Visualization of Zika Virus Infection via a Light-Initiated Bio-Orthogonal Cycloaddition Labeling Strategy
- 1Molecular Diagnosis and Treatment Center for Infectious Diseases, Dermatology Hospital, Southern Medical University, Guangzhou, China
- 2Department of Burn and Plastic Surgery, Guangzhou First People’s Hospital, South China University of Technology, Guangzhou, China
- 3Department of Clinical Laboratory, The First People’s Hospital of Foshan, Foshan, China
- 4Department of Burn Surgery & Department of Rheumatology, The First People’s Hospital of Foshan, Foshan, China
- 5Department of Infectious Disease, the Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, China
- 6NHC Key Laboratory of Metabolic Cardiovascular Diseases Research, Ningxia Key Laboratory of Vascular Injury and Repair Research, Ningxia Medical University, Yinchuan, China
Zika virus (ZIKV) is a re-emerging flavivirus that leads to devastating consequences for fetal development. It is crucial to visualize the pathogenicity activities of ZIKV ranging from infection pathways to immunity processes, but the accurate labeling of ZIKV remains challenging due to the lack of a reliable labeling technique. We introduce the photo-activated bio-orthogonal cycloaddition to construct a fluorogenic probe for the labeling and visualizing of ZIKV. Via a simple UV photoirradiation, the fluorogenic probes could be effectively labeled on the ZIKV. We demonstrated that it can be used for investigating the interaction between ZIKV and diverse cells and avoiding the autofluorescence phenomenon in traditional immunofluorescence assay. Thus, this bioorthogonal-enabled labeling strategy can serve as a promising approach to monitor and understand the interaction between the ZIKV and host cells.
1 Introduction
The mosquito-transmitted Zika virus (ZIKV), which belongs to the family Flaviviridae and genus flavivirus, can cause several Zika syndrome including ventriculomegaly and microcephaly (Miner and Diamond, 2017). Recent outbreaks of ZIKV have been reported in more than 30 countries or territories, emerging as a major threat to global health (Sampathkumar and Sanchez, 2016; Hills et al., 2017; Mengesha Tsegaye et al., 2018). The current diagnostic techniques available for ZIKV rely on the reverse transcriptase-polymerase chain reaction assay or ZIKV-specific IgM antibody testing, which play an important role in preventing the spread of disease (Ergünay et al., 2010; Barreiro, 2016; Kikuti et al., 2018; Low et al., 2021). The exploration of cellular signaling pathways on ZIKV infection has also attracted considerable research attention, greatly enriching the study of mechanisms of infection of RNA viruses (Grant et al., 2016; Chen et al., 2018; Chiramel and Best, 2018; Garcia et al., 2020). Among these, visualization of the Zika virus-host cell interactions is essential to comprehend the molecular mechanisms and pathogenesis of ZIKV disease.
Fluorescent dyes have been widely used in viral labeling and real-time imaging, which improve our understanding of the viral infection process (Zheng et al., 2019; Liao et al., 2020; Zhang et al., 2020). Thus, the photobleaching and spectral overlaps of fluorophores are inescapable, greatly affecting the efficacy of fluorophores, greatly affecting the efficacy of tracking dye-labeled viruses, and limiting the application in bioimaging. Fluorescent quantum dots (QDs) can be rationally chosen as an alternative candidate because they have distinguishing optical properties including narrow-band and tunable fluorescence emission, high fluorescence quantum yields, and photostability (Matea et al., 2017; Pleskova et al., 2018). Currently, numerous efforts have been devoted to constructing the QDs–virus imaging modality and demonstrating its capabilities in providing meaningful information (You et al., 2006; Zhang et al., 2012; Ribeiro et al., 2019; Kuang et al., 2020; Chen et al., 2021; Chung and Zhang, 2021; Lin et al., 2021; He et al., 2022; Yi et al., 2022). It is crucial to reveal the real virus–host cell interaction transversion by maintaining viral infiltration after riveting the virus on QDs (Cui et al., 2011; Liu et al., 2012; Hong et al., 2015; Ma et al., 2017). However, riveting virus on QDs via the unmild and uncontrollable physical-chemical process remains challenging.
To address this, we rationally designed a novel strategy with the photo-click cycloaddition-based QDs to tag and track the ZIKV (Scheme1). The light-initiated bio-orthogonal photo-click reaction has been widely applied in numerous biolabeling and bioimaging, enabling visualization of specific biomolecules with precise spatiotemporal control in their native environment (Lim and Lin, 2011; Huang et al., 2013; Herner and Lin, 2016). With this strategy, ZIKV was successfully tracked and visualized after cell entry in different cell lines, such as A549 and SNB19. Moreover, the ZIKV-QDs can map the ZIKV–host cell interactions under chlorpromazine hydrochloride (CPZ)- or nocodazole-treated conditions. This strategy would provide a reliable toolbox to elucidate the virus–host cell interactions and develop potential rapid diagnosis and therapeutic approaches.
2 Experimental Section
2.1 Materials and Reagents
Syto13 was obtained from Sigma. Amino-labeled QDs were obtained from Wuhan Jiayuan Quantum Dots Co., Ltd (Wuhan, China). The cell counting kit-8 (CCK-8) was obtained from Dojindo Laboratories (Kumamoto, Japan). 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDCI), N-hydroxysuccinimid esters (NHS), and N,N-4-dimethylaminopyridine (DMAP) were purchased from Energy Chemical (Shanghai, China).
2.2 Cell Lines
A549 cell and SNB19 cell were purchased from ATCC. The cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (Gibco, Ltd., Grand Island, NY, USA). The media were supplemented with 10% FBS (Gibco), 50 U mL−1 penicillin, and 50 μg ml−1 streptomycin. The cells were maintained in a humidified 37°C incubator in 5% CO2.
