A transgenic bacterial artificial chromosome approach to identify regulatory regions that direct Amhr2 and Osterix expression in Müllerian duct mesenchyme
- 1Department of Genetics, University of Texas MD Anderson Cancer Center, Houston, TX, United States
- 2Graduate Program in Genetics and Epigenetics, MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences, Houston, TX, United States
- 3Department of Reconstructive Sciences, School of Dental Medicine, University of Connecticut Health Center, Farmington, CT, United States
- 4Université de Rennes, Inserm, EHESP, Irset (Institut de Recherche en Santé, Environnement et Travail), Rennes, France
A transgenic mouse approach using bacterial artificial chromosomes (BAC) was used to identify regulatory regions that direct Müllerian duct expression for Amhr2 and Osterix (Osx, also known as Sp7). Amhr2 encodes the receptor that mediates anti-Müllerian hormone (AMH) signaling for Müllerian duct regression in male embryos. Amhr2 is expressed in the Müllerian duct mesenchyme of both male and female embryos. A ∼147-kb BAC clone containing the Amhr2 locus was used to generate transgenic mice. The transgene was able to rescue the block in Müllerian duct regression of Amhr2-null males, suggesting that the BAC clone contains regulatory sequences active in male embryos. Osx is expressed in the developing skeleton of male and female embryos but is also an AMH-induced gene that is expressed in the Müllerian duct mesenchyme exclusively in male embryos. Osx-Cre transgenic mice were previously generated using a ∼204-kb BAC clone. Crosses of Osx-Cre mice to Cre-dependent lacZ reporter mice resulted in reporter expression in the developing skeleton and in the Müllerian duct mesenchyme of male but not female embryos. Osx-Cherry transgenic mice were previously generated using a 39-kb genomic region surrounding the Osx locus. Osx-Cherry embryos expressed red fluorescence in the developing skeleton and Müllerian duct mesenchyme of male but not female embryos. In addition, female Osx-Cherry embryos ectopically expressing human AMH from an Mt1-AMH transgene activated red fluorescence in the Müllerian duct mesenchyme. These results suggest that the 39-kb region used to generate Osx-Cherry contains male-specific Müllerian duct mesenchyme regulatory sequences that are responsive to AMH signaling. These BAC transgenic mouse approaches identify two distinct regions that direct Müllerian duct mesenchyme expression and contribute fundamental knowledge to define a gene regulatory network for sex differentiation.
Introduction
Mammalian reproductive tract organ development begins with the formation of two genital ducts within the paired mesonephroi (Mullen and Behringer, 2014). The embryo, regardless of its sex genotype, first forms the Wolffian or mesonephric ducts, which have the potential to develop into male reproductive tract organs, including the epididymides, vasa deferentia, and seminal vesicles. Subsequently, the Müllerian or paramesonephric ducts form along the Wolffian ducts (Orvis and Behringer, 2007). The Müllerian ducts have the potential to develop into female reproductive tract organs, including the oviducts, uterus, cervix, and upper vagina. The elongation of the Müllerian ducts to the urogenital sinus in male and female mouse embryos is complete by E13.5 (Orvis and Behringer, 2007). During sex development, one pair of the genital ducts (Wolffian or Müllerian) will differentiate and the other will be eliminated. This is regulated by hormones and growth factors produced from the fetal testes or not produced by the fetal ovaries.
In male embryos, the testes send a signal to the Müllerian ducts, which triggers Müllerian duct regression, causing gaps in the Müllerian duct epithelium that lead to its elimination. This signal comes in the form of anti-Müllerian hormone (AMH) (Cate et al., 1986; Picard et al., 1986; Josso et al., 1993; Behringer, Finegold and Cate, 1994). The primary receptor that receives AMH in the Müllerian duct, anti-Müllerian hormone receptor 2 (AMHR2), forms a receptor complex with Type I receptors to trigger regression (Mishina et al., 1996, 1999; Gouedard et al., 2000; Clarke et al., 2001; Visser et al., 2001; Belville et al., 2005; Zhan et al., 2006; Orvis et al., 2008). Amhr2 is expressed in the Müllerian duct mesenchyme of both male and female embryos, indicating that the mesenchyme is the target tissue of AMH and that Müllerian duct regression results from mesenchyme-epithelial interactions (Baarends et al., 1994). Indeed, ectopic expression of AMH from an Mt1-AMH transgene induces Müllerian duct regression in females, resulting in the absence of oviducts and uterus (Behringer et al., 1990). The AMH receptor complex signals through redundant Smad genes (Orvis et al., 2008). Following this initial signal is a cascade of genetically regulated activity for Müllerian duct regression that has yet to be fully elucidated. However, a gene regulatory network has been described primarily based on genetic loss-of-function studies of Müllerian duct regression (Moses and Behringer, 2019).
A recent addition to the gene regulatory network for Müllerian duct regression is Osterix (Osx), formally known as Sp7 (Mullen et al., 2018). Osx encodes a zinc-finger transcription factor that is essential for osteoblast and odontoblast differentiation (Nakashima et al., 2002; Bae et al., 2018). Previously, we showed that Osx is an AMH-induced gene that is expressed in the Müllerian duct mesenchyme (Mullen et al., 2018). This places Osx downstream of Amhr2 in the gene regulatory network for Müllerian duct regression (Moses and Behringer, 2019). In contrast to Amhr2 which is expressed in the Müllerian duct mesenchyme of both male and female embryos, Osx is expressed in the Müllerian duct mesenchyme only in male embryos. Intriguingly, Amhr2 and Osx are located within 100 kb of each other.
Enhancers are DNA sequences that are bound by proteins to form complexes at basal promoters to direct cell type and tissue-specific transcription (Field and Adelman, 2020). These DNA sequences can reside close to a locus (within 1 kb) or can be located at relatively large distances (∼1 Mb) (Lettice et al., 2003). Enhancers have been identified 5′ to a locus, within introns, or 3’ to a locus (Ornitz et al., 1985; Behringer et al., 1987; Mukhopadhyay et al., 1995). Classically, enhancers have been identified using in vitro assays or in vivo using transgenic animals. More recently, analysis of chromatin modifications and open chromatin domains have identified candidate transcriptional enhancers (Vu and Ernst, 2022). Ultimately, enhancers that can direct tissue-specific transcription must be tested for activity in vivo. Although cis-regulatory elements in the gene regulatory network for Müllerian duct regression have been identified for the Amh gene expressed in the Sertoli cells of the testes, no cis elements have been identified for Müllerian duct epithelium or mesenchyme transcription. Previously, multiple lines of Amhr2-EGFP transgenic mice were generated using a 500-bp promoter region but reporter expression was not detected (Kimura et al., 2017). Thus, the immediate upstream region of Amhr2 is not sufficient for cell type-specific transcription.
