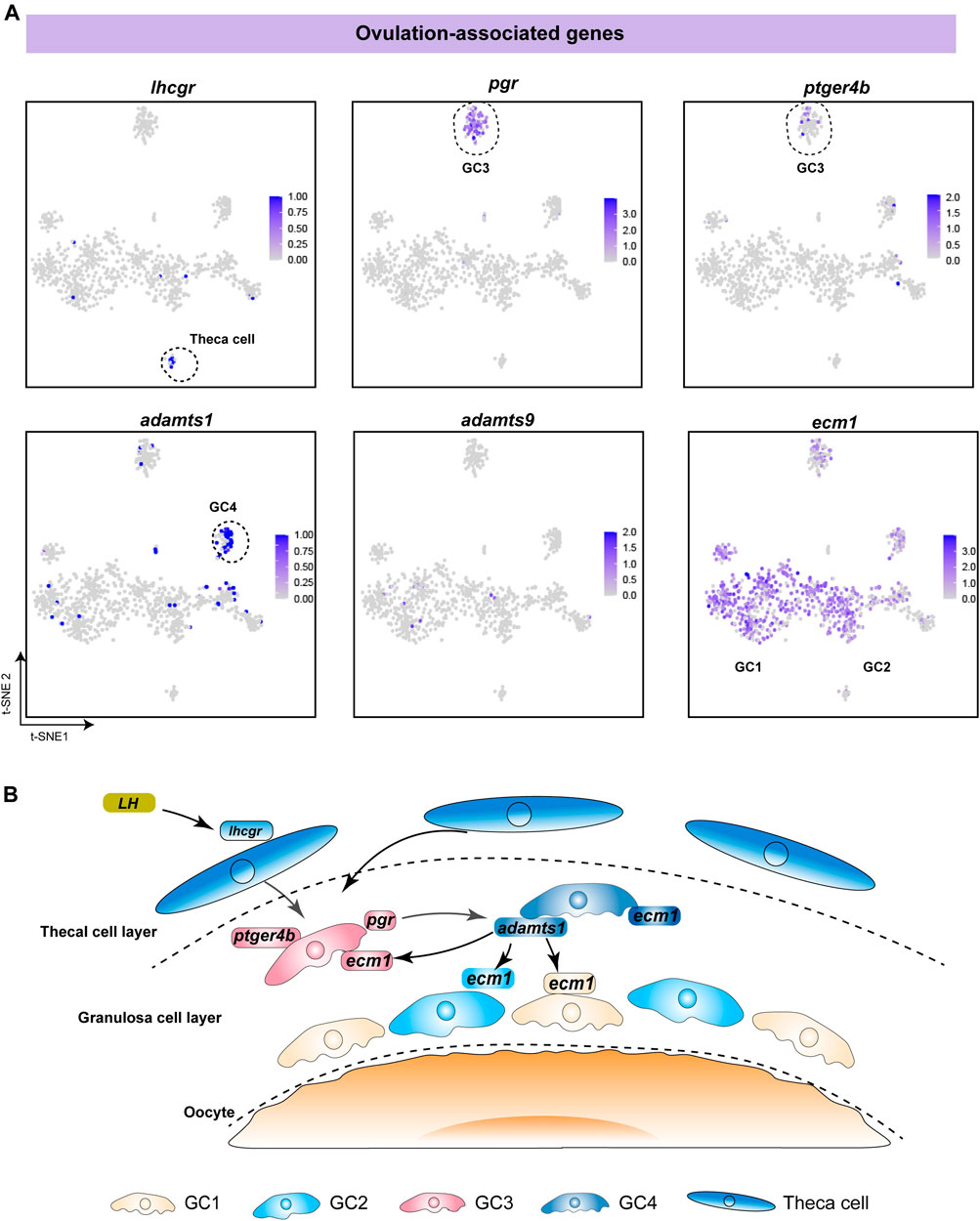
FIGURE 5. Analysis of specific expression patterns of ovulation-associated genes. (A) Feature plot of ovulation-associated genes in different follicle cells. The darker the colour, the higher the expression. Black dashed lines indicate the boundaries of the main clusters of interest. (B) Schematic diagram showing the regulatory mechanism of ovulation, involving the synergistic action of different follicle cells in fish.
Comparison of Germ Cell and Granulosa Cell Transcriptomic Profiles Across Species
To better understand the interspecies complexity of germ cell and granulosa cell transcriptomes in fish and mammals, we compared our data with the reported scRNA-seq datasets in macaques (Zhao et al., 2020a). To exclude the effects of abiotic factors, the raw data of the macaques and Chinese tongue sole were analysed via the same procedure. First, utilizing BLASTP, we identified 19,493 homologous genes shared by Chinese tongue sole and macaques. Focusing on germ cells, we identified 1,987 DEGs in Chinese tongue sole. Our results showed that there were 1738 genes with homologs in Chinese tongue sole and macaques (Figure 6A). Subsequently, 965 DEGs of Chinese tongue sole granulosa cells were analysed, 836 of which were homologous. Moreover, the homologous DEGs of the germ cells were enriched in the “metabolism of RNA”, “peptide biosynthetic process” and “cell cycle” terms, while the homologous DEGs of the granulosa cells were enriched in the “Wnt signaling pathway,” “VEGFA-VEGFR2 pathway,” “EGFR1 signaling pathway” and “focal adhesion” and “Delta-Notch signaling pathway” terms (Figure 6A). Notably, these terms are intimately associated with oogenesis. Additionally, several homologous DEGs that have been implicated in oogenesis, such as zar1, sall4, ybx2, dnd1, meiob, gdf9, foxl2, fshr, wt1, hsd17b1, cdh1 and bmpr2, showed similar expression patterns in Chinese tongue sole and macaque ovarian cells, suggesting the possibility that they have homologous functions in oogenesis (Figures 6B,C). Thus, these results demonstrate the evolutionarily conserved mechanisms involved in oogenesis across fish and mammals.
