Regulation of the Migration of Distinct Dendritic Cell Subsets
- Institute of Basic Medicine, Shandong First Medical University & Shandong Academy of Medical Science, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, China
Dendritic cells (DCs), a class of antigen-presenting cells, are widely present in tissues and apparatuses of the body, and their ability to migrate is key for the initiation of immune activation and tolerogenic immune responses. The importance of DCs migration for their differentiation, phenotypic states, and immunologic functions has attracted widespread attention. In this review, we discussed and compared the chemokines, membrane molecules, and migration patterns of conventional DCs, plasmocytoid DCs, and recently proposed DC subgroups. We also review the promoters and inhibitors that affect DCs migration, including the hypoxia microenvironment, tumor microenvironment, inflammatory factors, and pathogenic microorganisms. Further understanding of the migration mechanisms and regulatory factors of DC subgroups provides new insights for the treatment of diseases, such as infection, tumors, and vaccine preparation.
Introduction
Dendritic cells (DCs) are professional antigen-presenting cells that link innate and adaptive immune responses. In 1973, scientists isolated cells with unique dendritic processes from the peripheral lymphoid organs of mice and named them “dendritic cells” (Steinman and Cohn, 1973). Subsequently, Idoyaga and Steinman (2011) found that DCs participated in adaptive immune response after continuous migration and activation. The function of DCs, whether in maintaining immune tolerance or promoting immunity, require migration to a certain target destination. Recent studying has brought new ideas into the development of different DC subsets in immune responses. Herein, we reviewed the DC subsets that have been reported in recent years and discussed the regulatory factors and molecular mechanisms involved in DC migration. Elucidating the mechanisms underlying the migratory DCs would contribute to the development and function of different DC subsets and their role in diseases.
DC SUBSETS
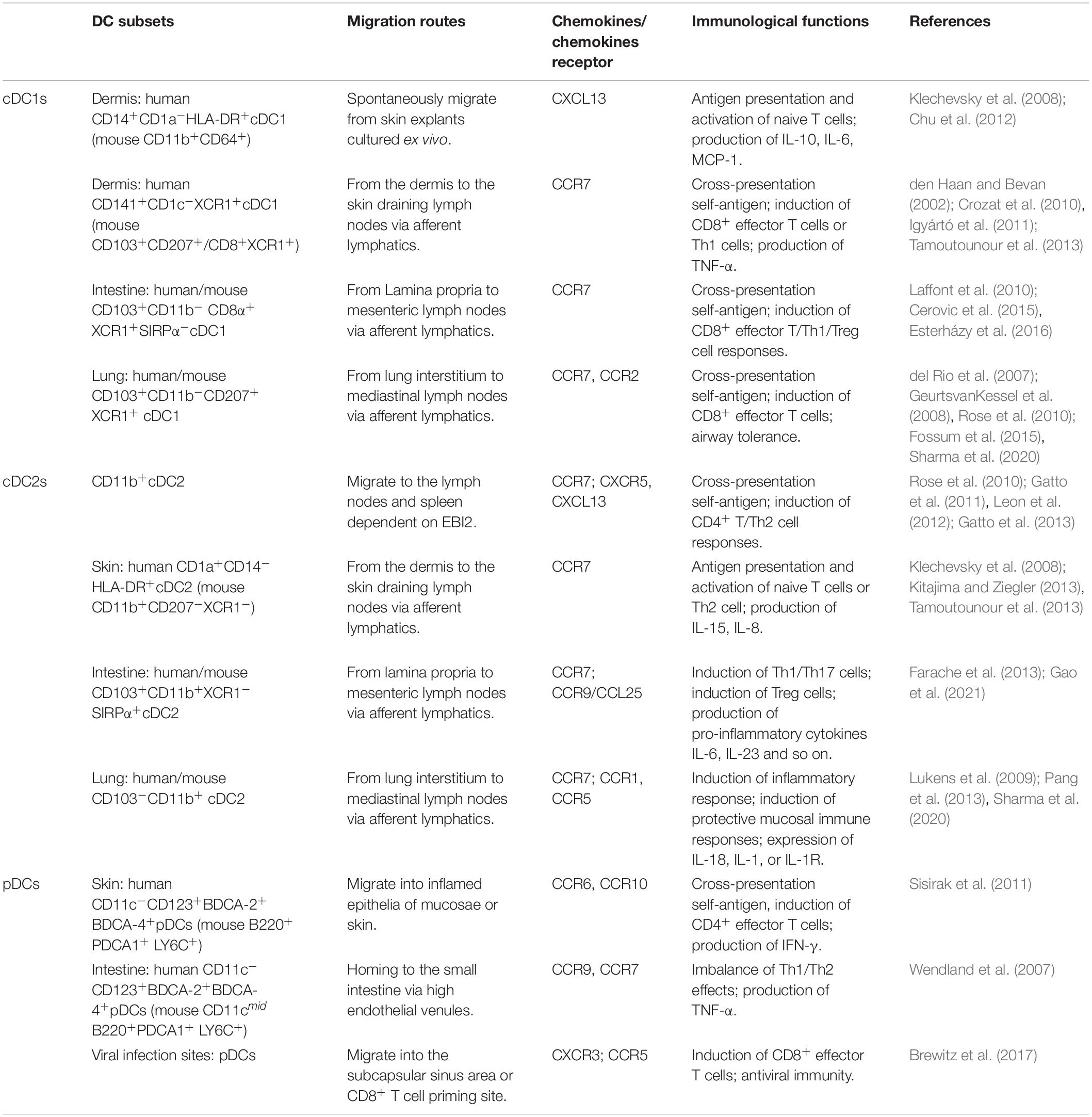
Dendritic cells are highly heterogeneous cells that have historically been categorized by phenotype, function, or location. DCs are unique hematopoietic cells that originate from precursor cells, such as monocytes and pre-DCs, in bone marrow (Naik et al., 2006; Liu et al., 2009; Liu and Nussenzweig, 2010). Precursor cells migrate to peripheral tissues and secondary lymphoid organs via blood circulation and/or lymphatic vessels where they differentiate into myeloid DCs and lymphoid DCs (Naik et al., 2007) (Figure 1). According to specific transcription factors and chemokines, these DCs are further differentiated into three classic subsets: conventional DC1s (cDC1s), conventional DC2s (cDC2s), and plasmacytoid DCs (pDCs). According to their states of maturity, DCs are divided into immature DCs (imDCs), mature DC (mDCs), semi-mature DCs (smDCs), and tolerogenic DCs (tol-DCs). Semi-mature DCs (smDCs), which are an activation state between immature and mature DC cells, are difficult to define (Lutz and Schuler, 2002). These classic DC subsets play a critical role in regulating immune response and immune tolerance.
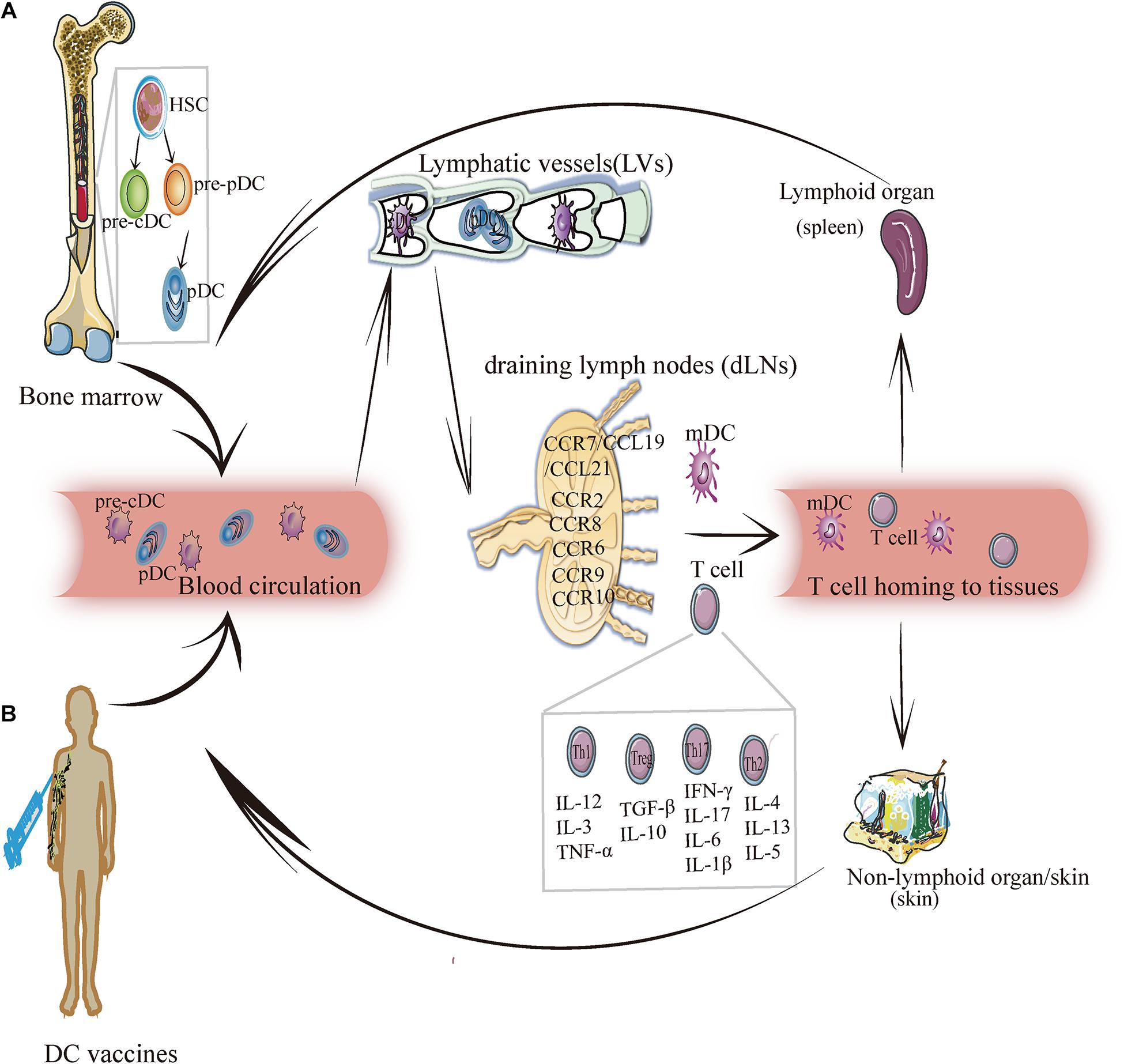
Figure 1. The migration of DC subsets. (A) DC endogenous migration: The precursor DC (pre-DC) develops from hematopoietic stem cells and gradually differentiates into pre-cDC and pDC. Then pDC and pre-cDC migrates from the bone marrow and enters blood circulation (in this case, DC is imDC and pre-cDC differentiates into cDC). Under the mediation of chemokines and cytokines, imDC enters lymphatic vessels, and then reaches draining lymph nodes. In this process, imDC relies upon chemokines (for example: CCR7/CCL19/CCL21, CCR8, CCR6, CCR9, CCR10, etc.) to migrate and transform into mDCs and induce T cells (Th1, Th2, Th17, and Treg) to migrate into lymphoid tissues (such as: the spleen) or non-lymphoid tissues (such as: skin) to exert an immunity effect. (B) DC exogenous migration: After the human body is injected with the DC vaccine, DCs loaded with specific antigens enter the blood circulation, and crawls along the blood vessel wall to reach the lymphatic vessels, and then enters the draining lymph nodes to activate the adaptive immune response, by which it exerts anti-tumorigenic or antiviral effect.