2.3 Modification of Zika Virus
9,10-phenanthrenequinone (PQ, 11 mg, 4.17*10–5 mol) was dispersed into 10 ml MES buffer, EDC (80 mg, 4.17*10–4 mol) and NHS (120 mg, 4.17*10–4 mol) were added, and then ultrasonic was applied for 15 s, the sealing film was sealed, and then it was shaken at 37°C for 15 min. Then, 100 µl of Zika virus (concentration is 2 mg/ml) was dispersed in 10 ml PBS. The liquid was added to the aforementioned MES buffer, shaken at 37°C overnight, and then purified by a NAP-5 desalting column to obtain Zika virus modified with the PQ group.
2.4 Modification of Quantum Dots
Take 1 µl of quantum dots (QDs) with the carboxyl group (molar concentration is 8*10–6), vinyl ether (VE) 1,000 μl, EDCI 15.3 mg, DAMP 9.8 mg, and put them into a 3-neck flask, respectively, then add 5 ml of dichloromethane to completely dissolve. Then, shake for 2 h at room temperature in a dark environment. Then, go through silica gel column chromatography to obtain quantum dots modified with vinyl ether group.
2.5 The Click Reaction of Zika Virus and Quantum Dots
The Zika viruses and the quantum dots were first modified with the PQ group and vinyl ether group, respectively. After that, these modified ZIKV and QDs were dissolved in a PBS solution and then irradiated with the LED lamp for 1 min to obtain the Zika virus modified with the quantum dots (ZIKV-QDs).
2.6 ZIKV-QD Internalization Assays
ZIKV-QDs and viral nucleic acid dye Syto13 were mixed and incubated for 1 h and then ultracentrifuged at 10,000 g for 0.5 h to remove the remaining dye. Then mixed with Vero or cells and shook 5 times with an interval of 15 min and then incubated in 5% carbon dioxide at 37°C for 24 h. Then, the cells were fixed with 4% paraformaldehyde, and the fluorescence imaging was observed under the confocal laser scanning microscope (CLSM).
2.7 Cell Culture and Cytotoxicity Evaluation In Vitro
SNB19 cells were cultured in DMEM containing 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a humidified 5% CO2 atmosphere. Cell density was determined using a hemocytometer before experimentation. Relative cell viabilities were determined by the standard CCK-8 assay. SNB19 cells were seeded into 96-well plates (104 cells per well). After cells were cultured for 12 h, they were added to a fresh culture medium and excited with a LED lamp (0.5 W/cm2) at different times. After incubation at 37°C for 12 h, those cells were added containing 10% CCK-8 DMEM (100 µl). After incubation for 2 h at 37°C, OD450, the absorbance value at 450 nm, was measured with a microplate reader to determine the cell viability.
2.8 Confocal Laser Scanning Microscope
The aforementioned fluorescent dye particles were photographed with a confocal laser scanning microscope (LSM880). The excitation wavelength of the quantum dots is 561 nm, and the emission wavelength is 605 nm. The excitation wavelength of Syto13 is 488 nm, and the emission wavelength is 509 nm.
3 Results and Discussion
3.1 Feasibility and Characterizations of QD-Modified Zika Virus
Photo-click reaction has been developed as an alternative strategy for spatiotemporally labeling and imaging (Lim and Lin, 2011; Li et al., 2018). Among them, 9,10-phenanthrenequinone (PQ) and electron-rich vinyl ether were widely chosen as photo-click substrates (Supplementary Figure S1, S2). As previous reports revealed that the ZIKV could be chemically labeled with the chemical proteomics strategies owning to the ZIKV surface E proteins to understand the early-stage entry of ZIKV into host cells (Srivastava et al., 2020; King and Irigoyen, 2021). To prepare a fluorescent tracking method, PQ and vinyl ether were functionalized in the ZIKV and QDs, respectively. Then, the ZIKV was labeled with fluorescent QDs via the cyclization under the irradiation (Scheme 1). As shown in Figure 1A, the primary absorbance peak of the as-prepared 9,10-phenanthrenequinone exhibited a significant blue shift that goes from 330 to 310 nm (Supplementary Figure S3), and the absorbance band at 425 nm disappeared along with the irradiation of LED lamp, which was in accordance with the 9,10-phenanthrenequinone analog reported by Zhang group (Li et al., 2018).
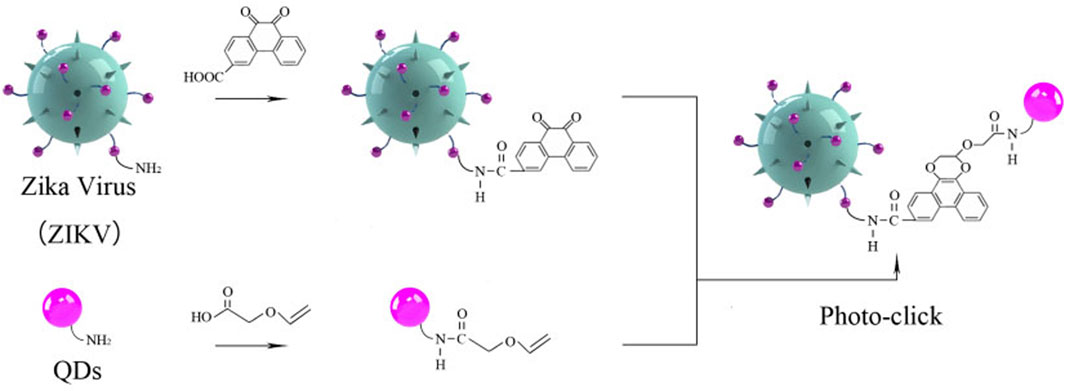
SCHEME 1. Schematic illustration of Zika virus labeling and imaging via light-initiated photo-click cycloaddition.