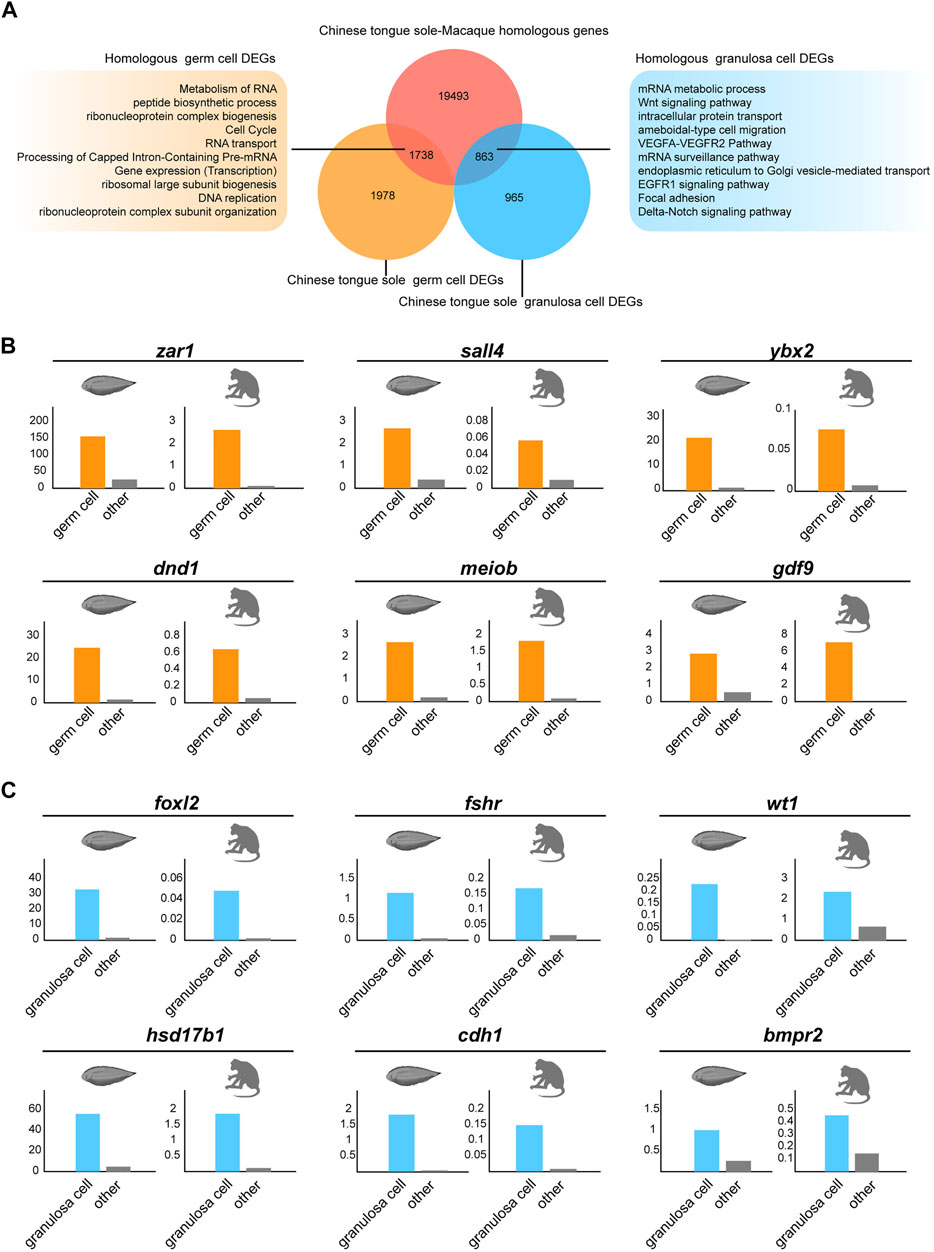
FIGURE 6. Comparison of germ cells and granulosa cells based on Chinese tongue sole and macaque transcriptomic profiles. (A) Venn diagram of DEGs in the germ cells and granulosa cells of Chinese tongue sole showing overlap with the Chinese tongue sole-macaque homologous genes (light red). The enriched terms of the homologous DEGs in germ cells and granulosa cells are shown. (B) Histogram of the normalized expression of the shared homologous genes between Chinese tongue sole and macaques related to oogenesis in germ cells relative to the average expression of the same genes in all other cell types. (C) Histogram of the normalized expression of the shared homologous genes between Chinese tongue sole and macaques related to oogenesis in granulosa cells relative to the average expression of the same genes in all other cell types.
