Profiling of Sexually Dimorphic Genes in Neural Cells to Identify Eif2s3y, Whose Overexpression Causes Autism-Like Behaviors in Male Mice
- 1Fujian Provincial Key Laboratory of Neurodegenerative Disease and Aging Research, Institute for Neuroscience, School of Medicine, Xiamen University, Xiamen, China
- 2Emergency Department, Xiang’an Hospital, Xiamen University, Xiamen, China
- 3Institute of Chemistry, University of Vienna, Vienna, Austria
- 4Xiamen Key Laboratory of Brain Center, The First Affiliated Hospital of Xiamen University, Xiamen, China
Many neurological disorders exhibit sex differences and sex-specific therapeutic responses. Unfortunately, significant amounts of studies investigating molecular and cellular mechanisms underlying these neurological disorders use primary cell cultures with undetermined sexes; and this may be a source for contradictory results among different studies and impair the validity of study conclusion. Herein, we comprehensively compared sexual dimorphism of gene expression in primary neurons, astrocytes, and microglia derived from neonatal mouse brains. We found that overall sexually dimorphic gene numbers were relatively low in these primary cells, with microglia possessing the most (264 genes), neurons possessing the medium (69 genes), and astrocytes possessing the least (30 genes). KEGG analysis indicated that sexually dimorphic genes in these three cell types were strongly enriched for the immune system and immune-related diseases. Furthermore, we identified that sexually dimorphic genes shared by these primary cells dominantly located on the Y chromosome, including Ddx3y, Eif2s3y, Kdm5d, and Uty. Finally, we demonstrated that overexpression of Eif2s3y increased synaptic transmission specifically in male neurons and caused autism-like behaviors specifically in male mice. Together, our results demonstrate that the sex of primary cells should be considered when these cells are used for studying the molecular mechanism underlying neurological disorders with sex-biased susceptibility, especially those related to immune dysfunction. Moreover, our findings indicate that dysregulation of sexually dimorphic genes on the Y chromosome may also result in autism and possibly other neurological disorders, providing new insights into the genetic driver of sex differences in neurological disorders.
Introduction
Numerous neurological disorders display a prominent sexual dimorphism, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), schizophrenia, autism, and multiple sclerosis (Aleman et al., 2003; Orton et al., 2006; Haaxma et al., 2007; Werling and Geschwind, 2013; Mielke et al., 2014; Maldonado Weng et al., 2019). For example, it is well-known that men are more vulnerable to autism and autism spectrum disorders (ASDs) than women, with a prevalence ratio of 4∼5:1 (Fombonne, 2009; Christensen et al., 2016). Elucidating the mechanistic underpinnings are important not only for understanding the sex-biased susceptibility of these devastating diseases but also for sex-specific treatment purposes.
Sex differences in disease prevalence and manifestation are rooted in the genetic sex differences, which are genetic females XX or genetic males XY in most mammals. The primary testicular/ovarian gonadal phenotype is determined by the sex-determining region of the Y chromosome (SRY) gene during embryonic development. Sex hormones (e.g., androgens and estrogens) and their receptors (e.g., androgen and estrogen receptors) have traditionally been considered the major cause of sex differences in the brain. However, emerging evidence has indicated that sex chromosome effects also contribute to the brain sex differences throughout the lifespan, independent of sex hormones (Arnold, 2017). Sex chromosome aneuploidies (SCAs) are characterized by an abnormal number of X or Y chromosome (Ngun et al., 2011; Hong and Reiss, 2014). Turner Syndrome (45,X), Klinefelter Syndrome (47,XXY), and XYY Syndrome (47,XYY) are the most common SCAs (Heard and Turner, 2011). Clinical characteristics of SCAs often include intellectual disability, motor impairments, and an increased risk of neurological and psychiatric disorders such as ASDs, demonstrating that genes on sex chromosomes are important for maintaining normal brain functions (Rovet et al., 1995; Visootsak and Graham, 2009; Leggett et al., 2010; Tartaglia et al., 2010; Bishop et al., 2011; Hong et al., 2011; Hong and Reiss, 2012, 2014; Printzlau et al., 2017; Green et al., 2019). Indeed, mutations in multiple X-chromosomal genes have been found to cause X-linked intellectual disability and ASDs (Schaafsma and Pfaff, 2014; Fung and Reiss, 2016). Patients carrying more than one Y chromosome also exhibit ASD symptoms (Demily et al., 2017; Tartaglia et al., 2017; Matsuzaki et al., 2019), suggesting that abnormality of Y-chromosomal genes may also result in ASD. However, the underlying mechanism remains largely elusive.
Much of our understanding of cellular and molecular mechanisms underlying neurological disorders are based on research using primary neural cells derived from animals at embryonic or neonatal developmental stages. Unfortunately, the sex of donors of these primary cells is not reported in most published studies. In many cases, the cells used in experiments are a mixture of both sexes. This could potentially jeopardize the conclusion of these studies, especially when genetic sex differences play a significant role in such disorders.
Therefore, a major aim of the present study is to determine whether the transcriptome of primary neurons, astrocytes, and microglia isolated from neonatal mice exhibit any significant sex difference. Herein, we found that overall sexually dimorphic gene numbers were low in these primary cells, and sexually dimorphic genes were enriched for the immune system and immune-related diseases. In addition, sexually dimorphic genes shared by all the three cell types dominantly located on sex chromosomes; and one of them is Eif2s3y.
Eif2s3y, a gene located on the non-recombining region of the mouse Y chromosome, is essential for normal spermatogonia proliferation and differentiation (Mazeyrat et al., 2001; Yamauchi et al., 2014; Matsubara et al., 2015). Eif2s3y is ubiquitously expressed in various tissues (Wolstenholme et al., 2013). In the brain, Eif2s3y is preferentially expressed in specific regions including the hippocampus, thalamus, striatum, and olfactory areas (Xu et al., 2006). Prior to gonadal differentiation (∼E12–E14), Eif2s3y is highly expressed in male mouse brain; and this sexual dimorphism expression can also be observed in the stage of gonadal hormone secretion phase (E18.5) and endures to adulthood (Wolstenholme et al., 2013). However, the function of Eif2s3y in the brain has not yet been reported. In the current study, we demonstrated that overexpression of Eif2s3y altered synaptic transmission specifically in male neurons and led to autism-like behaviors specifically in male mice.
Materials and Methods
Animals
All wild-type C57BL/6J mice were housed under Specific-Pathogen-Free (SPF) conditions in the core animal facility at the Xiamen University. Mice were maintained on an 8:00–20:00 light/dark cycle with ad libitum access to phytoestrogen-free chow and water. Pregnant females were housed separately. All animal procedures were approved by the Animal Ethics Committee of Xiamen University and were conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Antibodies
Anti-Myc (2276S), anti-β-actin (8457S), anti-GluR1 (13185S), anti-NR1 (5704S), anti-PSD-95 (3450S), and anti-Map2 (4542S) antibodies are from Cell Signaling Technology. Anti-GFAP (16825-1-AP), anti-GluR2 (11994-1-AP), anti-NR2A (19953-1-AP), and anti-NR2B (19954-1-AP) antibodies are from Proteintech. Anti-Iba1 (019-19741), anti-GFP (M20004L), anti-NeuN (ab177487), anti-GluR3 (MAB5416), and anti-synaptophysin (S5768) antibodies are from Wako, Abmart, Abcam, Millipore, and Sigma, respectively. HRP-conjugated goat anti-mouse IgG (H + L) secondary antibody (31430), HRP-conjugated goat anti-rabbit IgG (H + L) secondary antibody (31460), Alexa fluor 488-conjugated donkey anti-mouse IgG (H + L) secondary antibody (A-21202), Alexa fluor 488-conjugated goat anti-rabbit IgG (H + L) secondary antibody (A-11008), and Alexa fluor 594-conjugated goat anti-rabbit IgG (H + L) secondary antibody (A-11012) are from Thermo Fisher Scientific.
Primary Neuron, Astrocyte, and Microglia Cultures
Wild-type C57BL/6J mice at postnatal day 0 (P0) were euthanized within 12 h after birth. The brain was dissected and placed in ice-cold Hanks’ Balanced Salt Solution (HBSS; Gibco). After stripping off meninges, half of the cerebrum (including cortex and hippocampus) was used for preparing neuron cultures, and the other half of the cerebrum plus the entire cerebellum was used for preparing astrocyte and microglia cultures.
For neuron cultures, half of the cerebrum was dissociated in HBSS with a pipette and then incubated with an enzymatic digestion solution containing 0.25% trypsin-EDTA (Gibco) and DNase I (0.2 kU/mL, Worthington) at 37° for 15 min. After centrifuging at 1000 rpm for 5 min, cell pellets were re-suspended in neurobasal medium (Gibco) supplemented with 2% B-27 (Gibco), 2 mM L-glutamine (Gibco), and 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco). Cell density was counted using TC20 Automated Cell Counter (Bio-Rad). Primary neurons were seeded in an appropriate poly-L-lysine coated dish, cultured for 12 days in vitro (DIV) in a 37° incubator with 5% CO2, and used for RNA sequencing.
For astrocyte and microglia cultures, brain samples were triturated with a 5 mL pipette and then seeded in a poly-L-lysine coated T75 flask with microglia culture media containing DMEM (Gibco), 10% heat-inactivated fetal bovine serum (FBS; Gibco), and 100 U/mL penicillin and 100 μg/mL streptomycin. After incubating in a 37° incubator with 5% CO2 for 24 h, culture media were replaced with new microglia culture media and supplemented with 25 ng/mL mGM-CSF (R&D Systems). On DIV11, the flask was shaken on a rotating platform at 200 RPM for 20 min. Microglia floated in the media were collected by centrifugation and then re-suspended and cultured separately in microglia culture media for another day before subjected to RNA sequencing. The flask was refilled with culture media and incubated for another 3 days. After shaking off and removing newly proliferated microglia, the flask was refilled with fresh astrocyte culture medium (DMEM + 10% regular FBS + 100 U/mL penicillin + 100 μg/mL streptomycin) to prevent microglia proliferation. 3 days later, unattached cells were shaken off and discarded. Astrocytes were detached from the flask bottom by 0.25% trypsin digestion and then re-suspended and cultured in astrocyte culture media for another day before subjected to RNA sequencing.
Sex Identification
Mouse sex was identified by PCR. Briefly, genomic DNA was isolated from mouse tails using NaOH extraction and subjected to nested PCR using the primer pair: Sex-F (5′-ATGTGTTCCCGTGGTGAGAG-3′) and Sex-R (5′-GGTCGTCCAGTTTTGTTGAG-3′).