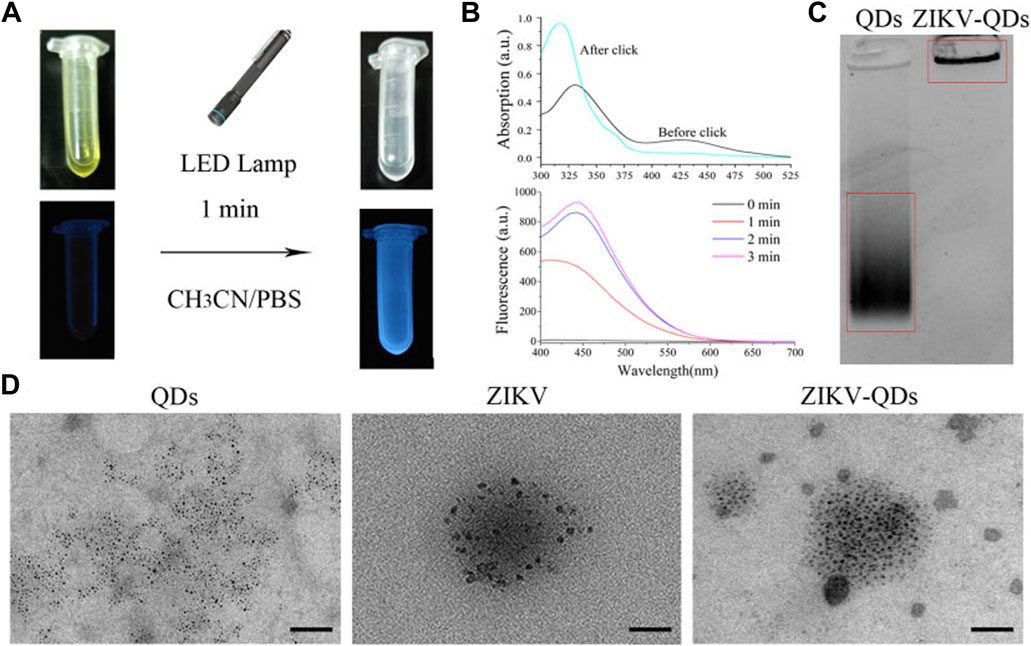
FIGURE 1. Characterizations of Zika virus labeling and imaging via light-initiated bio-orthogonal photo-click cycloaddition. (A) Images of ZIKV and QD solution without (left) or with (right) the irradiation of the LED lamp. (B) Absorbance and fluorescence changes of ZIKV and QDs in CH3CN/PBS solution upon irradiation treatment. (C) Polyacrylamide gel electrophoresis analysis of QDs and ZIKV-QDs. (D) Transmission electron microscopy (TEM) of QDs, ZIKV, and ZIKV-QDs.
Accordingly, the fluorescence intensity of PQ reacted with electron-rich vinyl ether at 450 nm was found to be greatly increased upon increasing irradiation time or concentration (Supplementary Figure S4), resulting in the photo-click reaction between PQ and electron-rich vinyl ether (Coelho et al., 2017). Meanwhile, the color of their mixed solution changed from yellow to colorless, and the blue light can be found, which was consistent with the antecedent results of absorbance and fluorescence (Figure 1B). In addition, the polyacrylamide gel electrophoresis analysis was chosen to validate whether the QDs were labeled on the ZIKV. As shown in Figure 1C, ZIKV-QDs held the original position compared to the QDs, which contributed to the macromolecular proteins of viruses conjugated with QDs by photo-click cycloaddition reaction. On the other hand, transmission electron microscopy (TEM) of ZIKV-QDs shows the typical ZIKV pattern functionalized with QDs, intuitively verifying that the QDs have been successfully decorated in the ZIKV (Figure 1D). Taken together, the photo-click cyclization between PQ and vinyl ether can be applied to label the ZIKV with QDs.
3.2 Quantitative Evaluation of QD-Modified Zika Virus
It is crucial to obtain high-purity viruses in various fields, ranging from virus pathogenesis to structure decomposition and vaccine development. Herein, the ZIKV particles were purified using a discontinuous sucrose gradient (Petruska et al., 2002), and all fractions were titrated by plaque assay. We observed the presence of ZIKV in all fractions, and the purified fraction has high titers above 108 PFU/ml (Supplementary Figure S5). In addition, a high RNA level was obtained at a sucrose concentration of 20%, resulting in 5.0 × 1010 RNA copies/ml (Supplementary Figure S6). To characterize the modification of QDs on ZIKV, we analyzed the particle sizes of ZIKV and ZIKV-QDs using the NanoSight NS300 equipment. As shown in Supplementary Figures S7 and S8, the average size increased from 108.9 ± 8.2 nm to 119.3 ± 11.5 nm, indicating that the QDs were successfully modified on ZIKV. Furthermore, the zeta potential data showed that the complex of ZIKV-QDs has a higher negative potential (−15.47 mV) than that of the individual QDs (−8.14 mV) and ZIKV (−12.03 mV) (Supplementary Figure S9). This result also indicated the successful modification of QDs on ZIKV through the amido bond between the carboxyl group and amino group. Additionally, no significant difference between the ZIKV and ZIKV-QDs viral titers was observed (Supplementary Figure S10), suggesting that the labeling process does not affect the infiltration performance.
