Two Types of Contact Between Lipid Droplets and Mitochondria
- 1National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China
- 2University of Chinese Academy of Sciences, Beijing, China
Lipid droplets (LDs) and mitochondria are essential organelles involved in cellular lipid metabolism and energy homeostasis. Accumulated studies have revealed that the physical contact between these two organelles is important for their functions. Current understanding of the contact between cellular organelles is highly dynamic, fitting a “kiss-and-run” model. The same pattern of contact between LDs and mitochondria has been reported and several proteins are found to mediate this contact, such as perilipin1 (PLIN1) and PLIN5. Another format of the contact has also been found and termed anchoring. LD-anchored mitochondria (LDAM) are identified in oxidative tissues including brown adipose tissue (BAT), skeletal muscle, and heart muscle, and this anchoring between these two organelles is conserved from mouse to monkey. Moreover, this anchoring is generated during the brown/beige adipocyte differentiation. In this review, we will summarize previous studies on the interaction between LDs and mitochondria, categorize the types of the contacts into dynamic and stable/anchored, present their similarities and differences, discuss their potential distinct molecular mechanism, and finally propose a working hypothesis that may explain why and how cells use two patterns of contact between LDs and mitochondria.
Introduction
The lipid droplet (LD) is a specialized cellular organelle with a neutral lipid core covered by a monolayer phospholipid membrane and associated proteins (Murphy, 2001; Tauchi-Sato et al., 2002; Fujimoto et al., 2008; Thiam et al., 2013). Starting with the discovery by Antony van Leeuwenhoek in 1674, LD biology has taken a sweeping 300 year-long developmental path to the modern era. Our understanding has accelerated in the last two decades, importantly marked by the discovery of marker proteins, development of methods for purification, and the application of omics studies (Ding et al., 2013; Roberts and Olzmann, 2020).
The LD is an ancient organelle with a new face and a recent recognition of its importance has attracted great attention in the fields of biology and life science. The newly discovered fact that LDs exist in some bacteria extends our view of the origin of membrane-bound organelles. The finding that LDs contain not only triacylglycerol (TAG), but also cholesterol ester, retinal ester, and monoalk(en)yl diacylglycerol in animal cells (Bartz et al., 2007a) suggests many potential roles for this organelle. In addition, the broad distribution of LDs from bacteria to humans implies its significance in all organisms (Murphy, 2012; Olzmann and Carvalho, 2019; Zhang and Liu, 2019).
The unique property of LDs in their sequestration of neutral lipids led to a perception of them merely as “a drop of oil,” negatively impacting scientific interest and slowing down its research. However, the description of their proteome and lipidome and their linkage to metabolic syndromes impacting human health provoked a renaissance in research interest and activity (Brasaemle et al., 2004; Liu et al., 2004; Bartz et al., 2007a). The LD differs from other membrane-bound organelles by containing (1) a hydrophobic core compared with an aqueous phase lumen for other membranous organelles, (2) a monolayer phospholipid membrane in contrast with the bilayer or double bilayer membrane for others, (3) a distinct protein complements termed LD resident proteins. With these differences, the LD is a unique cellular organelle. The functions of the LD are lipid storage, lipid transportation, lipid synthesis, and lipid hydrolysis, a suite of functions far beyond their image as a static oil reservoir. According to recent proteomic and other experimental lines of evidence, LD functions are also thought to include membrane trafficking, protein storage and degradation, signal transduction, detoxification, and nucleic acid handling. The multifunctional property of the LD endows the organelle with an irreplaceable position in almost all cellular activities.
The LD is integrated with other organelles in maintaining lipid homeostasis (Walther and Farese, 2012; Schuldiner and Bohnert, 2017), which is responsible for the ectopic storage of lipids that has been linked to many metabolic disorders (Greenberg et al., 2011; Shulman, 2014; Xu et al., 2018). LDs possess the ability for directed movement on the cytoskeleton, likely supporting the movement of neutral lipids within the cell and between organelles. Biochemical studies previously revealed the interaction between LD and early endosome (Liu et al., 2007), mitochondria (Pu et al., 2011), endoplasmic reticulum (ER) (Martin et al., 2005; Ozeki et al., 2005), and peroxisomes (Binns et al., 2006). More interactions between LDs and other organelles have been developed in the past decade (Olzmann and Carvalho, 2019). Therefore, LD-governed cellular lipid homeostasis is driven by lipid metabolism on the LD as well as the interaction between LD and other organelles. For example, TAG sequestered in the LD is converted to fatty acids (FAs) via lipolysis and lipophagy (Zechner et al., 2017; Ogasawara et al., 2020). The FAs in turn, are transported from LD to mitochondria, where they are oxidized as an energy source to produce ATP or heat, and provide building blocks for synthesis of biological molecules. Thus, the LD and mitochondrion form a functional organelle pair, managing cellular energy homeostasis.
Mitochondria are the site of β-oxidation, converting hydrophobic substrates into usable cellular energy and reducing potential. The fuel for this reaction is stored in the hydrophobic interior of LDs, necessitating a mechanism for the transfer of FAs between these two organelles. Free transfer of FA (or coenzyme A ligated FA) is restricted by the hydrophobic character of the molecules. In fact, the accumulation of FA or acyl-CoA under certain stress conditions leads to cellular toxicity (lipotoxicity). To facilitate the transfer, cells developed a specific interaction between these two organelles. An understanding of this interaction was initiated by the visualization of the physical contact between LDs and mitochondria. The best images depicting this interaction were electron micrographs taken by George Palade, in which the physical contact was clearly shown between LDs and mitochondria in guinea pig pancreas (Figure 1Aa). Since then, the development of high resolution light microscopy techniques and the availability of dyes and marker proteins for both LDs and mitochondria have greatly facilitated the visualization of the association of the two organelles.