cDC1s
Conventional DC1s widely exist in the blood and peripheral tissues of human and mouse, but their expression is very low in mouse blood. Mouse cDC1s have strong homogeneity in expressing CD8 and/or CD103 (Edelson et al., 2010). Mouse CD8+ cDC1s are identified as CD11chiCD45R–MHCII+ CD8α+DEC205+CD11bloSirpαlo and express C-type lectin Clec9A (DNGR1), Nectin-like protein 2 (Necl2; also called CADM1). Migratory CD103+ DCs in most non-lymphoid tissues are defined as CD11c+MHCII+CD103+CD11bloCX3CR1– F4/80–Sirpα– (McLellan et al., 2002; Bursch et al., 2007; Huysamen et al., 2008; Ginhoux et al., 2009). Both resident CD8a+ and migrating CD103+ cDCs express CD36, CD24, and XCR1 and play a critical role in immunity against intracellular pathogens, viruses, and cancer. In mouse blood, activated cDC1s secrete interleukin (IL)-12p70 and induce the T helper type 1 (Th1) response (Maldonado-Lopez et al., 1999; Farrand et al., 2009). Human CD141+/BDCA-3+ Conventional DC1s are primarily distributed in lymphoid tissues, express C-type lectin receptor 9 (Clec9) and X-C motif chemokine receptor 1 (XCR1), and contribute to antiviral immunity (Silvin et al., 2017), whereas human thymus CD141+cDC1 produces high levels of IL-12 and induces the Th17 response (Vandenabeele et al., 2001). A group of specific DC subgroup Langerin+(CD207+)CD103+CD8+cDC1 was found in the human spleen, and it was a key regulator of immune responses toward blood-borne antigens in the steady-state and during inflammation (Backer et al., 2019). In bacteria-infected human or mouse skin, a subset of CD59+EpCAM+Ly6D+ cDC1 promotes the infiltration of numerous neutrophils by producing the vascular endothelial growth factor (VEGF)-α (Janela et al., 2019). cDC1 contributes to antigen presentation, induces angiogenesis, and promotes inflammation.
The migration of cDC1 is primarily correlated with CXCR3 and CCR7 expression. CXCR3 expression is restricted to mice pre-cDC1 and pDC lineages and is specifically expressed in pre-cDC1 (Siglec-H–Ly6C–) but not pre-cDC2 (Siglec-H–Ly6C+). Trafficking to periphery CCR7-CCL21α–/– interactions guides the migration of pre-cDCs (Lin–CD11c+MHCII–Flt3+Sirpαlo), which accumulate in the thymus, where they may be important for T-cell tolerance (Cosway et al., 2018).
cDC2s
Conventional DC2s have high heterogeneity and play dual roles of immune activation and regulation in the immune response. In the blood, activated cDC2s secrete IL-1β, IL-6, and IL-23 and induce the Th17 response (Persson et al., 2013). In the mouse intestine and thymus, cDC2s can induce the production of regulatory T cells (Treg) (Proietto et al., 2008; Balan et al., 2019). Recently, human cDC2s have been divided into two subsets: (1) CD1cloCLEC10A–CLEC4hi cDC2A expresses a high level of amphiregulin (Areg) and matrix metalloproteinase-9 (MMP-9) but low levels of IL-23, IL-6, and tumor necrosis factor-α (TNF-α). This subset exhibits anti-inflammatory effects. (2) CD1c+CLEC10A+CLEC4lo cDC2B has pro-inflammatory effects with high expression levels of IL-6 and TNF-α (Brown et al., 2019). The corresponding two subgroups of cDC2s in mice are T-bet+ cDC2A and T-bet– cDC2B (Brown et al., 2019), which are different from the previously described cDC2 subsets. Notch2 targeting of CD11c+CD11b+ CD103+ IRF4+ cDC2s was associated with the induction of the Th17 cell response (Lewis et al., 2011), whereas Kruppel-like factor 4 (Klf4)-dependent CD11c+ IRF4+ cDC2s promote Th2, but not Th17 (Tussiwand et al., 2015). In addition, CD9 divided CD11b+cDC2s into two subgroups in B16-F10 tumor-bearing mice, namely CD9–(CD301–)/CD9+(CD301+)CD11b+cDC2s, which are required for activating antitumor CD4+ Tconv (Binnewies et al., 2019). Although cDC2s are divided into many subsets, migratory cDC2s subsets typically require CCR7, whereas extrathymic Sirpα+cDC2s enter the thymus primarily via CCR2 (Tomohisa et al., 2009).
pDCs
Plasmacytoid DCs were first discovered in human lymph nodes (LNs). Human CD11c–CD123+CD303+ pDCs are equivalent to mice PDCA-1+ pDCs. pDC differentiation depends on E2.2 and IRF7, and expresses the CD123/IL-3α chain, CD303 (BDCA-2), CD304 (BDCA-4), and immunosuppressive molecule ILT2, etc. (Dzionek et al., 2000; Swiecki et al., 2010; Mathan et al., 2013). A new subgroup, AXL+AS DCs (SIGLEC1+, SIGLEC6+), exists in human blood and expresses a similar marker as that of pDCs (Villani et al., 2017). Although these subgroups are incapable of proliferation, they can activate T cells and play an antiviral role. Furthermore, pDCs can be converted into cDCs. When transcription factor E2-2 is downregulated or ID2, PU.1, and BATF3 are significantly upregulated, CC-chemokine receptor 9 (CCR9)– pDCs in intestinal epithelial cells (IECs) migrate to peripheral tissues (Chen et al., 2015). Subsequently, they are transformed into CD11b+CD8+MHCII+ cDC-like cells under the stimulation of granulocyte-macrophage colony-stimulating factors (GM-CSF) or soluble factors produced by IEC (Schlitzer et al., 2011). This transformation leads to an imbalance or abnormal distribution of pDC and cDC subpopulations in the body, which induce autoimmune diseases (Chen et al., 2015; Qian and Cao, 2018). CCR4, CCR6, CCR7, CCR9, CCR10, and chemokine-like receptor 1/chemerin receptor 23 (CKLR1/ChemR23) are correlated with pDC migration (Penna et al., 2001; Vermi et al., 2005; Wendland et al., 2007; Sisirak et al., 2011).
tol-DCs
Tolerogenic DCs (tol-DCs) can be derived from monocytes or pre-DCs. GM-CSF and TGF-β1 stimulated mouse liver-derived pre-DCs into tol-DCs, which prolong the survival time of donors in organ transplantation (Bonham et al., 1996; Khanna et al., 2000). These tol-DCs induce Treg cells to exert immune tolerance by secreting large amounts of IL-10. In addition, skin-settled CD141+CD14+ DC are derived from colonized monocytes (Chu et al., 2012; Han et al., 2014), and inhibit the CD4+ T-cell response by secreting IL-10 and IDO. IDO+CD11b+ DC is a subset of tol-DCs (Park et al., 2012) and induces immune tolerance. Tol-DCs are classified as induced tolerogenic DCs (itDCs) and natural tolerogenic DCs (ntDCs). ItDCs contribute to the maintenance of homeostasis under potentially proinflammatory conditions. While under steady-state conditions, ntDCs facilitate the establishment of tolerance. These findings provide insights on a new framework for the use of DC-mediated mechanisms of tolerance to treat diseases (Iberg and Hawiger, 2020).
Mechanism Underlying DC Migration
Migration is the key process through which DCs exercise their uptake, processing, and presentation, and it runs throughout the entire process of DC differentiation and development. DC migration affects its phenotype and maturity, thus resulting in the different localization of different DC subgroups. DCs can directly pass through the blood vessel wall and migrate from peripheral tissues to a specific location or can enter lymph vessels from the bloodstream, from which they are passively transported to the subcapsular sinus (SCS) of the LNs through lymph flow and enter LNs to complete migration (Figure 1A).
Migration Kinetics of DCs
Differentiation and development of DCs occurs in four stages: (1) DC precursors, (2) imDCs, (3) migration DCs, and (4) mDCs. Previous studies have shown that only CD34+ DC precursor cells express E-cadherin, which promotes DC migration and maturity (Mackensen et al., 2000). During acute inflammation, DC precursors quickly mobilize to non-lymphoid tissues. Most DCs in the peripheral organs are imDCs, as immune response sentinel, which can take up antigens more efficiently. With exogenous antigens and inflammatory factor stimulation, imDCs migrate from peripheral tissues to secondary lymphoid areas. During this process, imDCs develop into mDCs, which present antigens and induce the T-cell response in LNs (Caux et al., 2000). Differentiation from imDCs to mDCs depends on migratory DCs. This type of DC exists mainly in lymphatic tissues, input lymphatic vessels, and peripheral blood. Through blood and lymphatic circulation, migratory DCs enter the secondary lymphatic organs from the input lymphatic vessels and drive the DCs to mature.
The dynamic migration process of DCs has an important guiding role in elucidating their homeostasis and pathology in tissues. However, different DC subsets display distinct migration kinetics during migration from skin to the draining LN (dLN). After photoconversion, self-antigens that are present on CD103– dermal DCs are rapidly transported from the skin to the dLN and are responsible for the transport of invading pathogens to the dLN. In contrast, CD103+ DCs reached a plateau on day 3 after photoconversion and participated in antigen cross-presentation (Tomura et al., 2014). Moreover, different DC subsets survey different regions of the spleen to induce specific T cell responses. For example, 33D1+ DC migrates to the periphery of the T cell zone of the spleen to induce CD4+ T cell responses, whereas XCR1+ DC migrates to the center of the T cell zone in the white pulp of spleen to induce CD8+ T cell responses (Calabro et al., 2016). In addition, the migration of localized skin CD1c+/CD14+/CD141+ DC subgroups to the inflammation site depends on CCR7/CXCL10 (Chu et al., 2012; Haniffa et al., 2012, 2015). However, research on the migration of a certain DC subgroup to a specific site in the tissues under steady-state and inflammatory conditions remain insufficient. Furthermore, whether independent DC subsets can selectively induce the T cell response in other immune organs warrants further research.
Essence and Mechanism of Migration
The migration of DCs is a complex and dynamic cyclical process. Cell migration occurs due to interaction between chemokines and chemokine receptors. Under the guidance of chemokines, DCs move to specific sites and exert corresponding biological functions. In addition, adhesion factors, integrins, semaphores, and cytoskeletal proteins play various roles in cell migration. Furthermore, the migration of DCs is essential for T cell responses.
Chemokines
Chemokines and chemokine receptors guide the positioning and chemotaxis effects of DCs at different developmental stages. Chemokines are a class of highly conserved small, secreted proteins that regulate DC migration by identifying chemokine receptors that bind to the DC surface. CCR7 plays an important role in DC migration from peripheral tissues to draining LNs and is a key factor that affects DC migration and function (Yanagihara et al., 1998; Hirao et al., 2000; Randolph et al., 2004). CCR7 ligands CCL19 and CCL21, which are expressed in lymphoid organs mainly, drive DC migration (Elke et al., 2004; Tiberio et al., 2018). CCL21 forms the “CCL21 gradient” by binding to heparan sulfates in the interstitium, thereby providing adhesion for DC migration and guiding DC migration into LNs (Weber et al., 2013). The discovery of chemokines and the chemokine–chemokine receptor axes facilitates research to elucidate the migration mechanism of DCs.