3.3 Evaluation of Infiltration Capability With QD-Modified Zika Virus
Next, we evaluated its infiltration capability using the QD-modified ZIKV. In comparison absence of evidence for ZIKV infection in cells, we thus applied QD-modified ZIKV in mapping the infection process. The fluorescence imaging of A549 cells incubated with ZIKV-QDs in the red channel was found to be much brighter compared to the ZIKV group without QDs, and no significant difference in the green emission of Syto13 dye was observed under both conditions (Supplementary Figure S11A). As expected, the fluorescence image of QDs overlaps well with that of a commercially available fluorescent dye of the nucleic acid tracking dye Syto13. Also, the corresponding Mander’s overlap coefficient, a well-established colocation parameter, was performed as 0.94 for the ZIKV-QDs group (Supplementary Figure S11B, S11C). In order to confirm that the QDs were labeled on the ZIKV, ZIKV-QDs were first incubated with the nucleic acid dye Syto13. After that, Syto13-pretreated ZIKV-QDs mixed with cells and subsequently observed interaction behavior between ZIKV and host cell under the confocal laser scanning microscope. To verify the wide application of Syto13-pretreated ZIKV-QDs targeted tracing performance, we also obtained significant fluorescence colocalization experiments on the SNB19 cancer cell line via confocal fluorescence microscopy (Figure 2A). The Pearson correlation coefficient had been drawn via the confocal image (Figure 2B), and the corresponding Mander’s overlap coefficient had also been calculated to be 0.97 (Figure 2C). These bioimaging results demonstrated that the ZIKV labeled with QDs could localize and trace the functions in virus infection procedures.
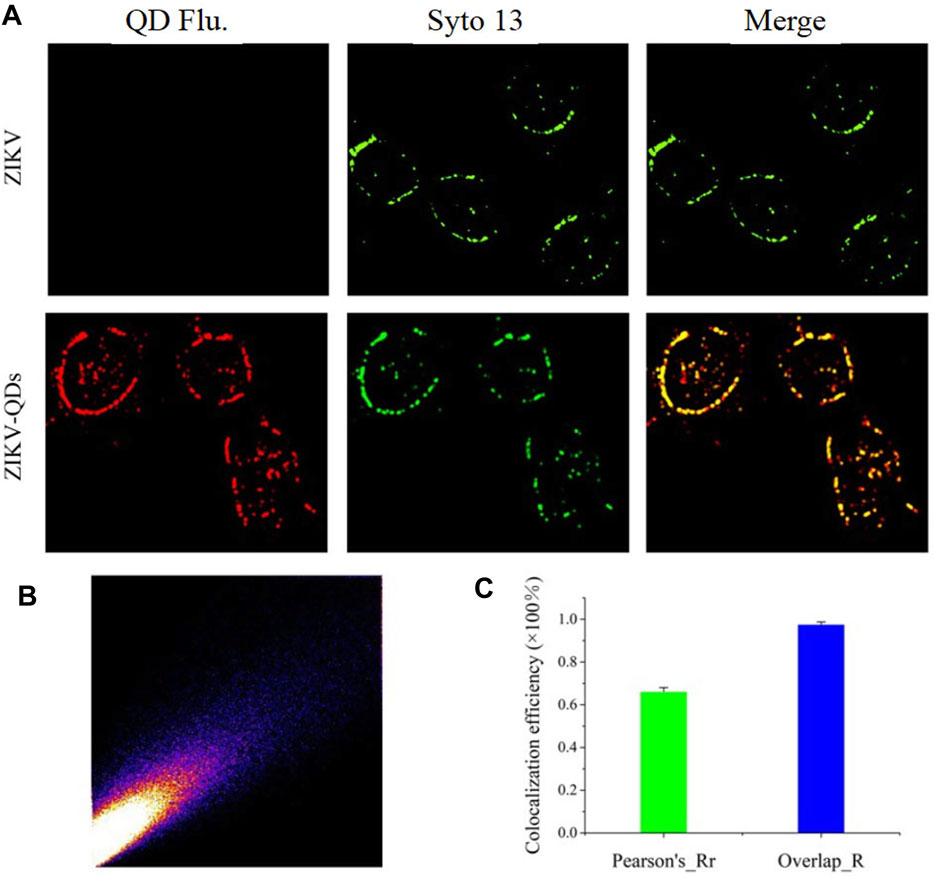
FIGURE 2. (A) Confocal microscopic images of ZIKV nucleic acid colocalized with the commercially available organelle trackers Syto13 in SNB19 cells (Flu is the abbreviation for fluorescence). (B) Colocalization scatterplots of (A). (C) Corresponding Pearson correlation coefficient of (A).
3.4 Evaluation of Drug Efficacy Through the QD-Modified Zika Virus
Furthermore, we demonstrated its applicability for assessing the efficacy of ZIKV-related treatments. Here, we introduced two typical kinds of antivirus drugs including CPZ and nocodazole, which could specifically inhibit clathrin-dependent endocytosis and formation of the microtubule, respectively (Xia and Liang, 2019; Sathyamoorthy et al., 2020). The cellular fluorescence intensity of QDs in the group treated with CPZ decreased compared to the control group (Figure 3). This was attributed to the less ZIKV crossing the monolayer of the barrier of Vero cells. In addition, the treatment of nocodazole also obviously abated the entry and infection of ZIKV at 2 h post-inoculation, providing direct evidence that the microtubule polymerization is required for intracellular trafficking of the ZIKV. These results prompt us to use the fluorescent nanoprobe QDs to trace the process of ZIKV infection and evaluate the treatment efficacy with the independent fluorescent signals.
