Exosome Mimetics-Loaded Hydrogel Accelerates Wound Repair by Transferring Functional Mitochondrial Proteins
- 1Department of Plastic and Reconstructive Surgery, Second Affiliated Hospital of Naval Medical University, Shanghai, China
- 2Department of Histology and Embryology, College of Basic Medicine, Naval Medical University, Shanghai, China
- 3Stem Cell and Regeneration Medicine Institute, Research Center of Translational Medicine, Naval Medical University, Shanghai, China
- 4Department of Histology and Embryology, Harbin Medical University, Harbin, China
- 5Shanghai Key Laboratory of Cell Engineering, Shanghai, China
- 6Department of Plastic and Reconstructive Surgery, Shanghai East Hospital, Tongji University, Shanghai, China
Loading human umbilical mesenchymal stem cell (hUMSC) derived exosomes onto hydrogel scaffolds is a strategy for rapid wound healing. The clinical application of exosomes is hindered by low production, and exosome mimetics could be substituted for exosomes. Here, the therapeutic effects of exosome-loaded hydrogels and exosome mimetic-loaded hydrogels on wounds are evaluated. Our results revealed that exosome mimetic-loaded hydrogels promote wound healing more efficiently than exosome-loaded hydrogels. Exosome mimetics can promote the proliferation and migration of dermal fibroblasts (hDF-a) cells in vitro. To investigate how exosome mimetics play a role, proteomics analysis was applied, and the obtained results suggested that exosome mimetics significantly enrich mitochondrial-derived oxidative phosphorylation-related proteins in comparison to exosomes. Overall, our work envisages the emerging potential of exosome mimetics, which take the advantage of exosomes and can be promising candidates for exosomes. It also suggests that hUMSC-derived exosome mimetic-loaded hydrogels have remarkable prospects for clinical application.
Introduction
Skin wound healing, which is essential for the survival of organisms, is a natural process that restores the integrity of normal skin and relieves injury (Han and Ceilley, 2017). However, inefficient treatments for skin wound healing have always attracted the community’s attention because they cause a tremendous burden on patients, families, and society (Singer and Clark, 1999; Brown et al., 2008). Skin wound healing is a multi-process cascade that involves inflammation, hemagglutination, cell proliferation, and extracellular matrix (ECM) remodeling (Gurtner et al., 2008; Rodrigues et al., 2019). Delayed wound healing may occur due to a disorder in one of the four stages described earlier. During different stages, rapid closure of the wound site through migration and proliferation of epithelial cells is essential to restore barrier function (Takeo et al., 2015). In relation to re-epithelialization, the restoration of the dermis occurs by the proliferation and migration of fibroblasts (Driskell et al., 2013).
Human umbilical mesenchymal stem cells (hUMSCs), which can differentiate into cells, migrate to injured tissues and secrete many factors, show special advantages in medical applications due to their multipotent ability (Ding et al., 2011; Biazar and Keshel, 2013; Nakamura et al., 2013; Huang et al., 2020). However, there are still many obstacles to stem cell-based therapy; for example, the allogenic source and the tumorigenicity of these cells hinder their clinical application (Han and Sidhu, 2011; Kim et al., 2017). Exosomes (Exos) are extracellular vesicles with a size of 30–150 nm. Exosomes contain different proteins, lipids, nucleic acids, and various molecules (Kalluri and LeBleu, 2020). Evidence suggests that exosomes, as a cell-free therapy, can stimulate the local healing process via paracrine secretion (Tonnard et al., 2013; Bernardini et al., 2015). Exosomes isolated from hUMSCs have previously been shown to be capable of promoting skin wound healing (Chen et al., 2018; Tao et al., 2018; Henriques-Antunes et al., 2019). However, exosome isolation is still difficult and costly (Gurunathan et al., 2019). Recently, studies revealed that high-yield generation of exosome mimetics (EMs) can be substituted for exosomes for use in clinical practice, but this approach has not been applied for skin wound healing and tissue regeneration (Nasiri Kenari et al., 2020). Moreover, whether MSC-prepared exosome mimetics could promote wound healing, and the underlying mechanism in the treatment of skin injuries remain unclear.