RNA Sequencing and Analysis
Cultured primary neurons, astrocytes, and microglia were soaked in the TRIzol reagent (Life Technologies) and transferred to Beijing Genomics Institute. RNA extraction, cDNA library preparation, and RNA sequencing using the BGISEQ-500 platform were conducted by Beijing Genomics Institute with standard protocols. Generated raw sequencing reads were filtered by using SOAPnuke1 to remove reads with adaptors, reads with more than 5% of unknown bases, and low-quality reads. Clean reads were aligned to Mus musculus reference genes (reference genome version: GCF_000001635.25_GRCm38.p5) using Bowtie2 (version 2.2.5) (Langmead and Salzberg, 2012). Gene expression levels were calculated and normalized to fragments per kilobase of transcript per million mapped reads (FPKM) using the RSEM software (version 1.2.12) (Li and Dewey, 2011) with default parameters. Principal component analysis (PCA), which converts the data into a few independent metagenes by dimensionality reduction analysis, was carried out using the princomp function in the R software package. Pearson’s correlation analysis between samples was carried out using the cor function in the R software package. Differentially expressed genes (DEGs) between males and females were determined using DEGseq (Wang et al., 2010), with a fold change ≥ 2 and false discovery rate (FDR) adjusted P-value ≤ 0.001 considered to be differentially expressed. DEG lists were annotated with the Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Quantitative Real-Time PCR (qRT-PCR)
Total RNAs were extracted using the TRIzol reagent. One microgram RNA was used for cDNA synthesis using Rever Tra Ace qPCR RT Master Mix (TOYOBO). qRT-PCR was performed on the LightCycler 480 II instrument (Roche) with FastStart Universal SYBR Green Master (ROX) (Roche). The primers used are shown in Supplementary Table 1. Expression levels of target genes were normalized to those of the control gene β-actin for comparison.
Adeno-Associated Virus (AAV) Infection
AAV2/9 (serotype 2/9) viruses carrying mouse Eif2s3y-Myc-P2A-EGFP or control EGFP were packaged by OBIO Technology (Shanghai, China). Cultured primary neurons were infected with AAV on DIV 5, and electrophysiology was carried out on DIV12-13.
For in vivo injection, P0 C57BL/6J wild-type mice were subjected to hypothermic anesthesia. Then one microliter virus (1.55 × 1013 V.G./mL) was slowly injected into each lateral ventricle (2 mm distance from ventral to skin and 2/5 from lambda suture to the eye). Injected mice were laid on a warming pad until the body temperature was fully recovered.
Immunostaining
Primary neurons, astrocytes, and microglia were fixed in 4% paraformaldehyde and permeabilized in 0.2% Triton X-100 before blocking in 5% BSA for 1 h at room temperature. Cells were incubated with indicated primary antibodies overnight at 4°C, and then incubated with appropriate secondary antibodies conjugated with fluorescence and DAPI for 1 h at room temperature. Images were captured using the A1R (Nikon) confocal microscope.
Mice injected with AAV were anesthetized and perfused with PBS from the heart. The brain was dissected quickly, post-fixed in 4% paraformaldehyde at 4°C for 24 h, and cryoprotected in 30% sucrose. After sectioning using a freezing microtome (Leica), brain sections (15 μm thickness) were blocked in 0.2% Triton X-100 and 5% BSA for 1 h at room temperature and then incubated with indicated primary antibodies overnight at 4°C, followed by incubation with appropriate secondary antibodies conjugated with fluorescence for 2 h at room temperature. Images were captured with the Aperio Versa 200 (Leica), the FV1000MPE-B (Olympus), or the LSM 880 (Carl Zeiss) microscope.
Western Blot
Mouse brain samples were lysed in the TNEN lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, and 1% NP-40, supplemented with protease inhibitors and phosphatase inhibitors). Protein lysates (30 μg) were subjected to SDS-polyacrylamide gel electrophoresis. After being transferred to PVDF membranes, protein samples were incubated with indicated primary antibodies overnight at 4°C, with appropriate HRP-conjugated secondary antibodies for 1 h at room temperature, and then with enhanced chemiluminescent reagent (Millipore) for protein band development. Protein band intensities were quantified by the Image J software for comparison.
Preparation of Synaptosomal and Postsynaptic Density Fractions
The hippocampal tissues of male mice injected with AAV were dissected and homogenized in ice-cold sucrose buffer (0.32 M sucrose, and 25 mM HEPES, pH 7.4, supplemented with protease inhibitors and phosphatase inhibitors). The homogenate was centrifuged at 300 g for 5 min to separate the supernatant (S1) and the pellet (P1, nuclei and large debris fraction). The P1 fraction was discarded and the S1 fraction was centrifuged for 12 min at 10,000 g to separate the supernatant (S2, light membrane and cytosolic fraction) and the pellet (P2, crude synaptosomal fraction). The P2 fraction was washed twice by ice-cold sucrose buffer and re-suspended in ice-cold HBS buffer (150 mM NaCl, and 25 mM HEPES, pH 7.4, supplemented with protease inhibitors and phosphatase inhibitors) to get the synaptosomal (Syn) fraction. Partial Syn fraction was supplemented with ice-cold 1% Triton, incubated at 4°C for 30 min, and centrifuging at 20,000 g for 30 min. The resulting pellet was re-dissolved in 3% SDS HBS buffer to get the postsynaptic density (PSD) fraction.
Whole-Cell Patch-Clamp Electrophysiology
Primary neurons infected with AAV containing Eif2s3y-Myc-P2A-EGFP or control EGFP were bathed in an artificial cerebrospinal fluid (aCSF) containing 126 mM NaCl, 2.5 mM KCl, 2.4 mM MgCl2⋅6H2O, 1.2 mM CaCl2, 1.2 mM NaH2PO4, 11 mM NaHCO3, and 18 mM glucose, pH 7.4, 290–300 mOsm, bubbled with 95% O2/5% CO2 (v/v). Infected neurons were selected with a microscope equipped with a GFP fluorescent filter set (Olympus), and visualized with differential interference contrast (DIC) at room temperature. Recording pipettes (5∼8 MΩ tip resistance) were filled with a solution containing 140 mM CsCH3SO3, 10 mM HEPES, 1 mM EGTA, 2 mM MgCl2⋅6H2O, 5 mM TEA-CL, 2.5 mM Mg-ATP, and 0.3 mM Na-GTP, pH 7.3, 280 mOsm. Spontaneous excitatory postsynaptic currents (sEPSCs) and spontaneous inhibitory postsynaptic currents (sIPSCs) were measured in the voltage-clamp mode at holding potential of -70 mV and 0 mV, respectively. Synaptic currents were amplified and filtered at 2 kHz (Multiclamp 700B, Molecular Devices) and digitized at 10 kHz (pClamp10.7/Axon Digidata 1550B, Molecular Devices). sEPSCs and sIPSCs were recorded for 5 min and analyzed with MiniAnalysis (Synaptosoft).
Behavioral Analysis
All behavioral tests, except nesting, were performed between 9: 00 A.M. and 4: 00 P.M. in a blinded manner. Mice were acclimated to handling for 3 days before tests and to the test room for 1 h prior to tests. All behavioral tests, except nesting and self-grooming, were recorded and analyzed by a Smart 3.0 video tracking system (Panlab, Harvard Apparatus).
Open-Field Test
Mice were placed in an open field box (40 cm (L) × 40 cm (W) × 40 cm (H)) and allowed to explore for 10 min freely. Their total travel distance and time spent in the center arena were analyzed.
Three-Chamber Social Interaction Test
The three-chamber social interaction test was performed as described previously (Zheng et al., 2017). Briefly, mice were tested in three steps: (1) their time spent in each of the three empty chambers (left, center, and right) during a 10 min habituation; (2) their time spent interacting with an empty wire cage (E) in the right chamber versus a mouse (S1) in one wire cage in the left chamber during a 10-min sociability test; and (3) their time spent interacting with the familiar mouse (S1) versus an unfamiliar mouse (S2) put into the wire cage in the right chamber during a 10-min social novelty test.
Nesting
Mice were individually placed in a home cage containing 2 cm bedding and cotton tissues (3 g) overnight. Nest quality was then scored on a 1-5 scale as described previously (Kim et al., 2017): 1 point, cotton tissue rarely touched; 2 points, cotton tissue partially ripped; 3 points, cotton tissue mostly torn; 4 points, cotton tissue shredded into flat nest shape (walls lower than the mouse); and 5 points, perfect nest shape (walls higher than the mouse). Nesting scores were manually analyzed in a blinded manner.
Self-Grooming
Mice were individually placed in a home cage with no bedding. Following a 5 min habituation, mouse activity was videotaped for another 10 min. The number of bouts and total grooming duration were manually analyzed in a blinded manner.
Elevated Plus-Maze Test
Mice were placed in the center area of an elevated plus-maze with two 30 cm (L) × 6 cm (W) open arms and two 30 cm (L) × 6 cm (W) × 15 cm (H) closed arms. The movement of mice was recorded for 5 min. The time spent in open arms and the number of open arm entries were analyzed.
Morris Water Maze Test
The Morris Water Maze test was performed as described previously (Zheng et al., 2017). In brief, mice were trained for 5 consecutive days in a circular tank (90 cm (D) × 35 cm (H)) filled with tap water (made opaque by TiO2) at a temperature of 22°C. Contrasted shapes were posted on the walls to serve as reference cues. A hidden platform was placed in the southeast quadrant 1 cm below the water surface. Mice were released into the pool at one of four points (northwest, northeast, southwest, and southeast) in a random order to let them find and climb onto the hidden platform within 60 s. If a mouse failed to reach the hidden platform within the timeline, it was gently guided to the platform and allowed to stay on the platform for 10 s. Two trials per day were performed with at least 1 h inter-trial interval. On the 6th day, the hidden platform was removed from the pool, and mice were released into the pool for a 60 seconds’ probe test. Mouse escape latency during the training phase and quadrant entry time during the probe test phase were analyzed.
Y-Maze Test
Mice were placed in the center area of a Y-shape maze with three symmetrical arms at 120° angles (30 cm (L) × 6 cm (W) × 15 cm (H)). Each mouse was allowed to explore the maze freely for 5 min. An alternation is defined as consecutive entries into three different arms. The alternations and the total arm entries were recorded. The percentage of alternation triplet was calculated using the following formula as previously described (Miedel et al., 2017; Lachance et al., 2019):
Tail Suspension Test
Mice were suspended by the tail in an acrylic bar (15 cm (L) × 30 cm (H)) for 7 min. Total immobility duration during the last 6 min period was analyzed.
Statistical Analysis
All statistical analysis was performed using Prism 8.3 (GraphPad Software, La Jolla, CA, USA). Statistical differences between two groups were assessed by unpaired t-test for normally distributed data or non-parametric Mann Whitney test for non-normally distributed data. Statistical differences between multiple groups were assessed by one-way ANOVA with post hoc Dunnett’s multiple comparisons test for non-normally distributed data, or two-way ANOVA with post hoc Sidak’s multiple comparisons test for normally distributed data. Data are plotted as mean ± standard error of mean (SEM). p < 0.05 was considered to be statistically significant.