In this study, we sought to identify regulatory regions associated with Amhr2 and Osx that direct Müllerian duct mesenchyme transcription, using a bacterial artificial chromosome (BAC) transgenic mouse approach. We screened BAC clones containing Amhr2 or Osx for Müllerian duct mesenchyme-specific transcriptional activity in transgenic mice. Using this assay, we have identified two genomic regions that direct Müllerian duct mesenchyme-specific activity. The subsequent identification of Müllerian duct-specific regulatory regions will further define the gene regulatory network for Müllerian duct regression during male differentiation.
Materials and methods
Mice
Swiss outbred mice were purchased from Taconic Biosciences. B6SJLF1/J and Tg(Sp7-tTA,tetO-EGFP/cre)1Amc (Osx-Cre) transgenic mice were obtained from the Jackson Laboratory. Tg(Sp7/mCherry)2Pmay/J (Osx-Cherry) transgenic mice were produced using a CD1 outbred stock (Strecker et al., 2013). Osx-Cre and Osx-Cherry mice were subsequently outcrossed to Swiss Webster mice (Taconic Biosciences). Amhr2ΔE1-6 (Amhr2-ΔE1-6; Mishina et al., 1996), Amhr2tm2Bhr (Amhr2-lacZ; Arango et al., 2008), Gt(ROSA)26Sortm1(lacZ)Sor (R26R-lacZ; Soriano 1999), and Mt1-AMH (Behringer et al., 1990) mice were maintained on a predominantly C57BL/6J genetic background. All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Texas MD Anderson Cancer Center. Studies were performed consistent with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Generation of Amhr2 BAC transgenic mice
High density BAC clone arrays on filters from the RPCI-22 129/SvEv female mouse genomic library (BACPAC Genomics, Emeryville, CA) were screened with a32P-labelled probe located ∼7 kb 5′ of Amhr2 exon 1, using a Megaprime DNA Labelling System kit (Amersham) (Mishina et al., 1996). Six positive clones were identified using the manufacturer’s grid key. Purified BAC clone DNA was digested with NotI, subjected to pulse-field agarose gel electrophoresis, and analyzed by Southern blot, using the same probe used in the initial screen. An additional probe within intron 6 of the Amhr2 locus was used (Mishina et al., 1996). This analysis suggested that all six BAC clones contained the Amhr2 locus. Sequencing of the ends of BAC clone RPCI-22 315D12 delineated the genomic region contained in the clone (NCBI37/mm9: Chr 15, 102,401,136–102,548,427). RPCI-22 315D12 was linearized with I-SceI. The linearized DNA (0.2 ng/ul) was injected into the pronuclei of B6SJLF2/J hybrid zygotes and then transferred into the oviducts of pseudopregnant Swiss surrogate mothers.
Genotyping
Osx-Cre, Osx-Cherry, Amhr2ΔE1-6, Amhr2-lacZ, R26R-lacZ were genotyped as previously described (Mishina et al., 1996; Soriano 1999; Rodda and Mcmahon, 2006; Arango et al., 2008; Strecker et al., 2013). Mt1-AMH transgenic mice were genotyped by PCR using the following primers: hMIS-Fw1: 5′ CCC TAG TGC TGT CTG CCC T 3′ and hMIS-Rv2: 5′ GGA GCT GCT GCC ATT GCT G, resulting in a 176 bp amplified DNA fragment. PCR genotyping for the Amhr2 BAC transgene was performed using the following pBACe3.6 vector primers: BAC-F: 5′ GTG ATA TCG CGG AAG GAA AA 3′ and BAC-R: 5′ AGG ATA TAC GGC AGG CAT TG 3’, resulting in a 499 bp amplified DNA fragment.
X-gal staining
Embryos were stained for lacZ expression as described (Arango et al., 2008).
Histology
Ten µm sections were cut from paraffin embedded lacZ-stained tissues and counterstained with 0.1% Nuclear Fast Red. 10 µm frozen sections were cut from OCT embedded Osx-Cherry embryos and stained with DAPI (4′,6-diamidino-2-phenylindole).
Fluorescent microscopy
Embryos were dissected in phosphate-buffered saline and visualized using a Leica MZ10F fluorescent dissecting microscope for Cherry fluorescence. Images were captured using a JENOPTIK GRYPHAX camera. Frozen sections of Osx-Cherry embryos were visualized for Cherry and DAPI fluorescence using an A1 Nikon confocal microscope.
Results
Rescue of Müllerian duct regression in Amhr2-null males by an Amhr2 BAC transgene
The lack of cell type-specific regulatory sequences in the 500-bp 5′ region of Amhr2 (Kimura et al., 2017) motivated us to screen a larger region surrounding the locus. BAC RPCI-22 315D12 has a ∼147 kb (147,291 bp) region of mouse chromosome 15 from a 129S6/SvEvTac inbred mouse, containing the Amhr2 locus (Figure 1). There is ∼35 kb of sequence 5′ and ∼94 kb 3′ of the Amhr2 locus. The BAC clone contains 7 other genes (Figure 1). We hypothesized that Amhr2 Müllerian duct mesenchyme-specific regulatory sequences were located within this BAC clone. To test this idea, we generated transgenic mice with this unmanipulated Amhr2-containing BAC clone. Two independent transgenic mouse lines (TgBAC-Amhr2) were established.