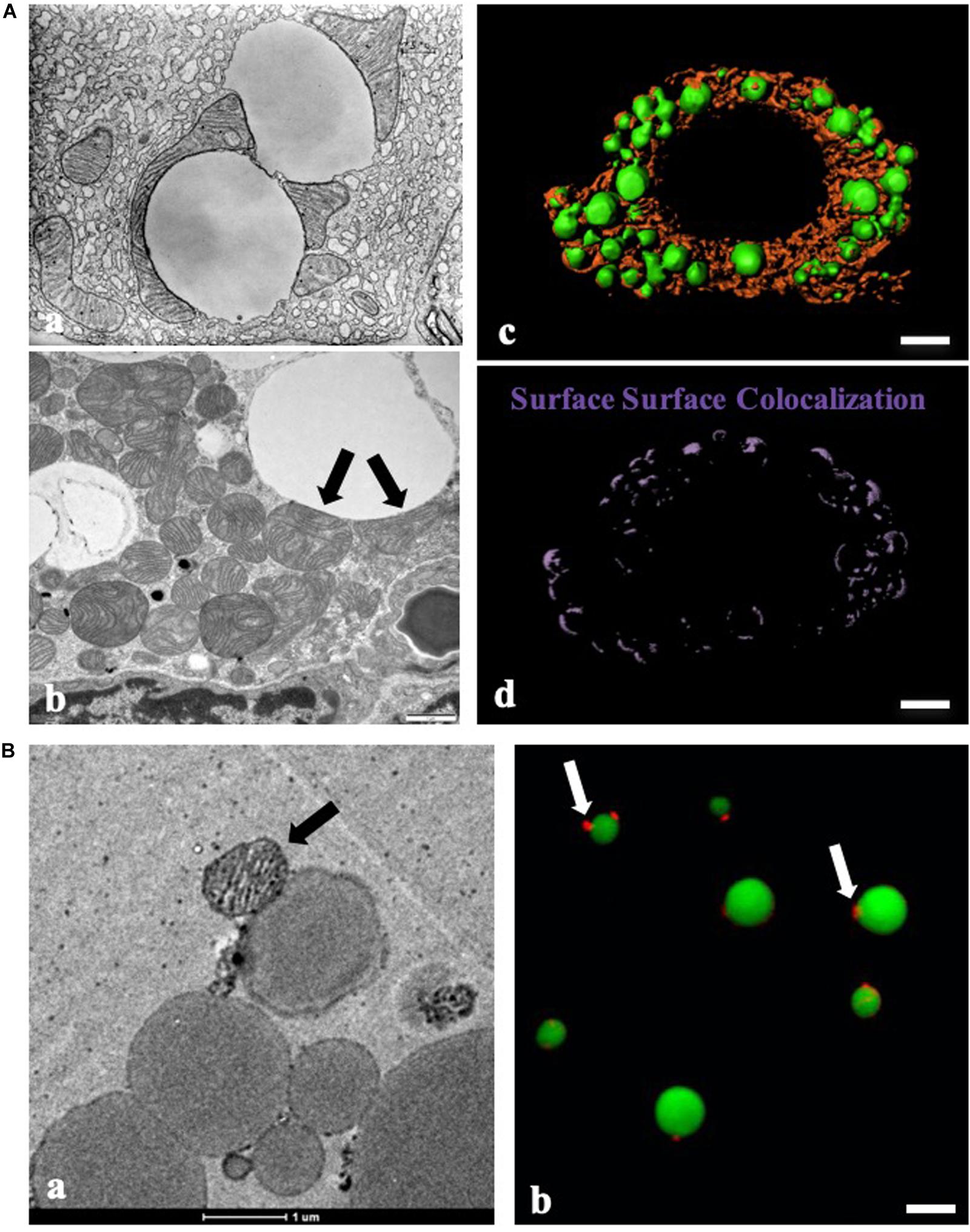
Figure 1. Contact between LDs and mitochondria by morphological studies. (A) Morphological study of contact between lipid droplets (LDs) and mitochondria. (a) Physical contact between LDs and mitochondria in guinea pig pancreas imaged with transmission electron microscopy (TEM) by George E. Palade (Cite from http://www.microscopy.info/Gallery/Details/58). (b) Physical contact between LDs and mitochondria in mouse BAT using TEM. Arrows point to the physical contact between LDs and mitochondria. (c,d) LDs and mitochondria in differentiated brown adipocytes (day 8) were stained with LipidTOX Green for LDs and MitoTracker Red for mitochondria, respectively. The image was analyzed using three-dimensional structured illumination microscopy (3D-SIM), and Imaris analysis was applied to detect surface-surface colocalization. Bar = 5 μm. (B) Morphologic analysis of isolated BAT LDs from mice housed at 23°C. (a) TEM of isolated BAT LDs, Bar = 1 μm. (b) LipidTOX Green staining for LDs and MitoTracker Red staining for mitochondria in isolated BAT LDs. Bar = 5 μm. Arrows indicate LD-anchored mitochondrion (LDAM) in isolated BAT LDs (cite from Cui et al., 2019).
Despite early, clear visual evidence of the contact, it has been a long journey toward a biochemical understanding of the interaction. Investigations were hampered by underdeveloped techniques for biochemical isolation and a lack of molecular tools. The discovery of LD marker proteins perilipin (PLIN1) and adipocyte differentiation-related protein (ADRP/PLIN2) in the early 1990s (Greenberg et al., 1991; Jiang and Serrero, 1992; Brasaemle et al., 1997) permitted the development of purification techniques and an evaluation of the purity of isolated organelle (Wu et al., 2000; Ding et al., 2013). These techniques coupled with the blossoming of proteomics opened a window into the functions, interactions, and underlying molecular machinery governing LD dynamics. These advances permitted the discovery that other organelles, like ER and mitochondria, co-isolate with LDs. Morphological studies provided further evidence for inter-organelle contact while proteomic results identified likely protein candidates regulating the contact. Interestingly, it has been observed that some organelles which are co-purified with LDs when isolated with low speed centrifugation can be separated from the LDs in higher centrifugal force, while other remain firmly associated (Yu et al., 2015; Benador et al., 2018; Cui et al., 2019; Freyre et al., 2019). This distinction probably reflects underlying functional relationships. For example, LDs isolated from heart muscle (Li et al., 2016), skeletal muscle (Zhang et al., 2011), and brown adipocytes (Yu et al., 2015; Cui et al., 2019) always contain many mitochondrial proteins that were once considered contamination. On the other side of the co-isolation, some LD-associated proteins have also been identified on mitochondria, such as PLIN5 and PLIN3 (Bosma et al., 2012; Ramos et al., 2014). However, these proteins cannot be removed by ultracentrifugation. With morphological and biochemical evidence, these proteins reflect a stable contact between LDs and mitochondria in these highly oxidative cells.
This review summarizes the evidence supporting the contact between LDs and mitochondria using morphological, biochemical, and biophysical data and focuses on interactions which cannot be separated by ultracentrifugation. This review brings forward a new point of view, proposing a distinction between stable and dynamic contact which can describe and explain some recent experimental findings. We propose that the stable interaction between LDs and mitochondria is maintained by rivet-like protein structures that prevent the separation between them during ultracentrifugation.
Identification of Membrane Trafficking Proteins on Lipid Droplets
The potential trafficking capability of LDs was revealed through the visualization of LDs moving along microtubules, mediated by the protein Klar (Welte et al., 1998) and other motor proteins (Xie et al., 2019). Proteomic studies of isolated LDs identified Rab and SNARE proteins (Fujimoto et al., 2004; Liu et al., 2004). The targeting of trafficking proteins to LDs has been studied through fluorescent fusion proteins and immunogold-labeled electron microscopy, providing further evidence that LDs are involved in membrane trafficking (Martin et al., 2005; Liu et al., 2007). In functional studies LD-associated Rabs have been demonstrated to regulate interactions between LDs and ER (Rab 18) (Martin et al., 2005; Ozeki et al., 2005), LDs and mitochondria (Rabs and SNARE) (Jägerström et al., 2009; Pu et al., 2011), LDs and early endosomes (Rabs) (Liu et al., 2007), and to drive LD fusion (Rab8) (Wu et al., 2014). The movement of LDs throughout the cell with regulated interactions with other organelles hints at an active and important role for the LD in cellular lipid homeostasis.