Discussion
Studies on fish reproduction assist the conservation of germplasm resources and the investigation of animal reproductive regulation mechanism in fish model. Most studies on fish oogenesis focus on the structural changes during oocyte maturation observed by light and electron micrograph sections and the regulation of endocrine factors (Selman et al., 1993; Tyler and Sumpter, 1996; Patiño et al., 2005; Kaviani et al., 2013). Conventional genome analysis approaches have been used to capture global gene expression patterns and chromatin conformation of fish ovaries or oocytes (Shao et al., 2014; Can et al., 2020; Dong et al., 2021). However, the bulk sequencing method does not account for cell-to-cell heterogeneity. The development of scRNA-seq technology in recent years can separate cells with different gene expression characteristics through principal component analysis rather than artificial definition, which can overcome these limitations (Tang et al., 2009; Kobak and Berens, 2019). Several studies have revealed unique transcriptional features and gene signatures of oogenesis across vertebrate and invertebrates with single-cell technology (Fan et al., 2019; Rust et al., 2020; Wang et al., 2020; Zhao et al., 2020b). The single-cell transcriptomic atlas of Asian sea bass has identified the specific gene signatures of granulosa cells and oocytes in the fish ovary, that is the first comprehensive analysis of fish ovary at single-cell resolution (Liu et al., 2021). Still, we are far from understanding the dynamic molecular regulation of germ cell development in fish oogenesis, the complex interactions between germ cells and somatic cells, and the transcriptional characteristics of specific cell populations. Thus, herein, we performed a single-cell survey of oogenesis in Chinese tongue sole. We identified eight ovarian cell types by using the known landmark genes and uncovered that pluripotency-related genes, mitosis-related genes and meiosis-related genes were dynamically expressed from oogonia to oocytes. When evaluating the different populations of somatic cells, we defined four granulosa cell subtypes and one theca cell type with diverse signature genes and TFs, which broke through the previous observations of the bilayer follicle cells of fish follicles. Moreover, we explored the bidirectional communication between germ cells and granulosa cells during fish oogenesis, and revealed the interaction of distinct granulosa cell subtypes in fish ovulation through the expression pattern of ovulation-associated genes. Besides, elucidating the conservation and divergence of oogenesis across vertebrates will help us to understand the advantages and disadvantages of various reproductive strategies from different angles and conduct the choice of infertility therapeutic strategies. In terms of ovarian cell types, we found that oogonia, oocytes, granulosa cells, endothelial cells and immune cells identified in mammalian ovaries were also present in fish. The signature genes of these cell types were similar to those of other vertebrates (Fan et al., 2019). Additionally, we found that most of the DEGs of ovarian germ cells and granulosa cells identified in Chinese tongue sole are homologous to macaques, and these homologous genes are significantly enriched in biological processes or pathways related to their functions. These results suggest that although fish and mammals represent different reproduction models, the process of gamete production is conserved between these taxa.
TFs are crucial for the differentiation of oocytes. Differentially expressed TFs such as arid3a, znf407, zeb1, pou2f2, ikzf2, tcf21, zhx1, gtf3ab, and znf706 were detected during early oogenesis, suggesting their possible roles in the development of germ cells. Two gtf3a gene paralogues (gtf3aa and gtf3ab) are present in teleost genomes due to the teleost-specific genome duplication event. Gtf3ab has been proved to be a marker of zebrafish oocyte differentiation, which facilitates massive oocyte stockpiling of 5S rRNAs. Notably, 5S rRNAs play essential roles in the developmental processes of newly formed embryos (Rojo-Bartolomé et al., 2020). Our data showed that gtf3ab was significantly upregulated from oogonia to oocytes, which suggests gtf3ab has the potential to serve as a broad marker of oocyte differentiation in diverse teleost species.
The granulosa cells of human follicles can be sorted into two functionally distinct subtypes: mural granulosa cells, which participate in cell differentiation and signal transduction; and cumulus cells, which participate in cell proliferation and metabolism (Harris et al., 2009; Roberts et al., 2002; Wigglesworth et al., 2015). In this study, we achieved the novel identification of five subtypes of follicle cells in Chinese tongue sole. Interestingly, we found that foxl2 and inhbb, which have been proved to be expressed in human cumulus cells, were specifically expressed in GC1 (Pisarska et al., 2004; Fan et al., 2019; Sadat Tahajjodi et al., 2019). Therefore, we speculate that GC1 is a differentiated granulosa cell type in mature follicles and its function is similar to that of mammalian cumulus cells. Gene expression correlation analysis argued that GC2 is a proliferating granulosa cell subtype of GC1, as GC1 showed a strong correlation with GC2, and cell-cycle phase prediction analysis indicated that most of cells in GC2 were in G2/M phase (Supplementary Figures S2A, S2B). Interestingly, GC4 uniquely expressed tnfaip6 whose orthologous gene in mammals is a signature gene in periovulatory cumulus cells playing a key role in the formation of the cumulus extracellular matrix (Fülöp et al., 2003). Notably, GC4 also specifically expressed adamts1, a gene required for the degradation of ECM in the cumulus oocyte complex to induce cumulus expansion and ovulation (Russell et al., 2003; Yang et al., 2021). These argues that GC4 is a differentiated granulosa cell type in preovulatory follicles and plays a key role in ovulation. In contrast, krt18, a marker gene of pre-granulosa cells and mural granulosa cells in human ovary (Fan et al., 2019), was expressed in GC3. Additionally, GC3 showed specific high activity of TFs related to pre-granulosa cells, such as lhx9, wt1, and irx3 (Fu et al., 2020; Knarston et al., 2020). Accordingly, GC3 might be pre-granulosa cells surrounding early follicles. Nevertheless, we also observed the expression of ovulation associated genes pgr and ptger4b showed high specificity in GC3, and pgr showed high regulatory activity synchronously. Pgr and ptger4b have been proved to be highly expressed in the preovulatory follicles of medaka to regulate the ovulation process (Hagiwara et al., 2014). Therefore, we speculate that GC3 may be the source of other granulosa cells; however, it is not only a granulosa cell subtype in primordial follicles, but has a broader function at any stage of oocytes, which is inconsistent with that observed in mammals. One problem is whether these different states of follicle cells are assigned to different cell types or different developmental stages. Our results are more inclined to the former. First, the DEGs of these subtypes hardly overlap and showed differential functional enrichment. Second, we observed that the signature genes of mammalian cumulus cells and mural granulosa cells were differentially expressed in the subtypes. Third, ovulation associated genes are assigned to different subtypes, which indicates that ovulation requires the combined action of different granulosa cell subtypes. For theca cells, we only observed a single cell type, but distinct populations of theca cells exist in different follicles in humans (Fan et al., 2019); however, the function of theca cells (i.e., steroidogenesis) is conserved across vertebrates. From an evolutionary perspective, inconsistent follicular structures indicate the different investment strategies of parents in the reproductive process. These structures undergo diversification during evolution; however, the core function of the follicle cell population remains similar.