As a biomaterial, hydrogels have already been widely used for various biomedical applications, such as drug delivery, tissue adhesion, and tissue regeneration (Pacelli et al., 2015). Gelatine methacryloyl (GelMA) hydrogels, with photo-crosslinking properties, resemble the properties of ECM based on their appropriate biological properties and tunable physical characteristics (Klotz et al., 2016). Drugs loaded on GelMA can be released slowly, improving the local drug administration concentration (Wang et al., 2019; Zhao et al., 2020). In this study, we investigated whether exosome mimetics could promote skin wound healing and suggested the underlying mechanism by defining proteomic differences (Figure 1A).
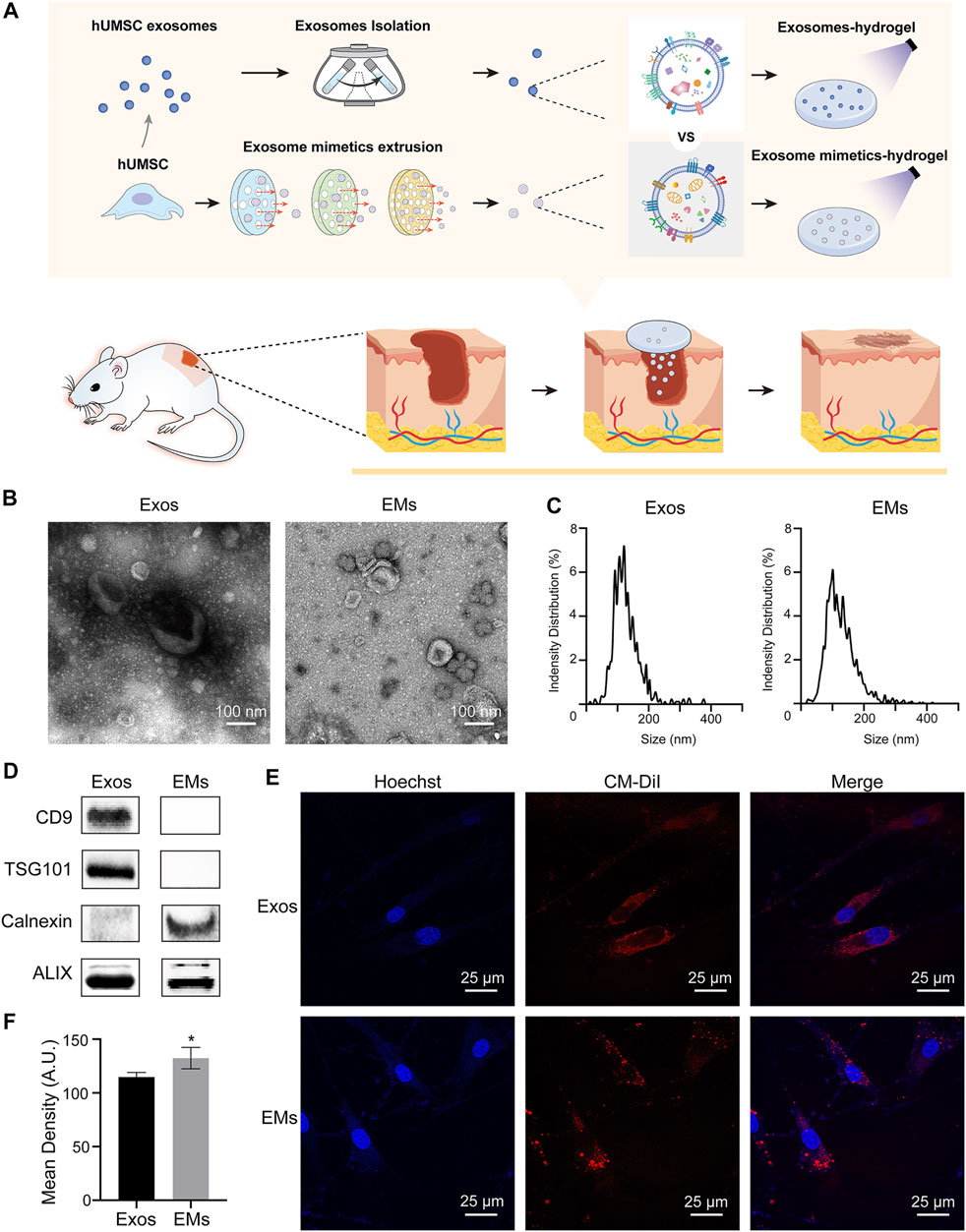
FIGURE 1. (A) Schematic overview of the development of hUMSC-Exos and hUMSC-EM hydrogels for skin wound healing and tissue regeneration. (B) TEM images of hUMSC-derived exosomes and exosome mimetics (hUMSC-derived exosomes: Exos; hUMSC-derived exosome mimetics: EMs). (C) Particle size distribution of hUMSC-derived exosomes and exosome mimetics measured by using nanoparticle tracking analysis (NTA). (D) Characteristic markers of hUMSC-derived exosomes and exosome mimetics. Membrane surface marker proteins (CD9), exosome biogenesis marker proteins (TSG101), endoplasmic reticulum related protein (calnexin), and endoplasmic marker proteins (ALIX) were analyzed by Western blotting. (E) Internalization of exosomes and EMs by fibroblasts. Exosomes and EMs are labeled in red, and fibroblast nuclei are labeled in blue. (F) The quantification of internalization. (n = 3, *p < 0.05)
Materials and Methods
Cell Culture and Synthesis of Human Umbilical Mesenchymal Stem Cell-Derived Exosomes and Exosome Mimetics
All the cells were incubated at 37°C with 5% CO2, and hUMSCs were grown in a mesenchymal stem cell basal medium (Cell Farm, Shanghai, China) with 10% exosome-free fetal bovine serum (FBS) (Gibco). FBS was managed by ultracentrifugation at × 120,000 g for 120 min to remove the exosomes from FBS. All of the supernatant was collected from hUMSCs from passages 4 to 8. After collecting the supernatant of hUMSCs, the exosomes were ultracentrifuged at ×300 g for 10 min, ×2000 g for 10 min, and ×15,000 g for 40 min to remove cells and cell debris. Next, the supernatant was ultra-centrifuged at ×1,00,000 g for 70 min twice in Ultra-Clear centrifuge tubes (Beckman Coulter, United States) for exosome purification. Finally, the precipitate was resuspended in PBS and stored at −80°C for identification and the following experiments.