Results
Identification of Sexually Dimorphic Genes in Neurons, Astrocytes, and Microglia of Neonatal Mouse Brains
To study gene expression profiling differences between sexes in neurons, astrocytes, and microglia, we collected 6 postnatal day 0 (P0) males and 6 P0 females from 2 C57BL/6J mouse litters (3 males and 3 females from each litter) (Figure 1A and Supplementary Figure 1A). These mice were euthanized within 12 h after birth. Primary neurons, astrocytes, and microglia were isolated from the same individual mouse brain and cultured in vitro (Figure 1A). Purities of each harvested cultures were determined to be over 90% (Supplementary Figure 1B). A total of 36 cell cultures were used for RNA sequencing, including 12 (6 males and 6 females) neuronal, 12 (6 males and 6 females) astrocytic, and 12 (6 males and 6 females) microglial cultures.
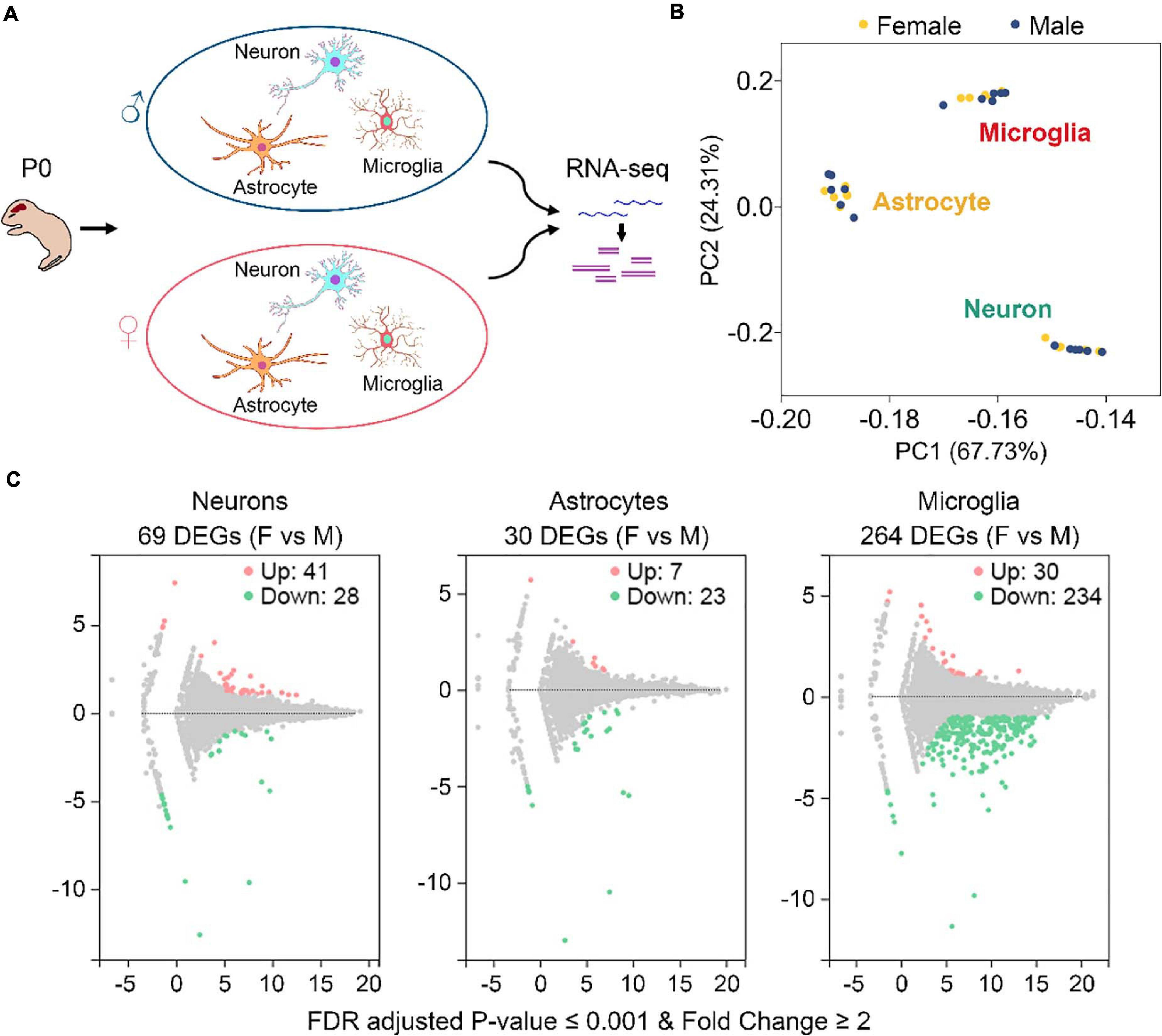
Figure 1. Identification of sexually dimorphic genes in neurons, astrocytes, and microglia. (A) Sample preparation strategy. Primary neurons, astrocytes, and microglia were isolated from the same postnatal day 0 (P0) male and female mice and cultured in vitro. RNAs were extracted from individual cultures and subjected to RNA sequencing. For each cell type, n = 6 males from two litters and n = 6 females from the same two litters. (B) PCA analysis of gene expression depicts three discrete subsets corresponding to neuron, astrocyte, and microglia samples, respectively. Yellow and blue dots represent female and male samples, respectively. (C) Volcano plots of gene expression change between female (F) and male (M) in neurons, astrocytes, and microglia. Pink and green dots represent genes that are expressed significantly higher and lower in females vs. males, respectively. Benjamini-Hochberg method, Storey and Tibshirani’s approach, false discovery rate (FDR) adjusted P-value ≤ 0.001, fold change ≥ 2. A total of 69, 30, and 264 differentially expressed genes (DEGs) were identified in neurons, astrocytes, and microglia, respectively.
RNA sequencing generated about 21.94 million raw reads for each sample (Supplementary Table 2). After data filtering using the SOAPnuke software, an average of 21.33 million clean reads for each sample was acquired. Mapping the clean reads onto the mouse reference genes using the Bowtie2 software (Langmead and Salzberg, 2012) revealed an average mapping ratio of 86.48% (Supplementary Table 2). A total of 20,073 genes were identified from the 36 samples.
Gene expression levels, described as fragments per kilobase of transcript per million mapped reads (FPKM), were determined using the RSEM software (Li and Dewey, 2011). As expected, principal component analysis (PCA) revealed three discrete expression clusters consistent with the three different cell types (Figure 1B). Pearson’s correlation analysis also showed high correlations for samples within the same cell type (all r > 0.91) (Supplementary Figure 1C). However, neither PCA nor Pearson’s analysis distinguished male and female samples within each cell type, suggesting that gene expression differences between males and females are minimal compared to those between different cell types.
By using the DEGseq software (Wang et al., 2010), we identified 69, 30, and 264 differentially expressed genes (DEGs, FDR adjusted P-value ≤ 0.001 and fold change ≥ 2) between males and females in neurons, astrocytes, and microglia, respectively (Figure 1C and Supplementary Table 3). Among them, 41 DEGs were upregulated and 28 DEGS were downregulated in female neurons versus male neurons, 7 DEGs were upregulated and 23 DEGS were downregulated in female astrocytes versus male astrocytes, and 30 DEGs were upregulated and 234 DEGS were downregulated in female microglia versus male microglia. We carried out Gene Ontology (GO) and KEGG pathway analysis and found that identified sexually dimorphic genes showed similar annotation patterns in neurons, astrocytes, and microglia. In GO analysis, sexually dimorphic genes in all the three cell types were strongly enriched for “binding” in the “molecular function” category, enriched for “cell” in the “cellular component” category, and enriched for “cellular process” in the “biological process” category (Figures 2A–C). In KEGG pathway analysis, sexually dimorphic genes in all the three cell types were strongly enriched for “immune system” in the “organismal system” category, enriched for “cancers: overview” and “infectious diseases: viral” in the “human diseases” category, and enriched for “signal transduction” in the “environmental information processing” category (Figures 2D–F).
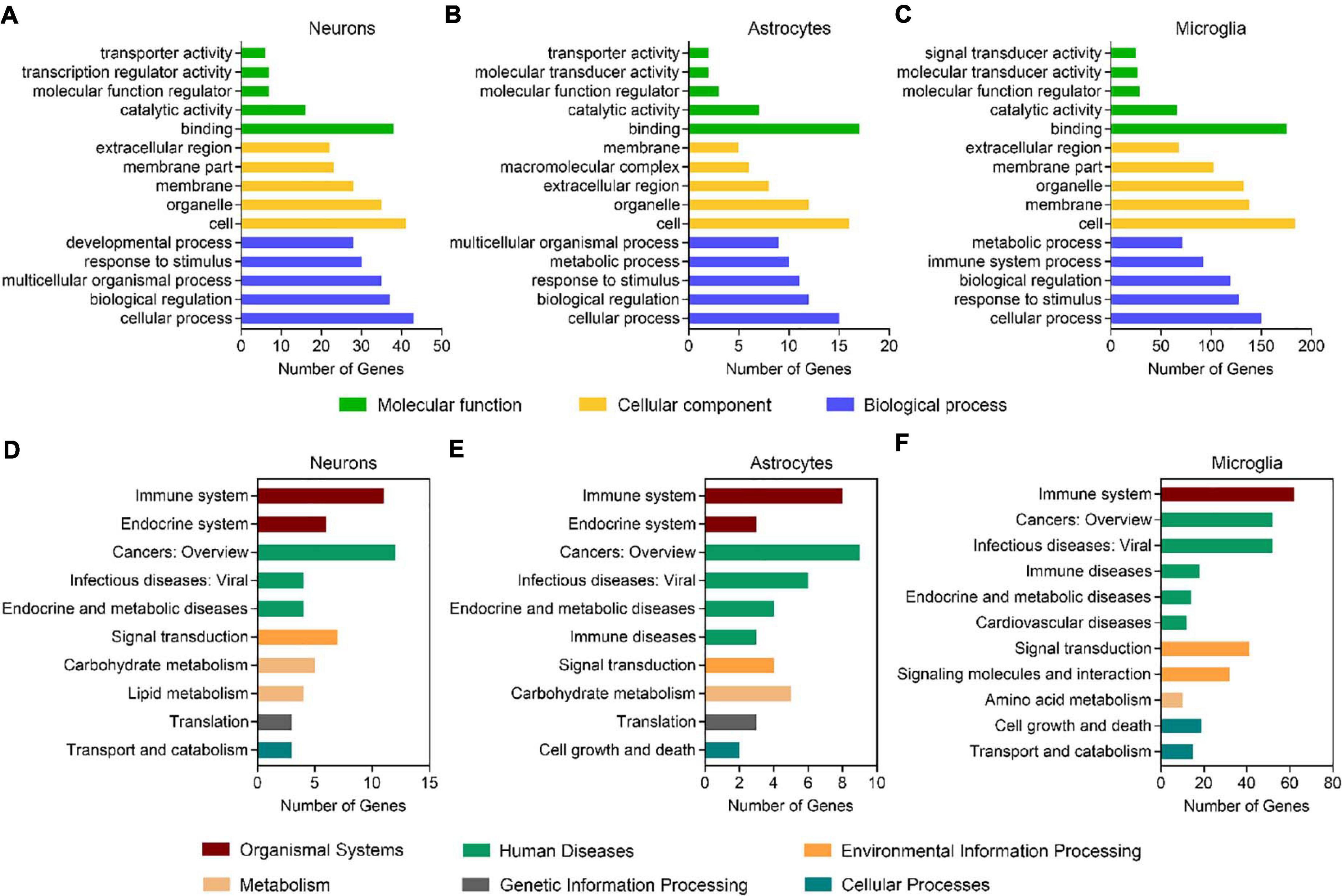
Figure 2. Sexually dimorphic genes exhibit similar annotation patterns in neurons, astrocytes, and microglia. (A–C) GO analysis of DEGs (male versus female) in neurons (A), astrocytes (B), and microglia (C). Green bars represent the molecular function, yellow bars represent the cellular component, and blue bars represent the biological process. (D–F) KEGG analysis of DEGs (male versus female) in neurons (D), astrocytes (E), and microglia (F). Red bars represent organismal systems, green bars represent human diseases, orange bars represent environmental information processing, flesh color bars represent metabolism, gray bars represent genetic information processing, and blue bars represent cellular processes.