Niche–germline bidirectional communication coordinately regulates oogenesis, but the knowledge of the relevant signaling pathway interactions in fish remains limited. In this study, by conducting an unbiased ligand–receptor interaction analysis between the germ and somatic cell subclusters, we have identified many bidirectional communications between germ cells and granulosa cells, but less in theca cells, which may be ascribed to the spatial distribution of germ cells, granulosa cells and theca cells.
Additionally, we analysed the expression patterns of two common oogenesis-associated signaling pathways. We noted that, in Chinese tongue sole, gdf9, and bmp15, the ligand of the TGF-β signaling pathway involved in follicular development, were specifically expressed in germ cells. Moreover, bmpr2, the receptor of bmp15/gdf9, was expressed in germ cells and granulosa cells. These findings are consistent with observations in other vertebrates, i.e., human (Zhang et al., 2018), short finned eel and European sea bass (Halm et al., 2008; Falahati et al., 2021), suggesting that the function of bmp15/gdf9-bmpr2 signaling in oogenesis is conserved across vertebrates. Moreover, our data revealed that the expression pattern of NOTCH signaling pathway components in fish was similar to that in human. This is consistent with the existing view that the NOTCH signaling pathway is highly conserved in vertebrates (Kopan and Ilagan, 2009).
Previous research demonstrated the ovulation mechanism of female mice, in which LH induces the expression of PGR and further stimulates granulosa cells to produce adamts1. Likewise, our study showed that pgr was specifically expressed in GC3, while adamts1 was mainly expressed in GC4. Moreover, ecm1, encoding the target protein of adamts1, was expressed in all granulosa cell subtypes. Therefore, we speculate that the ovulation mechanism mediated by adamts1 is conserved across vertebrates. Notably, ptger4b, a pge2 receptor isoform of ptger4, has been proven to be crucial for ovulation in medaka (Fujimori et al., 2012; Hagiwara et al., 2014) and is expressed in pgr-dependent cells of zebrafish (Tang et al., 2016; Takahashi et al., 2018). Of particular interest was the finding that the expression patterns of ptger4b and pgr were consistent. The observations led to the proposal that pge2/ptger4 signaling may have wide regulatory effects on ovulation across teleosts.
Based on the scRNA-seq method, this work provides several noteworthy contributions, as summarized below. We succeeded in identifying three germ cell types and five somatic cell types from fish ovary samples and revealed the gene expression signatures of each of these cell types. Especially, we identified four granulosa cell subtypes and one theca cell type in fish follicle, accompanied by different signature genes and transcriptional regulatory activities. Moreover, we found that the extensive niche–germline bidirectional communications regulate fish oogenesis, whereas ovulation in fish is accompanied by the coordination of simultaneous and tightly sequential processes across different granulosa cells. Altogether, these observations provide a global perspective on fish oogenesis, which may offer a valuable reference for future studies on fish resource propagation and contribute to the long-standing topics about the adaptability of reproductive strategies in lower vertebrates and common regularity of gametogenesis across vertebrates.
Data Availability Statement
According to national legislation/guidelines, specifically the Administrative Regulations of the People’s Republic of China on Human Genetic Resources (http://www.gov.cn/zhengce/content/2019-06/10/content_5398829.htm, http://english.www.gov.cn/policies/latest_releases/2019/06/10/content_281476708945462.htm), no additional raw data is available at this time. Data of this project can be accessed after an approval application to the China National Genebank (CNGB, https://db.cngb.org/cnsa/). Please refer to https://db.cngb.org/, or email: CNGBdb@cngb.org for detailed application guidance. The accession code CNP0002319 should be included in the application.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee at the Chinese Academy of Fishery Sciences.
Author Contributions
CS designed and supervised the research. XL, J-YC, XZ, and XZ performed all experiments, collected and interpreted the data. YH and FT did the bioinformatics analysis of single-cell transcriptomic data. H-YW, FP, SL, KL, QW, and GF provided essential reagents and suggestions. XL and H-YW collected the fish samples. XL and YH wrote the manuscript. XL, CS, and FP edited the manuscript and revised it critically. All authors took part in the interpretation of data.