The discovery of hormone-sensitive lipase (HSL) and PLIN1 on LDs, and the identification of the mechanism of activation and LD targeting of HSL via phosphorylation of both proteins, identified the catalytic function of this organelle. Additional proteomic studies found lipid synthetic enzymes on isolated LDs, pointing to an anabolic role (Fujimoto et al., 2004; Liu et al., 2004). In addition, some non-coding RNA-coded proteins are recently identified in isolated LDs (Huang et al., 2020). Functional studies of isolated LDs using in vitro assays confirmed that LD is a site for cellular lipid synthesis (Fujimoto et al., 2007; Zhang et al., 2016).
Thus, the LD is a site of neutral lipid storage, but is also metabolically active, can move rapidly throughout the cell, and possesses the molecular machinery to regulate interactions with other organelles. Collectively, these observations point to a central role for LDs in cellular lipid homeostasis.
The Morphological Studies of Contact Between Lipid Droplets and Mitochondria
The interaction between LDs and mitochondria was first observed more than a half-century ago. George E. Palade was one of the major contributors who visualized the physical contact between LDs and mitochondria in guinea pig pancreas using transmission electron microscopy (TEM) (Figure 1Aa). EM also revealed the contact between these two organelles in mammary gland cells (Stemberger et al., 1984) and brown adipose tissue (BAT) (Figure 1Ab). Since these discoveries, many biologists have contributed to this field, accumulated visual evidence documenting the interactions between LDs and mitochondria by EM (Peute et al., 1978; Pu et al., 2011; Yu et al., 2015; Cui et al., 2019).
Advances in biological dyes, such as JC-1 and MitoTracker for mitochondria and Nile red, Bodipy, Oil Red O, and LipidTOX for LDs, have permitted the application of fluorescence microscopy to the study of LD-mitochondrion interactions (Figures 1Ac,d). The discovery of LD marker proteins enabled the use of immunolabeling and fluorescent fusion proteins. Together these advances have greatly simplified the study of the organellar interactions. Furthermore, the recent development of high resolution light microscopy techniques has facilitated finer observations, significantly aiding functional studies of the interactions between LDs and mitochondria (Valm et al., 2017).
Morphological studies have described highly dynamic interactions between LDs and mitochondria. For example, it was observed by EM that endurance exercise training enhances the physical contact between LDs and mitochondria in muscle cells. This finding is fully consistent with the upregulation of FA β-oxidation in mitochondria to meet the energetic needs during exercise (Tarnopolsky et al., 2007). In another study using live-imaging light microscopy, the distance between LDs and mitochondria was found to change in response to various experimental manipulations with close co-localization interpreted as direct contact (Valm et al., 2017).
A wide range of terminology has been applied to direct the contact between LDs and mitochondria. These include, but are not limited to, approximation, association, interaction, targeting, tethering, contact, binding, and anchoring. In one instance, the interaction site was called recognition site (Zehmer et al., 2009). Table 1 summarizes the most common terms used to describe inter-organelle interaction. Mainly based on morphologic observations, the current well-accepted term for the interaction between organelles, including LDs and mitochondria, is “contact” and the site for the interaction is termed as the “contact site.” Contact is characterized by: (1) co-localization of organelles, (2) the distance between organelles approximately 10–70 nm, (3) a contact site composed of tethering proteins, and (4) the ability to transfer ions, lipids, and other molecules (Schuldiner and Bohnert, 2017).
The contact between LDs and mitochondria has been studied and visualized in many types of cells using a variety of technologies. However, molecular techniques are required to push our understanding further. As in other fields of biology, the powerful omics tools have been applied with great success. The first interactomic study screened LD and mitochondrial surface proteins, searching for interacting protein pairs using a bimolecular fluorescence complementation assay in Saccharomyces cerevisiae (Pu et al., 2011). In the same work an in vitro assay to study the interaction between isolated LDs and mitochondria was established, in which GTP was found to play a key role for this interaction (Pu et al., 2011). The most recent organelle-level interactomic study using high resolution microscopy visualized not only the interaction between LDs and mitochondria but also four other membranous organelles using a multispectral image acquisition method (Valm et al., 2017). Interactomic studies of this type provide a broad, system wide view of the network for interactions. However, insight into the mechanisms enabling these interactions requires other experimental techniques.
The Biochemical and Biophysical Studies of Contact Between Lipid Droplets and Mitochondria
Biochemical and biophysical studies have been used to probe the molecular basis of inter-organelle contact. Contact has been dissected into discrete phases: recognition/targeting, tethering, and binding/anchoring, with distinct proteins playing key roles in each step. While the Rab proteins previously described are key candidates for recognition and tethering, other proteins including PLIN1, PLIN5, MFN2, and MIGA2 have also been implicated in these phases (Wasilewski et al., 2012; Boutant et al., 2017; Freyre et al., 2019; Varghese et al., 2019). Some proteins serve multiple functions. For example, VPS13C functions both as a binding component and plays a role in lipid transport (Kumar et al., 2018). Some proteins involved in LD-mitochondrion contact have been found to mediate the contact between other organelles as well. Studies using live-cell imaging have shown that these proteins are involved in highly dynamic contact, fitting a “kiss-and-run” model. Beyond morphologic observation, experiments using co-isolation (Figures 1Ba,b) and in vitro reconstitution have been used to further dissect the mechanisms of this contact (Pu et al., 2011).
Distinct from other membrane-bound organelles, the LD uniquely possesses a density less than 1 g/cm3 due to the TAG forming its core. This enables the method of floatation in a centrifugal field for the isolation/purification of LDs (Ding et al., 2013). All other membrane-bound organelles with densities higher than 1 g/cm3 are driven in the opposite direction into a pellet below the aqueous phase media. Therefore, a simple centrifugation can easily separate LDs from other membrane-bound organelles. Strong binding between LDs and other organelles could result in co-migration in the centrifugal field, either to the low-density region or the pellet.
In fact, other membranous organelles, such as ER and mitochondria, are commonly co-isolated with LDs and this co-fractionation historically was considered as contamination. Indeed, some membranous structures can be stripped from LDs through ultracentrifugation, but other structures remain tightly bound, suggesting that these co-fractionated organelles are bound physiologically in a type of organelle complex. The analysis of isolated and re-isolated LD fractions by EM and light microscopy provides visual evidence of intact membranous organelles bound to the surface of LDs, demonstrating the physiological and physical binding of LDs with other membranous organelles. The strength of the centrifugal field can be used to measure the binding strength, distinguishing weak contact (association/kiss-run) from strong contact (stable binding/anchoring). Therefore, this experimental approach can be used to classify types of contact.
This property of LDs can also be studied using in vitro assays (Pu et al., 2011). After incubation with controlled components, LDs can be re-isolated by floatation. Membranous structures induced to bind to the LDs in the in vitro system are co-floated. Utilizing this unique property of LDs, isolated LDs were incubated with isolated early endosomes in the presence or absence of GTP, the reaction mixture was centrifuged, and the re-isolated LDs were analyzed. Through this experiment, the physiological binding of early endosomes to LDs was identified to be regulated by GTP. Stripping Rab proteins from both isolated LDs and early endosomes totally blocked the binding, demonstrating that Rab protein(s) are key players governing the physiological binding between early endosomes and LDs (Liu et al., 2007).