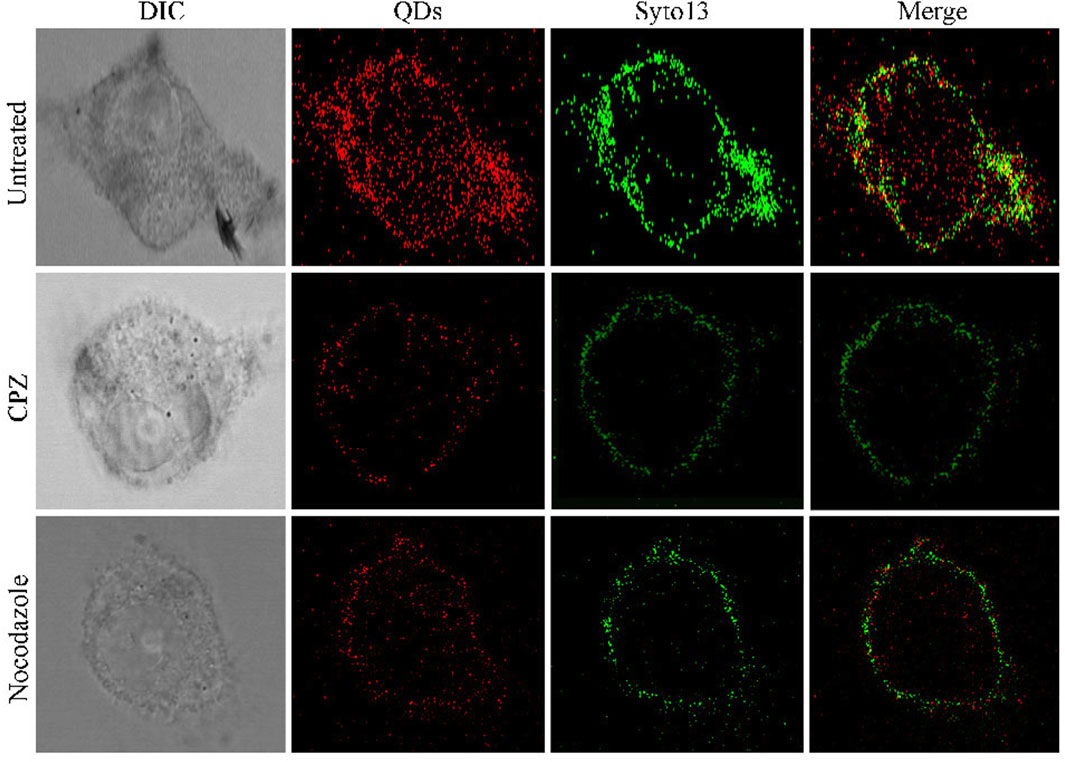
FIGURE 3. Confocal microscopic images of ZIKV nucleic acid colocalized with the commercially available organelle trackers Syto13 in SNB19 cells treated with different drugs including chlorpromazine and nocodazole.
To further validate the infiltration activity of the QD-labeled ZIKV, we chose the level of nascent protein synthesis to evaluate the ZIKV infection after being treated with CPZ or nocodazole. ZIKV envelope protein E (ZIKV E) is the major structural protein exposed on the cell surface of the particle, which is engaged in viral attachment, penetration, and membrane fusion. Thus, we chose the ZIKV E to study and evaluate the reason why hydrochloride and nocodazole could induce the infection of ZIKV. Western blot experiments indicate that ZIKV infection had a dramatic effect on the synthesis of ZIKV E (Figure 4A, top), similar to the case that cells infected with the translation inhibitor nocodazole (Figure 4A, bottom). At the same time, the CPZ and nocodazole exhibited some side effects (Figures 4B, C), which means these two drugs could not only inhibit the microtubule polymerization but also lead to the cellular apoptosis due to the disruption of intracellular trafficking (Liao et al., 2018). Furthermore, to test the biological application ability of pre-QDs, the cytotoxicity of PQ and VE was tested with Vero cells. After incubation with different concentrations of PQ and VE, there was no significant change in SNB19 cells (Figures 4D, E). In order to evaluate the influence of light on cells, the effect of light was investigated by the standard CCK-8 assay. Following incubation of various times of light ranging from 0 to 5 min, the cellular viabilities were determined to exceed 80% (Figure 4F), indicating its low effect with the LED lamp laser. Taken together, these aforementioned findings indicated that ZIKV-QDs could accurately map and localize the ZIKV via monitoring the fluorescence of QDs under various conditions.
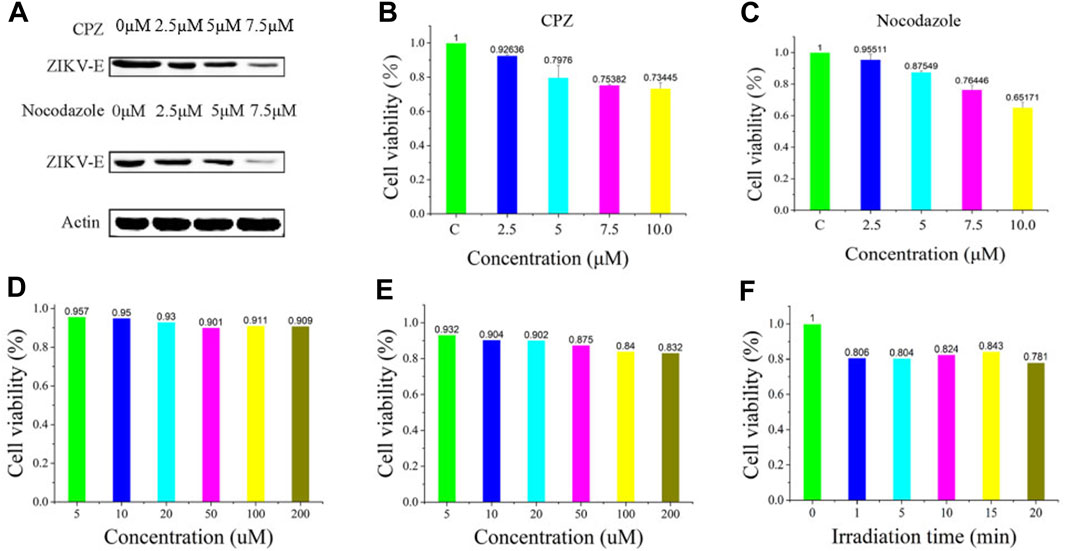
FIGURE 4. (A) Levels of β-actin and ZIKV envelope protein E (ZIKV E) of SNB19 cells treated with CPZ or nocodazole. Cytotoxicity studies of (B) CPZ and (C) nocodazole. Cytotoxicity studies of (D) PQ and (E) VE, or (F) irradiation time.