(1) The CCR7-CCL21/CCL19 axis: The CCR7-CCL19/CCL21 chemokine axis is vital for the regulation of adaptive immunity and tolerance by affecting mDC migration from the peripheral tissue to lymphatic vessels and LNs (Forster et al., 2008). Depending on this axis, human skin CD141+CD1c–XCR1+ cDC1 (Igyártó et al., 2011)/CD1a+CD1c+cDC2 (Kitajima and Ziegler, 2013) migrates from the dermis to skin-draining LNs (Tamoutounour et al., 2013) or intestinal CD103+ CD11b–XCR1+ SIRPα–CD141+DNGR1+cDC1 (Olga et al., 2009)/ CD103+ CD11b+ XCR1– SIRPα+ CD141–DNGR–cDC2 (Persson et al., 2013) migrate from the lamina propria to mesenteric LNs (Farache et al., 2013). CCR7-CCL19/CCL21 promote the migration of corneal mDCs to intraocular lymphatic vessels and mediate the CD4+ T cell immune response (Wang et al., 2019). Migrations of Newcastle disease virus-like particles (NDV-VLP)-treated DCs to draining LNs or the spleen rely upon the CCR7-CCL19/CCL21 axis, thus leading to CD4+ T cell activation (Qu et al., 2005). CCR7-CCL19/CCL21-dependent DC migration is involved in the coordination of the activation of specific Tregs, which is beneficial for maintaining peripheral tolerance (Leventhal et al., 2016). This provides new insights for further understanding the role of the CCR7-CCL19/CCL21 axis in maintaining a balance between the adaptive immune response and immune tolerance. Inflammatory factors CCRL1 (called ACKR4) (Ulvmar et al., 2014), transcription factor PU.1 (Yashiro et al., 2019), and IL-18-driven human helper NK cells (Wong et al., 2014) participated in the regulation of DC migration, which may contribute to adaptive immune responses that are associated with infection, cancer, or vaccination.
(2) The leukotriene B4 (LTB4)-BLT1 axis: This axis is critical for regulating DC transport and inducing an adaptive immune response (Del Prete et al., 2006). DCs can be stimulated by LTB4 in vitro and upregulate the expression of CCR7 and CCL19 while promoting chemokines CCL19 and CCL21 to induce DC migration to LNs (Del Prete et al., 2006). This indicates that LTB4 plays an important role in regulating DC migration and inducing adaptive immune responses.
(3) The CXCR4-CXCL12 axis: This axis relies on CCR7 to promote the migration of DCs from peripheral organs to LNs and participate in the migration of DCs across lymphatic endothelial cells and lymphatic vessels, as well as the migration of epidermal DCs to the dermis (Kabashima et al., 2007; Villablanca and Mora, 2008). Thus, CXCR4-CXCL12 is a key axis for DC migration during skin inflammation.
(4) The CCR8-CCL21/CCL8 axis: CCR8 and its ligand CCL21/CCL8 promote DC homing toward LNs (Sokol et al., 2018). In addition, CCR8 and CCL21 coordinate the promotion of CCR7-mediated CD301b+ DC migration from the SCS to LNs and induce Th2 effects. Th2 immunization specifically induces CCL8 expression by CD169+SIGN-R1+ macrophages. CCL8, and CCL21 synergistically promote CD301b+ DC migration (Sokol et al., 2018). These factors may contribute to adaptive immune deviation and cancer cell metastasis associated with DC migration.
Some chemotactic signals can directly activate DC migration or promote the production of chemokines (CXCL12, CXCL14, CCL19, CCL3, etc.), thereby causing secondary recruitment of cells (Majumdar et al., 2014; Tiberio et al., 2018). New paradigms have emerged in the establishment and maintenance of gradients during directed cell migration. Such chemotactic signals include bacterial components, lipid mediators, signaling proteins, and proinflammatory cytokines. For example, cathelin-related antimicrobial peptide (CRAMP), platelet-activating factor (PAF), Activin A, serum amyloid A (SAA), and leukotriene B4 (LTB4). The formylpeptide receptor (Fpr2) expressed on the surface of DCs and CRAMP is jointly involved in the activation and aggregation of DCs involved in allergic airway inflammation (Chen et al., 2014). SAA can directly induce the migration of imDCs via the secondary release of CXCL12 and CXCL14 (Gouwy et al., 2015). Furthermore, the chemokine signals induce faster migration of DCs.
Adhesion Molecules and Proteins
The acquisition of DC migration capacity also depends on the change of its adhesion state. During inflammation, ICAM-1, ICAM-2, Mac-1 (αMβ2), and LFA-1 (αLβ2) play crucial roles in regulating DC migration. The expression of intracellular chemokine CXCR3 promote Mac-1 and LFA-1 binding to their ligand ICAM-1/2, thereby targeting cell adhesion (Springer, 1994). L/E/P-selectin on activated endothelial cells is required for the DC migration process and is involved in the DC homing of lymphoid and peripheral tissues (Tedder et al., 1995; Pendl et al., 2002; Lekakis et al., 2010).
Tetraspanins are expressed on the surface of DCs and control DC migration by coordinating the expression and aggregation of cytokines, selectins, integrins, or other cell–cell proteins on the DC surface (Charrin et al., 2009). P-selectin-independent rolling decreases in the absence of tetraspanin CD63 (Doyle et al., 2011). Tetraspanin CD53 stabilizes L-selectin surface expression and promotes lymphocyte recirculation (Demaria et al., 2020), which indicates that tetraspanin pairs with its partner protein of selectin in co-participation in DC migration.
Rho-associated protein kinases (ROCKs) affect the migration of DCs to draining LNs by mediating the activation of actin nuclear contraction (Nitschké et al., 2012), rapid reconstruction of F-actin throughout DC migration, cell polarity formation, and interaction between cell proteins (Tang and Gerlach, 2017). The actin-related protein 2/3 (Arp2/3) complex mediated F-actin formation of pseudopodia at the front end of this movement and boosted the CCR7-CCL19/CCL21 response axis to induce chemotactic migration of mDCs (Leithner et al., 2016). Under the regulation of the Rho-GTPase signaling pathway, Arp2/3-mediated actin nucleation weakened at the front end of the migration movement (Suraneni et al., 2015), whereas morphogenetic formin-protein-mediated actin nucleation increased at the end of the migration movement (Vargas et al., 2016), which resulted in the rapid migration of mDCs. Calcium ions maintain cell polarity and stabilize the actin cytoskeleton. DC migration requires the participation of a variety of adhesion factors or proteins, which provides a more comprehensive explanation of the mechanism underlying DC migration.
Migration Patterns of Different Cell Subsets
Migrating cDCs and pDCs recruited from the blood to LNs can promote peripheral Treg cells to induce immune tolerance, thereby linking migrating DCs as potential markers for the treatment of autoimmune diseases (Bonasio et al., 2006; Hadeiba et al., 2012). The development of cDCs and pDCs depends on the expression of CCR6/7 but relies upon CCR1/4/8 or CCR2/9/10, respectively (Figure 2).
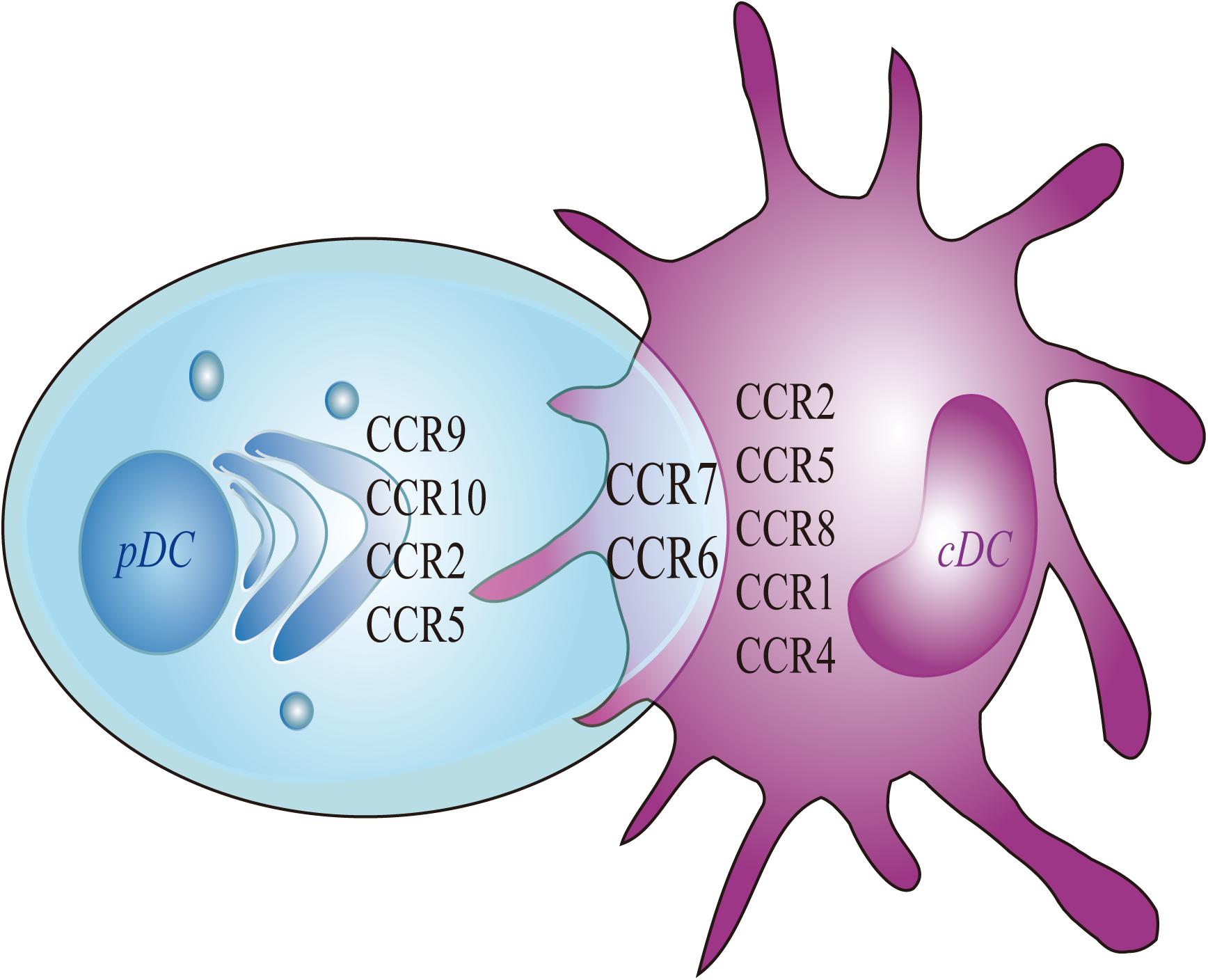
Figure 2. Chemokines of pDC and cDC. Migratory pDC and cDC depends on the expression of CCR7 and CCR6. More specifically, the migration of cDC depends on the expression of CCR8, whereas the migration of pDC depends on the expressions of CCR9 and CCR10. The current study found that the migration of mouse cDC depends on CCR1 and CCR4. The migration of human cDC is dependent on CCR2 and CCR5. At the same time, the expressions of CCR2 and CCR5 are also involved in mediating the migration of mouse pDC.