A similar experiment was conducted to investigate factors that govern the binding between LDs and mitochondria. The two organelles, isolated separately, were mixed and incubated in the presence or absence of GTP or cytosol, followed by the re-isolation of the LDs. Mitochondria were co-fractionated with the floating LD fraction under the GTP condition, identifying GTP as a key factor regulating the binding between these two organelles (Pu et al., 2011).
Many mitochondrial proteins were identified in LDs isolated from mouse skeletal muscle using ultracentrifugation, indicating the strong contact between these two organelles (Zhang et al., 2011). Another study of LDs isolated from rat heart with ultracentrifugation also found numerous mitochondrial proteins (Li et al., 2016). A summary of finding from proteomic studies of isolated LDs shows that mitochondrial proteins co-fractionate with LDs isolated from a broad range of cell types (Table 2). Most of these studies used ultracentrifugation conditions, demonstrating the tight contact between these two organelles. It is also quite interesting that this phenomenon primarily is present in cells from highly oxidative tissues (Cui et al., 2019). The LD-mitochondrion contact is especially easy to visualize in BAT, an observation that is consistent with the role of these cells in heat production and the resulting high energy demand of mitochondria (Géloën et al., 1990; Ohue and Makita, 1992). Therefore, brown adipocytes represent a uniquely suitable experimental system for these investigations.
Recent studies of isolated BAT LDs have suggested a distinction between two types of contact (Cui et al., 2019). The first is tight/stable contact (Cui et al., 2019), also called anchoring since it cannot be separated by ultracentrifugation, similar to the results from isolated LDs of muscle and heart (Zhang et al., 2011; Li et al., 2016). The other is dynamic contact that can be separated using high speed centrifugation (9,000g). The term peridroplet mitochondria (PDM) was proposed to describe these loosely associated mitochondria (Benador et al., 2018). After centrifugation with 9,000g, about 50% of LD-associated were still remained on re-isolated LD fraction. In addition, PDM are functionally different with cytosolic mitochondria and more toward to fatty acid synthesis (Benador et al., 2018). These differences can be understood physiologically due to the different demands placed on oxidative systems of the cell, depending on the role of the tissue.
Dynamic and Stable Contacts Between Lipid Droplets and Mitochondria
The empirical distinction between weak and strong contact revealed by centrifugal force is likely to reflect important physiological complexity. The contact likely functions to permit efficient signaling and transport of hydrophobic molecules. For oxidative tissues such as heart muscle, skeletal muscle, and BAT, rapid delivery of FAs from LDs to mitochondria is an essential element for efficient ATP and heat production. The contact between these two organelles not only makes this delivery fast but also avoids insolubility and cytotoxicity of free FAs. Stable/anchoring contact found in these tissues (Cui et al., 2019) well represents the necessity for the constant production of ATP or heat with high efficiency (Figure 3). The term of LD-anchored mitochondria (LDAM) has been proposed for the mitochondria that stably associate with LDs (Cui et al., 2019). Both LDAM and PDM co-fractionate with LDs that are isolated with the low centrifugal force while only LDAM remain in the following centrifugation at speeds generating over 9,000g.