Funding
This work was supported by the National Nature Science Foundation of China (31722058, 31802275 and 31472269); the National Key R&D Program of China (2018YFD0900301); the AoShan Talents Cultivation Program Supported by Qingdao National Laboratory for Marine Science and Technology (2017ASTCP-ES06); the Taishan Scholar Project Fund of Shandong of China to C.S.; the National Ten-Thousands Talents Special Support Program to C.S.; the Central Public-interest Scientific Institution Basal Research Fund, CAFS (No.2020TD19); and the China Agriculture Research System (CARS-47-G03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer ML declared shared affiliation with one of the authors YH to the handling editor.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.828124/full#supplementary-material
References
Bachvarova, R. F., Masi, T., Drum, M., Parker, N., Mason, K., Patient, R., et al. (2004). Gene Expression in the Axolotl Germ Line: Axdazl, Axvh, Axoct-4, and Axkit. Dev. Dyn. 231, 871–880. doi:10.1002/dvdy.20195
Bertho, S., Clapp, M., Banisch, T. U., Bandemer, J., Raz, E., and Marlow, F. L. (2021). Zebrafish Dazl Regulates Cystogenesis and Germline Stem Cell Specification during the Primordial Germ Cell to Germline Stem Cell Transition. Development 148 (7), dev187773. doi:10.1242/dev.187773
Biase, F. H. (2017). Oocyte Developmental Competence: Insights from Cross-Species Differential Gene Expression and Human Oocyte-Specific Functional Gene Networks. OMICS: A J. Integr. Biol. 21, 156–168. doi:10.1089/omi.2016.0177
Brown, H. M., Dunning, K. R., Robker, R. L., Pritchard, M., and Russell, D. L. (2006). Requirement for ADAMTS-1 in Extracellular Matrix Remodeling during Ovarian Folliculogenesis and Lymphangiogenesis. Develop. Biol. 300, 699–709. doi:10.1016/j.ydbio.2006.10.012
Can, H., Chanumolu, S. K., Gonzalez-Muñoz, E., Prukudom, S., Otu, H. H., and Cibelli, J. B. (2020). Comparative Analysis of Single-Cell Transcriptomics in Human and Zebrafish Oocytes. BMC Genom. 21, 471. doi:10.1186/s12864-020-06860-z
Chen, C. F., Wen, H. S., Wang, Z. P., He, F., Zhang, J. R., Chen, X. Y., et al. (2010). Cloning and Expression of P450c17-I (17α-Hydroxylase/17,20-Lyase) in Brain and Ovary during Gonad Development in Cynoglossus Semilaevis. Fish. Physiol. Biochem. 36, 1001–1012. doi:10.1007/s10695-009-9378-7
Chen, S., Zhang, G., Shao, C., Huang, Q., Liu, G., Zhang, P., et al. (2014). Whole-Genome Sequence of a Flatfish Provides Insights into ZW Sex Chromosome Evolution and Adaptation to a Benthic Lifestyle. Nat. Genet. 46, 253–260. doi:10.1038/ng.2890
Curry, T. E. (2010). ADAMTS1 and Versican: Partners in Ovulation and Fertilization. Biol. Reprod. 83, 505–506. doi:10.1095/biolreprod.110.087056
Dong, Y., Lyu, L., Zhang, D., Li, J., Wen, H., and Shi, B. (2021). Integrated lncRNA and mRNA Transcriptome Analyses in the Ovary of Cynoglossus Semilaevis Reveal Genes and Pathways Potentially Involved in Reproduction. Front. Genet. 12, 671729. doi:10.3389/fgene.2021.671729
Eppig, J. J., Wigglesworth, K., Pendola, F., and Hirao, Y. (1997). Murine Oocytes Suppress Expression of Luteinizing Hormone Receptor Messenger Ribonucleic Acid by Granulosa Cells. Biol. Reprod. 56, 976–984. doi:10.1095/biolreprod56.4.976
Falahati, A., Ozaki, Y., Damsteegt, E. L., Zadmajid, V., Freeman, K. J., and Lokman, P. M. (2021). Spatiotemporal Expression of Activin Receptor-Like Kinase-5 and Bone Morphogenetic Protein Receptor Type II in the Ovary of Shortfinned Eel, Anguilla Australis. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 251, 110509. doi:10.1016/j.cbpb.2020.110509
Fan, X., Bialecka, M., Moustakas, I., Lam, E., Torrens-Juaneda, V., Borggreven, N. V., et al. (2019). Single-Cell Reconstruction of Follicular Remodeling in the Human Adult Ovary. Nat. Commun. 10, 3164. doi:10.1038/s41467-019-11036-9
Fu, A., Koth, M. L., Brown, R. M., Shaw, S. A., Wang, L., Krentz, K. J., et al. (2020). IRX3 and IRX5 Collaborate during Ovary Development and Follicle Formation to Establish Responsive Granulosa Cells in the Adult Mouse†. Biol. Reprod. 103, 620–629. doi:10.1093/biolre/ioaa100
Fujimori, C., Ogiwara, K., Hagiwara, A., and Takahashi, T. (2012). New Evidence for the Involvement of Prostaglandin Receptor EP4b in Ovulation of the Medaka, Oryzias L. Mol. Cell Endocrinol. 362, 76–84. doi:10.1016/j.mce.2012.05.013
Fülöp, C., Szántó, S., Mukhopadhyay, D., Bárdos, T., Kamath, R. V., Rugg, M. S., et al. (2003). Impaired Cumulus Mucification and Female Sterility in Tumor Necrosis Factor-Induced Protein-6 Deficient Mice. Development 130, 2253–2261. doi:10.1242/dev.00422