Sexually Dimorphic Genes Shared by Different Neural Cell Types Locate Dominantly on Sex Chromosomes
We next used Venn diagrams to determine which genes show consistent sexually dimorphic expression in neurons, astrocytes, and microglia and identified five such genes as Ddx3y, Eif2s3y, Kdm5d, Uty, and Gm21975 (Figure 3A). The formal four identified genes locate on the Y chromosome and have relatively medium (FPKM > 1) to high (FPKM > 10) expressions in all three cell types of male origin (Figure 3B and Supplementary Figure 2). Protein-protein interaction (PPI) network analysis showed a robust interaction within the four Y-chromosomal genes (Figure 3C). The Gm21975 gene (also known as Evi2) locates on chromosome 11. Gm21975 expresses a naturally occurring read-through transcript covering the neighboring Evi2a (ecotropic viral integration site 2a) and Evi2b (ecotropic viral integration site 2b) genes and encodes a protein identical to the Evi2b gene product. However, although RNA sequencing results showed that Gm21975 was expressed significantly higher in male samples than female samples, quantitative real-time PCR (qRT-PCR) validation found lower expression of Gm21975 in male astrocytes than female astrocytes, and no Gm21975 expression differences between male and female samples of neurons and microglia (Supplementary Figures 3A–C). Therefore, we excluded Gm21975 from subsequent analysis.
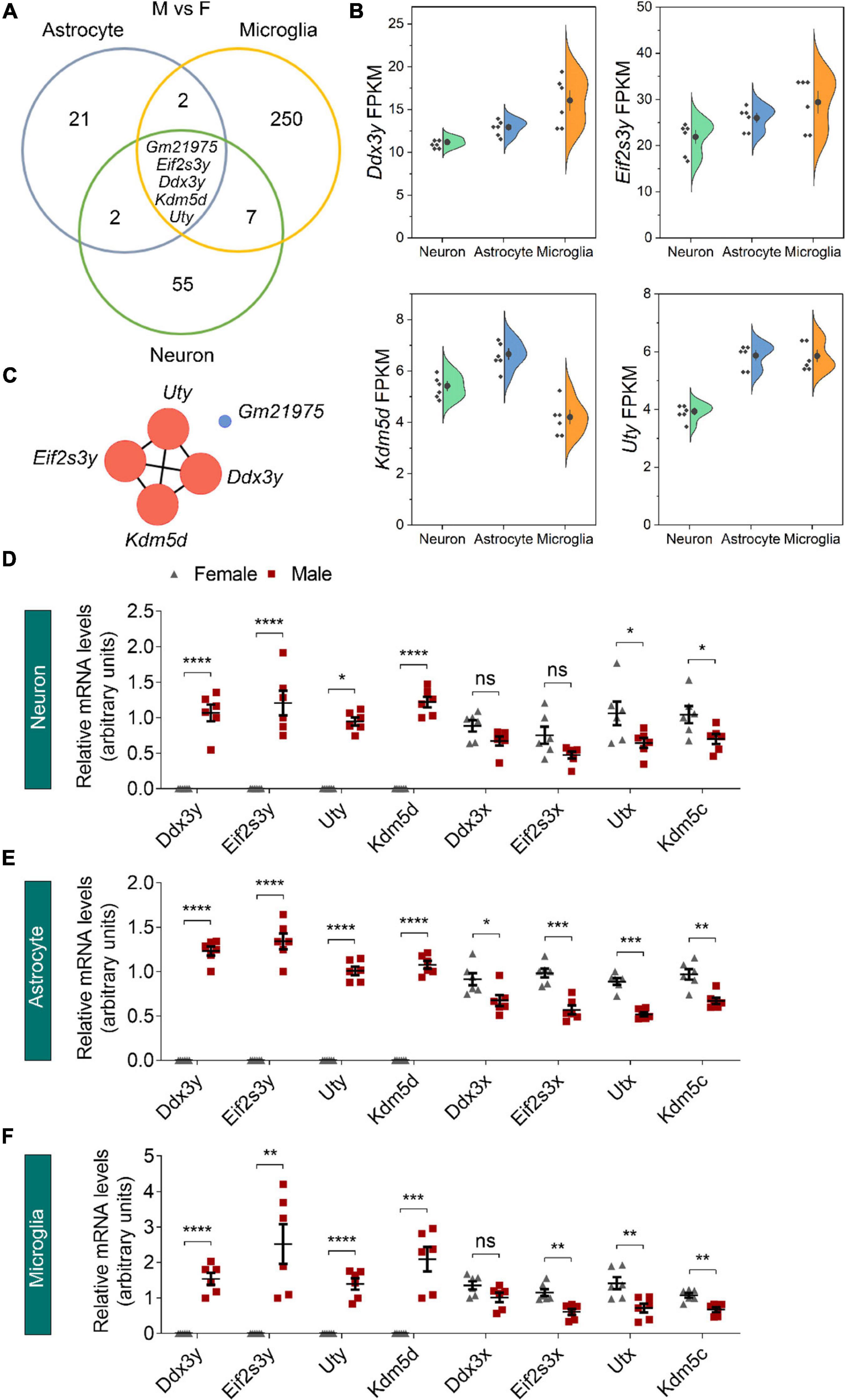
Figure 3. Identification of sexually dimorphic genes shared by different neural cell types. (A) Venn diagram analysis revealed five DEGs (male vs. female, M vs. F) shared by neurons, astrocytes, and microglia. (B) RNA sequencing identified Ddx3y, Eif2s3y, Kdm5d, and Uty expression levels (represented using FPKM) in male neurons, astrocytes, and microglia. n = 6 per group. (C) Protein-protein interaction analysis depicted a robust interaction within the four Y-linked genes: Ddx3y, Eif2s3y, Kdm5d, and Uty. (D–F) Relative mRNA levels of the four Y-linked genes and respective homologous genes on the X chromosome in neurons (D), astrocytes (E), and microglia (F) were determined by qRT-PCR and compared between male and female samples. n = 6 per group. Unpaired t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, not significant.
We also validated the four identified Y-chromosomal genes by qRT-PCR. Consistent with RNA sequencing data, Ddx3y, Eif2s3y, Kdm5d, and Uty were solely expressed in neurons, astrocytes, and microglia of male origin, at levels dramatically higher in males than females (Figures 3D–F). Since all these genes have respective homologs on the X chromosome (Ddx3x for Ddx3y, Eif2s3x for Eif2s3y, Kdm5c for Kdm5d, and Utx for Uty), we speculated that their X chromosomal homologs might have a biased expression in females for functional compensation purpose. Indeed, qRT-PCR analysis revealed that Kdm5c and Utx were expressed significantly higher in all the three cell types of female origin than those of male origin (Figures 3D–F). However, Ddx3x was expressed only significantly higher in female astrocytes than male astrocytes; and Eif2s3x was expressed only significantly higher in astrocytes and microglia of female origin than those of male origin. Together, these results suggest that sexually dimorphic genes dominantly locate on sex chromosomes.
Overexpression of Eif2s3y Increases sEPSC and sIPSC Amplitudes Specifically in Male Neurons
Given that Eif2s3y was relatively highly expressed compared to the other three Y-chromosomal genes in male neurons (Supplementary Figure 2) and that its X-linked homolog, Eif2s3x showed no significant differences in neurons between males and females (Figures 3B,D), we hypothesize that Eif2s3y plays a unique role in neuronal functions. Therefore, we studied whether overexpression of Eif2s3y affects neuronal synaptic transmission.
We packaged AAVs that express the mouse Eif2s3y protein with a Myc-tag on the C-terminus (AAV-Eif2s3y) or an EGFP control (AAV-NC) (Figure 4A). Primary neurons from P0 male and female mice were infected with AAV-Eif2s3y or AAV-NC. Eif2s3y overexpression was confirmed by western blotting (Figure 4B) and qRT-PCR (Supplementary Figure 4). We performed whole-cell recordings in these primary neurons. We found that compared to controls, the amplitude but not the frequency of spontaneous excitatory postsynaptic currents (sEPSCs) (Figures 4C–E) and spontaneous inhibitory postsynaptic currents (sIPSCs) (Figures 4I–K) were significantly increased in male neurons overexpressing Eif2s3y. While in female neurons, neither the frequency nor the amplitude of sEPSCs (Figures 4F–H) and sIPSCs (Figures 4L–N) was affected by Eif2s3y overexpression.