The current definition of contact is mainly based on morphological studies and includes four key characteristics that describe the dynamic contact well but seem to lack a criterion that defines the strength of contact. The nature of the molecules bridging these two organelles may explain the different strengths of contacts. Identifying the proteins mediating the different types of contact is important to understand the underlying physiology. Since LDAM are not affected in PLIN1 or PLIN5 deleted mouse BAT (Cui et al., 2019), the interaction between PLIN1 and MFN2 may be a good example of the protein pair involved in dynamic contact (Boutant et al., 2017; Olzmann and Carvalho, 2019). The proteins forming cellular tight junctions and the proteins with a nail (Havaux, 1998) or rivet-like (Iacovache et al., 2006) conformation may provide a useful model for understanding the stable contact (Figure 3). It is reasonable to include a fifth criterion classifying the contact based on its stability in a centrifugal field.
The LDAM have also been identified in the oxidative tissues of Rhesus monkeys (Macaca mulatta), including BAT (BAT), heart (Heart), and muscles (MG and MS) (Figure 2, lanes 1–12), but not in liver (Liver) (Figure 2, lanes 13–15), suggesting that the LDAM are conserved from mice to monkeys. To further understand the physiological significance of this stable contact (or LDAM), the monkeys with different metabolic conditions, such as normal (CK), obese (OB), and diabetes (TM), were utilized and the significant differences are found not only in LD-associated proteins but also in LDAM proteins (Figure 2).
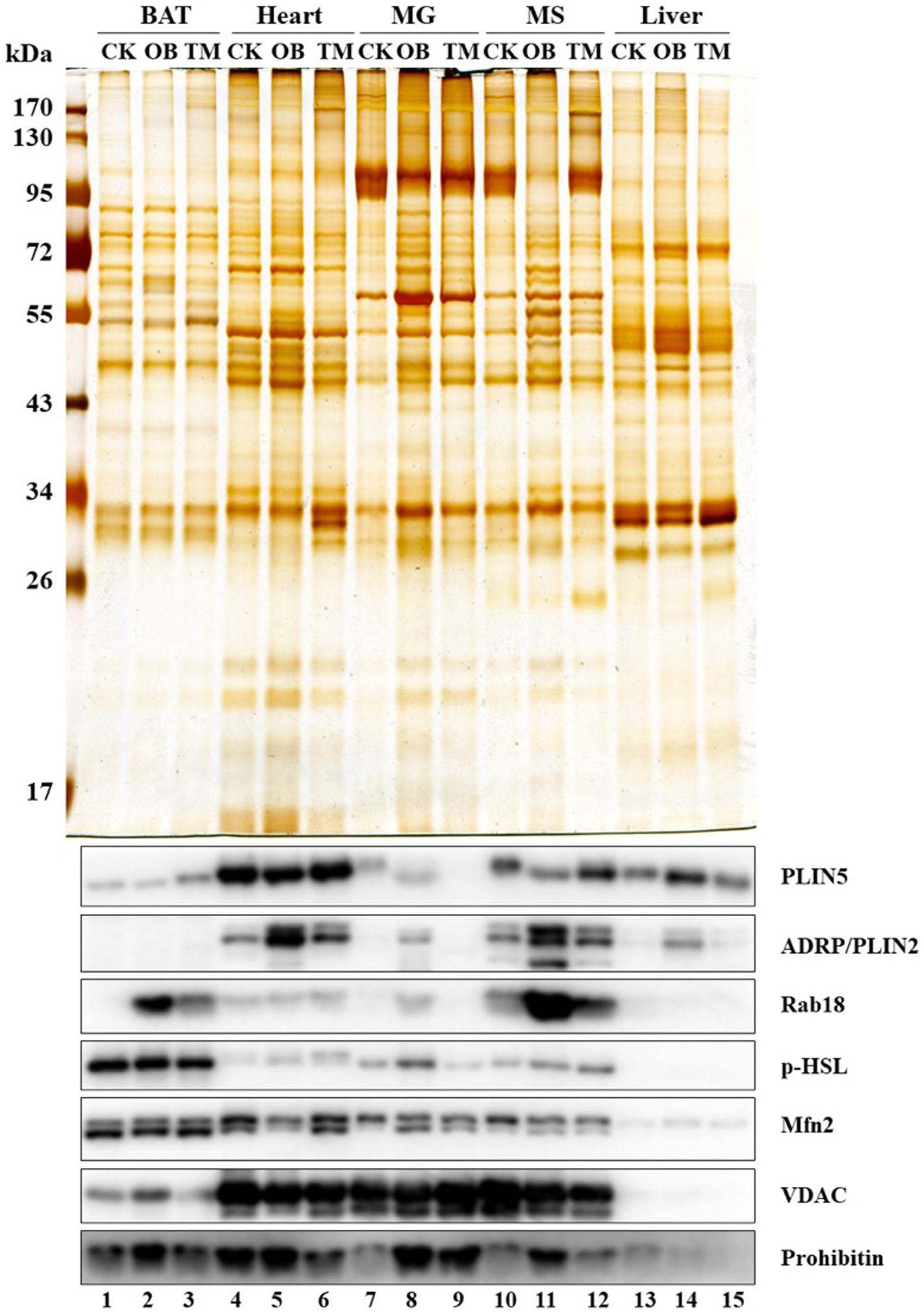
Figure 2. Co-isolation of LDs and mitochondria. Rhesus monkeys (Macaca mulatta) including Normal (CK), Obese (OB), and Type 2 diabetes mellitus (TM) were obtained from Kunming Institute of Zoology, Chinese Academy of Sciences. The LDs from different tissues of these monkeys were isolated using indicated homogenization methods and centrifugal forces as following: (1) A dounce type glass-teflon homogenizer and a speed of 247,000g were used for heart, musculus gastrocnemius (MG), and musculus soleus (MS). (2) A dounce type loose-fitting potter-elvehjem tissue grinder and a speed of 247,000g were used for liver. (3) A 200-mesh screen with plastic syringe piston and speeds of 1,000g and 247,000g were used for BAT. The proteins from isolated LDs were separated by SDS-PAGE and visualized by silver staining (upper panel). The marker proteins for LDs (PLIN5, ADRP/PLIN2, Rab18, and p-HSL) and for mitochondria (Mfn2, VDAC, and Prohibitin) were determined by Western blots (lower panel) (cite from Cui et al., 2019).
Another fundamental question remaining to be answered for this anchoring hypothesis (Figure 3), is whether two types of contact are interchangeable based on physiological requirements. If this is possible, dynamic contact could transition to stable contact and back as energetic requirements fluctuate. Alternatively, if these are permanent states of association, there may be other important structural and functional differences between these two states. It is possible that mitochondria and LDs locked in stable contact coordinate in a manner similar to some protein complexes. In this case, it may be useful to think of LD and mitochondrion engaged in a stable contact as a distinct cellular structure, an organelle complex. Nevertheless, we suggest that both morphological and biochemical methods should be used together to study the contact between LDs and mitochondria as well as other organelle contacts.
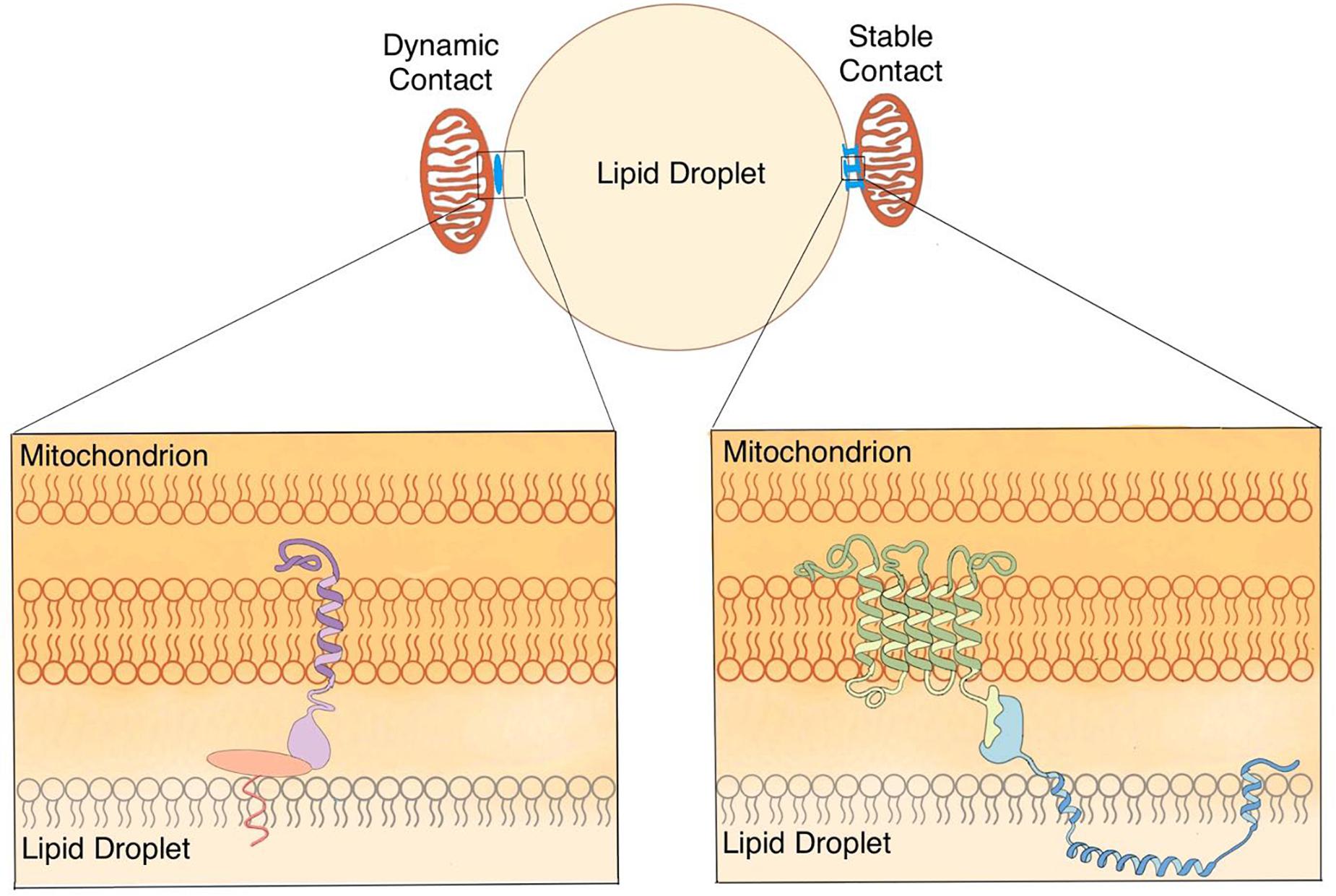
Figure 3. Anchoring hypothesis of the contact between LDs and mitochondria. Based on previous studies from ours and other laboratories, the working hypothesis is proposed that there are two types of contact (dynamic and stable) between LDs and mitochondria in oxidative cells. Dynamic contact can tether LDs and mitochondria through protein complexes. Stable contact can anchor mitochondria on LDs by nail- or rivet-like proteins.