Although cDCs usually upregulate the expression of CCR7 to induce migration, during inflammation, IRF4–/–CD11b+ cDCs could not upregulate CCR7 expression to induce migration to inflamed skin (Bajana et al., 2012; Plantinga et al., 2013). When Staphylococcus epidermidis infects skin tissue, dermal CD103+ cDC1s carry bacterial antigens that migrate to skin LNs, promote IL-17 secretion, and induce the recruitment of CD8+ T cells to the skin to resist pathogen infection (Farache et al., 2013). The chemotactic receptor Epstein–Barr virus-induced 2 (EBI2) can guide the migration of CD11b+ cDC2 to the LNs and spleen by up-regulating CCR7, CXCR5, and CXCL13 and inducing CD4+ T-cell effects (Gatto et al., 2011; Leon et al., 2012; Gatto et al., 2013). pDCs usually enter LNs through high endothelial veins and assist other DC subsets in performing antigen presentation functions. Upon viral infection, pDCs were directed toward two different sites in the LN, they either migrated to infected macrophages residing in the SCS area in a CXCR3-dependent manner or to CD8+ T cell priming sites in a CCR5-dependent manner. This may be essential to induce antiviral immunity (Brewitz et al., 2017). Moreover, CCR9 mediates pDC migration to the intestine (Wendland et al., 2007). CKLR1/ChemR23 mediates pDC migration to LNs or inflamed skin (Zabel et al., 2005; Albanesi et al., 2009). pDCs also depend on CCR6, CCR7, and CCR10 to complete homing from the blood to inflamed skin (Sisirak et al., 2011). pDCs and cDCs express different chemokine and chemokine receptors which lead them to have different migrate route and functions (Table 1). However, the specific mechanism needs to be further studied.
During inflammation, the migration of imDCs mainly depends on the mediation of E/P-selectin (Pendl et al., 2002). Endothelial selectins are involved in the rolling, extravasation, and migration of imDC in the vascular endothelium. The ChemR23 ligand chemerin can increase the migration of imDCs to endothelial cells with the participation of CCL7 (Gouwy et al., 2013). imDCs and mDCs may have the opposite reactivity to the same chemokine. For example, imDCs have weak reactivity to CC-chemokine-MIP-1b and CXC-chemokine-SDF-1a, but the reverse occurs when imDCs are mature (Lore et al., 1998). In addition, the Rho-mDia1-dependent actin pool is involved in the forward movement of imDCs and the migration of mDCs to lymphatic vessels (Vargas et al., 2016). The migration patterns of different subgroups affect the progress of immune regulation, but the migration mechanism of each subgroup is unclear.
Regulatory Factors Affecting DC Migration
The Hypoxic Microenvironment
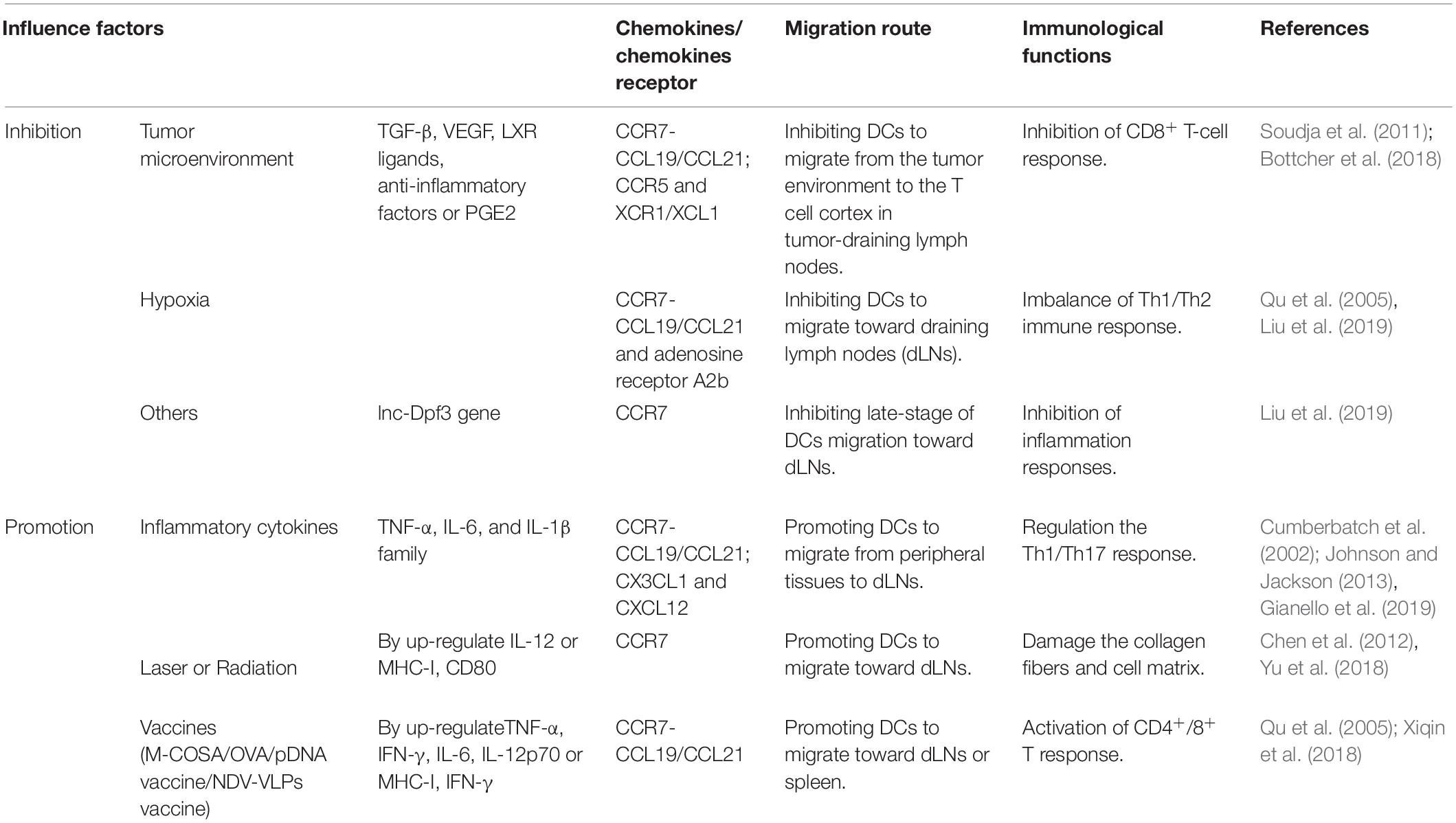
A sufficient oxygen environment is required to maintain the normal development and metabolism of cells. Hypoxia can downregulate the expression of CCR7 and DC surface adenosine receptor A2b, whereas the cyclic AMP/protein kinase A signaling pathway reduces the inhibition of MMP-9/TIMP gene secretion during hypoxia by acting on the A2b receptor (Qu et al., 2005). The downregulation of CCR7 and the change in MMP-9/TIMP gene expression are the main factors that inhibit DC migration, which cause an imbalance in the Th1/Th2 immune response.
Tumor Microenvironment
Elucidating the migration processes of DCs in the tumor microenvironment (TME) can explain how DC-derived cancer vaccines will effectively work in the human body, thus leading to the development of effective vaccines. However, knowledge of the exogenous migration pathway of DC is limited. When tumors occur, increased secretion of TGF-β, VEGF, and LXR ligands and anti-inflammatory factors may recruit DC precursors and convert them into tol-DCs, thereby inhibiting DC maturation and migration to LNs (Soudja et al., 2011). Likely, TGF-β may be involved in DC migration under phosphatidylinositol 3-kinase/Akt activation (Bakin et al., 2000) and increase cell tolerance (Lee et al., 1998). This confirms that the TME can inhibit DC migration. The occurrence of ectopic LNs in tumors can also induce DC migration via CCL21 (Chen et al., 2002; Di Caro et al., 2014). NK cells promote cDC1 accumulation in incipient tumors by producing CCL5 and XCL1/2 (Bottcher et al., 2018). Immunoregulatory factor PEG2 can downregulate the expression of chemokines CCL5, XCL1, XCR1, and CCR5 on cDC1 to inhibit DC accumulation and CD8+ T cell action in the TME (Bottcher et al., 2018). Regulating the expression of chemokines and/or chemokine receptors may interfere with the accumulation of DC in tumors or tumor-draining LNs.
A nano-vaccine containing M-COSA/OVA/pDNA can promote the expression of MHC-I and cytokines (such as IFN-γ) after injection into the human body, enhance antigen presentation as an immune adjuvant, induce DC migration to LNs, and activate CD8+ T-cell effects to inhibit tumor growth (Xiqin et al., 2018). The key to effective DC vaccines involves the migration of DC-carrying antigens to T-cell-rich LN regions. The use of magnetic resonance imaging, fluorescent labeling, and other technical methods to track the migration route of DC in the body enables DC vaccines to target their effects on cancer (de Chickera et al., 2011). In-depth understanding of DC migration routes are conducive to the preparation of targeted DC vaccines.
Inflammation Cytokines
Inflammatory cytokines promote DC migration through paracrine or autocrine signaling and induce the expression of CCR7 and its ligand CCL19/CCL21 in DCs, thereby promoting DC migration (MartIn-Fontecha et al., 2003; Del Prete et al., 2006). The TNF-α, IL-6, and IL-1β families are involved in DC migration to inflammation sites that are under mediation by CCR7, and this process may be related to the Toll-like receptor/transcription factor nuclear factor-κB (TLR/NF-κB) pathway, which can modulate the Th1/Th17 polarization effect (Cumberbatch et al., 2002; Gianello et al., 2019). In addition, CX3CL1 and CXCL12 may participate in DC migration in the inflammatory environment and promote DC migration through the vascular endothelium to lymphatic vessels (Johnson and Jackson, 2013). In the non-inflammatory environment, the atypical Iκβ-dependent pathway activated by NF-κB appears to regulate CCR7 and co-stimulatory molecule expression (Baratin et al., 2015). At the same time, TLR ligands can enhance DCs to express CCR7 and CCL19 and promote DC migration from peripheral tissues to draining LNs (González et al., 2014). Thus, the inflammatory environment or inflammatory signals (TGF-α, IL-1β, IL-6, and IL-12) promote DC maturation and migration depending on CCR7 expression.
Pathogenic Microbes
Invasion by pathogenic microbes affects the migration and location of DC subgroups. In the gut, CD103+CD11b+ cDC2 in the intestinal lamina propria of Salmonella infection upregulate CCR7 and migrate to the IEC layer, which helps epithelial DCs acquire bacterial infections (Farache et al., 2013). A substantial number of CCR2-dependent LY6Chi monocytes that secrete proinflammatory factors and accumulate in the intestine may transform into inf-DCs and then migrate to mesenteric LNs and induce T-cell effects (Zigmond et al., 2012). However, acute intestinal bacterial infection may cause a substantial number of migrating DCs to converge in the adipose tissue area of mesenteric LNs, thereby preventing transfer to mesenteric LNs (Fonseca et al., 2015).