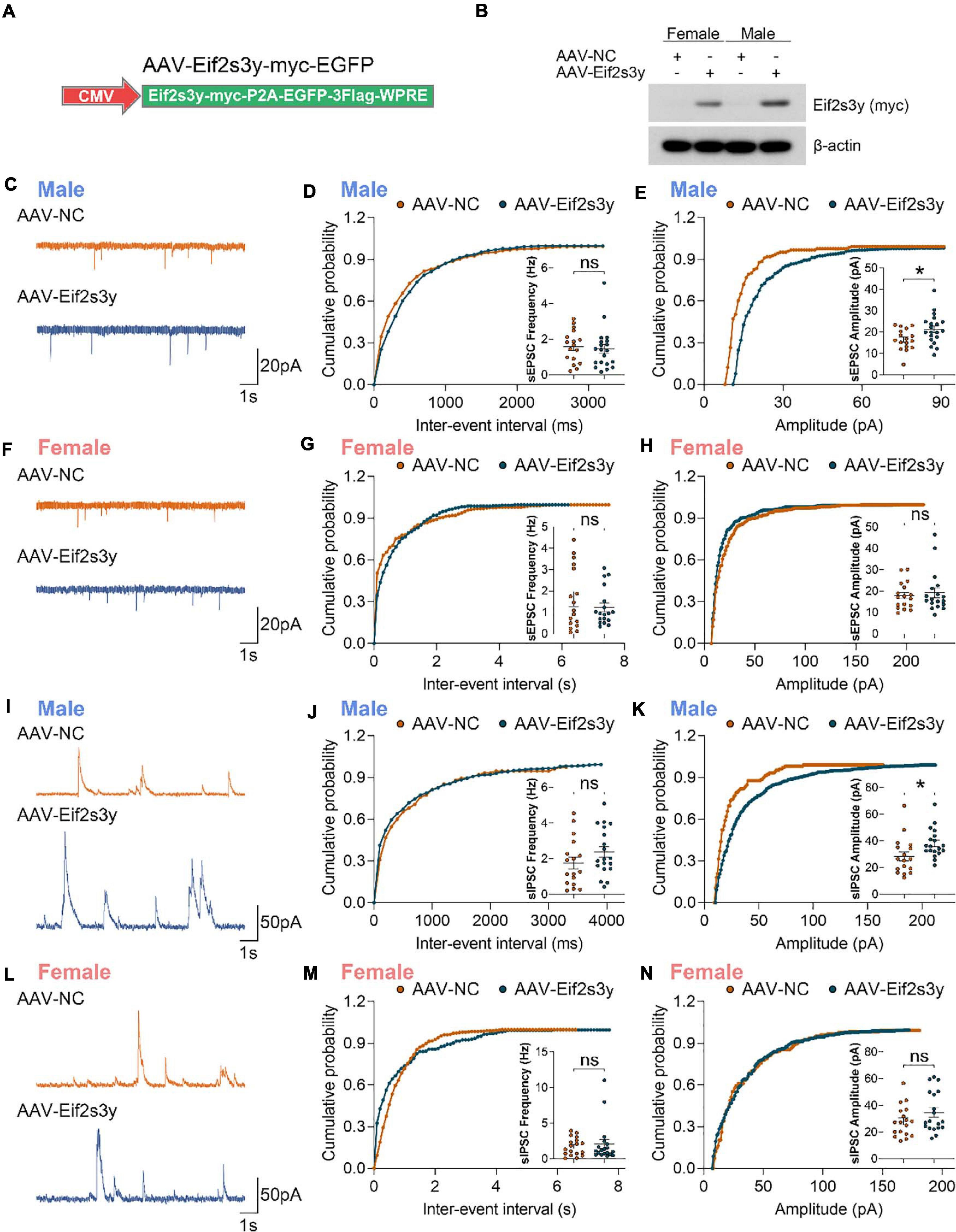
Figure 4. Overexpression of Eif2s3y promotes excitatory and inhibitory synaptic transmission specifically in male neurons. (A) Scheme of the Eif2s3y-Myc expressing construct used for AAV packaging. (B) Primary neurons from E16.5 male and female mice were infected with AAV expressing Eif2s3y (AAV-Eif2s3y) or EGFP (AAV-NC). Exogenous Eif2s3y protein was detected by western blotting using an anti-Myc antibody. (C) Representative traces of sEPSCs recorded from primary male neurons infected with AAV-Eif2s3y (n = 20 cells) and AAV-NC (n = 17 cells). Scale bars, 20 pA (vertical) and 1 s (horizontal). (D,E) Cumulative probabilities of sEPSC frequency (D) and amplitude (E) and respective quantification and comparison in male neurons. (F) Representative traces of sEPSCs recorded from primary female neurons infected with AAV-Eif2s3y (n = 18 cells) and AAV-NC (n = 17 cells). Scale bars, 20 pA (vertical) and 1 s (horizontal). (G,H) Cumulative probabilities of sEPSC frequency (G) and amplitude (H) and respective quantification and comparison in female neurons. (I) Representative traces of sIPSCs recorded from primary male neurons infected with AAV-Eif2s3y (n = 19 cells) and AAV-NC (n = 17 cells). Scale bars, 50 pA (vertical) and 1 s (horizontal). (L,K) Cumulative probabilities of sIPSC frequency (L) and amplitude (K) and respective quantification and comparison in male neurons. (L) Representative traces of sIPSCs recorded from primary female neurons infected with AAV-Eif2s3y (n = 19 cells) and AAV-NC (n = 18 cells). Scale bars, 50 pA (vertical) and 1 s (horizontal). (M,N) Cumulative probabilities of sIPSC frequency (M) and amplitude (N) and respective quantification and comparison in female neurons. Mann Whitney test was used for (G,H,M). Unpaired t-test was used for (D,E,J,K,N). *p < 0.05, ns, not significant.
Overexpression of Eif2s3y Causes Autism-Like Behaviors Specifically in Male Mice
We next investigated whether Eif2s3y overexpression affects mouse behaviors. Both male and female neonatal C57BL/6 mice received a bilateral injection in the lateral ventricle of AAV-Eif2s3y or AAV-NC (Figure 5A). EGFP fluorescence observation revealed that AAVs infected largely the hippocampal and nearby regions (Figure 5B and Supplementary Figures 5A,B). Exogenous Eif2s3y protein expression was also detected in the hippocampus of both female and male mice injected with AAV-Eif2s3y compared to controls (Supplementary Figure 5C). In addition, we found that EGFP colocalized well with the neuronal marker NeuN but not with the astrocytic marker GFAP or the microglial marker Iba1 (Supplementary Figures 5D,E), indicating that AAVs mostly infected neurons, i.e., exogenous Eif2s3y was dominantly expressed in neurons.
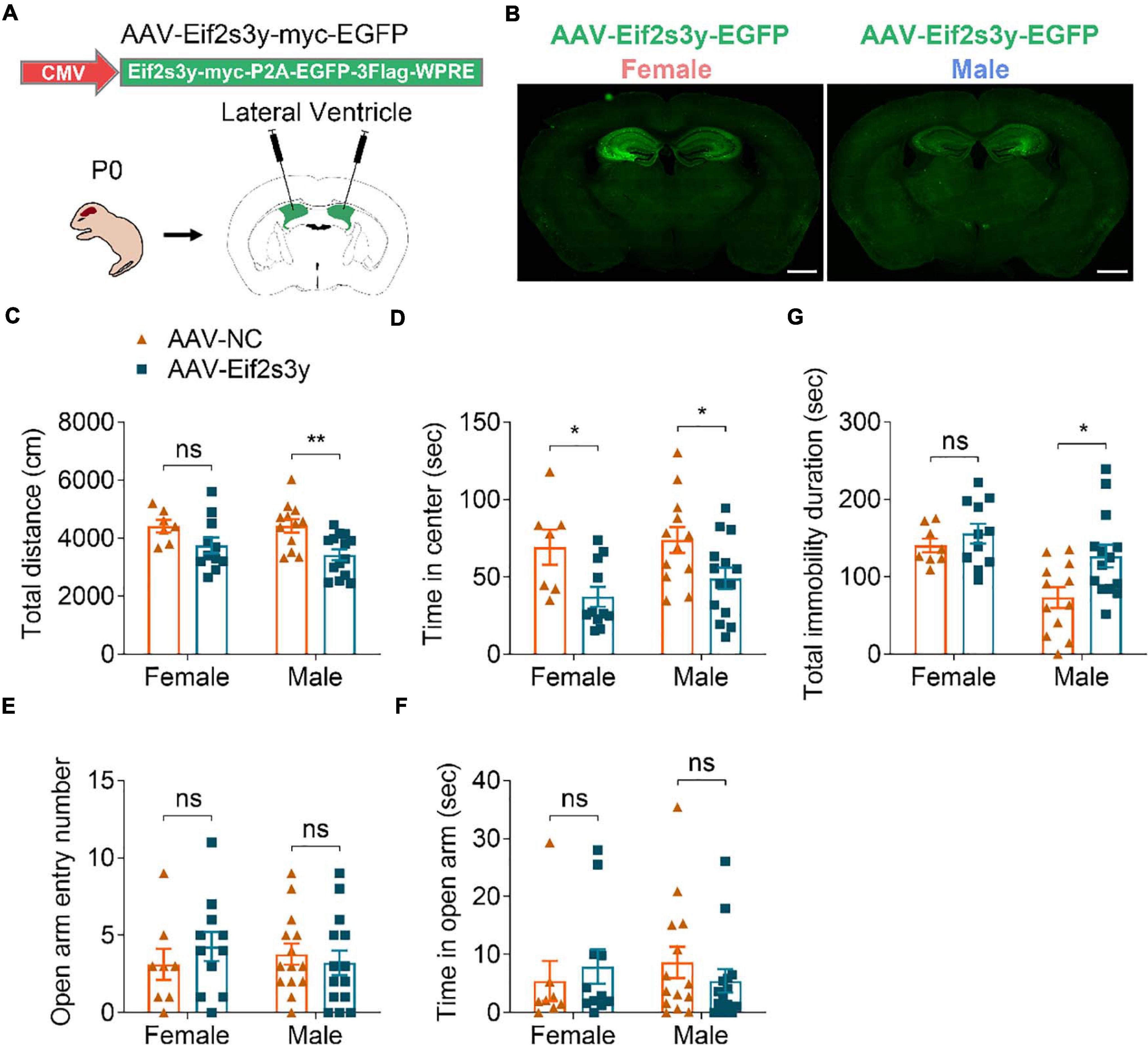
Figure 5. Overexpression of Eif2s3y causes male-specific depression-like behavior. (A) Scheme of the AAV injection paradigm. (B) Representative images of the EGFP expression localization in male and female mice injected with AAV-Eif2s3y. Scale bar, 1 mm. (C,D) In the open-field test, total travel distance (C) and time in the center (D) were analyzed. n = 14 AAV-Eif2s3y male mice, n = 12 AAV-NC male mice, n = 11 AAV-Eif2s3y female mice, and n = 7 AAV-NC female mice. (E,F) In the elevated plus-maze test, time spent in the open arm (E) and open arm entry numbers (F) were analyzed. n = 14 AAV-Eif2s3y male mice, n = 14 AAV-NC male mice, n = 11 AAV-Eif2s3y female mice, and n = 8 AAV-NC female mice. (G) In the tail suspension test, mouse immobility duration was analyzed. n = 14 AAV-Eif2s3y male mice, n = 12 AAV-NC male mice, n = 11 AAV-Eif2s3y female mice, and n = 8 AAV-NC female mice. Unpaired t-test. *p < 0.05, **p < 0.01, ns, not significant.
Six weeks after AAV injection, we carried out various behavioral tests. In the open field test, Eif2s3y overexpression reduced total travel distance in male mice (Figure 5C) and decreased the time spent in the center region for both male and female mice (Figure 5D). However, in the elevated open arm test, Eif2s3y overexpression had no effect on the time spent in the open arm and open arm entries in either male or female mice (Figures 5E,F). These results suggest that Eif2s3y overexpression has a marginal effect on animal anxiety in both sexes. In the tail suspension test, Eif2s3y overexpression significantly increased immobility duration in male mice but not in female mice (Figure 5G), implying that Eif2s3y overexpression may specifically cause depression in male mice.