To conduct the biochemical and biophysical study for the contact between LDs and mitochondria using isolation method, two critical factors should be addressed for isolation of LDs from BAT and brown adipocytes since they contain supersize LDs that are fragile and easy to be broken. These factors are (1) stringency of homogenization and (2) centrifugal force of floatation. Three homogenization methods that have been applied on BAT and brown adipocytes are listed by stringency (low to high): Nitrogen Cavitation technology (N2 bomb) (Cui et al., 2019), grinding tissue on 200-mesh screen using plastic syringe piston (Yu et al., 2015; Cui et al., 2019), and dounce homogenization (Benador et al., 2018). The centrifugal force has been utilized for sample from BAT and brown adipocytes ranging from 500 to 100,000g depending on the processes of isolation, washing, and separation. The high homogenization stringency and high centrifugal force can remove nonspecific bound contaminations but result missing of supersize LDs. But low centrifugal force cannot float small LDs efficiently (Zhang et al., 2016). Therefore, homogenization methods and centrifugal forces should be selected properly to control the quality, the size, and the type of LDs isolated from different tissues and cells.
Finally, based on previous studies from ours and other laboratories, the working hypothesis is proposed that there are two types of contact (dynamic and stable) between LDs and mitochondria in oxidative cells (Figure 3). The co-isolation method is introduced to study the contact between LDs and mitochondria, as well as other bilayer membrane-bound organelles. Combined with morphological and biochemical approaches, the co-isolation study is going to uncover new aspects toward to better dissect and understand the contact between these two organelles.
Author Contributions
PL conceived the project. LC carried out the experiments and data analysis. LC and PL wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (Grant Nos. 2018YFA0800700, 2016YFA0500100, and 2018YFA0800900), National Natural Science Foundation of China (Grant Nos. 91857201, 91954108, 31671402, 31671233, 31701018, and U1702288). This work was also supported by the “Personalized Medicines-Molecular Signature-based Drug Discovery and Development,” Strategic Priority Research Program of the Chinese Academy of Sciences, Grant No. XDA12040218.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. John Zehmer for his critical reading and useful suggestions. We also thank Ms. Siyu Wang for her artwork of Figure 3.
References
Barbosa, A. D., Savage, D. B., and Siniossoglou, S. (2015). Lipid droplet–organelle interactions: emerging roles in lipid metabolism. Curr. Opin. Cell Biol. 35, 91–97. doi: 10.1016/j.ceb.2015.04.017
Bartz, R., Li, W.-H., Venables, B., Zehmer, J. K., Roth, M. R., Welti, R., et al. (2007a). Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 48, 837–847. doi: 10.1194/jlr.m600413-jlr200
Bartz, R., Zehmer, J. K., Zhu, M., Chen, Y., Serrero, G., Zhao, Y., and Liu, P. (2007b). Dynamic Activity of Lipid Droplets: Protein Phosphorylation and GTP-Mediated Protein Translocation. J. Proteome. Res. 6, 3256–3265. doi: 10.1021/pr070158j
Benador, I. Y., Veliova, M., Mahdaviani, K., Petcherski, A., Wikstrom, J. D., Assali, E. A., et al. (2018). Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab. 27, 869.e6–885.e6.
Binns, D., Januszewski, T., Chen, Y., Hill, J., Markin, V. S., Zhao, Y., et al. (2006). An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173, 719–731. doi: 10.1083/jcb.200511125
Bosma, M., Minnaard, R., Sparks, L. M., Schaart, G., Losen, M., De Baets, M. H., et al. (2012). The lipid droplet coat protein perilipin 5 also localizes to muscle mitochondria. Histochem. Cell Biol. 137, 205–216. doi: 10.1007/s00418-011-0888-x
Boutant, M., Kulkarni, S. S., Joffraud, M., Ratajczak, J., Valera-Alberni, M., Combe, R., et al. (2017). Mfn2 is critical for brown adipose tissue thermogenic function. EMBO J. 36, 1543–1558. doi: 10.15252/embj.201694914
Brasaemle, D., Barber, T., Wolins, N. E., Serrero, G., Blanchette-Mackie, E. J., and Londos, C. (1997). Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J. Lipid Res. 38, 2249–2263.
Brasaemle, D. L., Dolios, G., Shapiro, L., and Wang, R. (2004). Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 279, 46835–46842. doi: 10.1074/jbc.m409340200
Cui, L., Mirza, A. H., Zhang, S., Liang, B., and Liu, P. (2019). Lipid droplets and mitochondria are anchored during brown adipocyte differentiation. Protein Cell 10, 921–926. doi: 10.1007/s13238-019-00661-1
Ding, Y., Zhang, S., Yang, L., Na, H., Zhang, P., Zhang, H., et al. (2013). Isolating lipid droplets from multiple species. Nat. Protoc. 8:43. doi: 10.1038/nprot.2012.142
Freyre, C. A. C., Rauher, P. C., Ejsing, C. S., and Klemm, R. W. (2019). MIGA2 links mitochondria, the ER, and lipid droplets and promotes de novo Lipogenesis in Adipocytes. Mol. Cell. 76, 811.e4–825.e4.
Fujimoto, T., Ohsaki, Y., Cheng, J., Suzuki, M., and Shinohara, Y. (2008). Lipid droplets: a classic organelle with new outfits. Histochem. Cell Biol. 130, 263–279. doi: 10.1007/s00418-008-0449-0
Fujimoto, Y., Itabe, H., Kinoshita, T., Homma, K. J., Onoduka, J., Mori, M., et al. (2007). Involvement of ACSL in local synthesis of neutral lipids in cytoplasmic lipid droplets in human hepatocyte HuH7. J. Lipid Res. 48, 1280–1292. doi: 10.1194/jlr.m700050-jlr200
Fujimoto, Y., Itabe, H., Sakai, J., Makita, M., Noda, J., Mori, M., et al. (2004). Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta 1644, 47–59. doi: 10.1016/j.bbamcr.2003.10.018
Géloën, A., Collet, A., Guay, G., and Bukowiecki, L. J. (1990). In vivo differentiation of brown adipocytes in adult mice: an electron microscopic study. Am. J. Anat. 188, 366–372. doi: 10.1002/aja.1001880404
Gemmink, A., Daemen, S., Kuijpers, H. J. H., Schaart, G., Duimel, H., Lopez-Iglesias, C., et al. (2018). Super-resolution microscopy localizes perilipin 5 at lipid droplet-mitochondria interaction sites and at lipid droplets juxtaposing to perilipin 2. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 1423–1432. doi: 10.1016/j.bbalip.2018.08.016
Greenberg, A. S., Coleman, R. A., Kraemer, F. B., Mcmanaman, J. L., Obin, M. S., Puri, V., et al. (2011). The role of lipid droplets in metabolic disease in rodents and humans. J. Clin. Investig. 121, 2102–2110. doi: 10.1172/jci46069
Greenberg, A. S., Egan, J. J., Wek, S. A., Garty, N. B., Blanchette-Mackie, E., and Londos, C. (1991). Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem. 266, 11341–11346.