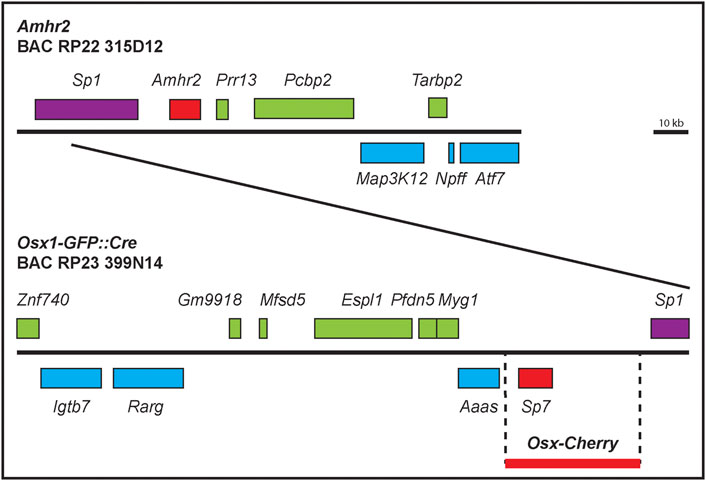
FIGURE 1. Amhr2-and Osx-containing BAC clone maps. Diagram of the genomic regions contained within BAC clones RPCI-22 315D12 and RP23-399N14. Genes (introns and exons) are represented by boxes above or below the line, representing the location of the genes on the two strands of DNA. The ∼39 kb region used to create Osx-Cherry mice is also shown (thick red line) that lacks coding sequences from neighboring genes.
We next bred male mice from the two Amhr2 BAC transgenic lines to female mice heterozygous for an Amhr2-lacZ allele that is also a null allele (Arango et al., 2008). The Amhr2-lacZ allele encodes a convenient visual marker of the uterus and oviduct because lacZ is expressed in the myometrium and myosalpinx, respectively (Arango et al., 2008). From the progeny of that cross, TgBAC-Amhr2; Amhr2lacZ/+ male mice were identified and then bred to female mice homozygous for an Amhr2 deletion allele that is also a null allele (ΔE1-6; Mishina et al., 1996) to generate TgBAC-Amhr2 transgenic males on an Amhr2 compound heterozygous null (ΔE1-6/lacZ) genetic background. Amhr2ΔE1-6/lacZ males served as controls for persistent Müllerian duct derivatives, exploiting the lacZ allele to label Müllerian duct derivatives, i. e. uterus and oviducts (Mishina et al., 1996; Arango et al., 2008). At postnatal day 0 (P0) there was complete regression of the Müllerian system in control males (Figure 2A). Positive lacZ staining was detected in the persistent uterus/oviduct of an Amhr2lacZ/ΔE1-6 male (Figure 2B). We found that TgBAC-Amhr2; Amhr2ΔE1-6/lacZ males were negative for lacZ staining because they lacked a uterus and oviduct (Figure 2C). The complete rescue of the Amhr2 mutant phenotype was observed for both Amhr2 BAC transgenic lines (Line 1, n = 2; Line 2, n = 2). This suggests that the ∼147 kb Amhr2-containing BAC clone contains all of the required regulatory regions for Müllerian duct mesenchyme expression to mediate AMH signaling for regression.
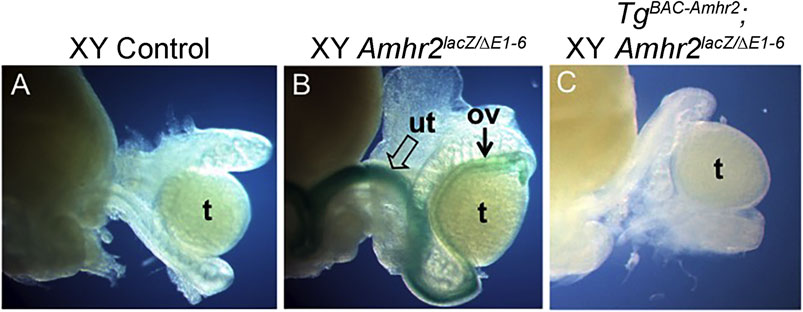
FIGURE 2. An Amhr2-containing BAC transgene rescues the block in Müllerian duct regression of Amhr2-null male mice. (A) Control male mouse possesses only male reproductive tract organs (epididymis, vas deferens) seen adjacent to the testis. (B) Amhr2ΔE1-6/lacZ (null) male mouse carry one deletion allele and one lacZ knock-in/knockout allele. lacZ expression marks the uterus (open arrow) and oviduct (arrow) that forms in these mutants. (C) Male mouse with the Amhr2 BAC transgene (TgBAC-Amhr2) on an Amhr2ΔE1-6/lacZ background lacks lacZ expression because the block in Müllerian duct regression has been rescued preventing uterus/oviduct differentiation (Line 1, n = 2; Line 2, n = 2). Ov, oviduct; t, testis; ut, uterus.
An Osx-Cre BAC transgene activates Cre reporter expression in the Müllerian duct mesenchyme of male embryos
We next used a different BAC transgenic mouse approach to screen for Müllerian duct mesenchyme-specific regulatory regions for Osx. BAC RP23-399N14 has a ∼204 kb (204,096 bp) portion of mouse chromosome 15 from a female C57BL/6J mouse containing the Osx locus (Figure 1). There is ∼145 kb of sequence 5′ and ∼50 kb 3′ of the Osx locus. The BAC clone contains 10 other genes (Figure 1). Like our strategy to identify a Müllerian duct mesenchyme-specific regulatory regions for Amhr2, we tested the idea that an Osx Müllerian duct mesenchyme-specific regulatory sequences were located within BAC clone RP23-399N14. Transgenic mice carrying this Osx-containing BAC have been previously generated (Rodda and Mcmahon, 2006). The Osx-containing BAC was previously modified by recombineering to insert a Tet-off regulatable GFP:Cre fusion protein under the control of the Osx promoter (Osx-Cre). In the absence of doxycycline, Cre expressed from the Osx locus mediates recombination of floxed alleles in mice. The Osx-Cre BAC transgene was found to be active in the osteoblast lineage throughout embryonic and early postnatal development (Rodda and Mcmahon, 2006).