Gershon, E., and Dekel, N. (2020). Newly Identified Regulators of Ovarian Folliculogenesis and Ovulation. Int. J. Mol. Sci. 21, 4565. doi:10.3390/ijms21124565
Hagiwara, A., Ogiwara, K., Katsu, Y., and Takahashi, T. (2014). Luteinizing Hormone-Induced Expression of Ptger4b, a Prostaglandin E2 Receptor Indispensable for Ovulation of the Medaka Oryzias L, Is Regulated by a Genomic Mechanism Involving Nuclear Progestin Receptor. Biol. Reprod. 90, 126. doi:10.1095/biolreprod.113.115485
Halm, S., Ibañez, A. J., Tyler, C. R., and Prat, F. (2008). Molecular Characterisation of Growth Differentiation Factor 9 (Gdf9) and Bone Morphogenetic Protein 15 (Bmp15) and Their Patterns of Gene Expression during the Ovarian Reproductive Cycle in the European Sea Bass. Mol. Cell Endocrinol. 291, 95–103. doi:10.1016/j.mce.2008.03.002
Harris, S. E., Leese, H. J., Gosden, R. G., and Picton, H. M. (2009). Pyruvate and Oxygen Consumption throughout the Growth and Development of Murine Oocytes. Mol. Reprod. Dev. 76, 231–238. doi:10.1002/mrd.20945
Hunyady, B., Mezey, É., Pacak, K., and Palkovits, M. (1996). Identification of Endogenous Peroxidase-Containing Cells as Eosinophils in the Gastrointestinal System. Histochem. Cel Biol. 106, 447–456. doi:10.1007/bf02473306
Hurley, D. J., Wilson, R. A., Baldwin, C. L., Liu, J.-Y., and Mastro, A. M. (1994). Characterization of Resting and Phorbol Ester or Concanavalin A Activated Bovine Lymph Node Cells with Leukocyte Specific Monoclonal Antibodies. Vet. Immunol. Immunopathology 40, 49–61. doi:10.1016/0165-2427(94)90014-0
Kaviani, E. F., Shabanipour, N., and Mirnategh, S. B. (2013). Light and Electron Microscope Structural Study of the Zona Radiata in the Oocyte of Zebrafish (Danio R). Microscopy (Tokyo) 62, 377–382. doi:10.1093/jmicro/dfs086
Knarston, I. M., Pachernegg, S., Robevska, G., Ghobrial, I., Er, P. X., Georges, E., et al. (2020). An In Vitro Differentiation Protocol for Human Embryonic Bipotential Gonad and Testis Cell Development. Stem Cel Rep. 15, 1377–1391. doi:10.1016/j.stemcr.2020.10.009
Kobak, D., and Berens, P. (2019). The Art of Using T-SNE for Single-Cell Transcriptomics. Nat. Commun. 10, 5416. doi:10.1038/s41467-019-13056-x
Kopan, R., and Ilagan, M. X. G. (2009). The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell 137, 216–233. doi:10.1016/j.cell.2009.03.045
Kostyrko, K., Bosshard, S., Urban, Z., and Mermod, N. (2015). A Role for Homologous Recombination Proteins in Cell Cycle Regulation. Cell Cycle 14, 2853–2861. doi:10.1080/15384101.2015.1049784
Li, M., Zhu, F., Li, Z., Hong, N., and Hong, Y. (2016). Dazl Is a Critical Player for Primordial Germ Cell Formation in Medaka. Sci. Rep. 6, 28317. doi:10.1038/srep28317
Li, R., and Albertini, D. F. (2013). The Road to Maturation: Somatic Cell Interaction and Self-Organization of the Mammalian Oocyte. Nat. Rev. Mol. Cel Biol. 14, 141–152. doi:10.1038/nrm3531
Li, Z., Wang, J., Zhao, Y., Ma, D., Zhao, M., Li, N., et al. (2021). scRNA‐Seq of Ovarian Follicle Granulosa Cells from Different Fertility Goats Reveals Distinct Expression Patterns. Reprod. Dom Anim. 56, 801–811. doi:10.1111/rda.13920
Liu, L., and Ge, W. (2007). Growth Differentiation Factor 9 and its Spatiotemporal Expression and Regulation in the Zebrafish Ovary1. Biol. Reprod. 76, 294–302. doi:10.1095/biolreprod.106.054668
Liu, X., Li, W., Yang, Y., Chen, K., Li, Y., Zhu, X., et al. (2021). Transcriptome Profiling of the Ovarian Cells at the Single-Cell Resolution in Adult Asian Seabass. Front. Cel Dev. Biol. 9, 647892. doi:10.3389/fcell.2021.647892
Lodé, T. (2012). Oviparity or Viviparity? That Is the Question. Reprod. Biol. 12, 259–264. doi:10.1016/j.repbio.2012.09.001
Lundholm, M., Mayans, S., Motta, V., Löfgren-Burström, A., Danska, J., and Holmberg, D. (2010). Variation in the Cd3 Zeta (Cd247) Gene Correlates with Altered T Cell Activation and Is Associated with Autoimmune Diabetes. J. Immunol. 184, 5537–5544. doi:10.4049/jimmunol.0904012
Luo, W., Gu, L., Li, J., and Gong, Y. (2020). Transcriptome Sequencing Revealed that Knocking Down FOXL2 Affected Cell Proliferation, the Cell Cycle, and DNA Replication in Chicken Pre-Ovulatory Follicle Cells. PLoS One 15, e0234795. doi:10.1371/journal.pone.0234795
Moolhuijsen, L. M. E., and Visser, J. A. (2020). Anti-Müllerian Hormone and Ovarian Reserve: Update on Assessing Ovarian Function. J. Clin. Endocrinol.Metab. 105, 3361–3373. doi:10.1210/clinem/dgaa513
Munday, A. D., Berndt, M. C., and Mitchell, C. A. (2000). Phosphoinositide 3-Kinase Forms a Complex with Platelet Membrane Glycoprotein Ib-IX-V Complex and 14-3-3ζ. Blood 96, 577–584. doi:10.1182/blood.v96.2.577.014k15_577_584