Respiratory syncytial virus (RSA) infection promoted CD11b+ DCs to carry allergens to mediastinal LNs by CCR2/CCL2 and CCR7, which induced Th2 cell immunity and caused allergic asthma (Plantinga et al., 2013). During RSV infection, the G protein receptor EOS1 caused the lung CD103+CD11b+ DC subgroup to migrate to mediastinal LNs (Lukens et al., 2009), thereby up-regulating the expressions of IL-18, IL-1, and IL-1R, which promoted respiratory DC migration and increased the inflammatory response (Pang et al., 2013). NDV-VLPs, an emerging virus vaccine (Qu et al., 2005), up-regulates MHC-II, co-stimulatory molecules, and proinflammatory cytokines TNF-α, IFN-γ, IL-6, and IL-12p70 through the TLR4/NF-κB pathway, thereby effectively activating DC maturity. In addition, NDV-VLPs induce the expression of CCR7 on DCs and cooperate with CCL19/CCL21 to mediate the migration of DCs to draining LNs or the spleen to activate CD4+ T-cell response. These discoveries provides new insight toward the development of similar VLP vaccines.
Notably, the coronavirus disease 2019 (COVID-19) infection also produces a large number of chemokines (CCL2, CCL3, CCL5, CXCL8, CXCL9, CXCL10, etc.), that might promote DCs and/or T cell infiltration into infected sites, thereby causing cytokine storms that destroy lung function and cause a systemic inflammatory response that leads to organ failure (Huang et al., 2020; Rivellese and Prediletto, 2020; Xu et al., 2020). Recently, it was found that inflammatory disease-inflammatory type 2 cDCs (inf-cDC2s) (Bosteels et al., 2020), which are structurally similar to DCs but have the combined functional advantages of monocytes, macrophages, and cDC functionality, exert an anti-inflammatory effect in COVID-19 patients.
Others
Proteomic and transcriptome analyses confirmed that the lnc-Dpf3 gene can negatively inhibit CCR7-mediated HIF-1α activation and glycolysis gene LDHA expression, ultimately negatively regulating CCR7-mediated DC migration and inflammation (Liu et al., 2019). This study shows that deletion of lnc-Dpf3 gene can enhance CCR7-mediated activation of HIF-1α and DC migration and provides direction for research on the expression or role of non-coding long-chain RNAs in DC migration and inflammatory diseases.
In addition, laser irradiation or radiation may damage collagen fibers and the cell matrix of cells, thus causing collagen fibers to become disordered or broken, which affects the local recruitment of DC and promotes the migration of DCs to LNs (Chen et al., 2012; Yu et al., 2018). Laser-irradiated DCs may be accompanied by an increase in MHC-I and CD80 (Chen et al., 2012). It was established that, depending on the ATM/NF-κB signaling pathway, low-dose radiation may increase CCR7-mediated DC migration and is accompanied by an increased secretion of IL-12. Whether external infection factors or internal genetic factors affect DC migration by regulating the expression of chemotactic or adhesion factors in DCs (Table 2), further study of the mechanisms regulating the migration of DCs will elucidate important factors underlying the pathogenesis and immune status of disease.
Conclusion and Outlook
The essence of DC migration involves chemokines, adhesion factors, integrins, and contributing biological activities. Different migration modes eventually lead to differences in the DC phenotype, location, and function. DC migration has a guiding role in the development and functions of DC-tumor or DC-virus vaccines, which are injected and migrated through blood vessels or lymphatic vessels, and eventually to the site of infection or tumor to play a role in antiviral or anti-tumorigenic effects (Figure 1B). Further studies are needed to determine whether the DC-tumor vaccine can effectively reach the local tumor to induce an anti-tumorigenic effect and whether DC-virus vaccine can effectively reach the infected site and elicit antiviral response. The development of transcriptomes, proteomics, and other technologies will provide technical support for more precise expression and regulation of DC migration to achieve a more effective treatment.
Migration from non-lymphoid to lymphoid tissue is a key feature of DCs that regulates immune response. Chemokine/chemokine receptors, integrins, protein receptors, and transcription factors, promote DC migration and specific intra-organ localization. In tumor tissues, removing inhibitory factors on DC migration may activate immunity and anti-tumorgenicity. However, inhibiting DC migration may be related to the inhibition of excessive activation in autoimmune diseases. Targeting CCR7 or other key mediators of DC trafficking may represent more suitable approaches for targeting DCs in diseases. Research on the migration and function of specific DC subgroups in diseases requires further study.
At present, research on genomes that affect DC migration and the migration modes of pDC, cDC, and other DC subgroups are unclear, and there is still a question about how to precisely target the direction of DC migration to make DC vaccines effective in antitumor and antiviral therapies. For the development and clinical application of an effective DC antiviral vaccine for COVID-19, which has rapidly spread around the world since December 2019, an in-depth exploration of changes in DC migration during the immune response to infection by pathogenic microorganisms is key, but greater elucidation is urgently needed.
Author Contributions
WL and MF contributed toward the concept and manuscript writing. SZ, YY, QS, and XL participated in the literature search and discussion. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Natural Science Foundation of China (81500710), Shandong Key Research and Development Project (2019GSF108189), projects of medical and health technology development program in Shandong province (2019WS186), the science and technology program from Shandong Academy of Medical Sciences (2015–2025), and the Innovation Project of Shandong Academy of Medical Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Albanesi, C., Scarponi, C., Pallotta, S., Daniele, R., Bosisio, D., Madonna, S., et al. (2009). Chemerin expression marks early psoriatic skin lesions and correlates with plasmacytoid dendritic cell recruitment. J. Exp. Med. 206, 249–258. doi: 10.1084/jem.20080129
Backer, R. A., Diener, N., and Clausen, B. E. (2019). Langerin(+)CD8(+) dendritic cells in the splenic marginal zone: not so marginal after all. Front. Immunol. 10:741. doi: 10.3389/fimmu.2019.00741
Bajana, S., Roach, K., Turner, S., Paul, J., and Kovats, S. (2012). IRF4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. J. Immunol. 189, 3368–3377. doi: 10.4049/jimmunol.1102613
Bakin, A. V., Tomlinson, A. K., Bhowmick, N. A., Moses, H. L., and Arteaga, C. L. (2000). Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J. Biol. Chem. 275, 36803–36810. doi: 10.1074/jbc.M005912200
Balan, S., Saxena, M., and Bhardwaj, N. (2019). Dendritic cell subsets and locations. Int. Rev. Cell Mol. Biol. 348, 1–68. doi: 10.1016/bs.ircmb.2019.07.004
Baratin, M., Foray, C., Demaria, O., Habbeddine, M., Pollet, E., Maurizio, J., et al. (2015). Homeostatic NF-κB signaling in steady-state migratory dendritic cells regulates immune homeostasis and tolerance. Immunity 42, 627–639. doi: 10.1016/j.immuni.2015.03.003
Binnewies, M., Mujal, A. M., Pollack, J. L., Combes, A. J., Hardison, E. A., Barry, K. C., et al. (2019). Unleashing type-2 dendritic cells to drive protective antitumor CD4(+) T cell. Immunity Cell 177, 556–571. doi: 10.1016/j.cell.2019.02.005
Bonasio, R., Scimone, M. L., Schaerli, P., Grabie, N., Lichtman, A. H., and von Andrian, U. H. (2006). Clonal deletion of thymocytes by circulating dendritic cells homing to the thymus. Nat. Immunol. 7, 1092–1100. doi: 10.1038/ni1385
Bonham, C. A., Lu, L., Banas, R. A., Fontes, P., Rao, A. S., Starzl, T. E., et al. (1996). TGF-beta 1 pretreatment impairs the allostimulatory function of human bone marrow-derived antigen-presenting cells for both naive and primed T cells. Transpl. Immuno. 4, 186–191. doi: 10.1016/s0966-3274(96)80015-3
Bosteels, C., Neyt, K., Vanheerswynghels, M., van Helden, M. J., Sichien, D., Debeuf, N., et al. (2020). Inflammatory Type 2 cDCs Acquire Features of cDC1s and macrophages to orchestrate immunity to respiratory virus infection. Immunity 52, 1039.e9–1056.e9. doi: 10.1016/j.immuni.2020.04.005
Bottcher, J. P., Bonavita, E., Chakravarty, P., Blees, H., Cabeza-Cabrerizo, M., Sammicheli, S., et al. (2018). NK cells stimulate recruitment of cdc1 into the tumor microenvironment promoting cancer immune control. Cell 172, 1022–1037. doi: 10.1016/j.cell.2018.01.004
Brewitz, A., Eickhoff, S., Dähling, S., Quast, T., Bedoui, S., Kroczek, R. A., et al. (2017). CD8+ T cells orchestrate pDC-XCR1+ dendritic cell spatial and functional cooperativity to optimize priming. Immunity 46, 205–219. doi: 10.1016/j.immuni.2017.01.003
Brown, C. C., Gudjonson, H., Pritykin, Y., Deep, D., Lavallée, V., Mendoza, A., et al. (2019). Transcriptional basis of mouse and human dendritic cell heterogeneity. Cell 179, 846.e24–863.e24. doi: 10.1016/j.cell.2019.09.035
Bursch, L. S., Wang, L., Igyarto, B., Kissenpfennig, A., Malissen, B., Kaplan, D. H., et al. (2007). Identification of a novel population of Langerin+ dendritic cells. J. Exp. Med. 204, 3147–3156. doi: 10.1084/jem.20071966
Calabro, S., Liu, D., Gallman, A., Nascimento, M. S., Yu, Z., Zhang, T. T., et al. (2016). Differential intrasplenic migration of dendritic cell subsets tailors adaptive immunity. Cell Rep. 16, 2472–2485. doi: 10.1016/j.celrep.2016.07.076