In the three-chamber social interaction test, we found that Eif2s3y overexpression in male mice had no effect on their chamber exploration pattern during the habituation stage (Figure 6A) and their sociability to a mouse (S1) rather than to an empty cage (Figure 6B). However, Eif2s3y overexpression impaired the social novelty recognition of male mice to a familiar mouse (S1) versus an unfamiliar mouse (S2) (Figure 6C). In contrast, Eif2s3y overexpression in female mice had no effect on animal habituation, sociability, or social novelty recognition (Figures 6D–F). In the nest-building test, we also found that Eif2s3y overexpression specifically impaired nest building ability in male but not female mice (Figure 6G). Moreover, male but not female mice with Eif2s3y overexpression exhibited significantly increased bout numbers and grooming duration compared to controls (Figures 6H,I). Together, these results indicate that overexpression of Eif2s3y causes autism-like behaviors specifically in male mice.
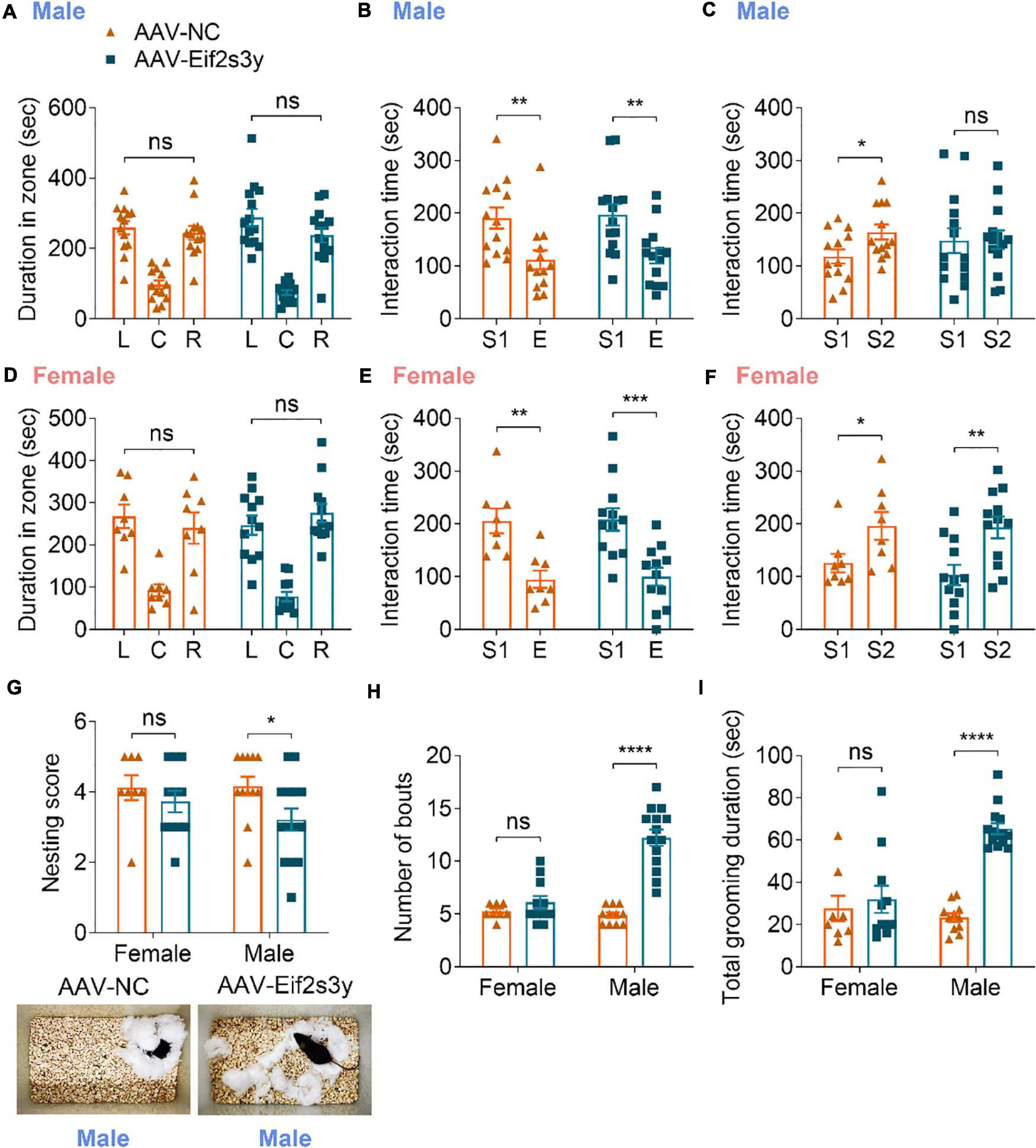
Figure 6. Overexpression of Eif2s3y causes autism-like behaviors specifically in male mice. (A–F) In the three-chamber social interaction test, male mice (A–C) injected with AAV-Eif2s3y (n = 14) or AAV-NC (n = 13), and female mice (D–F) injected with AAV-Eif2s3y (n = 12) or AAV-NC (n = 8) were tested in three steps. For habituation (A,D), mice were compared for their time spent in each empty chamber (L, left; C, center; R, right). For sociability (B,E), mice were compared for their interaction time with an empty wire cage (E) and with a mouse (S1). For social novelty recognition (C,F), mice were compared for their interaction time with a familiar mouse (S1) and with an unfamiliar mouse (S2). (G) Both male and female mice were compared for their nest-building ability. Representative images of nests built by male mice are shown. n = 14 AAV-Eif2s3y male mice, n = 12 AAV-NC male mice, n = 11 AAV-Eif2s3y female mice, and n = 8 AAV-NC female mice. (H,I) During free movement, mice were analyzed for bout numbers (H) and total grooming duration (I). n = 14 AAV-Eif2s3y male mice, n = 11 AAV-NC male mice, n = 11 AAV-Eif2s3y female mice, and n = 8 AAV-NC female mice. Unpaired t-test was used for (A–F,I). Mann Whitney test was used for (G,H). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, not significant.
We also studied whether Eif2s3y overexpression affects animal learning and memory. However, in the Morris water maze test, both male (Figures 7A,B) and female (Figures 7C,D) mice with Eif2s3y overexpression showed similar escape latencies during the training phase and similar time spent in the target quadrant during the probe test phase compared to respective controls. In the Y maze test, both male and female mice with Eif2s3y overexpression had similar alternation triplet and total arm entry numbers compared to respective controls (Figures 7E,F). Therefore, Eif2s3y overexpression may not affect learning and memory in mice.
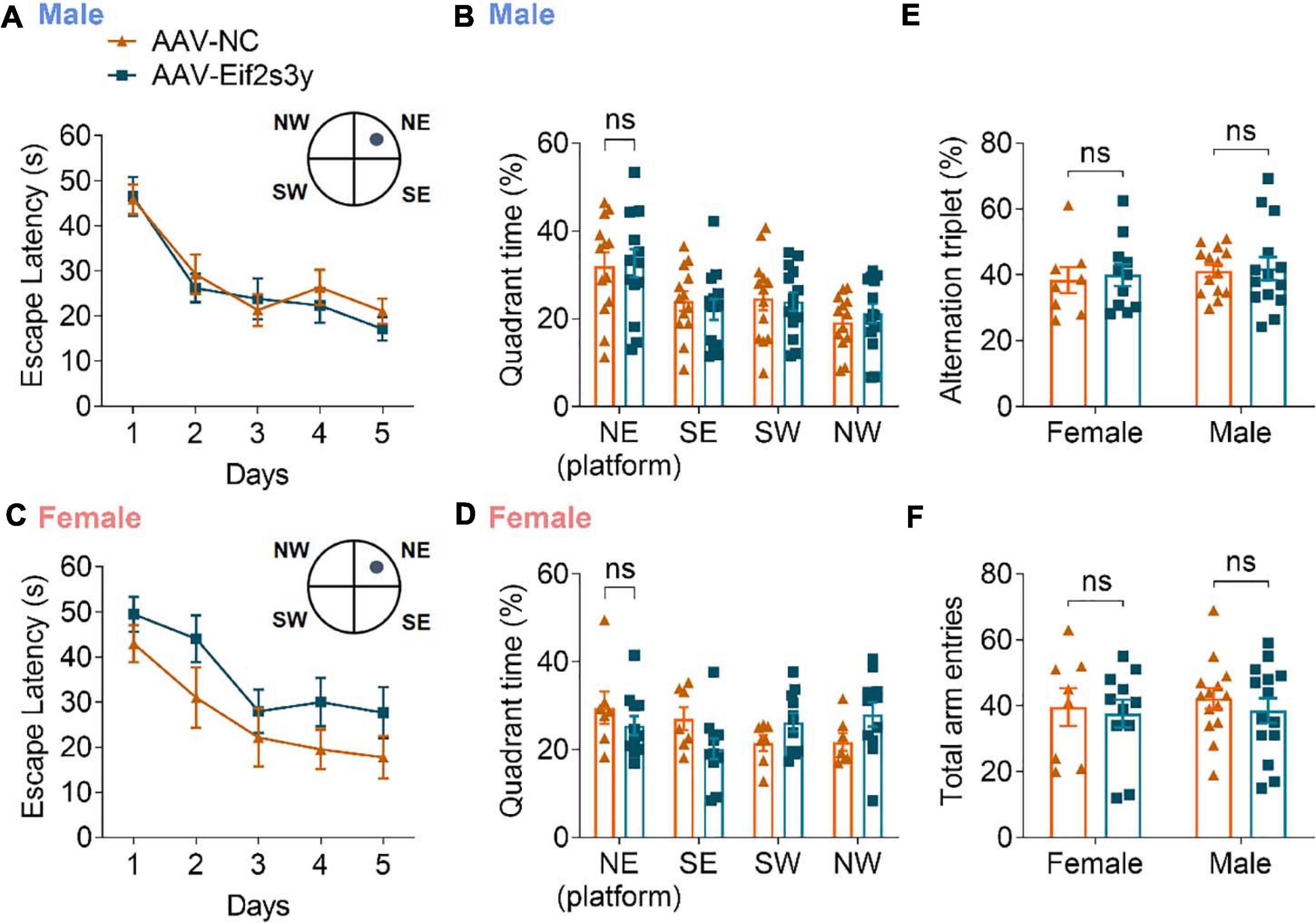
Figure 7. Overexpression of Eif2s3y has no effect on mouse learning and memory. (A–D) In the Morris water maze test, escape latency (A,C) in the training phase and time spent in each quadrant during the probe test phase (B,D) were analyzed for male mice (A,B) injected with AAV-Eif2s3y (n = 14) or AAV-NC (n = 13), and female mice (C,D) injected with AAV-Eif2s3y (n = 11) or AAV-NC (n = 7). Two-way ANOVA followed by post hoc Sidak’s multiple comparisons test was used for (A,C). Unpaired t-test was used for (B,D). ns, not significant. (E,F) In the Y maze test, alternation triplet (E) and total arm entries (F) were analyzed. n = 14 AAV-Eif2s3y male mice, n = 14 AAV-NC male mice, n = 11 AAV-Eif2s3y female mice, and n = 8 AAV-NC female mice. Unpaired t-test was used for (E,F). ns, not significant.