Nagahama, Y., and Yamashita, M. (2008). Regulation of Oocyte Maturation in Fish. Dev. Growth Differ. 50 (Suppl. 1), S195–S219. doi:10.1111/j.1440-169X.2008.01019.x
Nagahama, Y., Yoshikuni, M., Yamashita, M., Tokumoto, T., and Katsu, Y. (1995). 4 Regulation of Oocyte Growth and Maturation in Fish. Curr. Top. Dev. Biol. 30, 103–145. doi:10.1016/s0070-2153(08)60565-7
Nagai, N., Ohguchi, H., Nakaki, R., Matsumura, Y., Kanki, Y., Sakai, J., et al. (2018). Downregulation of ERG and FLI1 Expression in Endothelial Cells Triggers Endothelial-To-Mesenchymal Transition. Plos Genet. 14, e1007826. doi:10.1371/journal.pgen.1007826
Patiño, R., Bolamba, D., Thomas, P., and Kumakura, N. (2005). Effects of External pH on Hormonally Regulated Ovarian Follicle Maturation and Ovulation in Atlantic Croaker. Gen. Comp. Endocrinol. 141, 126–134. doi:10.1016/j.ygcen.2004.12.006
Pisarska, M. D., Bae, J., Klein, C., and Hsueh, A. J. W. (2004). Forkhead L2 Is Expressed in the Ovary and Represses the Promoter Activity of the Steroidogenic Acute Regulatory Gene. Endocrinology 145, 3424–3433. doi:10.1210/en.2003-1141
Pisarska, M. D., Barlow, G., and Kuo, F.-T. (2011). Minireview: Roles of the Forkhead Transcription Factor FOXL2 in Granulosa Cell Biology and Pathology. Endocrinology 152, 1199–1208. doi:10.1210/en.2010-1041
Roberts, R., Franks, S., and Hardy, K. (2002). Culture Environment Modulates Maturation and Metabolism of Human Oocytes. Hum. Reprod. 17, 2950–2956. doi:10.1093/humrep/17.11.2950
Rojo-Bartolomé, I., Santana de Souza, J. E., Diaz de Cerio, O., and Cancio, I. (2020). Duplication and Subfunctionalisation of the General Transcription Factor IIIA (Gtf3a) Gene in Teleost Genomes, with Ovarian Specific Transcription of Gtf3ab. PLoS One 15, e0227690. doi:10.1371/journal.pone.0227690
Russell, D. L., Doyle, K. M. H., Ochsner, S. A., Sandy, J. D., and Richards, J. S. (2003). Processing and Localization of ADAMTS-1 and Proteolytic Cleavage of Versican during Cumulus Matrix Expansion and Ovulation. J. Biol. Chem. 278, 42330–42339. doi:10.1074/jbc.M300519200
Rust, K., Byrnes, L. E., Yu, K. S., Park, J. S., Sneddon, J. B., Tward, A. D., et al. (2020). A Single-Cell Atlas and Lineage Analysis of the Adult Drosophila Ovary. Nat. Commun. 11, 5628. doi:10.1038/s41467-020-19361-0
Sadat Tahajjodi, S., Farashahi Yazd, E., Agha-Rahimi, A., Aflatoonian, R., Ali Khalili, M., Mohammadi, M., et al. (2019). Biological and Physiological Characteristics of Human Cumulus Cell in Adherent Culture Condition. Int. J. Reprod. Biomed. (Yazd) 18, 1–10. doi:10.18502/ijrm.v18i1.6189
Sánchez, F., and Smitz, J. (2012). Molecular Control of Oogenesis. Biochim. Biophys. Acta (Bba) - Mol. Basis Dis. 1822, 1896–1912. doi:10.1016/j.bbadis.2012.05.013
Selman, K., Wallace, R. A., Sarka, A., and Qi, X. (1993). Stages of Oocyte Development in the Zebrafish, Brachydanio Rerio. J. Morphol. 218, 203–224. doi:10.1002/jmor.1052180209
Sendler, M., Weiss, F.-U., Golchert, J., Homuth, G., van den Brandt, C., Mahajan, U. M., et al. (2018). Cathepsin B-Mediated Activation of Trypsinogen in Endocytosing Macrophages Increases Severity of Pancreatitis in Mice. Gastroenterology 154, 704–718. doi:10.1053/j.gastro.2017.10.018
Shao, C., Li, Q., Chen, S., Zhang, P., Lian, J., Hu, Q., et al. (2014). Epigenetic Modification and Inheritance in Sexual Reversal of Fish. Genome Res. 24, 604–615. doi:10.1101/gr.162172.113
Shozu, M., Minami, N., Yokoyama, H., Inoue, M., Kurihara, H., Matsushima, K., et al. (2005). ADAMTS-1 Is Involved in normal Follicular Development, Ovulatory Process and Organization of the Medullary Vascular Network in the Ovary. J. Mol. Endocrinol. 35, 343–355. doi:10.1677/jme.1.01735
Sylvestre, E.-L., Robert, C., Pennetier, S., Labrecque, R., Gilbert, I., Dufort, I., et al. (2013). Evolutionary Conservation of the Oocyte Transcriptome Among Vertebrates and its Implications for Understanding Human Reproductive Function. Mol. Hum. Reprod. 19, 369–379. doi:10.1093/molehr/gat006
Takahashi, T., Hagiwara, A., and Ogiwara, K. (2018). Prostaglandins in Teleost Ovulation: A Review of the Roles with a View to Comparison with Prostaglandins in Mammalian Ovulation. Mol. Cel. Endocrinol. 461, 236–247. doi:10.1016/j.mce.2017.09.019
Tang, F., Barbacioru, C., Wang, Y., Nordman, E., Lee, C., Xu, N., et al. (2009). mRNA-Seq Whole-Transcriptome Analysis of a Single Cell. Nat. Methods 6, 377–382. doi:10.1038/nmeth.1315