We used the R26R-lacZ Cre reporter mouse (Soriano 1999) to screen for Osx-Cre expression in the developing Müllerian ducts. Timed matings were established and E13.5 to 16.5 embryos were dissected and stained with X-gal. X-gal staining was detected initially at E14.5 in the developing Müllerian ducts of male embryos (n = 6) (Figure 3). Although Osx is expressed throughout the male Müllerian duct mesenchyme (Mullen et al., 2018), we observed strong lacZ expression in the anterior Müllerian duct but reduced lacZ expression in the posterior Müllerian duct (Figure 3B). X-gal staining was also observed at E15.5 in the male Müllerian duct, in the region adjacent to the testes (n = 4) (data not shown). A very small population of lacZ-expressing cells was observed in the Müllerian ducts of female embryos (Figures 3A,C). Histological sections demonstrated that X-gal staining in male embryos was restricted to the Müllerian duct mesenchyme (Figure 3D). This suggests that the ∼204 kb Osx-containing BAC contains regulatory information for male-specific Müllerian duct mesenchyme expression.
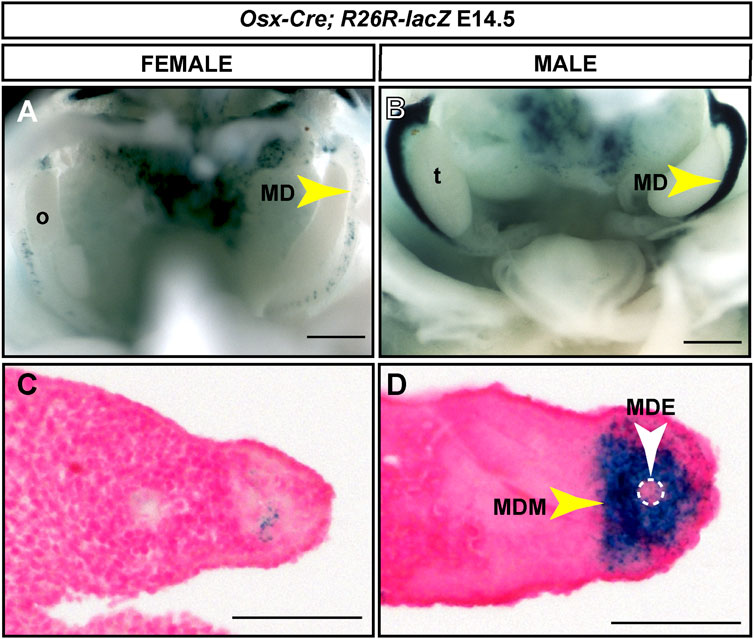
FIGURE 3. Male-specific Müllerian duct mesenchyme expression induced by an Osx-Cre transgene. E14.5 reproductive tract organs from Osx-Cre tg/0; R26R-lacZ/+ mouse embryos stained for lacZ expression. (A and B) Whole mount images for female (A) and male (B) embryos. Scale bar = 500um. (C and D) Cross sections through Müllerian ducts counterstained with Nuclear Fast Red. Scale bar = 50 um. t, testis; o, ovary; MD, Müllerian duct; MDE, Müllerian duct epithelium; MDM, Müllerian duct mesenchyme. Osx-Cre tg/0; R26R-lacZ/+ male embryos, n = 6.
A 39 kb region surrounding Osx directs male-specific Müllerian duct mesenchyme cherry fluorescent reporter expression in transgenic mice
Transgenic mice have previously been generated using ∼39 kb of genomic sequence containing the Osx gene derived from BAC clone RP24-362M3 (C57BL/6J) (Strecker et al., 2013). This ∼39 kb region excluded other gene coding sequences and encompassed ∼20% of the BAC cloned used to generate Osx-Cre transgenic mice. A Cherry Fluorescent Protein reporter was inserted just upstream of the second translational start site of Osx. Cherry fluorescence was detected in developing and adult skeletal tissues (Strecker et al., 2013). To determine if the ∼39 kb region also contained Müllerian duct mesenchyme regulatory sequences, we examined E14.5 male and female embryos for Cherry fluorescence in the Müllerian ducts (Figure 4). At E14.5, we initially identified Osx-Cherry transgenic embryos because they express Cherry fluorescence in the developing skeleton. Male and female Osx-Cherry transgenic embryos were identified by brightfield assessment of the gonads. Although Cherry fluorescence was observed in the developing skeleton tissues of female Osx-Cherry transgenic embryos, no Cherry fluorescence was detected in the Müllerian ducts (n = 4) (Figures 4A,C). Likewise, Cherry fluorescence was detected in the developing skeleton tissues of male Osx-Cherry transgenic embryos (Figure 4B). In addition, Cherry fluorescence was detected in the Müllerian ducts (n = 6) (Figures 4B,D). Histological sections demonstrated that Cherry fluorescence was present in the male but not female Müllerian duct mesenchyme (Figures 4E,F). We also examined AMH-responsive organs, including testes, ovaries and uteri of adult Osx-Cherry transgenic and control mice but no Cherry fluorescence was detected compared to controls (Supplemental Figure S1). These results suggest that the ∼39 kb genomic region included in the Osx-Cherry transgene contains a male-specific, Müllerian duct mesenchyme regulatory sequences that direct Osx transcription.
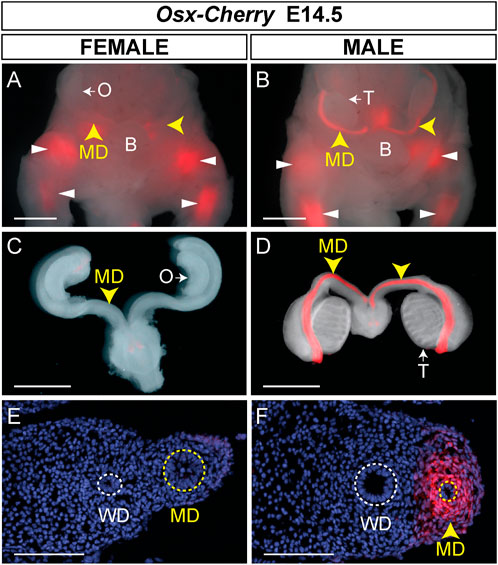
FIGURE 4. Male-specific Müllerian duct mesenchyme expression in Osx-Cherry transgenic mice. (A–D) E14.5 reproductive tract organs from Osx-Cherry tg/0 mouse embryos visualized for Cherry fluorescence. (A and B) Ventral views of whole mount brightfield and fluorescent images merged for female (A) and male (B) embryos. Yellow arrowheads, Müllerian ducts (MD); white arrowheads, skeletal tissues. O, ovary; T, testis. (C and D) Brightfield and fluorescent images merged of isolated reproductive tracts at E14.5. (C) Female, (D) male. (E and F) Cross sections through Müllerian ducts, visualized for Cherry fluorescence and counterstained with DAPI. (E) Female, (F) male. WD, Wolffian ducts. Scale bars (A–D) = 1000 um; (E and F) = 100 um. Osx-Cherry tg/0 female embryos, n = 4; male embryos, n = 6.