4 Conclusion
In this work, we developed a novel QD-based probe for reliable labeling and visualization of the Zika virus. This probe leverages photo-activated bio-orthogonal cycloaddition for high-efficient conjugation of ZIKV and QDs, exhibiting a simple labeling process and no influence on the infiltration performance. We verified that such a conjugation of ZIKV and QD probe can localize and trace the functions in the virus infection procedure. Moreover, the infiltration activity of the QD-labeled ZIKV is validated using the anti-ZIKV drugs including CPZ or nocodazole by monitoring the level of nascent protein synthesis. These findings suggest that the proposed QD probe could accurately map and localize the ZIKV via monitoring the fluorescence. Thus, this bioorthogonal-enabled QD probe might be a promising approach for monitoring the pathogenicity activities of ZIKV.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JZ and RY designed and performed the experiment. RHY, QW, and YW were responsible for data collation and drafting the manuscript. MH, XC, and JH contributed to synthetic designs. WL, CX, and YJ, BY edited and revised the manuscript draft. JZ and YL were responsible for providing funds and unified management of work. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81972019, 21904145, 82002253, 81671970, 81772136, and 82102444), the Special Fund of Foshan Summit Plan (2020B019, 2020B012, 2020A015, 2019A006, 2019C002, and 2019D008), the Training Project of National Science Foundation for Outstanding/Excellent Young Scholars of Southern Medical University (C620PF0217), the China Postdoctoral Science Foundation (2020M682783 and 2021M693638), the Regional Joint Fund of Natural Science Foundation of Guangdong Province (2020A1515110529), the Guangdong Basic and Applied Basic Research Foundation (2020A1515010754), the Medical Research Fund of Guangdong Province (A2020322), Special Fund for Science and Technology Innovation Strategy of Guangdong Province (2020A1515011402), the Foundation of Foshan City (FS0AA-KJ218-1301-0034 and 2018AB003411), and the Young Scientists Project of the National Key Research and Development Program (2021YFC2302200).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.940511/full#supplementary-material
References
Barreiro, P. (2016). Evolving RNA Virus Pandemics: HIV, HCV, Ebola, Dengue, Chikunguya, and Now Zika. AIDS Rev. 18 (1), 54–55.
Chen, J., Yang, Y.-f., Yang, Y., Zou, P., Chen, J., He, Y., et al. (2018). AXL Promotes Zika Virus Infection in Astrocytes by Antagonizing Type I Interferon Signalling. Nat. Microbiol. 3 (3), 302–309. doi:10.1038/s41564-017-0092-4
Chen, X., Zhu, J., Song, W., and Xiao, L.-P. (2021). Integrated Cascade Biorefinery Processes to Transform Woody Biomass into Phenolic Monomers and Carbon Quantum Dots. Front. Bioeng. Biotechnol. 9, 803138. doi:10.3389/fbioe.2021.803138
Chiramel, A. I., and Best, S. M. (2018). Role of Autophagy in Zika Virus Infection and Pathogenesis. Virus Res. 254, 34–40. doi:10.1016/j.virusres.2017.09.006
Chung, S., and Zhang, M. (2021). Microwave-Assisted Synthesis of Carbon Dot - Iron Oxide Nanoparticles for Fluorescence Imaging and Therapy. Front. Bioeng. Biotechnol. 9, 711534. doi:10.3389/fbioe.2021.711534
Coelho, S. V. A., Neris, R. L. S., Papa, M. P., Schnellrath, L. C., Meuren, L. M., Tschoeke, D. A., et al. (2017). Development of Standard Methods for Zika Virus Propagation, Titration, and Purification. J. Virol. Methods 246, 65–74. doi:10.1016/j.jviromet.2017.04.011
Cui, Z.-Q., Ren, Q., Wei, H.-P., Chen, Z., Deng, J.-Y., Zhang, Z.-P., et al. (2011). Quantum Dot-Aptamer Nanoprobes for Recognizing and Labeling Influenza A Virus Particles. Nanoscale 3 (6), 2454–2457. doi:10.1039/c1nr10218d
Ergünay, K., Saygan, M. B., Aydoğan, S., Litzba, N., Niedrig, M., Pınar, A., et al. (2010). Investigation of Dengue Virus and Yellow Fever Virus Seropositivities in Blood Donors from Central/Northern Anatolia, Turkey. Mikrobiyol. Bul. 44 (3), 415–424.