After hUMSCs reached a confluence of approximately 70%–80%, the cells were resuspended in a PBS solution of 1 × 106 cells per 1 ml. The EMs were collected using serial extrusive approaches through polycarbonate membranes (Whatman, United States) of 10 nm, 5 nm, 1 nm, 800 μm, 400 μm, and 200 μm using a mini-extruder (Morgec, Shanghai, China). Exosome mimetics were centrifuged at × 100,000 g for 70 min twice and purified by a 100 kDa centrifugal filter (Millipore, CA, United States) at ×4,000 g for 20 min. The final EM sample was stored at −80°C.
Identification of Human Umbilical Mesenchymal Stem Cell-Derived Exosomes and Exosome Mimetics
Transmission electron microscopy (TEM), ZetaView, and Western blotting analysis were applied to identify the characteristics of the isolated hUMSC-Exos. The morphology and microstructure of exosomes were observed using TEM (Jeol 1230, Japan). The size distribution and concentration of exosomes were measured at VivaCell Biosciences with ZetaView PMX 110 (Particle Metrix, Meerbusch, Germany) and the corresponding software ZetaView 8.04.02. Exosomal characteristic surface markers, including ALIX, CD9, TSG101, and calnexin, were analyzed by Western blotting analysis.
Western Blotting Analysis
Exosome and EM solutions were used to isolate total cellular protein, and a BCA kit (Beyotime Biotechnology, Shanghai, China) was used for the quantification of protein levels. Proteins were separated by 10% SDS-PAGE, transferred to PVDF membranes, and finally probed with the appropriate primary antibodies specific for ALIX (#ab275377, Abcam, Cambridge, United Kingdom, 1:1000), CD9 (#ab236630, Abcam, Cambridge, United Kingdom, 1:1000), TSG101 (#ab125011, Abcam, Cambridge, United Kingdom, 1:1000), and calnexin (#133615, Abcam, Cambridge, United Kingdom, 1:1000). The hUMSC and EM solutions were used to isolate total cellular protein, and a BCA kit was used for the quantification of protein levels. Proteins were separated by 10% SDS-PAGE, transferred to PVDF membranes, and finally probed with the appropriate primary antibodies specific for cardiotin (sc-53002, Biocompare, 1:1000) and β-actin (#3700, CST, 1:1000). Then, we used secondary antibodies to probe the blots, and the proteins were detected with a Tanon 5200 scanner. The AllDoc_x software was used to quantify the protein band density.