Finally, we explored whether Eif2s3y overexpression affects synaptic proteins. However, the results showed that Eif2s3y overexpression in male mice did not affect total levels (Supplementary Figures 6A,B) and synaptic distribution (Supplementary Figures 6C-F) of proteins studied here, including AMPA receptor subunits (GluR1, GluR2, and GluR3), NMDA receptor subunits (NR1, NR2A, and NR2B), the presynaptic marker synaptophysin (SYP), and the postsynaptic marker PSD-95.
Discussion
Sex differences have been documented in multiple neurological disorders, but underlying mechanisms remain largely elusive. Primary neural cells have been widely used as in vitro models for investigating cellular and molecular mechanisms of neurological disorders. However, the sex of donors of these primary cells is neglected in significant amounts of such studies; and this could potentially affect the correctness of their conclusions and be a source for contradictory results of different studies. Therefore, to determine whether a significant gene expression difference exists between male and female neural cells, we comprehensively compared the transcriptome of primary neurons, astrocytes and microglia derived from neonatal mice of the two sexes. We identified 69, 30, and 264 sexually dimorphic genes in primary neurons, astrocytes, and microglia, respectively. Compared to a total of 20,073 genes identified in this study, the numbers of sexually dimorphic genes in these cells are relatively low. Among the three cell types, microglia exhibit the most divergent expression differences between the two sexes, which is consistent with previous reports showing that microglia display sex differences both in their transcriptome and function (Hanamsagar et al., 2017; Guneykaya et al., 2018; Thion et al., 2018; Villa et al., 2018).
Gene ontology and KEGG analysis revealed that identified sexually dimorphic genes in neurons, astrocytes, and microglia exhibited similar annotation patterns. Interestingly, sexually dimorphic genes were strongly enriched for the immune system and immune-related diseases in all three cell types, suggesting that immune systems are the most divergent between males and females in the central nervous system (CNS). This finding is in agreement with many previous studies suggesting that immune systems are involved in the sex-specific etiology of multiple neurological disorders (Crain et al., 2009, 2013; Schwarz et al., 2012; Lenz et al., 2013; Dunn et al., 2015; Guneykaya et al., 2018; Villa et al., 2018).
Our RNA sequencing results identified five genes showing consistent sexually dimorphic expression in neurons, astrocytes, and microglia. Among them, four genes (Ddx3y, Eif2s3y, Kdm5d, and Uty) locate on the short arm of the mouse Y chromosome. This region is part of the Y-chromosomal non-recombining region so that genes on this region are present and expressed exclusively in males (Armoskus et al., 2014). Previous studies also identified sexually dimorphic expression of Ddx3y, Eif2s3y, Kdm5d, and Uty in the mouse brain at different ages by using northern blotting and RT-PCR (Xu et al., 2002), and of Ddx3y, Eif2s3y, and Kdm5d in the neonatal mouse cortex/hippocampus by using microarray analysis (Armoskus et al., 2014). Ddx3y encodes an RNA helicase containing a DEAD-box motif and has been implicated in RNA metabolism, neuronal regulation, apoptosis, and cell cycle progression (Vakilian et al., 2015). Eif2s3y is suggested to play a crucial role in mouse spermatogenesis (Mazeyrat et al., 2001; Yamauchi et al., 2014; Matsubara et al., 2015). Kdm5d encodes a histone demethylase and participates in H3K4 demethylation (Bernstein et al., 2002). Uty and its X-homolog Utx (also known as Kdm6a) belong to the KDM6 subfamily. Utx encodes a histone H3 lysine 27 (H3K27) demethylase. However, human UTY has been reported to lose enzymatic activity due to sequence divergence (Shpargel et al., 2012).
Ddx3y, Eif2s3y, Kdm5d, and Uty have their homologs on the X chromosome as Ddx3x, Eif2s3x, Kdm6a, and Utx, respectively. We found that Kdm5c and Kdm6a were expressed higher in all three female cell types than male cells, Eif2s3x was expressed higher in female astrocytes and microglia than male cells, and Ddx3x was expressed only significantly higher in female astrocytes than male astrocytes. These results imply that these X-chromosomal genes escape from X chromosome inactivation in certain female cell types, possibly as compensation for balancing the function of their Y-chromosomal homologous genes.
Eif2s3y was found to have a high expression in male neurons. However, the function of Eif2s3y in the CNS remains unknown. To study whether dysregulation of Eif2s3y affects neuronal functions in a sex-specific manner, we infected primary mouse neurons with AAVs expressing mouse Eif2s3y and found that overexpression of Eif2s3y increased the amplitude of both sEPSCs and sIPSCs in male but not female neurons. These results indicate that overexpression of Eif2s3y promotes both excitatory and inhibitory synaptic transmission specifically in males.
Moreover, we overexpressed Eif2s3y in the mouse brain and found that overexpression of Eif2s3y led to autism-like behaviors specifically in male mice, including impaired social novelty recognition and nest-building abilities and increased repetitive behavior. Since the protein sequence of mouse Eif2s3y is highly conserved (over 97% similarity) with those of mouse Eif2s3x and human X-linked EIF2S3, and there is no Y-linked homolog of Eif2s3y in humans (Supplementary Figures 7A,B), overexpression of Eif2s3y in male and female mice may partially resemble patients with Klinefelter Syndrome (47,XXY) and XXX Syndrome (47,XXX), respectively. Indeed, Klinefelter Syndrome patients have been reported to have an increased risk of ASDs (Visootsak and Graham, 2009; Tartaglia et al., 2010; Bishop et al., 2011; Hong and Reiss, 2014; Green et al., 2019); and our results suggest that an increased expression of EIF2S3 may at least partially contribute to this.
The human homolog of Eif2s3y, EIF2S3 encodes eukaryotic translation initiation factor 2 subunit γ (eIF2γ) that plays a regulatory role in Met-tRNAiMet binding and thus is involved in early protein synthesis (Young-Baird et al., 2019). Since dysregulation of protein synthesis has been found in ASD patients and animal models and proposed as a common mechanism underlying disease pathogenesis (Levitt and Campbell, 2009; Lo and Lai, 2020; Lu and Hsueh, 2021), it is possible that a dysregulation of Eif2s3y may promote susceptibility to ASD through interfering with the synthesis of proteins critically involved in ASD pathogenesis; and this warrants further investigation.
In summary, our present study provides a novel and unbiased sexually dimorphic gene expression dataset in primary neurons, astrocytes, and microglia derived from female and male neonatal mice. Our study also indicates that sexually dimorphic genes on the Y chromosome may be a genetic driver of neurological disorders, which deserves further scrutiny.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (https://ftp.cngb.org/pub/CNSA/data3/CNP0001675/), accession number CNP0001675.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of Xiamen University.
Author Contributions
MZ and Y-WZ designed the research. MZ performed most experiments. YZ contributed to virus experiments. YJ, ZL, XX, and JN helped with animal experiments. XX helped with sample preparation for RNA sequencing. HS helped with electrophysiological experiments. XZ, HL, DC, JL, and HX made intellectual contributions. MZ, ZL, and Y-WZ wrote the manuscript. Y-WZ supervised the project. All authors reviewed the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81771377 and U1705285 to Y-WZ) and National Key Research and Development Program of China (2018YFC2000400 and 2016YFC1305903 to Y-WZ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jingru Huang and Xiang You for helping with the Laser Scanning Confocal Microscope operation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.669798/full#supplementary-material
Footnotes
References
Aleman, A., Kahn, R. S., and Selten, J. P. (2003). Sex differences in the risk of schizophrenia: evidence from meta-analysis. Arch. Gen. Psychiatry 60, 565–571. doi: 10.1001/archpsyc.60.6.565
Armoskus, C., Moreira, D., Bollinger, K., Jimenez, O., Taniguchi, S., and Tsai, H. W. (2014). Identification of sexually dimorphic genes in the neonatal mouse cortex and hippocampus. Brain Res. 1562, 23–38. doi: 10.1016/j.brainres.2014.03.017
Arnold, A. P. (2017). A general theory of sexual differentiation. J. Neurosci. Res. 95, 291–300. doi: 10.1002/jnr.23884
Bernstein, B. E., Humphrey, E. L., Erlich, R. L., Schneider, R., Bouman, P., Liu, J. S., et al. (2002). Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. U.S.A. 99, 8695–8700. doi: 10.1073/pnas.082249499
Bishop, D. V., Jacobs, P. A., Lachlan, K., Wellesley, D., Barnicoat, A., Boyd, P. A., et al. (2011). Autism, language and communication in children with sex chromosome trisomies. Arch. Dis. Child. 96, 954–959. doi: 10.1136/adc.2009.179747
Christensen, D. L., Baio, J., Braun, K. V., Bilder, D., Charles, J., Constantino, J. N., et al. (2016). Prevalence and characteristics of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2012. MMWR Surveill. Summ. 65, 1–23. doi: 10.15585/mmwr.ss6802a1
Crain, J. M., Nikodemova, M., and Watters, J. J. (2009). Expression of P2 nucleotide receptors varies with age and sex in murine brain microglia. J. Neuroinflammation 6:24. doi: 10.1186/1742-2094-6-24
Crain, J. M., Nikodemova, M., and Watters, J. J. (2013). Microglia express distinct M1 and M2 phenotypic markers in the postnatal and adult central nervous system in male and female mice. J. Neurosci. Res. 91, 1143–1151. doi: 10.1002/jnr.23242
Demily, C., Poisson, A., Peyroux, E., Gatellier, V., Nicolas, A., Rigard, C., et al. (2017). Autism spectrum disorder associated with 49,XYYYY: case report and review of the literature. BMC Med. Genet. 18:9.
Dunn, S. E., Lee, H., Pavri, F. R., and Zhang, M. A. (2015). Sex-based differences in multiple sclerosis (Part I): biology of disease incidence. Curr. Top. Behav. Neurosci. 26, 29–56. doi: 10.1007/7854_2015_371
Fung, L. K., and Reiss, A. L. (2016). Moving toward integrative, multidimensional research in modern psychiatry: lessons learned from fragile X syndrome. Biol. Psychiatry 80, 100–111. doi: 10.1016/j.biopsych.2015.12.015
Green, T., Flash, S., and Reiss, A. L. (2019). Sex differences in psychiatric disorders: what we can learn from sex chromosome aneuploidies. Neuropsychopharmacology 44, 9–21. doi: 10.1038/s41386-018-0153-2
Guneykaya, D., Ivanov, A., Hernandez, D. P., Haage, V., Wojtas, B., Meyer, N., et al. (2018). Transcriptional and translational differences of microglia from male and female brains. Cell Rep. 24, 2773–2783 e2776.