Caux, C., Ait-Yahia, S., Chemin, K., de Bouteiller, O., Dieu-Nosjean, M. C., Homey, B., et al. (2000). Dendritic cell biology and regulation of dendritic cell trafficking by chemokines. Springer Semin. Immunopathol. 22, 345–369. doi: 10.1007/s002810000053
Cerovic, V., Houston, S. A., Westlund, J., Utriainen, L., Davison, E. S., Scott, C. L., et al. (2015). Lymph-borne CD8α+ dendritic cells are uniquely able to cross-prime CD8+ T cells with antigen acquired from intestinal epithelial cells. Mucosal. Immunol. 8, 38–48. doi: 10.1038/mi.2014.40
Charrin, S., Le Naour, F., Silvie, O., Milhiet, P. E., Boucheix, C., and Rubinstein, E. (2009). Lateral organization of membrane proteins: tetraspanins spin their web. Biochem. J. 420, 133–154. doi: 10.1042/BJ20082422
Chen, K., Xiang, Y., Huang, J., Gong, W., Yoshimura, T., Jiang, Q., et al. (2014). The formylpeptide receptor 2 (Fpr2) and its endogenous ligand cathelin-related antimicrobial peptide (CRAMP) promote dendritic cell maturation. J. Biol. Chem. 289, 17553–17563. doi: 10.1074/jbc.M113.535674
Chen, P., Denniston, A. K., Hirani, S., Hannes, S., and Nussenblatt, R. B. (2015). Role of dendritic cell subsets in immunity and their contribution to noninfectious uveitis. Surv. Ophthalmol. 60, 242–249. doi: 10.1016/j.survophthal.2015.01.003
Chen, S. C., Vassileva, G., Kinsley, D., Holzmann, S., Manfra, D., Wiekowski, M. T., et al. (2002). Ectopic expression of the murine chemokines CCL21a and CCL21b induces the formation of lymph node-like structures in pancreas, but not skin, of transgenic mice. J. Immunol. 168, 1001–1008. doi: 10.4049/jimmunol.168.3.1001
Chen, X., Zeng, Q., and Wu, M. X. (2012). Improved efficacy of dendritic cell-based immunotherapy by cutaneous laser illumination. Clin. Cancer Res. 18, 2240–2249. doi: 10.1158/1078-0432.CCR-11-2654
Chu, C. C., Ali, N., Karagiannis, P., Di Meglio, P., Skowera, A., Napolitano, L., et al. (2012). Resident CD141 (BDCA3)+ dendritic cells in human skin produce IL-10 and induce regulatory T cells that suppress skin inflammation. J. Exp. Med. 209, 935–945. doi: 10.1084/jem.20112583
Cosway, E. J., Ohigashi, I., Schauble, K., Parnell, S. M., Jenkinson, W. E., Luther, S., et al. (2018). Formation of the intrathymic dendritic cell pool requires ccl21-mediated recruitment of ccr7(+) progenitors to the thymus. J. Immunol. 201, 516–523. doi: 10.4049/jimmunol.1800348
Crozat, K., Guiton, R., Contreras, V., Feuillet, V., Dutertre, C. A., Ventre, E., et al. (2010). The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J. Exp. Med. 207, 1283–1292. doi: 10.1084/jem.20100223
Cumberbatch, M., Dearman, R. J., Groves, R. W., Antonopoulos, C., and Kimber, I. (2002). Differential regulation of epidermal langerhans cell migration by interleukins (IL)-1alpha and IL-1beta during irritant- and allergen-induced cutaneous immune responses. Toxicol. Appl. Pharmacol. 182, 126–135. doi: 10.1006/taap.2002.9442
de Chickera, S., Christy, W., Christiane, M., Ronan, F., Paula, F., and Dekaban, G. A. (2011). Cellular MRI as a suitable, sensitive non-invasive modality for correlating in vivo migratory efficiencies of different dendritic cell populations with subsequent immunological outcomes. Int. Immunol. 24, 29–41. doi: 10.1093/intimm/dxr095
Del Prete, A., Shao, W. H., Mitola, S., Santoro, G., Sozzani, S., and Haribabu, B. (2006). Regulation of dendritic cell migration and adaptive immune response by leukotriene B4 receptors: a role for LTB4 in up-regulation of CCR7 expression and function. Blood 109, 626–631. doi: 10.1182/blood-2006-02-003665
del Rio, M. L., Rodriguez-Barbosa, J. I., Kremmer, E., and Förster, R. (2007). CD103– and CD103+ bronchial lymph node dendritic cells are specialized in presenting and cross-presenting innocuous antigen to CD4+ and CD8+ T cells. J. Immunol. 178, 6861–6866. doi: 10.4049/jimmunol.178.11.6861
Demaria, M. C., Yeung, L., Peeters, R., Wee, J. L., Mihaljcic, M., Jones, E. L., et al. (2020). Tetraspanin CD53 promotes lymphocyte recirculation by stabilizing L-Selectin surface expression. iScience 23:101104. doi: 10.1016/j.isci.2020.101104
den Haan, J. M., and Bevan, M. J. (2002). Constitutive versus activation-dependent cross-presentation of immune complexes by CD8(+) and CD8(-) dendritic cells in vivo. J. Exp. Med. 196, 817–827. doi: 10.1084/jem.20020295
Di Caro, G., Bergomas, F., Grizzi, F., Doni, A., Bianchi, P., Malesci, A., et al. (2014). Occurrence of tertiary lymphoid tissue is associated with T-Cell infiltration and predicts better prognosis in early-stage colorectal cancers. Clin. Cancer Res. 20, 2147–2158. doi: 10.1158/1078-0432.CCR-13-2590
Doyle, E. L., Ridger, V., Ferraro, F., Turmaine, M., Saftig, P., and Cutler, D. F. (2011). CD63 is an essential cofactor to leukocyte recruitment by endothelial P-selectin. Blood 118, 4265–4273. doi: 10.1182/blood-2010-11-321489
Dzionek, A., Fuchs, A., Schmidt, P., Cremer, S., Zysk, M., Miltenyi, S., et al. (2000). BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J. Immunol. 165, 6037–6046. doi: 10.4049/jimmunol.165.11.6037
Edelson, B. T., Kc, W., Juang, R., Kohyama, M., Benoit, L. A., Klekotka, P. A., et al. (2010). Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med. 207, 823–836. doi: 10.1084/jem.20091627
Elke, S., Ying, M., Legler, D. F., Silke, G., Ladislav, P., Burkhard, L., et al. (2004). CCL19/CCL21-triggered signal transduction and migration of dendritic cells requires prostaglandin E2. Blood 103, 1595–1601. doi: 10.1182/blood-2003-05-1643
Esterházy, D., Loschko, J., London, M., Jove, V., Oliveira, T. Y., Mucida, D., et al. (2016). Classical dendritic cells are required for dietary antigen-mediated induction of peripheral T(reg) cells and tolerance. Nat. Immunol. 17, 545–555. doi: 10.1038/ni.3408
Farache, J., Koren, I., Milo, I., Gurevich, I., Kim, K. W., Zigmond, E., et al. (2013). Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity 38, 581–595. doi: 10.1016/j.immuni.2013.01.009
Farrand, K. J., Dickgreber, N., Stoitzner, P., Ronchese, F., Petersen, T. R., and Hermans, I. F. (2009). Langerin+ CD8alpha+ dendritic cells are critical for cross-priming and IL-12 production in response to systemic antigens. J. Immunol. 183, 7732–7742. doi: 10.4049/jimmunol.0902707
Fonseca, D. M. D., Hand, T. W., Han, S., Gerner, M. Y., Zaretsky, A. G., Byrd, A. L., et al. (2015). Microbiota-Dependent sequelae of acute infection compromise tissue-specific immunity. Cell 163, 354–366. doi: 10.1016/j.cell.2015.08.030
Forster, R., Davalos-Misslitz, A. C., and Rot, A. (2008). CCR7 and its ligands: balancing immunity and tolerance. Nat. Rev. Immunol. 8, 362–371. doi: 10.1038/nri2297
Fossum, E., Grødeland, G., Terhorst, D., Tveita, A. A., Vikse, E., Mjaaland, S., et al. (2015). Vaccine molecules targeting Xcr1 on cross-presenting DCs induce protective CD8+ T-cell responses against influenza virus. Eur. J. Immunol. 45, 624–635. doi: 10.1002/eji.201445080
Gao, W., Wang, Y., Bi, J., Chen, X., Li, N., Wang, Y., et al. (2021). Impaired CCR9/CCL25 signalling induced by inefficient dendritic cells contributes to intestinal immune imbalance in nonalcoholic steatohepatitis. Biochem. Biophys. Res. Commun. 534, 34–40. doi: 10.1016/j.bbrc.2020.12.007
Gatto, D., Wood, K., and Brink, R. (2011). EBI2 operates independently of but in cooperation with CXCR5 and CCR7 to direct B cell migration and organization in follicles and the germinal center. J. Immunol. 187, 4621–4628. doi: 10.4049/jimmunol.1101542
Gatto, D., Wood, K., Caminschi, I., Murphy-Durland, D., Schofield, P., Christ, D., et al. (2013). The chemotactic receptor EBI2 regulates the homeostasis, localization and immunological function of splenic dendritic cells. Nat. Immunol. 14, 446–453. doi: 10.1038/ni.2555
GeurtsvanKessel, C. H., Willart, M. A., van Rijt, L. S., Muskens, F., Kool, M., Baas, C., et al. (2008). Clearance of influenza virus from the lung depends on migratory langerin+CD11b– but not plasmacytoid dendritic cells. J. Exp. Med. 205, 1621–1634. doi: 10.1084/jem.20071365
Gianello, V., Salvi, V., Parola, C., Moretto, N., Facchinetti, F., Civelli, M., et al. (2019). The PDE4 inhibitor CHF6001 modulates pro-inflammatory cytokines, chemokines and Th1- and Th17-polarizing cytokines in human dendritic cells. Biochem. Pharmacol. 163, 371–380. doi: 10.1016/j.bcp.2019.03.006
Ginhoux, F., Liu, K., Helft, J., Bogunovic, M., Greter, M., Hashimoto, D., et al. (2009). The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206, 3115–3130. doi: 10.1084/jem.20091756
González, F. E., Ortiz, C., Reyes, M., Dutzan, N., Patel, V., Pereda, C., et al. (2014). Melanoma cell lysate induces CCR7 expression andin vivo migration to draining lymph nodes of therapeutic human dendritic cells. Immunology 142, 396–405. doi: 10.1111/imm.12264
Gouwy, M., De Buck, M., P Rtner, N. M., Opdenakker, G., Proost, P., Struyf, S., et al. (2015). Serum amyloid a chemoattracts immature dendritic cells and indirectly provokes monocyte chemotaxis by induction of cooperating CC and CXC chemokines. Eur. J. Immunol. 45, 101–112. doi: 10.1002/eji.201444818
Gouwy, M., Struyf, S., Leutenez, L., Pörtner, N., Sozzani, S., and Van Damme, J. (2013). Chemokines and other GPCR ligands synergize in receptor-mediated migration of monocyte-derived immature and mature dendritic cells. Immunobiology 219, 218–229. doi: 10.1016/j.imbio.2013.10.004
Hadeiba, H., Lahl, K., Edalati, A., Oderup, C., Habtezion, A., Pachynski, R., et al. (2012). Plasmacytoid dendritic cells transport peripheral antigens to the thymus to promote central tolerance. Immunity 36, 438–450. doi: 10.1016/j.immuni.2012.01.017
Han, Y., Chen, Z., Yang, Y., Jiang, Z., Gu, Y., Liu, Y., et al. (2014). Human CD14+ CTLA-4+ regulatory dendritic cells suppress T-cell response by cytotoxic T-lymphocyte antigen-4-dependent IL-10 and indoleamine-2,3-dioxygenase production in hepatocellular carcinoma. Hepatology 59, 567–579. doi: 10.1002/hep.26694