Exosome and Exosome Mimetics Uptake by Human Dermal Fibroblasts-a
To examine the internalization of hUMSC-derived exosomes by hDF-a, exosomes were labeled with a red fluorescent dye, CM-DiI (Sigma, United States). Then, labeled exosomes were incubated with hDF-a for 24 h. After that, the cells were fixed in 4% paraformaldehyde for 15 min and washed with PBS. The nuclei were stained with 1 μg/ml Hoechst (Invitrogen, United States) for 10 min before observation by using TEM.
Preparation of GelMA Loaded With Human Umbilical Mesenchymal Stem Cell-Exosomes and Human Umbilical Mesenchymal Stem Cell-Exosome Mimetics
GelMA hydrogels (30% in LAP solution, EFL, Suzhou, China) were prepared according to the manufacturer’s instructions. Briefly, 3 g of gelatin was dissolved in 10 ml of LAP solution in a 55°C water bath for 30 min. Next, another 200 μg of exosomes or EMs were added to the mixed solution at room temperature. The mixed solution was irradiated under UV light for 1 min for crosslinking. To detect the distribution of exosomes in GelMA, exosomes and EMs were labeled with CM-DiI and then observed by TEM.
Cell Counting Kit 8 Assay
CCK-8 assays were used to detect the effects of hUMSC-derived Exos and EMs on the proliferation of hDF-a cells. Briefly, hDF-a cells were seeded into 96-well plates at a density of 5 × 103 cells per well and divided into seven groups when the cells were incubated until an exponential growth period. A complete high-glucose DMEM was used as the control (CN) group. The concentrations of exosomes and EMs were as follows: exosome 5 μg/ml (E5), exosome 20 μg/ml (E20), exosome 50 μg/ml (E50), EMs 5 μg/ml (EM5), EMs 20 μg/ml (EM20), and EMs 50 μg/ml (EM50). After being treated with different concentrations of exosomes and EMs for 1 day, 90 μl of culture medium mixed with 10 μl of CCK-8 reagent (Dojindo, Japan) was added into each well and incubated with 5% CO2 at 37°C for 2 h in a humidified incubator. Then, the optical density (OD) value at 450 nm was measured by the microplate reader (Tecan, Thermo Scientific, United States).
EdU Proliferation Assay
An EdU kit (Riobo, Guangzhou, China) was used to detect the effects of hUMSC-derived Exos and EMs on the proliferation of hDF-a cells. The hDF-a cells were seeded at 1 × 105 cells per well in 24-well plates and were cultured until the exponential growth period. Then, the cells were divided into seven groups as follows: the control (CN) group, exosomes 5 μg/ml (E5), exosomes 20 μg/ml (E20), exosomes 50 μg/ml (E50), EMs 5 μg/ml (EM5), EMs 20 μg/ml (EM20), and EMs 50 μg/ml (EM50). The control group of cells was cultured in high-glucose DMEM. Moreover, the exosomes and EM groups were prepared with a concentration gradient. After the cells were cultured for 24 h, the EdU experiment was performed following the manufacturer’s instructions. Cell proliferation was determined by counting the number of EdU-positive cells and total cells from randomly selected image fields. Then, the ratio of EdU-positive cells to total cells was calculated. Images were taken with a Zeiss fluorescence microscope (Axio Observer.D1, Shanghai, China).
Scratch Wound Assay
Briefly, 1 × 105 cells per well were seeded into 6-well plates and incubated for 24 h. Then, the confluent layer of cells was scratched using a sterile 20–200 μL pipette tip. After washing the cells with PBS, 0, 20, or 50 μg/ml exosomes or EMs were added. Images were recorded at 0, 6, and 12 h after the monolayers were scratched. Scratched areas were measured by ImageJ software.