Haaxma, C. A., Bloem, B. R., Borm, G. F., Oyen, W. J., Leenders, K. L., Eshuis, S., et al. (2007). Gender differences in Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 78, 819–824.
Hanamsagar, R., Alter, M. D., Block, C. S., Sullivan, H., Bolton, J. L., and Bilbo, S. D. (2017). Generation of a microglial developmental index in mice and in humans reveals a sex difference in maturation and immune reactivity. Glia 65, 1504–1520. doi: 10.1002/glia.23176
Heard, E., and Turner, J. (2011). Function of the sex chromosomes in mammalian fertility. Cold Spring Harb. Perspect. Biol. 3:a002675. doi: 10.1101/cshperspect.a002675
Hong, D. S., and Reiss, A. L. (2012). Cognition and behavior in turner syndrome: a brief review. Pediatr. Endocrinol. Rev. 9(Suppl. 2), 710–712.
Hong, D. S., and Reiss, A. L. (2014). Cognitive and neurological aspects of sex chromosome aneuploidies. Lancet Neurol. 13, 306–318. doi: 10.1016/s1474-4422(13)70302-8
Hong, D. S., Dunkin, B., and Reiss, A. L. (2011). Psychosocial functioning and social cognitive processing in girls with turner syndrome. J. Dev. Behav. Pediatr. 32, 512–520. doi: 10.1097/dbp.0b013e3182255301
Kim, H. J., Cho, M. H., Shim, W. H., Kim, J. K., Jeon, E. Y., Kim, D. H., et al. (2017). Deficient autophagy in microglia impairs synaptic pruning and causes social behavioral defects. Mol. Psychiatry 22, 1576–1584. doi: 10.1038/mp.2016.103
Lachance, V., Wang, Q., Sweet, E., Choi, I., Cai, C. Z., Zhuang, X. X., et al. (2019). Autophagy protein NRBF2 has reduced expression in Alzheimer’s brains and modulates memory and amyloid-beta homeostasis in mice. Mol. Neurodegener 14:43.
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Leggett, V., Jacobs, P., Nation, K., Scerif, G., and Bishop, D. V. (2010). Neurocognitive outcomes of individuals with a sex chromosome trisomy: XXX, XYY, or XXY: a systematic review. Dev. Med. Child. Neurol. 52, 119–129. doi: 10.1111/j.1469-8749.2009.03545.x
Lenz, K. M., Nugent, B. M., Haliyur, R., and McCarthy, M. M. (2013). Microglia are essential to masculinization of brain and behavior. J. Neurosci. 33, 2761–2772. doi: 10.1523/jneurosci.1268-12.2013
Levitt, P., and Campbell, D. B. (2009). The genetic and neurobiologic compass points toward common signaling dysfunctions in autism spectrum disorders. J. Clin. Invest. 119, 747–754. doi: 10.1172/jci37934
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.
Lo, L. H., and Lai, K. O. (2020). Dysregulation of protein synthesis and dendritic spine morphogenesis in ASD: studies in human pluripotent stem cells. Mol. Autism 11:40.
Lu, M. H., and Hsueh, Y. P. (2021). Protein synthesis as a modifiable target for autism-related dendritic spine pathophysiologies. FEBS J. [Epub ahead of print]. doi: 10.1111/febs.15733
Maldonado Weng, J., Parikh, I., Naqib, A., York, J., Green, S. J., Estus, S., et al. (2019). Synergistic effects of APOE and sex on the gut microbiome of young EFAD transgenic mice. Mol. Neurodegener. 14:47.
Matsubara, Y., Kato, T., Kashimada, K., Tanaka, H., Zhi, Z., Ichinose, S., et al. (2015). TALEN-mediated gene disruption on Y chromosome reveals critical role of EIF2S3Y in mouse spermatogenesis. Stem Cells Dev. 24, 1164–1170. doi: 10.1089/scd.2014.0466
Matsuzaki, J., Bloy, L., Blaskey, L., Miller, J., Kuschner, E. S., Ku, M., et al. (2019). Abnormal auditory mismatch fields in children and adolescents with 47,XYY syndrome. Dev. Neurosci. 41, 123–131. doi: 10.1159/000500799
Mazeyrat, S., Saut, N., Grigoriev, V., Mahadevaiah, S. K., Ojarikre, O. A., Rattigan, A., et al. (2001). A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat. Genet. 29, 49–53. doi: 10.1038/ng717
Miedel, C. J., Patton, J. M., Miedel, A. N., Miedel, E. S., and Levenson, J. M. (2017). Assessment of spontaneous alternation, novel object recognition and limb clasping in transgenic mouse models of amyloid-beta and tau neuropathology. J. Vis. Exp. 123:55523.
Mielke, M. M., Vemuri, P., and Rocca, W. A. (2014). Clinical epidemiology of Alzheimer’s disease: assessing sex and gender differences. Clin. Epidemiol. 6, 37–48. doi: 10.2147/clep.s37929
Ngun, T. C., Ghahramani, N., Sanchez, F. J., Bocklandt, S., and Vilain, E. (2011). The genetics of sex differences in brain and behavior. Front. Neuroendocrinol. 32, 227–246.
Orton, S.-M., Herrera, B. M., Yee, I. M., Valdar, W., Ramagopalan, S. V., Sadovnick, A. D., et al. (2006). Sex ratio of multiple sclerosis in Canada: a longitudinal study. Lancet Neurol. 5, 932–936. doi: 10.1016/s1474-4422(06)70581-6
Printzlau, F., Wolstencroft, J., and Skuse, D. H. (2017). Cognitive, behavioral, and neural consequences of sex chromosome aneuploidy. J. Neurosci. Res. 95, 311–319. doi: 10.1002/jnr.23951
Rovet, J., Netley, C., Bailey, J., Keenan, M., and Stewart, D. (1995). Intelligence and achievement in children with extra X aneuploidy: a longitudinal perspective. Am. J. Med. Genet. 60, 356–363. doi: 10.1002/ajmg.1320600503
Schaafsma, S. M., and Pfaff, D. W. (2014). Etiologies underlying sex differences in autism spectrum disorders. Front. Neuroendocrinol. 35, 255–271. doi: 10.1016/j.yfrne.2014.03.006
Schwarz, J. M., Sholar, P. W., and Bilbo, S. D. (2012). Sex differences in microglial colonization of the developing rat brain. J. Neurochem. 120, 948–963.
Shpargel, K. B., Sengoku, T., Yokoyama, S., and Magnuson, T. (2012). UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 8:e1002964. doi: 10.1371/journal.pgen.1002964
Tartaglia, N. R., Wilson, R., Miller, J. S., Rafalko, J., Cordeiro, L., Davis, S., et al. (2017). Autism spectrum disorder in males with sex chromosome aneuploidy: XXY/klinefelter syndrome, XYY, and XXYY. J. Dev. Behav. Pediatr. 38, 197–207. doi: 10.1097/dbp.0000000000000429
Tartaglia, N., Cordeiro, L., Howell, S., Wilson, R., and Janusz, J. (2010). The spectrum of the behavioral phenotype in boys and adolescents 47,XXY (Klinefelter syndrome). Pediatr. Endocrinol. Rev. 8(Suppl. 1), 151–159.
Thion, M. S., Low, D., Silvin, A., Chen, J., Grisel, P., Schulte-Schrepping, J., et al. (2018). Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell 172, 500–516 e516.
Vakilian, H., Mirzaei, M., Sharifi Tabar, M., Pooyan, P., Habibi Rezaee, L., and Parker, L. (2015). DDX3Y a male-specific region of Y chromosome gene may modulate neuronal differentiation. J. Proteome Res. 14, 3474–3483. doi: 10.1021/acs.jproteome.5b00512
Villa, A., Gelosa, P., Castiglioni, L., Cimino, M., Rizzi, N., Pepe, G., et al. (2018). Sex-specific features of microglia from adult mice. Cell Rep. 23, 3501–3511. doi: 10.1016/j.celrep.2018.05.048
Visootsak, J., and Graham, J. M. (2009). Social function in multiple X and Y chromosome disorders: XXY, XYY, XXYY, XXXY. Dev. Disabil. Res. Rev. 15, 328–332. doi: 10.1002/ddrr.76
Wang, L., Feng, Z., Wang, X., Wang, X., and Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Werling, D. M., and Geschwind, D. H. (2013). Sex differences in autism spectrum disorders. Curr. Opin. Neurol 26, 146–153.
Wolstenholme, J. T., Rissman, E. F., and Bekiranov, S. (2013). Sexual differentiation in the developing mouse brain: contributions of sex chromosome genes. Genes Brain Behav. 12, 166–180. doi: 10.1111/gbb.12010
Xu, J., Burgoyne, P. S., and Arnold, A. P. (2002). Sex differences in sex chromosome gene expression in mouse brain. Hum. Mol. Genet. 11, 1409–1419. doi: 10.1093/hmg/11.12.1409
Xu, J., Watkins, R., and Arnold, A. P. (2006). Sexually dimorphic expression of the X-linked gene Eif2s3x mRNA but not protein in mouse brain. Gene Expr. Patterns 6, 146–155. doi: 10.1016/j.modgep.2005.06.011
Yamauchi, Y., Riel, J. M., Stoytcheva, Z., and Ward, M. A. (2014). Two Y genes can replace the entire Y chromosome for assisted reproduction in the mouse. Science 343, 69–72. doi: 10.1126/science.1242544
Young-Baird, S. K., Shin, B. S., and Dever, T. E. (2019). MEHMO syndrome mutation EIF2S3-I259M impairs initiator Met-tRNAiMet binding to eukaryotic translation initiation factor eIF2. Nucleic Acids Res. 47, 855–867. doi: 10.1093/nar/gky1213
Keywords: autism, Eif2s3y, immune system, sex differences, sexually dimorphic genes, Y chromosome
Citation: Zhang M, Zhou Y, Jiang Y, Lu Z, Xiao X, Ning J, Sun H, Zhang X, Luo H, Can D, Lu J, Xu H and Zhang Y-w (2021) Profiling of Sexually Dimorphic Genes in Neural Cells to Identify Eif2s3y, Whose Overexpression Causes Autism-Like Behaviors in Male Mice. Front. Cell Dev. Biol. 9:669798. doi: 10.3389/fcell.2021.669798
Received: 19 February 2021; Accepted: 12 April 2021;
Published: 06 July 2021.
Edited by:
Zhifang Dong, Chongqing Medical University, ChinaCopyright © 2021 Zhang, Zhou, Jiang, Lu, Xiao, Ning, Sun, Zhang, Luo, Can, Lu, Xu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun-wu Zhang, yunzhang@xmu.edu.cn
 Muxian Zhang
Muxian Zhang Yunqiang Zhou1
Yunqiang Zhou1  Xian Zhang
Xian Zhang Huaxi Xu
Huaxi Xu Yun-wu Zhang
Yun-wu Zhang