Previously, we showed that an Osx-lacZ knock-in allele was expressed in male but not female Müllerian duct mesenchyme, reflecting the sex-specific pattern of Osx expression in the Müllerian ducts (Mullen et al., 2018). We also showed that ectopic expression of human AMH from an Mt1-AMH transgene could activate the Osx-lacZ knock-in allele in female embryos, indicating that Osx is an AMH-induced gene (Mullen et al., 2018). To determine if the Osx-Cherry transgene could also respond to ectopic human AMH in female embryos, we crossed Osx-Cherry females to Mt1-AMH males to generate E14.5 Mt1-AMH; Osx-Cherry double transgenic female embryos (Figure 5). We observed Cherry fluorescence in the Müllerian ducts of the Mt1-AMH; Osx-Cherry double transgenic female embryos but not in Osx-Cherry transgenic female embryos (n = 5) (Figures 5A,B). The Cherry fluorescence in the Müllerian ducts of the Mt1-AMH; Osx-Cherry double transgenic female embryos was localized to the mesenchyme (Figure 5D). These results demonstrate that the 39 kb Osx region contains a Müllerian duct mesenchyme regulatory sequences that are responsive to AMH signaling.
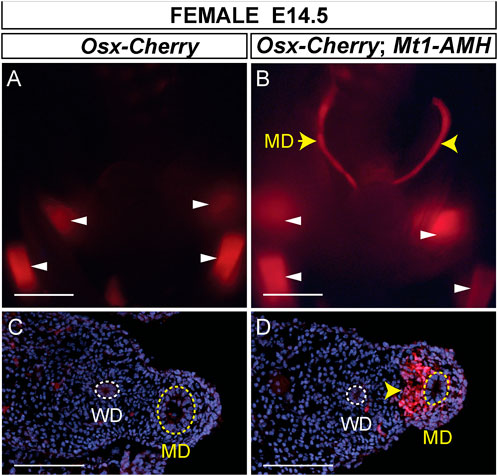
FIGURE 5. Mt1-AMH induces Osx-Cherry in the Müllerian ducts of female embryos. (A and B) Whole mount images of E14.5 reproductive tract organs from female mouse embryos visualized for Cherry fluorescence. (A) Osx-Cherry tg/0 (B) Osx-Cherry tg/0; Mt1-AMH tg/0 female embryos. White arrowheads point to Cherry-expressing bone-forming tissues. Yellow arrowheads point to Cherry expression in the Müllerian ducts. (C and D) Cross sections through Müllerian ducts, visualized for Cherry fluorescence and counterstained with DAPI. (C) Osx-Cherry tg/0 (D) Osx-Cherry tg/0; Mt1-AMH tg/0 female embryos. Arrowhead in D points to Cherry-expressing Müllerian duct mesenchyme. MD, Müllerian duct (yellow dotted line); WD, Wolffian duct (white dotted line). Scale bars (A and B) = 1000 um; (C and D) = 100 um. Cherry tg/0; Mt1-AMH tg/0 female embryos, n = 5.
Discussion
AMH is secreted by Sertoli cells of the fetal testes and subsequently interacts with AMHR2 expressed by the Müllerian duct mesenchyme to induce the elimination of the ductal epithelium, blocking the formation of female reproductive organs in males (Mullen and Behringer, 2014). Previously, we described a gene regulatory network (GRN) for AMH-induced Müllerian duct regression primarily based on in vivo genetic data (Moses and Behringer, 2019). Multiple cis regulatory elements have been identified in the 5′ region of the Amh gene with binding sites for SOX9, NR5A1, and GATA4 that are required for testis-specific transcription (Shen et al., 1994; De Santa Barbara et al., 1998; Arango et al., 1999; Lasala et al., 2004; Bouchard, et al., 2019). Amhr2 and Osx are expressed in the Müllerian duct mesenchyme for Müllerian duct regression during male differentiation (Baarends et al., 1994; Mishina et al., 1996; Mullen et al., 2018). Although cis regulatory elements have been identified for fetal Sertoli cell-specific expression of Amh, no transcriptional enhancers have been identified in the rest of this GRN, notably for genes expressed in the Müllerian duct epithelium or mesenchyme. In this study, we used BAC transgenic mouse approaches to identify genomic regions that contain Amhr2 and Osx sequences that direct Müllerian duct mesenchyme expression.
Localization of a Müllerian duct mesenchyme regulatory regions for Amhr2
We found that transgenic mice generated with a mouse BAC clone containing the entire Amhr2 locus rescued the persistent Müllerian duct-derived tissues found in Amhr2-null males (Mishina et al., 1996; Arango et al., 2008). This suggests that the BAC clone contains regulatory sequences sufficient for Müllerian duct mesenchyme expression. It is also possible that Amhr2 expression from the BAC clone was expressed ubiquitously. However, this could lead to engagement of Type I receptors for AMH signaling in ectopic tissues that might lead to mutant phenotypes. The Amhr2 BAC transgenic mice in both lines were normal and fertile.
We also tested the 6.2 kb region 5′ of Amhr2 including the endogenous promoter for tissue-specific activity using a lacZ reporter. However, in 9 independent lines of transgenic mice there was no lacZ expression in E14.5 Müllerian ducts of both sexes, adult testes, ovary and uterus (unpublished observations). Thus, the immediate upstream region of Amhr2 is not sufficient for cell type-specific transcription (Kimura et al., 2017). While our current results are a step forwards, the BAC clone is ∼147 kb in length, creating a challenge to localize the Müllerian duct mesenchyme regulatory sequence to a discreet location.