In Vivo Cutaneous Wound Healing Model of Mice
C57 mice (22 ± 2 g) at 8 weeks old were purchased from Shanghai Jihui Laboratory Animal Technology Co., Ltd. (Shanghai, China). Animals were housed in an SPF-class laboratory and allowed access to water and food. All animal care and experimental procedures were supported by the Committee on Ethics of Biomedicine, Naval Medical University (Approval No: 20180309020). Before the surgery, the mice were anesthetized by an intraperitoneal injection of 2% pentobarbital (50 mg/kg). After shaving the dorsal hair and disinfecting the skin, two 10 mm × 10 mm bilateral symmetry round full-thickness incisions were made by a 10-mm punch biopsy on the upper back. The mice were randomly divided into five groups of six animals as follows: 1) Control group: the wounds were merely covered by gauze and Tegaderm™ film (3M, United States) without any treatment. 2) Exos group: the 20 μg/ml hUSMC-Exos were injected subcutaneously around the wound area. 3) Exosome mimetics group: the 20 μg/ml hUSMC-EMs were injected subcutaneously around the wound area. 4) GelMA-Exos group: the wounds were covered by GelMA loaded with 20 μg/ml hUSMC-Exos. 5) GelMA-EMs group: the wounds were covered by GelMA loaded with 20 μg/ml hUSMC-EMs. The wounds were photographed at 0, 4, 7, and 14 days after surgery. The wound size was measured by ImageJ software (NIH, United States). At 4, 7, and 14 days after surgery, two mice in each group were sacrificed for further analysis.
Histology Analysis
The wounded skin in each group was removed at days 4, 7, and 14 postsurgery, fixed in 4% paraformaldehyde for 48 h, gradually dehydrated with a series of graded ethanol solutions, embedded in paraffin, and sectioned into 5 μm-thick sections. Then, the sections were stained with hematoxylin and eosin (H&E) and Masson’s trichrome. H&E staining was used to evaluate the extent of wound healing, while Masson’s trichrome staining was performed to determine the maturity of collagen. Cell proliferation was tested by immunohistochemical staining of Ki67 (AF0198, Affinity Biosciences, United States). The sections were rehydrated and heated in a microwave in citrate buffer (0.01 M; pH 6.0) for 15 min to retrieve the antigen. Then, they were blocked in 1% BSA for 30 min at room temperature, incubated with the primary antibody anti-ki67 overnight at 4°C, and finally, they were incubated with the respective secondary antibodies at room temperature for 1 h in the dark. Images were acquired with a fluorescence microscope.
Proteomic Profiling
Liquid chromatography–tandem mass spectrometry (LC-MS/MS)-based proteomic technologies were used to ionize and separate eluted peptides by mass spectrometry according to their unique mass-to-charge ratio (m/z). The data were analyzed in Proteome Discoverer (v2.4.1.15) software (Thermo Fisher) for the main search. The differential expression threshold was set to a 1.5 fold change. After the t-test analysis, significantly differentially enriched proteins were identified through volcano plot filtering between the two experimental groups. Finally, hierarchical clustering was performed to show distinguishable enriched protein profiles among the samples using the GO and KEGG databases (http://eggnog5.embl.de).
Statistical Analysis
The data are presented as the mean ± standard deviation (SD). Two groups were compared by Student’s t-test using GraphPad Prism 8.0 software. A two-sided t-test with p < 0.05 was defined as statistically significant.
Results
Characterization of Human Umbilical Mesenchymal Stem Cell-Derived Exosomes and Exosome Mimetics
hUMSC exosomes were collected using ultracentrifugation as previously described (Livshits et al., 2015; Li et al., 2017). Then, hUMSCs were extruded to form EMs, which were collected by using ultracentrifugation (Huang et al., 2020; Nasiri Kenari et al., 2020).
hUMSC-derived exosomes and EMs were characterized by TEM, NTA, and Western blotting analysis. TEM showed the morphologies of hUMSC-derived exosomes and EMs as shown in Figure 1B. We can see that EMs, which are the same as exosomes, are round cup-shaped vesicles with lipid membranes (Kalluri and LeBleu, 2020). The NTA results (Figure 1C) of hUMSC-derived exosomes and EMs demonstrated an approximate Gaussian distribution of size with a peak diameter of approximately 100 nm, which is consistent with the TEM results. From Western blotting analysis, vesicle membrane markers (CD9), multivesicular body proteins (TSG101), and cell generation-related protein (ALIX) (Kalluri and LeBleu, 2020) are used as identification proteins for exosomes. In addition, EMs had endoplasmic reticulum-related protein (calnexin) which exosomes did not have (Figure 1D) (Chen et al., 2020).