Havaux, M. (1998). Carotenoids as membrane stabilizers in chloroplasts. Trends Plant Sci. 3, 147–151. doi: 10.1016/s1360-1385(98)01200-x
Herms, A., Bosch, M., Reddy, B. J. N., Schieber, N. L., Fajardo, A., Ruperez, C., et al. (2015). AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nat. Commun. 6:7176. doi: 10.1038/ncomms8176
Huang, T., Bamigbade, A. T., Xu, S., Deng, Y., Xie, K., Ogunsade, O. O., et al. (2020). Identification of noncoding RNA-encoded proteins on lipid droplets. Sci. Bull. doi: 10.1016/j.scib.2020.09.022 [Epub ahead of print].
Iacovache, I., Paumard, P., Scheib, H., Lesieur, C., Sakai, N., Matile, S., et al. (2006). A rivet model for channel formation by aerolysin-like pore-forming toxins. EMBO J. 25, 457–466. doi: 10.1038/sj.emboj.7600959
Jägerström, S., Polesie, S., Wickström, Y., Johansson, B. R., Schröder, H. D., Højlund, K., et al. (2009). Lipid droplets interact with mitochondria using SNAP23. Cell Biol. Int. 33, 934–940. doi: 10.1016/j.cellbi.2009.06.011
Jiang, H.-P., and Serrero, G. (1992). Isolation and characterization of a full-length cDNA coding for an adipose differentiation-related protein. Proc. Natl. Acad. Sci. U.S.A. 89, 7856–7860. doi: 10.1073/pnas.89.17.7856
Kim, S. C., Chen, Y., Mirza, S., Xu, Y., Lee, J., Liu, P., et al. (2006). A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J. Proteome Res. 5, 3446–3452. doi: 10.1021/pr0603396
Krahmer, N., Najafi, B., Schueder, F., Quagliarini, F., Steger, M., Seitz, S., et al. (2018). Organellar proteomics and phospho-proteomics reveal subcellular reorganization in diet-induced hepatic steatosis. Dev. Cell 47, 205–221.e207.
Kumar, N., Leonzino, M., Hancock-Cerutti, W., Horenkamp, F. A., Li, P., Lees, J. A., et al. (2018). VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell. Biol. 217, 3625–3639. doi: 10.1083/jcb.201807019
Li, L., Zhang, H., Wang, W., Hong, Y., Wang, J., Zhang, S., et al. (2016). Comparative proteomics reveals abnormal binding of ATGL and dysferlin on lipid droplets from pressure overload-induced dysfunctional rat hearts. Sci. Rep. 6:19782.
Liu, P., Bartz, R., Zehmer, J. K., Ying, Y.-S., Zhu, M., Serrero, G., et al. (2007). Rab-regulated interaction of early endosomes with lipid droplets. Biochim. Biophys. Acta Mol. Cell Res. 1773, 784–793. doi: 10.1016/j.bbamcr.2007.02.004
Liu, P., Ying, Y., Zhao, Y., Mundy, D. I., Zhu, M., and Anderson, R. G. (2004). Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279, 3787–3792. doi: 10.1074/jbc.m311945200
Lupette, J., Jaussaud, A., Seddiki, K., Morabito, C., Brugiere, S., Schaller, H., et al. (2019). The architecture of lipid droplets in the diatom Phaeodactylum tricornutum. Algal Res. 38:101415. doi: 10.1016/j.algal.2019.101415
Martin, S., Driessen, K., Nixon, S. J., Zerial, M., and Parton, R. G. (2005). Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 280, 42325–42335. doi: 10.1074/jbc.m506651200
Milakovic, I., Jeseta, M., Hanulakova, S., Knitlova, D., Hanzalova, K., Hulinska, P., et al. (2015). Energy status characteristics of porcine oocytes during in vitro maturation is influenced by their meiotic competence. Reprod. Domest Anim. 50, 812–819. doi: 10.1111/rda.12592
Murphy, D. J. (2001). The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40, 325–438. doi: 10.1016/s0163-7827(01)00013-3
Murphy, D. J. (2012). The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 249, 541–585. doi: 10.1007/s00709-011-0329-7
Murphy, S., Martin, S., and Parton, R. G. (2009). Lipid droplet-organelle interactions; sharing the fats. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1791, 441–447. doi: 10.1016/j.bbalip.2008.07.004
Nguyen, T. B., Louie, S. M., Daniele, J. R., Tran, Q., Dillin, A., Zoncu, R., et al. (2017). DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev. Cell 42, 9–21.e25. doi: 10.1016/Fj.devcel.2017.06.003
Ogasawara, Y., Tsuji, T., and Fujimoto, T. (2020). Multifarious roles of lipid droplets in autophagy – Target, product, and what else? Semin. Cell Dev. Biol. [Epub ahead of print].
Ohue, M., and Makita, T. (1992). Interrelationship between lipid droplets and mitochondria in brown adipocytes of the hamster. J. Vet. Med. Sci. 54, 1131–1135. doi: 10.1292/jvms.54.1131
Olzmann, J. A., and Carvalho, P. (2019). Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155. doi: 10.1038/s41580-018-0085-z
Ozeki, S., Cheng, J., Tauchi-Sato, K., Hatano, N., Taniguchi, H., and Fujimoto, T. (2005). Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 118, 2601–2611. doi: 10.1242/jcs.02401
Peute, J., Van Der Gaag, M. A., and Lambert, J. G. D. (1978). Ultrastructure and lipid content of the liver of the zebrafish, Brachydanio rerio, related to vitellogenin synthesis. Cell Tissue Res. 186, 297–308. doi: 10.1007/bf00225539
Pribasnig, M., Kien, B., Pusch, L., Haemmerle, G., Zimmermann, R., and Wolinski, H. (2018). Extendedresolution imaging of the interaction of lipid droplets and mitochondria. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 1285–1296.
Pu, J., Ha, C. W., Zhang, S., Jung, J. P., Huh, W.-K., and Liu, P. (2011). Interactomic study on interaction between lipid droplets and mitochondria. Protein Cell 2, 487–496. doi: 10.1007/s13238-011-1061-y
Rambold, A. S., Cohen, S., and Lippincott-Schwartz, J. (2015). Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678–692. doi: 10.1016/j.devcel.2015.01.029
Ramos, S. V., Macpherson, R. E., Turnbull, P. C., Bott, K. N., Leblanc, P., Ward, W. E., et al. (2014). Higher PLIN5 but not PLIN3 content in isolated skeletal muscle mitochondria following acute in vivo contraction in rat hindlimb. Physiol. Rep. 2:e12154. doi: 10.14814/phy2.12154
Roberts, M. A., and Olzmann, J. A. (2020). Protein quality control and lipid droplet metabolism. Annu. Rev. Cell Dev.Biol. 36, 115–139. doi: 10.1146/annurev-cellbio-031320-101827
Schuldiner, M., and Bohnert, M. (2017). A different kind of love – lipid droplet contact sites. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1188–1196. doi: 10.1016/j.bbalip.2017.06.005
Shiozaki, M., Hayakawa, N., Shibata, M., Koike, M., Uchiyama, Y., and Gotow, T. (2011). Closer association of mitochondria with lipid droplets in hepatocytes and activation of Kupffer cells in resveratrol-treated senescence-accelerated mice. Histochem. Cell Biol. 136, 475–489.