Garcia, G., Paul, S., Beshara, S., Ramanujan, V. K., Ramaiah, A., Nielsen-Saines, K., et al. (2020). Hippo Signaling Pathway Has a Critical Role in Zika Virus Replication and in the Pathogenesis of Neuroinflammation. Am. J. Pathol. 190 (4), 844–861. doi:10.1016/j.ajpath.2019.12.005
Grant, A., Ponia, S. S., Tripathi, S., Balasubramaniam, V., Miorin, L., Sourisseau, M., et al. (2016). Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe 19 (6), 882–890. doi:10.1016/j.chom.2016.05.009
He, C., Lin, X., Mei, Y., Luo, Y., Yang, M., Kuang, Y., et al. (2022). Recent Advances in Carbon Dots for In Vitro/Vivo Fluorescent Bioimaging: A Mini-Review. Front. Chem. 10, 905475. doi:10.3389/fchem.2022.905475
Herner, A., and Lin, Q. (2016). Photo-Triggered Click Chemistry for Biological Applications. Top. Curr. Chem. (Z) 374 (1), 77–107. doi:10.1007/s41061-015-0002-2
Hills, S. L., Fischer, M., and Petersen, L. R. (2017). Epidemiology of Zika Virus Infection. J. Infect. Dis. 216 (Suppl. l_10), S868–s874. doi:10.1093/infdis/jix434
Hong, Z.-Y., Lv, C., Liu, A.-A., Liu, S.-L., Sun, E.-Z., Zhang, Z.-L., et al. (2015). Clicking Hydrazine and Aldehyde: The Way to Labeling of Viruses with Quantum Dots. ACS Nano 9 (12), 11750–11760. doi:10.1021/acsnano.5b03256
Huang, L.-L., Lu, G.-H., Hao, J., Wang, H., Yin, D.-L., and Xie, H.-Y. (2013). Enveloped Virus Labeling via Both Intrinsic Biosynthesis and Metabolic Incorporation of Phospholipids in Host Cells. Anal. Chem. 85 (10), 5263–5270. doi:10.1021/ac4008144
Kikuti, M., Tauro, L. B., Moreira, P. S. S., Campos, G. S., Paploski, I. A. D., Weaver, S. C., et al. (2018). Diagnostic Performance of Commercial IgM and IgG Enzyme-Linked Immunoassays (ELISAs) for Diagnosis of Zika Virus Infection. Virol. J. 15 (1), 108. doi:10.1186/s12985-018-1015-6
King, E. L., and Irigoyen, N. (2021). Zika Virus and Neuropathogenesis: The Unanswered Question of Which Strain Is More Prone to Causing Microcephaly and Other Neurological Defects. Front. Cell. Neurosci. 15, 1–14. doi:10.3389/fncel.2021.695106
Kuang, Y., Zhang, J., Xiong, M., Zeng, W., Lin, X., Yi, X., et al. (2020). A Novel Nanosystem Realizing Curcumin Delivery Based on Fe3O4@Carbon Dots Nanocomposite for Alzheimer's Disease Therapy. Front. Bioeng. Biotechnol. 8, 614906–614916. doi:10.3389/fbioe.2020.614906
Li, J., Kong, H., Huang, L., Cheng, B., Qin, K., Zheng, M., et al. (2018). Visible Light-Initiated Bioorthogonal Photoclick Cycloaddition. J. Am. Chem. Soc. 140 (44), 14542–14546. doi:10.1021/jacs.8b08175
Liao, Y., Fan, Z., Deng, H., Yang, Y., Lin, J., Zhao, Z., et al. (2018). Zika Virus Liquid Biopsy: A Dendritic Ru(bpy)(3) (2+)-Polymer-Amplified ECL Diagnosis Strategy Using a Drop of Blood. ACS Cent. Sci. 4 (10), 1403–1411. doi:10.1021/acscentsci.8b00471
Liao, Y., Li, B., Zhao, Z., Fu, Y., Tan, Q., Li, X., et al. (2020). Targeted Theranostics for Tuberculosis: A Rifampicin-Loaded Aggregation-Induced Emission Carrier for Granulomas Tracking and Anti-infection. ACS Nano 14 (7), 8046–8058. doi:10.1021/acsnano.0c00586
Lim, R. K. V., and Lin, Q. (2011). Photoinducible Bioorthogonal Chemistry: a Spatiotemporally Controllable Tool to Visualize and Perturb Proteins in Live Cells. Acc. Chem. Res. 44 (9), 828–839. doi:10.1021/ar200021p
Lin, X., Xiong, M., Zhang, J., He, C., Ma, X., Zhang, H., et al. (2021). Carbon Dots Based on Natural Resources: Synthesis and Applications in Sensors. Microchem. J. 160, 105604. doi:10.1016/j.microc.2020.105604
Liu, S.-L., Zhang, Z.-L., Tian, Z.-Q., Zhao, H.-S., Liu, H., Sun, E.-Z., et al. (2012). Effectively and Efficiently Dissecting the Infection of Influenza Virus by Quantum-Dot-Based Single-Particle Tracking. ACS Nano 6 (1), 141–150. doi:10.1021/nn2031353
Low, S. L., Leo, Y. S., Lai, Y. L., Lam, S., Tan, H. H., Wong, J. C. C., et al. (2021). Evaluation of Eight Commercial Zika Virus IgM and IgG Serology Assays for Diagnostics and Research. PLoS One 16 (1), e0244601. doi:10.1371/journal.pone.0244601
Ma, Y., Wang, M., Li, W., Zhang, Z., Zhang, X., Tan, T., et al. (2017). Live Cell Imaging of Single Genomic Loci with Quantum Dot-Labeled TALEs. Nat. Commun. 8, 15318. doi:10.1038/ncomms15318