Previous work verified that exosomes and EMs have similar characteristics and extensive biological effects (Lu and Huang, 2020). After that, the uptake efficacy of exosomes and EMs by recipient cells, such as dermal fibroblasts, was also evaluated. We labeled the hUMSC membrane with CM-DiI in red and then introduced it into polycarbonate filters with pores to produce EMs. At the same time, the exosomes were also labeled with CM-DiI. Exosomes and EMs were incubated with the human dermis-immortalized fibroblast cell line hDF-a for 24 h separately (Zhang et al., 2016). After Hoechst staining of the nuclei in blue, the TEM merging graphics showed that these two kinds of nanoparticles could be internalized into the cells (Figure 1E). The obtained results indicate that hDF-a has effective internalization capabilities for both hUMSC-Exos and EMs.
Human Umbilical Mesenchymal Stem Cell-Derived Exosome Mimetics Can Promote Proliferation and Migration of Human Dermal Fibroblasts-a In Vitro
Next, the regulation of hDF-a behavior by exosomes and EMs was evaluated. The EdU incorporation assay and CCK-8 assays were used to evaluate hDF-a proliferation ability, and the scratch test was used to evaluate hDF-a migration ability (Liu et al., 2018). With increasing concentrations of exosomes and EMs, the number of EdU-probe-labeled proliferating cells per field was gradually increased in a dose-dependent manner compared to the exosome group and control group (Figure 2A). Moreover, the EMs group showed higher proliferative capability than the exosome group at a certain concentration (Figure 2B). After 4 h of incubation, as shown by the OD values at 450 nm, the results of the CCK-8 assays (Figure 2C) also showed the same tendency as the EdU incorporation assays. This result indicated that the proliferation-promoting effects of exosomes and EMs on hDF-a were equal at the same concentration.
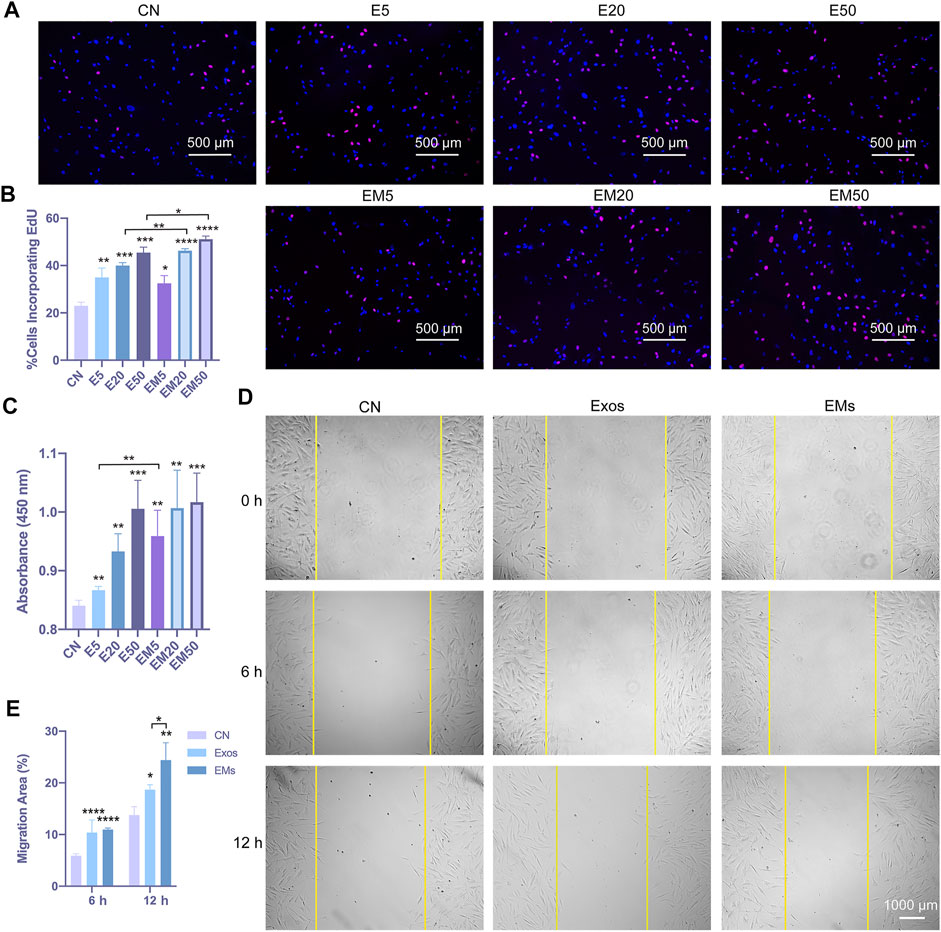