Tang, H., Liu, Y., Li, J., Yin, Y., Li, G., Chen, Y., et al. (2016). Gene Knockout of Nuclear Progesterone Receptor Provides Insights into the Regulation of Ovulation by LH Signaling in Zebrafish. Sci. Rep. 6, 28545. doi:10.1038/srep28545
Tsang, J. C. H., Vong, J. S. L., Ji, L., Poon, L. C. Y., Jiang, P., Lui, K. O., et al. (2017). Integrative Single-Cell and Cell-free Plasma RNA Transcriptomics Elucidates Placental Cellular Dynamics. Proc. Natl. Acad. Sci. U.S.A. 114, E7786–e7795. doi:10.1073/pnas.1710470114
Tyler, C. R., and Sumpter, J. P. (1996). Oocyte Growth and Development in Teleosts. Rev. Fish. Biol. Fish. 6, 287–318. doi:10.1007/BF00122584
Viana, I. K. S., Gonçalves, L. A. B., Ferreira, M. A. P., Mendes, Y. A., and Rocha, R. M. (2018). Oocyte Growth, Follicular Complex Formation and Extracellular-Matrix Remodeling in Ovarian Maturation of the imperial Zebra Pleco Fish Hypancistrus Zebra. Sci. Rep. 8, 13760. doi:10.1038/s41598-018-32117-7
Völkening, L., Vatselia, A., Asgedom, G., Bastians, H., Lavin, M., Schindler, D., et al. (2020). RAD50 Regulates Mitotic Progression Independent of DNA Repair Functions. FASEB j. 34, 2812–2820. doi:10.1096/fj.201902318R
Wang, S., Zheng, Y., Li, J., Yu, Y., Zhang, W., Song, M., et al. (2020). Single-Cell Transcriptomic Atlas of Primate Ovarian Aging. Cell 180, 585–600. doi:10.1016/j.cell.2020.01.009
Wang, Y., Wang, R., Zhang, S., Song, S., Jiang, C., Han, G., et al. (2019). iTALK: An R Package to Characterize and Illustrate Intercellular Communication. bioRxiv, 507871. doi:10.1101/507871
Wen, H., Si, Y., Zhang, Y., He, F., and Li, J. (2015). Cloning and Expression of Follistatin Gene in Half-Smooth Tongue Sole Cynoglossus Semilaevis during the Reproduction Cycle. Chin. J. Ocean. Limnol. 33, 299–308. doi:10.1007/s00343-015-4051-8
Wigglesworth, K., Lee, K.-B., Emori, C., Sugiura, K., and Eppig, J. J. (2015). Transcriptomic Diversification of Developing Cumulus and Mural Granulosa Cells in Mouse Ovarian Follicles. Biol. Reprod. 92, 23. doi:10.1095/biolreprod.114.121756
Wu, H., Liu, K., and Zhang, J. (2021). Excess Fibronectin 1 Participates in Pathogenesis of Pre-eclampsia by Promoting Apoptosis and Autophagy in Vascular Endothelial Cells. Mol. Hum. Reprod. 27 (6), gaab030. doi:10.1093/molehr/gaab030
Yang, C.-X., Wright, E. C., and Ross, J. W. (2012). Expression of RNA-Binding Proteins DND1 and FXR1 in the Porcine Ovary, and during Oocyte Maturation and Early Embryo Development. Mol. Reprod. Dev. 79, 541–552. doi:10.1002/mrd.22059
Yang, G., Yao, G., Xu, Z., Fan, H., Liu, X., He, J., et al. (2021). Expression Level of ADAMTS1 in Granulosa Cells of PCOS Patients Is Related to Granulosa Cell Function, Oocyte Quality, and Embryo Development. Front. Cel Dev. Biol. 9, 647522. doi:10.3389/fcell.2021.647522
Zhang, C.-P., Yang, J.-L., Zhang, J., Li, L., Huang, L., Ji, S.-Y., et al. (2011). Notch Signaling Is Involved in Ovarian Follicle Development by Regulating Granulosa Cell Proliferation. Endocrinology 152, 2437–2447. doi:10.1210/en.2010-1182
Zhang, Y., Yan, Z., Qin, Q., Nisenblat, V., Chang, H.-M., Yu, Y., et al. (2018). Transcriptome Landscape of Human Folliculogenesis Reveals Oocyte and Granulosa Cell Interactions. Mol. Cel 72, 1021–1034. doi:10.1016/j.molcel.2018.10.029
Zhao, Z. H., Li, C.-Y., Meng, T.-G., Wang, Y., Liu, W.-B., Li, A., et al. (2020a). Single-cell RNA Sequencing Reveals Regulation of Fetal Ovary Development in the Monkey (Macaca F). Cell Discov 6, 97. doi:10.1038/s41421-020-00219-0
Keywords: scRNA-seq, oogenesis, Chinese tongue sole, germ cell, granulosa cell
Citation: Liu X, Huang Y, Tan F, Wang H-Y, Chen J-Y, Zhang X, Zhao X, Liu K, Wang Q, Liu S, Piferrer F, Fan G and Shao C (2022) Single-Cell Atlas of the Chinese Tongue Sole (Cynoglossus semilaevis) Ovary Reveals Transcriptional Programs of Oogenesis in Fish. Front. Cell Dev. Biol. 10:828124. doi: 10.3389/fcell.2022.828124
Received: 03 December 2021; Accepted: 07 February 2022;
Published: 01 March 2022.
Edited by:
Zhao-Jia Ge, Qingdao Agricultural University, ChinaReviewed by:
Jie Mei, Huazhong Agricultural University, ChinaMingyou Li, Shanghai Ocean University, China
Andrea Tangherloni, University of Bergamo, Italy
Copyright © 2022 Liu, Huang, Tan, Wang, Chen, Zhang, Zhao, Liu, Wang, Liu, Piferrer, Fan and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangyi Fan, fanguangyi@genomics.cn; Changwei Shao, shaocw@ysfri.ac.cn
†These authors have contributed equally to this work
 Xiang Liu
Xiang Liu Yingyi Huang1,2†
Yingyi Huang1,2†  Qian Wang
Qian Wang Francesc Piferrer
Francesc Piferrer Guangyi Fan
Guangyi Fan Changwei Shao
Changwei Shao