Haniffa, M., Gunawan, M., and Jardine, L. (2015). Human skin dendritic cells in health and disease. J. Dermatol. Sci. 77, 85–92. doi: 10.1016/j.jdermsci.2014.08.012
Haniffa, M., Shin, A., Bigley, V., McGovern, N., Teo, P., See, P., et al. (2012). Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity 37, 60–73. doi: 10.1016/j.immuni.2012.04.012
Hirao, M., Onai, N., Hiroishi, K., Watkins, S. C., Matsushima, K., Robbins, P. D., et al. (2000). CC chemokine receptor-7 on dendritic cells is induced after interaction with apoptotic tumor cells: critical role in migration from the tumor site to draining lymph nodes. Cancer Res. 60, 2209–2217. doi: 10.1109/TPAS.1984.318706
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet 395, 497–506. doi: 10.1016/S0140-6736(20)30183-5
Huysamen, C., Willment, J. A., Dennehy, K. M., and Brown, G. D. (2008). CLEC9A is a novel activation C-type lectin-like receptor expressed on BDCA3+ dendritic cells and a subset of monocytes. J. Biol. Chem. 283, 16693–16701. doi: 10.1074/jbc.M709923200
Iberg, C. A., and Hawiger, D. (2020). Natural and induced tolerogenic dendritic cells. J. Immunol. 204, 733–744. doi: 10.4049/jimmunol.1901121
Idoyaga, J., and Steinman, R. M. (2011). SnapShot: dendritic cells. Cell 146:660. doi: 10.1016/j.cell.2011.08.010
Igyártó, B. Z., Haley, K., Ortner, D., Bobr, A., Gerami-Nejad, M., Edelson, B. T., et al. (2011). Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity 35, 260–272. doi: 10.1016/j.immuni.2011.06.005
Janela, B., Patel, A. A., Lau, M. C., Goh, C. C., Msallam, R., Kong, W. T., et al. (2019). A subset of Type I conventional dendritic cells controls cutaneous bacterial infections through vegfalpha-mediated recruitment of neutrophils. Immunity 50, 1069–1083. doi: 10.1016/j.immuni.2019.03.001
Johnson, L. A., and Jackson, D. G. (2013). The chemokine CX3CL1 promotes trafficking of dendritic cells through inflamed lymphatics. J. Cell Sci. 126, 5259–5270. doi: 10.1242/jcs.135343
Rose, C. E. Jr., Lannigan, J. A., Kim, P., Lee, J. J., Fu, S. M., Sung, S. S., et al. (2010). Murine lung eosinophil activation and chemokine production in allergic airway inflammation. Cell Mol. Immunol. 7, 361–374. doi: 10.1038/cmi.2010.31
Kabashima, K., Shiraishi, N., Sugita, K., Mori, T., Onoue, A., Kobayashi, M., et al. (2007). CXCL12-CXCR4 engagement is required for migration of cutaneous dendritic cells. Am. J. Pathol. 171, 1249–1257. doi: 10.2353/ajpath.2007.070225
Khanna, A., Morelli, A. E., Zhong, C., Takayama, T., Lu, L., and Thomson, A. W. (2000). Effects of liver-derived dendritic cell progenitors on Th1- and Th2-like cytokine responses in vitro and in vivo. J. Immunol. 164, 1346–1354. doi: 10.4049/jimmunol.164.3.1346
Kitajima, M., and Ziegler, S. F. (2013). Cutting edge: identification of the thymic stromal lymphopoietin-responsive dendritic cell subset critical for initiation of type 2 contact hypersensitivity. J. Immunol. 191, 4903–4907. doi: 10.4049/jimmunol.1302175
Klechevsky, E., Morita, R., Liu, M., Cao, Y., Coquery, S., Thompson-Snipes, L., et al. (2008). Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity 29, 497–510. doi: 10.1016/j.immuni.2008.07.013
Laffont, S., Siddiqui, K. R., and Powrie, F. (2010). Intestinal inflammation abrogates the tolerogenic properties of MLN CD103+ dendritic cells. Eur. J. Immunol. 40, 1877–1883. doi: 10.1002/eji.200939957
Lee, W. C., Zhong, C., Qian, S., Wan, Y., Gauldie, J., Mi, Z., et al. (1998). Phenotype, function, and in vivo migration and survival of allogeneic dendritic cell progenitors genetically engineered to express TGF-beta. Transplantation 66, 1810–1817. doi: 10.1097/00007890-199812270-00040
Leithner, A., Eichner, A., Muller, J., Reversat, A., Brown, M., Schwarz, J., et al. (2016). Diversified actin protrusions promote environmental exploration but are dispensable for locomotion of leukocytes. Nat. Cell. Biol. 18, 1253–1259. doi: 10.1038/ncb3426
Lekakis, J., Ikonomidis, I., Papoutsi, Z., Moutsatsou, P., Nikolaou, M., Parissis, J., et al. (2010). Selective serotonin re-uptake inhibitors decrease the cytokine-induced endothelial adhesion molecule expression, the endothelial adhesiveness to monocytes and the circulating levels of vascular adhesion molecules. Int. J. Cardiol. 139, 150–158. doi: 10.1016/j.ijcard.2008.10.010
Leon, B., Ballesteros-Tato, A., Browning, J. L., Dunn, R., Randall, T. D., and Lund, F. E. (2012). Regulation of T(H)2 development by CXCR5+ dendritic cells and lymphotoxin-expressing B cells. Nat. Immunol. 13, 681–690. doi: 10.1038/ni.2309
Leventhal, D. S., Gilmore, D. C., Berger, J. M., Nishi, S., Lee, V., Malchow, S., et al. (2016). Dendritic cells coordinate the development and homeostasis of organ-specific regulatory T cells. Immunity 44, 847–859. doi: 10.1016/j.immuni.2016.01.025
Lewis, K. L., Caton, M. L., Bogunovic, M., Greter, M., Grajkowska, L. T., Ng, D., et al. (2011). Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity 35, 780–791. doi: 10.1016/j.immuni.2011.08.013
Liu, J., Zhang, X., Chen, K., Cheng, Y., Liu, S., Xia, M., et al. (2019). CCR7 chemokine receptor-inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1α-mediated glycolysis. Immunity 50, 600–615. doi: 10.1016/j.immuni.2019.01.021
Liu, K., and Nussenzweig, M. C. (2010). Origin and development of dendritic cells. Immunol. Rev. 234, 45–54. doi: 10.1111/j.0105-2896.2009.00879.x
Liu, K., Victora, G. D., Schwickert, T. A., Guermonprez, P., Meredith, M. M., Yao, K., et al. (2009). In vivo analysis of dendritic cell development and homeostasis. Science 324, 392–397. doi: 10.1126/science.1170540
Lore, K., Sonnerborg, A., Spetz, A. L., Andersson, U., and Andersson, J. (1998). Immunocytochemical detection of cytokines and chemokines in Langerhans cells and in vitro derived dendritic cells. J. Immunol. Methods 214, 97–111. doi: 10.1016/s0022-1759(98)00040-4
Lukens, M. V., Kruijsen, D., Coenjaerts, F. E. J., Kimpen, J. L. L., and van Bleek, G. M. (2009). Respiratory syncytial virus-induced activation and migration of respiratory dendritic cells and subsequent antigen presentation in the lung-draining lymph node. J. Virol. 83, 7235–7243. doi: 10.1128/JVI.00452-09
Lutz, M. B., and Schuler, G. (2002). Immature, semi-mature and fully mature dendritic cells: which signals induce tolerance or immunity? Trends Immunol. 23, 445–449. doi: 10.1016/s1471-4906(02)02281-0
Mackensen, A., Herbst, B., Chen, J. L., Kohler, G., Noppen, C., Herr, W., et al. (2000). Phase I study in melanoma patients of a vaccine with peptide-pulsed dendritic cells generated in vitro from CD34(+) hematopoietic progenitor cells. Int. J. Cancer 86, 385–392. doi: 10.1002/(sici)1097-0215(20000501)86:3<385::aid-ijc13<3.0.co;2-t
Majumdar, R., Sixt, M., and Parent, C. A. (2014). New paradigms in the establishment and maintenance of gradients during directed cell migration. Curr. Opin Cell Biol. 30, 33–40. doi: 10.1016/j.ceb.2014.05.010
Maldonado-Lopez, R., De Smedt, T., Michel, P., Godfroid, J., Pajak, B., Heirman, C., et al. (1999). CD8alpha+ and CD8alpha– subclasses of dendritic cells direct the development of distinct T helper cells in vivo. J. Exp. Med. 189, 587–592. doi: 10.1084/jem.189.3.587
MartIn-Fontecha, A., Sebastiani, S., Hopken, U. E., Uguccioni, M., Lipp, M., Lanzavecchia, A., et al. (2003). Regulation of dendritic cell migration to the draining lymph node: impact on T lymphocyte traffic and priming. J. Exp. Med. 198, 615–621. doi: 10.1084/jem.20030448
Mathan, T. S., Figdor, C. G., and Buschow, S. I. (2013). Human plasmacytoid dendritic cells: from molecules to intercellular communication network. Front. Immunol. 4:372. doi: 10.3389/fimmu.2013.00372
McLellan, A. D., Kapp, M., Eggert, A., Linden, C., Bommhardt, U., Brocker, E. B., et al. (2002). Anatomic location and T-cell stimulatory functions of mouse dendritic cell subsets defined by CD4 and CD8 expression. Blood 99, 2084–2093. doi: 10.1182/blood.v99.6.2084
Naik, S. H., Metcalf, D., van Nieuwenhuijze, A., Wicks, I., Wu, L., O’Keeffe, M., et al. (2006). Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat. Immunol. 7, 663–671. doi: 10.1038/ni1340
Naik, S. H., Sathe, P., Park, H., Metcalf, D., Proietto, A. I., Dakic, A., et al. (2007). Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8, 1217–1226. doi: 10.1038/ni1522
Nitschké, M., Aebischer, D., Abadier, M., Haener, S., Lucic, M., Vigl, B., et al. (2012). Differential requirement for ROCK in dendritic cell migration within lymphatic capillaries in steady-state and inflammation. Blood 120, 2249–2258. doi: 10.1182/blood-2012-03-417923
Olga, S., Elin, J., Emma, K. P., Xiaosun, L., Tim, W., William, A. W., et al. (2009). Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 206, 3101–3114. doi: 10.1084/jem.20091925
Pang, I. K., Ichinohe, T., and Iwasaki, A. (2013). IL-1R signaling in dendritic cells replaces pattern recognition receptors to promote CD8+ T cell responses to influenza A virus. Nat. Immunol. 14, 246–253. doi: 10.1038/ni.2514
Park, M. J., Park, K. S., Park, H. S., Cho, M. L., Hwang, S. Y., Min, S. Y., et al. (2012). A distinct tolerogenic subset of splenic IDO(+)CD11b(+) dendritic cells from orally tolerized mice is responsible for induction of systemic immune tolerance and suppression of collagen-induced arthritis. Cell Immunol. 278, 45–54. doi: 10.1016/j.cellimm.2012.06.009
Pendl, G. G., Robert, C., Steinert, M., Thanos, R., Eytner, R., Borges, E., et al. (2002). Immature mouse dendritic cells enter inflamed tissue, a process that requires E- and P-selectin, but not P-selectin glycoprotein ligand 1. Blood 99:946. doi: 10.1182/blood.v99.3.946
Penna, G., Sozzani, S., and Adorini, L. (2001). Cutting edge: selective usage of chemokine receptors by plasmacytoid dendritic cells. J. Immunol. 167, 1862–1866. doi: 10.4049/jimmunol.167.4.1862