FIGURE 2. The effect of hUMSC-derived exosomes and EMs on the proliferation and migration of hDF-a cells in vitro (CN = control, E5 = exosome 5 μg/ml, E20 = exosome 20 μg/ml, E50 = exosome 50 μg/ml, EM5 = EMs 5 μg/ml, EM20 = EMs 20 μg/ml, EM50 = EMs 50 μg/ml). (A) hUMSC-derived exosomes and EMs promoted proliferation of hDF-a by EdU incorporation assays. Cell nuclei were stained blue. After the images are merged, proliferating cells were counterstained purple with an EdU probe. (B) Quantitative analysis of the EdU incorporation assays (n = 4, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). (C) hUMSC-derived exosomes and EMs promoted the proliferation of hDF-a cells as shown in CCK-8 assays. (n = 4, *p < 0.05, **p < 0.01) (D) A scratch assay verified the migration of hDF-a cells in vitro by hUMSC-derived exosomes and EMs. (E) Quantitative analysis of scratch assays (n = 4, *p < 0.05, **p < 0.01, ****p < 0.0001).
We also applied the scratch assay to verify the migration of hDF-a cells in vitro after hUMSC-derived exosome or EM treatments, and the result indicated that exosomes and EMs could promote the migration of hDF-a (Figure 2D). The quantitative analysis by ImageJ suggested that the percentage of migration area (%) of EMs was slightly smaller than that of exosomes (Figure 2E). In this part, the promoting effects of exosomes and EMs on the proliferation and migration of fibroblasts were tested, and EMs showed equal effects compared with exosomes.
Human Umbilical Mesenchymal Stem Cell-Derived Exosome Mimetics Loaded on GelMA Accelerate Wound Healing In Vivo
The effects of exosomes and EMs on wound healing were subsequently evaluated. hUMSC-derived exosomes and EMs were labeled with CM-DiI and then loaded on GelMA. Exosomes and EMs were both evenly incorporated into GelMA (Supplementary Figure S1). In addition, we investigated whether the hUMSC-derived EM-loaded GelMA hydrogel could improve the closure and tissue repair of full-thickness skin wounds (Supplementary Figure S2). Figure 3A shows optical images of the wound healing on days 4, 7, and 14 after surgery. While the wound sizes in all five groups decreased with time, especially on day 4 after injury, the wound sizes in the GelMA-EMs group were clearly smaller than those in the GelMA-Exos group, and the wounds had almost closed by 14 days. In addition, the EMs group had a repair effect similar to that of the exosomes group. The exosomes group showed a better repair effect than the control group as previously described. Quantification of the wound sizes confirmed the aforementioned results (Figure 3B).