Amhr2 transcription in the Müllerian duct mesenchyme is induced by WNT7a secreted by the Müllerian duct epithelium (Parr and McMahon, 1998). Wnt7a-null males do not express Amhr2 in the Müllerian duct mesenchyme, leading to a block in regression, resulting in the formation of a uterus (Parr and McMahon, 1998). Thus, WNT signaling lies upstream of Amhr2 transcription in the GRN (Moses and Behringer, 2019). In addition, beta-catenin is required in the ductal mesenchyme for Müllerian duct regression during male differentiation (Kobayashi et al., 2011). These observations suggest that there may be cis elements related to WNT signaling that are required for Amhr2 transcription.
Amhr2 is also expressed in Sertoli, granulosa cells and smooth muscle of the uterine myometrium (Arango et al., 2008). Amhr2 expression in these cell types have been shown to be regulated by multiple factors. A binding site for NR5A1 (also known as SF-1) located within ∼300 bp upstream of the transcription start site of the human AMHR2 gene was shown to be required for reporter expression in the NT2/D1 teratocarcinoma cell line (De Santa Barbara et al., 1998). Knockdown of Dmrt1 in male chick embryos caused a down-regulation of Amhr2 (Cutting et al., 2014). BMP4 and BMP15 were shown to enhance AMHR2 transcript levels in human and sheep ovarian granulosa cells in vitro (Pierre et al., 2016). This response to BMP signaling was mediated by a ∼2.2 kb region 5′ of the AMHR2 promoter (Pierre et al., 2016). A long non-coding (lnc) RNA, lnc-Amhr2 has been identified that enhanced Amhr2 promoter activity in a mouse ovarian granulosa cell line, OV3121 (Kimura et al., 2017). The factors that regulate Amhr2 transcription in Müllerian duct mesenchyme cells remain to be discovered.
Localization of a Müllerian duct mesenchyme regulatory region for Osx that is responsive to AMH signaling
The BAC used to generate the Osx-Cre line spans 204 kb of chromosome 15, from the 3′ end of Znf40 to the 5′ end of Sp1. Using the Osx-Cre BAC transgenic mouse line, we discovered male-specific Cre reporter gene expression in the Müllerian duct mesenchyme. Although the endogenous Osx gene is expressed throughout the entire male Müllerian duct mesenchyme (Mullen et al., 2018), we found that Osx-Cre directed reporter gene expression predominantly in the anterior Müllerian duct. It is possible that there are separate cis elements that direct anterior and posterior expression. However, these findings may be unique to this one transgenic mouse line.
The Osx-Cherry construct encompasses 39 kb within the 204 kb Osx-Cre BAC region, spanning Osx and intergenic sequences on both the 5′ and 3’ ends of the gene. Using the Osx-Cherry transgenic mouse line, we found male-specific Cherry fluorescence along the entire length of the Müllerian ducts, indicating that the 39 kb region contains a Müllerian duct mesenchyme-specific regulatory sequences. AMH signaling is necessary and sufficient for Osx expression in the Müllerian duct mesenchyme (Mullen et al., 2018). We found that ectopic AMH provided by an Mt1-AMH transgene could induce Osx-Cherry expression in the Müllerian duct mesenchyme of female embryos. This suggests that the 39 kb Osx region contains sequences that respond to AMH signaling. The sequences that direct Müllerian duct mesenchyme-specific transcription and those that mediate AMH signaling may be the same but await further study.
Two distinct Müllerian duct mesenchyme regulatory regions
In the mouse genome, Osx and Amhr2 are linked, residing on chromosome 15 within 100 kb of each other. There is one gene, namely Sp1, located between Osx and Amhr2. Sp1 encodes a transcription factor that is expressed in most tissues (O’connor et al., 2016). Interestingly, the 39 kb Osx-Cherry region (NCBI37/mm9: Chr 15, 102,182,823–102,220,102) does not overlap with the Amhr2 BAC clone (NCBI37/mm9: Chr 15, 102,401,136–102,548,427). Therefore, the Müllerian duct mesenchyme regulatory region we identified for Osx-Cherry is distinct from the one identified within the Amhr2 BAC. The identification of two distinct Müllerian duct mesenchyme regulatory regions, one for Amhr2 and one for Osx, seems reasonable because Amhr2 is expressed in the mesenchyme of both male and female Müllerian ducts that is dependent on epithelial-derived WNT7A, whereas Osx expression is restricted to male Müllerian ducts that depends on Amhr2 and AMH secreted by the fetal testes (Baarends et al., 1994; Parr and McMahon, 1998; Arango et al., 2008; Mullen et al., 2018). Thus, Amhr2 is upstream of Osx in the GRN. Analysis of the chromatin landscape of E14.5 male and female Müllerian duct mesenchyme could point to Müllerian duct-specific regulatory elements. However, these types of studies have not been reported perhaps because there are very few cells per embryo and single cell approaches still require cell numbers greater than practically available. The identification of genomic regions that contain regulatory sequences for Müllerian duct mesenchyme expression contributes to the definition of this GRN of sexual development (Moses and Behringer, 2019).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of the University of Texas MD Anderson Cancer Center.
Author contributions
MM, SJ, and RB conceived the study and designed experiments. MM, SJ, RM, and DI performed experiments and analyzed data. PM provided transgenic mice and guidance. MM and RB wrote the paper with input from all authors.
Funding
This research was supported by National Institutes of Health (NIH) grant HD030284 and the Ben F. Love Endowment to RB, Agence Nationale de la Recherche (ANR) grant ANR-08-JCJC-0059 to SJ, and NIH grant AR60899 to PM. MM was supported by NIH F31HD106600. DI was supported by NIH R25CA240137. Confocal microscopy was supported by NIH shared instrument grant OD024976. Veterinary resources and DNA sequencing were supported by NIH Grant CA16672.