Persson, E. K., Uronen-Hansson, H., Semmrich, M., Rivollier, A., Hagerbrand, K., Marsal, J., et al. (2013). IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity 38, 958–969. doi: 10.1016/j.immuni.2013.03.009
Plantinga, M., Guilliams, M., Vanheerswynghels, M., Deswarte, K., Branco-Madeira, F., Toussaint, W., et al. (2013). Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity 38, 322–335. doi: 10.1016/j.immuni.2012.10.016
Proietto, A. I., van Dommelen, S., Zhou, P., Rizzitelli, A., D’Amico, A., Steptoe, R. J., et al. (2008). Dendritic cells in the thymus contribute to T-Regulatory cell induction. Proc. Natl. Acad. Sci. U.S.A. 105, 19869–19874. doi: 10.1073/pnas.0810268105
Qian, C., and Cao, X. (2018). Dendritic cells in the regulation of immunity and inflammation. Semin. Immunol. 35, 3–11. doi: 10.1016/j.smim.2017.12.002
Qu, X., Mei-Xiang, Y., Bei-Hua, K., Lan, Q., Queenie Lai, K. L., Shi, Y., et al. (2005). Hypoxia inhibits the migratory capacity of human monocyte-derived dendritic cells. Immunol. Cell Bio. 83, 668–673. doi: 10.1111/j.1440-1711.2005.01383.x
Randolph, G. J., Sanchez-Schmitz, G., and Angeli, V. (2004). Factors and signals that govern the migration of dendritic cells via lymphatics: recent advances. Springer Semin. Immunopathol. 26, 273–287. doi: 10.1007/s00281-004-0168-0
Rivellese, F., and Prediletto, E. (2020). ACE2 at the centre of COVID-19 from paucisymptomatic infections to severe pneumonia. Autoimmun. Rev. 19:102536. doi: 10.1016/j.autrev.2020.102536
Schlitzer, A., Loschko, J., Mair, K., Vogelmann, R., and Krug, A. (2011). Identification of CCR9- murine plasmacytoid DC precursors with plasticity to differentiate into conventional DCs. Blood 117, 6562–6570. doi: 10.1182/blood-2010-12-326678
Sharma, P., Levy, O., and Dowling, D. J. (2020). The TLR5 agonist flagellin shapes phenotypical and functional activation of lung mucosal antigen presenting cells in neonatal mice. Front. Immunol. 11:171. doi: 10.3389/fimmu.2020.00171
Silvin, A., Yu, C. I., Lahaye, X., Imperatore, F., and Manel, N. (2017). Constitutive resistance to viral infection in human CD141+ dendritic cells. Sci. Immunol. 2:aai8071. doi: 10.1126/sciimmunol.aai8071
Sisirak, V., Vey, N., Vanbervliet, B., Duhen, T., Puisieux, I., Homey, B., et al. (2011). CCR6/CCR10-mediated plasmacytoid dendritic cell recruitment to inflamed epithelia after instruction in lymphoid tissues. Blood 118, 5130–5140. doi: 10.1182/blood-2010-07-295626
Sokol, C. L., Camire, R. B., Jones, M. C., and Luster, A. D. (2018). The chemokine receptor CCR8 promotes the migration of Dendritic Cells into the Lymph Node Parenchyma to initiate the allergic immune response. Immunity 49, 449–463. doi: 10.1016/j.immuni.2018.07.012
Soudja, S. M., Henri, S., Mello, M., Chasson, L., Mas, A., Wehbe, M., et al. (2011). Disrupted lymph node and splenic stroma in mice with induced inflammatory melanomas is associated with impaired recruitment of T and dendritic cells. PLoS One 6:e22639. doi: 10.1371/journal.pone.0022639
Springer, T. A. (1994). Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell 76, 301–314. doi: 10.1016/0092-8674(94)90337-9
Steinman, R. M., and Cohn, Z. A. (1973). Identification of a novel cell type in peripheral lymphoid organs of mice. I. morphology, quantitation, tissue distribution. J. Exp. Med. 137, 1142–1162. doi: 10.1084/jem.137.5.1142
Suraneni, P., Fogelson, B., Rubinstein, B., Noguera, P., Volkmann, N., Hanein, D., et al. (2015). A mechanism of leading-edge protrusion in the absence of Arp2/3 complex. Mol. Biol. Cell 26, 901–912. doi: 10.1091/mbc.E14-07-1250
Swiecki, M., Gilfillan, S., Vermi, W., Wang, Y., and Colonna, M. (2010). Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity 33, 955–966. doi: 10.1016/j.immuni.2010.11.020
Tamoutounour, S., Guilliams, M., Montanana, S. F., Liu, H., Terhorst, D., Malosse, C., et al. (2013). Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 39, 925–938. doi: 10.1016/j.immuni.2013.10.004
Tang, D. D., and Gerlach, B. D. (2017). The roles and regulation of the actin cytoskeleton, intermediate filaments and microtubules in smooth muscle cell migration. Respir Res. 18, 54. doi: 10.1186/s12931-017-0544-7
Tedder, T. F., Steeber, D. A., Chen, A., and Engel, P. (1995). The selectins: vascular adhesion molecules. FASEB J. 9, 866–873. doi: 10.1006/excr.1995.1233
Tiberio, L., Del Prete, A., Schioppa, T., Sozio, F., Bosisio, D., and Sozzani, S. (2018). Chemokine and chemotactic signals in dendritic cell migration. Cell Mol. Immunol. 15, 346–352. doi: 10.1038/s41423-018-0005-3
Tomohisa, B., Yasunari, N., and Naofumi, M. (2009). Crucial contribution of thymic Sirp alpha+ conventional dendritic cells to central tolerance against blood-borne antigens in a CCR2-dependent manner. J. Immunol. 183, 3053–3063. doi: 10.4049/jimmunol.0900438
Tomura, M., Hata, A., Matsuoka, S., Shand, F. H., Nakanishi, Y., Ikebuchi, R., et al. (2014). Tracking and quantification of dendritic cell migration and antigen trafficking between the skin and lymph nodes. Sci. Rep. 4:6030. doi: 10.1038/srep06030
Tussiwand, R., Everts, B., Grajales-Reyes, G. E., Kretzer, N. M., Iwata, A., Bagaitkar, J., et al. (2015). Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 42, 916–928. doi: 10.1016/j.immuni.2015.04.017
Ulvmar, M. H., Werth, K., Braun, A., Kelay, P., Hub, E., Eller, K., et al. (2014). The atypical chemokine receptor CCRL1 shapes functional CCL21 gradients in lymph nodes. Nat. Immunol. 15, 623–630. doi: 10.1038/ni.2889
Vandenabeele, S., Hochrein, H., Mavaddat, N., Winkel, K., and Shortman, K. (2001). Human thymus contains 2 distinct dendritic cell populations. Blood 97, 1733–1741. doi: 10.1182/blood.v97.6.1733
Vargas, P., Maiuri, P., Bretou, M., Saez, P. J., Pierobon, P., Maurin, M., et al. (2016). Innate control of actin nucleation determines two distinct migration behaviours in dendritic cells. Nat. Cell Biol. 18, 43–53. doi: 10.1038/ncb3284
Vermi, W., Riboldi, E., Wittamer, V., Gentili, F., Luini, W., Marrelli, S., et al. (2005). Role of ChemR23 in directing the migration of myeloid and plasmacytoid dendritic cells to lymphoid organs and inflamed skin. J. Exp. Med. 201, 509–515. doi: 10.1084/jem.20041310
Villablanca, E. J., and Mora, J. R. (2008). A two-step model for Langerhans cell migration to skin-draining LN. Eur. J. Immunol. 38, 2975–2980. doi: 10.1002/eji.200838919
Villani, A. C., Satija, R., Reynolds, G., Sarkizova, S., Shekhar, K., Fletcher, J., et al. (2017). Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356:eaah4573. doi: 10.1126/science.aah4573
Wang, T., Li, W., Cheng, H., Zhong, L., Deng, J., and Ling, S. (2019). The important role of the chemokine axis CCR7-CCL19 and CCR7-CCL21 in the Pathophysiology of the Immuno-inflammatory response in Dry Eye Disease. Ocul. Immunol. Inflamm. [Online ahead of print] 1–12. doi: 10.1080/09273948.2019.1674891
Weber, M., Hauschild, R., Schwarz, J., Moussion, C., de Vries, I., Legler, D., et al. (2013). Interstitial dendritic cell guidance by haptotactic chemokine gradients. Science 339, 328–332.
Wendland, M., Czeloth, N., Mach, N., Malissen, B., Kremmer, E., Pabst, O., et al. (2007). CCR9 is a homing receptor for plasmacytoid dendritic cells to the small intestine. Proc. Natl. Acad. Sci. U.S.A. 104, 6347–6352. doi: 10.1073/pnas.0609180104
Wong, J. L., Muthuswamy, R., Bartlett, D. L., and Kalinski, P. (2014). IL-18-based combinatorial adjuvants promote the intranodal production of CCL19 by NK cells and dendritic cells of cancer patients. OncoImmunology 2:e26245. doi: 10.4161/onci.26245
Xiqin, Y., Keke, L., Tingting, M., Xuan, L., Jing, M., Yanan, T., et al. (2018). Immune adjuvant targeting micelles allow efficient dendritic cell migration to lymph nodes for enhanced cellular immunity. ACS Appl. Mater. Interfaces 10, 33532–33544. doi: 10.1021/acsami.8b10081
Xu, H., Zhong, L., Deng, J., Peng, J., Dan, H., Zeng, X., et al. (2020). High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 12, 77–81. doi: 10.1038/s41368-020-0074-x
Yanagihara, S., Komura, E., Nagafune, J., Watarai, H., and Yamaguchi, Y. (1998). EBI1/CCR7 is a new member of dendritic cell chemokine receptor that is up-regulated upon maturation. J. Immunol. 161, 3096–3102.
Yashiro, T., Takeuchi, H., Nakamura, S., Tanabe, A., Hara, M., Uchida, K., et al. (2019). PU.1 plays a pivotal role in dendritic cell migration from the periphery to secondary lymphoid organs via regulating CCR7 expression. FASEB J. 33:11481. doi: 10.1096/fj.201900379RR
Yu, N., Wang, S., Song, X., Gao, L., Li, W., Yu, H., et al. (2018). Low-Dose radiation promotes Dendritic Cell Migration and IL-12 Production via the ATM/NF-KappaB Pathway. Radiat. Res. 189:409. doi: 10.1667/RR14840.1
Zabel, B. A., Silverio, A. M., and Butcher, E. C. (2005). Chemokine-like receptor 1 expression and chemerin-directed chemotaxis distinguish plasmacytoid from myeloid dendritic cells in human blood. J. Immunol. 174, 244–251. doi: 10.4049/jimmunol.174.1.244
Keywords: dendritic cells, migration, conventional dendritic cells, plasmacytoid dendritic cells, chemokines, adhesion molecules
Citation: Feng M, Zhou S, Yu Y, Su Q, Li X and Lin W (2021) Regulation of the Migration of Distinct Dendritic Cell Subsets. Front. Cell Dev. Biol. 9:635221. doi: 10.3389/fcell.2021.635221
Received: 30 November 2020; Accepted: 01 February 2021;
Published: 19 February 2021.
Edited by:
Zhichao Fan, UCONN Health, United StatesCopyright © 2021 Feng, Zhou, Yu, Su, Li and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Lin, linw1978@163.com; weilin11@fudan.edu.cn
 Meng Feng
Meng Feng Shuping Zhou
Shuping Zhou Yong Yu
Yong Yu Qinghong Su
Qinghong Su Xiaofan Li
Xiaofan Li Wei Lin
Wei Lin