Shulman, G. I. (2014). Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N. Engl. J. Med. 371, 1131–1141. doi: 10.1056/nejmra1011035
Stemberger, B. H., Walsh, R. M., and Patton, S. (1984). Morphometric evaluation of lipid droplet associations with secretory vesicles, mitochondria and other components in the lactating cell. Cell Tissue Res. 236, 471–475.
Song, Y.-F., Hogstrand, C., Ling, S.-C., Chen, G.-H., and Luo, Z. (2020). Creb-Pgc1α pathway modulates the interaction between lipid droplets and mitochondria and influences high fat diet-induced changes of lipid metabolism in the liver and isolated hepatocytes of yellow catfish. J. Nutr. Biochem. 80:108364. doi: 10.1016/j.jnutbio.2020.108364
Sturmey, R. G., O’toole, P. J., and Leese, H. J. (2006). Fluorescence resonance energy transfer analysis of mitochondrial:lipid association in the porcine oocyte. Reproduction 132, 829–837. doi: 10.1530/rep-06-0073
Tarique, I., Vistro, W. A., Bai, X., Yang, P., Hong, C., Huang, Y., et al. (2019). LIPOPHAGY: a novel form of steroidogenic activity within the LEYDIG cell during the reproductive cycle of turtle. Reprod. Biol. Endocrinol. 17:19. doi: 10.1186/s12958-019-0462-2
Tarnopolsky, M. A., Rennie, C. D., Robertshaw, H. A., Fedak-Tarnopolsky, S. N., Devries, M. C., and Hamadeh, M. J. (2007). Influence of endurance exercise training and sex on intramyocellular lipid and mitochondrial ultrastructure, substrate use, and mitochondrial enzyme activity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, R1271–R1278.
Tauchi-Sato, K., Ozeki, S., Houjou, T., Taguchi, R., and Fujimoto, T. (2002). The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277, 44507–44512. doi: 10.1074/jbc.m207712200
Thiam, A. R., Farese, R. V. Jr., and Walther, T. C. (2013). The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14, 775–786. doi: 10.1038/nrm3699
Valm, A. M., Cohen, S., Legant, W. R., Melunis, J., Hershberg, U., Wait, E., et al. (2017). Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546:162. doi: 10.1038/nature22369
Varghese, M., Kimler, V. A., Ghazi, F. R., Rathore, G. K., Perkins, G. A., Ellisman, M. H., et al. (2019). Adipocyte lipolysis affects Perilipin 5 and cristae organization at the cardiac lipid droplet-mitochondrial interface. Sci. Rep. 9:4734.
Vock, R., Hoppeler, H., Claassen, H., Wu, D. X., Billeter, R., Weber, J. M., et al. (1996). Design of the oxygen and substrate pathways. VI. structural basis of intracellular substrate supply to mitochondria in muscle cells. J. Exp. Biol. 199:1689.
Walther, T. C., and Farese, R. V. (2012). Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81, 687–714. doi: 10.1146/annurev-biochem-061009-102430
Wang, H., Lei, M., Hsia, R.-C., and Sztalryd, C. (2013). “Chapter 8–analysis of lipid droplets in cardiac muscle,” in Methods in Cell Biology, eds H. Yang and P. Li (Cambridge, MA: Academic Press), 129–149. doi: 10.1016/b978-0-12-408051-5.00008-5
Wang, H., Sreenivasan, U., Hu, H., Saladino, A., Polster, B. M., Lund, L. M., et al. (2011). Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 52, 2159–2168. doi: 10.1194/jlr.m017939
Wasilewski, M., Semenzato, M., Rafelski, S. M., Robbins, J., Bakardjiev, A. I, and Scorrano, L. (2012). Optic atrophy 1-dependent mitochondrial remodeling controls steroidogenesis in trophoblasts. Curr. Biol. 22, 1228–1234. doi: 10.1016/j.cub.2012.04.054
Welte, M. A., Gross, S. P., Postner, M., Block, S. M., and Wieschaus, E. F. (1998). Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell 92, 547–557. doi: 10.1016/s0092-8674(00)80947-2
Wu, C. C., Howell, K. E., Neville, M. C., Yates Iii, J. R., and Mcmanaman, J. L. (2000). Proteomics reveal a link between the endoplasmic reticulum and lipid secretory mechanisms in mammary epithelial cells. Electrophoresis Int. J. 21, 3470–3482. doi: 10.1002/1522-2683(20001001)21:16<3470::aid-elps3470>3.0.co;2-g
Wu, L., Xu, D., Zhou, L., Xie, B., Yu, L., Yang, H., et al. (2014). Rab8a-AS160-MSS4 regulatory circuit controls lipid droplet fusion and growth. Dev. Cell 30, 378–393. doi: 10.1016/j.devcel.2014.07.005
Xie, K., Zhang, P., Na, H., Liu, Y., Zhang, H., and Liu, P. (2019). MDT-28/PLIN-1 mediates lipid droplet-microtubule interaction via DLC-1 in Caenorhabditis elegans. Sci. Rep. 9:14902.
Xu, S., Zhang, X., and Liu, P. (2018). Lipid droplet proteins and metabolic diseases. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 1968–1983. doi: 10.1016/j.bbadis.2017.07.019
Yu, J., Zhang, S., Cui, L., Wang, W., Na, H., Zhu, X., et al. (2015). Lipid droplet remodeling and interaction with mitochondria in mouse brown adipose tissue during cold treatment. Biochim. Biophys. Acta 1853, 918–928. doi: 10.1016/j.bbamcr.2015.01.020
Zechner, R., Madeo, F., and Kratky, D. (2017). Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat. Rev. Mol. Cell Biol. 18:671. doi: 10.1038/nrm.2017.76
Zehmer, J. K., Huang, Y., Peng, G., Pu, J., Anderson, R. G. W., and Liu, P. (2009). A role for lipid droplets in inter-membrane lipid traffic. Proteomics 9, 914–921. doi: 10.1002/pmic.200800584
Zhang, C., and Liu, P. (2019). The new face of the lipid droplet: lipid droplet proteins. Proteomics 19:1700223. doi: 10.1002/pmic.201700223
Zhang, H., Wang, Y., Li, J., Yu, J., Pu, J., Li, L., et al. (2011). Proteome of skeletal muscle lipid droplet reveals association with mitochondria and apolipoprotein a-I. J. Proteome Res. 10, 4757–4768. doi: 10.1021/pr200553c
Keywords: contact, lipid droplets, mitochondria, anchor, isolation, centrifugal force
Citation: Cui L and Liu P (2020) Two Types of Contact Between Lipid Droplets and Mitochondria. Front. Cell Dev. Biol. 8:618322. doi: 10.3389/fcell.2020.618322
Received: 16 October 2020; Accepted: 25 November 2020;
Published: 15 December 2020.
Edited by:
Dan Zhang, Temasek Life Sciences Laboratory, SingaporeReviewed by:
Mark Christian, Nottingham Trent University, United KingdomWanzhu Jin, Chinese Academy of Sciences (CAS), China
Copyright © 2020 Cui and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pingsheng Liu, pliu@ibp.ac.cn
 Liujuan Cui
Liujuan Cui Pingsheng Liu
Pingsheng Liu