Acknowledgments
We thank members of our labs for helpful comments on the manuscript. We thank George Adebayo for zygote injections and Adriana Paulucci-Holthauzen for confocal microscope training.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.1006087/full#supplementary-material
References
Arango, N. A., Kobayashi, A., Wang, Y., Jamin, S. P., Lee, H. H., Orvis, G. D., et al. (2008). A mesenchymal perspective of Mullerian duct differentiation and regression in Amhr2-lacZ mice. Mol. Reprod. Dev. 75, 1154–1162. doi:10.1002/mrd.20858
Arango, N. A., Lovell-Badge, R., and Behringer, R. R. (1999). Targeted mutagenesis of the endogenous mouse mis gene promoter: In vivo definition of genetic pathways of vertebrate sexual development. Cell 99, 409–419. doi:10.1016/s0092-8674(00)81527-5
Baarends, W. M., Van Helmond, M. J., Post, M., Van Der Schoot, P. J., Hoogerbrugge, J. W., De Winter, J. P., et al. (1994). A novel member of the transmembrane serine/threonine kinase receptor family is specifically expressed in the gonads and in mesenchymal cells adjacent to the mullerian duct. Development 120, 189–197. doi:10.1242/dev.120.1.189
Bae, J. M., Clarke, J. C., Rashid, H., Adhami, M. D., Mccullough, K., Scott, J. S., et al. (2018). Specificity protein 7 is required for proliferation and differentiation of ameloblasts and odontoblasts. J. Bone Min. Res. 33, 1126–1140. doi:10.1002/jbmr.3401
Behringer, R. R., Cate, R. L., Froelick, G. J., Palmiter, R. D., and Brinster, R. L. (1990). Abnormal sexual development in transgenic mice chronically expressing mullerian inhibiting substance. Nature 345, 167–170. doi:10.1038/345167a0
Behringer, R. R., Finegold, M. J., and Cate, R. L. (1994). Mullerian-inhibiting substance function during mammalian sexual development. Cell 79, 415–425. doi:10.1016/0092-8674(94)90251-8
Behringer, R. R., Hammer, R. E., Brinster, R. L., Palmiter, R. D., and Townes, T. M. (1987). Two 3' sequences direct adult erythroid-specific expression of human beta-globin genes in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 84, 7056–7060. doi:10.1073/pnas.84.20.7056
Belville, C., Jamin, S. P., Picard, J. Y., Josso, N., and Di Clemente, N. (2005). Role of type I receptors for anti-Mullerian hormone in the SMAT-1 Sertoli cell line. Oncogene 24, 4984–4992. doi:10.1038/sj.onc.1208686
Bouchard, M. F., Bergeron, F., Grenier Delaney, J., Harvey, L. M., and Viger, R. S. (2019). In vivo ablation of the conserved GATA-binding motif in the Amh promoter impairs Amh expression in the male mouse. Endocrinology 160, 817–826. doi:10.1210/en.2019-00047
Cate, R. L., Mattaliano, R. J., Hession, C., Tizard, R., Farber, N. M., Cheung, A., et al. (1986). Isolation of the bovine and human genes for Mullerian inhibiting substance and expression of the human gene in animal cells. Cell 45, 685–698. doi:10.1016/0092-8674(86)90783-x
Clarke, T. R., Hoshiya, Y., Yi, S. E., Liu, X., Lyons, K. M., and Donahoe, P. K. (2001). Mullerian inhibiting substance signaling uses a bone morphogenetic protein (BMP)-like pathway mediated by ALK2 and induces SMAD6 expression. Mol. Endocrinol. 15, 946–959. doi:10.1210/mend.15.6.0664
Cutting, A. D., Ayers, K., Davidson, N., Oshlack, A., Doran, T., Sinclair, A. H., et al. (2014). Identification, expression, and regulation of anti-Mullerian hormone type-II receptor in the embryonic chicken gonad. Biol. Reprod. 90, 106. doi:10.1095/biolreprod.113.116491
De Santa Barbara, P., Bonneaud, N., Boizet, B., Desclozeaux, M., Moniot, B., Sudbeck, P., et al. (1998). Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Mullerian hormone gene. Mol. Cell. Biol. 18, 6653–6665. doi:10.1128/mcb.18.11.6653
Field, A., and Adelman, K. (2020). Evaluating enhancer function and transcription. Annu. Rev. Biochem. 89, 213–234. doi:10.1146/annurev-biochem-011420-095916
Gouedard, L., Chen, Y. G., Thevenet, L., Racine, C., Borie, S., Lamarre, I., et al. (2000). Engagement of bone morphogenetic protein type IB receptor and Smad1 signaling by anti-Mullerian hormone and its type II receptor. J. Biol. Chem. 275, 27973–27978. doi:10.1074/jbc.M002704200
Josso, N., Cate, R. L., Picard, J. Y., Vigier, B., Di Clemente, N., Wilson, C., et al. (1993). Anti-mullerian hormone: The jost factor. Recent Prog. Horm. Res. 48, 1–59. doi:10.1016/b978-0-12-571148-7.50005-1
Kimura, A. P., Yoneda, R., Kurihara, M., Mayama, S., and Matsubara, S. (2017). A long noncoding RNA, lncRNA-amhr2, plays a role in Amhr2 gene activation in mouse ovarian granulosa cells. Endocrinology 158, 4105–4121. doi:10.1210/en.2017-00619
Kobayashi, A., Stewart, A. C., Wang, Y., Fujioka, K., Thomas, N. C., and Jamin, S. P. (2011). β-Catenin is essential for Mullerian duct regression during male sexual differentiation. Development 138 (10), 1967–1975. doi:10.1242/dev.056143
Lasala, C., Carre-Eusebe, D., Picard, J. Y., and Rey, R. (2004). Subcellular and molecular mechanisms regulating anti-Mullerian hormone gene expression in mammalian and nonmammalian species. DNA Cell Biol. 23, 572–585. doi:10.1089/dna.2004.23.572
Lettice, L. A., Heaney, S. J., Purdie, L. A., Li, L., De Beer, P., Oostra, B. A., et al. (2003). A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12, 1725–1735. doi:10.1093/hmg/ddg180
Mishina, Y., Rey, R., Finegold, M. J., Matzuk, M. M., Josso, N., Cate, R. L., et al. (1996). Genetic analysis of the Mullerian-inhibiting substance signal transduction pathway in mammalian sexual differentiation. Genes Dev. 10, 2577–2587. doi:10.1101/gad.10.20.2577
Mishina, Y., Whitworth, D. J., Racine, C., and Behringer, R. R. (1999). High specificity of Mullerian-inhibiting substance signaling in vivo. Endocrinology 140, 2084–2088. doi:10.1210/endo.140.5.6705
Moses, M. M., and Behringer, R. R. (2019). A gene regulatory network for Mullerian duct regression. Environ. Epigenet. 5, dvz017. doi:10.1093/eep/dvz017
Mukhopadhyay, K., Lefebvre, V., Zhou, G., Garofalo, S., Kimura, J. H., and De Crombrugghe, B. (1995). Use of a new rat chondrosarcoma cell line to delineate a 119-base pair chondrocyte-specific enhancer element and to define active promoter segments in the mouse pro-alpha 1(II) collagen gene. J. Biol. Chem. 270, 27711–27719. doi:10.1074/jbc.270.46.27711
Mullen, R. D., and Behringer, R. R. (2014). Molecular genetics of Mullerian duct formation, regression and differentiation. Sex. Dev. 8, 281–296. doi:10.1159/000364935
Mullen, R. D., Wang, Y., Liu, B., Moore, E. L., and Behringer, R. R. (2018). Osterix functions downstream of anti-Mullerian hormone signaling to regulate Mullerian duct regression. Proc. Natl. Acad. Sci. U. S. A. 115, 8382–8387. doi:10.1073/pnas.1721793115
Nakashima, K., Zhou, X., Kunkel, G., Zhang, Z., Deng, J. M., Behringer, R. R., et al. (2002). The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108, 17–29. doi:10.1016/s0092-8674(01)00622-5
O'connor, L., Gilmour, J., and Bonifer, C. (2016). The role of the ubiquitously expressed transcription factor Sp1 in tissue-specific transcriptional regulation and in disease. Yale J. Biol. Med. 89, 513–525.