Matea, C., Mocan, T., Tabaran, F., Pop, T., Mosteanu, O., Puia, C., et al. (2017). Quantum Dots in Imaging, Drug Delivery and Sensor Applications. Int. J. Nanomed. 12, 5421–5431. doi:10.2147/ijn.S138624
Mengesha Tsegaye, M., Beyene, B., Ayele, W., Abebe, A., Tareke, I., Sall, A., et al. (2018). Sero-prevalence of Yellow Fever and Related Flavi Viruses in Ethiopia: a Public Health Perspective. BMC Public Health 18 (1), 1011. doi:10.1186/s12889-018-5726-9
Miner, J. J., and Diamond, M. S. (2017). Zika Virus Pathogenesis and Tissue Tropism. Cell Host Microbe 21 (2), 134–142. doi:10.1016/j.chom.2017.01.004
Petruska, J. M., Frank, D. W., Freeman, G. B., Evans, E. W., and MacDonald, J. S. (2002). Toxicity and Carcinogenicity Studies of Chlorpromazine Hydrochloride and P-Cresidine in the P53 Heterozygous Mouse Model. Toxicol. Pathol. 30 (6), 696–704. doi:10.1080/01926230290166788
Pleskova, S., Mikheeva, E., and Gornostaeva, E. (2018). Using of Quantum Dots in Biology and Medicine. Adv. Exp. Med. Biol. 1048, 323–334. doi:10.1007/978-3-319-72041-8_19
Ribeiro, J. F. F., Pereira, M. I. A., Assis, L. G., Cabral Filho, P. E., Santos, B. S., Pereira, G. A. L., et al. (2019). Quantum Dots-Based Fluoroimmunoassay for Anti-Zika Virus IgG Antibodies Detection. J. Photochem. Photobiol. B Biol. 194, 135–139. doi:10.1016/j.jphotobiol.2019.03.019
Sampathkumar, P., and Sanchez, J. L. (2016). Zika Virus in the Americas: A Review for Clinicians. Mayo Clin. Proc. 91 (4), 514–521. doi:10.1016/j.mayocp.2016.02.017
Sathyamoorthy, N., Chintamaneni, P. K., and Chinni, S. (2020). Plausible Role of Combination of Chlorpromazine Hydrochloride and Teicoplanin against COVID-19. Med. Hypotheses 144, 110011. doi:10.1016/j.mehy.2020.110011
Srivastava, M., Zhang, Y., Chen, J., Sirohi, D., Miller, A., Zhang, Y., et al. (2020). Chemical Proteomics Tracks Virus Entry and Uncovers NCAM1 as Zika Virus Receptor. Nat. Commun. 11 (1), 3896. doi:10.1038/s41467-020-17638-y
Xia, Y., and Liang, T. J. (2019). Development of Direct-Acting Antiviral and Host-Targeting Agents for Treatment of Hepatitis B Virus Infection. Gastroenterology 156 (2), 311–324. doi:10.1053/j.gastro.2018.07.057
Yi, X., Zeng, W., Wang, C., Chen, Y., Zheng, L., Zhu, X., et al. (2022). A Step-by-step Multiple Stimuli-Responsive Metal-Phenolic Network Prodrug Nanoparticles for Chemotherapy. Nano Res. 15, 1205–1212. doi:10.1007/s12274-021-3626-2
You, J.-O., Liu, Y.-S., Liu, Y.-C., Joo, K.-I., and Peng, C.-A. (2006). Incorporation of Quantum Dots on Virus in Polycationic Solution. Int. J. Nanomed. 1 (1), 59–64. doi:10.2147/nano.2006.1.1.59
Zhang, P., Liu, S., Gao, D., Hu, D., Gong, P., Sheng, Z., et al. (2012). Click-functionalized Compact Quantum Dots Protected by Multidentate-Imidazole Ligands: Conjugation-Ready Nanotags for Living-Virus Labeling and Imaging. J. Am. Chem. Soc. 134 (20), 8388–8391. doi:10.1021/ja302367s
Zhang, R., Zheng, J., and Zhang, T. (2020). In Vivo selective Imaging of Metabolic Glycosylation with a Tetrazine-Modified Upconversion Nanoprobe. RSC Adv. 10, 15990–15996. doi:10.1039/d0ra01832e
Keywords: Zika virus, quantum dots (DQs), light-initiated cycloaddition, fluorescent probe, phenanthrenequinone
Citation: Zheng J, Yue R, Yang R, Wu Q, Wu Y, Huang M, Chen X, Lin W, Huang J, Chen X, Jiang Y, Yang B and Liao Y (2022) Visualization of Zika Virus Infection via a Light-Initiated Bio-Orthogonal Cycloaddition Labeling Strategy. Front. Bioeng. Biotechnol. 10:940511. doi: 10.3389/fbioe.2022.940511
Received: 10 May 2022; Accepted: 30 May 2022;
Published: 08 July 2022.
Edited by:
Qitong Huang, Gannan Medical University, ChinaReviewed by:
Liewei Wen, Jinan University, ChinaZhixiong Wang, National Institutes of Health (NIH), United States
Xiaofeng Lin, Gannan Medical University, China
Copyright © 2022 Zheng, Yue, Yang, Wu, Wu, Huang, Chen, Lin, Huang, Chen, Jiang, Yang and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodong Chen, cxd234@163.com; Yideng Jiang, jwcjyd@163.com; Bin Yang, yangbin1@smu.edu.cn; Yuhui Liao, liaoyh8@mail.sysu.edu.cn
†These authors have contributed equally to this work
 Judun Zheng
Judun Zheng Rui Yue
Rui Yue Ronghua Yang
Ronghua Yang Qikang Wu3,4†
Qikang Wu3,4†  Mingxing Huang
Mingxing Huang Xiaodong Chen
Xiaodong Chen Bin Yang
Bin Yang Yuhui Liao
Yuhui Liao