Ornitz, D. M., Palmiter, R. D., Hammer, R. E., Brinster, R. L., Swift, G. H., and Macdonald, R. J. (1985). Specific expression of an elastase-human growth hormone fusion gene in pancreatic acinar cells of transgenic mice. Nature 313, 600–602. doi:10.1038/313600a0
Orvis, G. D., and Behringer, R. R. (2007). Cellular mechanisms of Mullerian duct formation in the mouse. Dev. Biol. 306, 493–504. doi:10.1016/j.ydbio.2007.03.027
Orvis, G. D., Jamin, S. P., Kwan, K. M., Mishina, Y., Kaartinen, V. M., Huang, S., et al. (2008). Functional redundancy of TGF-beta family type I receptors and receptor-Smads in mediating anti-Mullerian hormone-induced Mullerian duct regression in the mouse. Biol. Reprod. 78, 994–1001. doi:10.1095/biolreprod.107.066605
Parr, B. A., and McMahon, A. P. (1998). Sexually dimorphic development of the mammalian reproductive tract requires Wnt-7a. Nature 395 (6703), 707–710. doi:10.1038/27221
Picard, J. Y., Benarous, R., Guerrier, D., Josso, N., and Kahn, A. (1986). Cloning and expression of cDNA for anti-mullerian hormone. Proc. Natl. Acad. Sci. U. S. A. 83, 5464–5468. doi:10.1073/pnas.83.15.5464
Pierre, A., Estienne, A., Racine, C., Picard, J. Y., Fanchin, R., Lahoz, B., et al. (2016). The bone morphogenetic protein 15 up-regulates the anti-mullerian hormone receptor expression in granulosa cells. J. Clin. Endocrinol. Metab. 101, 2602–2611. doi:10.1210/jc.2015-4066
Rodda, S. J., and Mcmahon, A. P. (2006). Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133, 3231–3244. doi:10.1242/dev.02480
Shen, W. H., Moore, C. C., Ikeda, Y., Parker, K. L., and Ingraham, H. A. (1994). Nuclear receptor steroidogenic factor 1 regulates the mullerian inhibiting substance gene: A link to the sex determination cascade. Cell 77, 651–661. doi:10.1016/0092-8674(94)90050-7
Soriano, P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71. doi:10.1038/5007
Strecker, S., Fu, Y., Liu, Y., and Maye, P. (2013). Generation and characterization of Osterix-Cherry reporter mice. Genesis 51, 246–258. doi:10.1002/dvg.22360
Visser, J. A., Olaso, R., Verhoef-Post, M., Kramer, P., Themmen, A. P., and Ingraham, H. A. (2001). The serine/threonine transmembrane receptor ALK2 mediates Mullerian inhibiting substance signaling. Mol. Endocrinol. 15, 936–945. doi:10.1210/mend.15.6.0645
Vu, H., and Ernst, J. (2022). Universal annotation of the human genome through integration of over a thousand epigenomic datasets. Genome Biol. 23, 9. doi:10.1186/s13059-021-02572-z
Zhan, Y., Fujino, A., Maclaughlin, D. T., Manganaro, T. F., Szotek, P. P., Arango, N. A., et al. (2006). Mullerian inhibiting substance regulates its receptor/SMAD signaling and causes mesenchymal transition of the coelomic epithelial cells early in Mullerian duct regression. Development 133, 2359–2369. doi:10.1242/dev.02383
Keywords: osterix, osx-cre, Sp7/osterix, anti-müllerian hormone receptor 2, transgenic mice, sex diffentiation, bacterial artificial chromosome (BAC)
Citation: Moses MM, Mullen RD, Idowu DI, Maye P, Jamin SP and Behringer RR (2022) A transgenic bacterial artificial chromosome approach to identify regulatory regions that direct Amhr2 and Osterix expression in Müllerian duct mesenchyme. Front. Cell Dev. Biol. 10:1006087. doi: 10.3389/fcell.2022.1006087
Received: 28 July 2022; Accepted: 21 September 2022;
Published: 12 October 2022.
Edited by:
Fei Gao, Institute of Zoology (CAS), ChinaReviewed by:
Yi Athena Ren, Cornell University, United StatesIsabelle Stévant, Bar-Ilan University, Israel
Blanche Capel, Duke University Medical Center, United States
Nitzan Gonen, Bar-Ilan University, Israel
Francis Poulat, Institut National de la Santé et de la Recherche Médicale (INSERM), France
Copyright © 2022 Moses, Mullen, Idowu, Maye, Jamin and Behringer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soazik P. Jamin, soazik.jamin@inserm.fr; Richard R. Behringer, rrb@mdanderson.org
†These authors have contributed equally to this work and share senior authorship
 Malcolm M. Moses
Malcolm M. Moses Rachel D. Mullen
Rachel D. Mullen Daniel I. Idowu
Daniel I. Idowu Peter Maye
Peter Maye Soazik P. Jamin
Soazik P. Jamin Richard R. Behringer
Richard R. Behringer