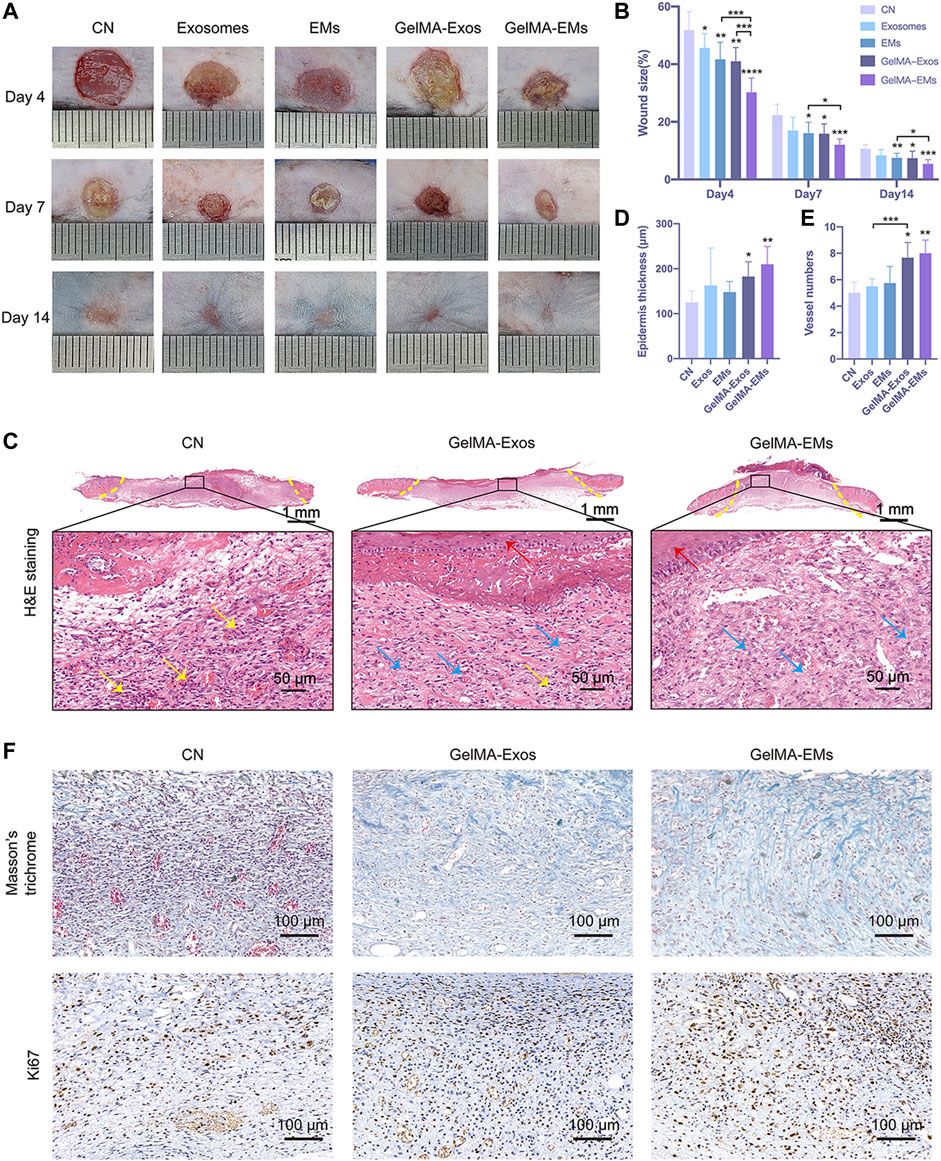
FIGURE 3. The effect of hUMSC-derived exosome mimetics on skin wound healing in vivo. Representative images (A) and quantification (B) of full-thickness cutaneous wound sizes showed that the hUMSC-derived exosome mimetics loaded on GelMA could accelerate the wound healing process. The data are shown as the mean ± SD. The significant difference between the untreated and hydrogel-treated groups was analyzed by the t-test (n = 6, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). Epidermis thickness (C) and vessel numbers (D) per random field were analyzed by the t-test. (n = 4, *p < 0.05, **p < 0.01) H&E staining (E) of the wound sites is shown. The yellow dotted lines indicate the boundary of the wound sites. The blue arrows indicate granulation tissues. The yellow arrows indicate inflammatory neutrophils. The red arrows label the newly formed epidermis. (F) Masson’s trichrome staining and ki67 immunohistochemistry staining of wound sites.
Hematoxylin and eosin (H&E) staining showed the histologic changes of the wounds at day 7 (Figure 3C). The yellow dotted lines indicate the boundary of the wound site. Compared to the control group, we can see that the GelMA-EMs group showed a reduced inflammatory response, and the granulation tissues were restored much more quickly. The area of granulation tissues in random sights differs remarkably. The newly formed epidermis was nearly intact and stretched under hydrogel residues in the GelMA-Exos and GelMA-EMs groups. The thickness (Figure 3D) of the neo-epithelium of GelMA-EMs was significantly increased. The vessel numbers in random 0.04 mm2 fields (Figure 3E) indicate that the wound bed of the experimental groups has higher angiogenesis activity than that of the control group. It is known that the fibroblasts showed higher proliferation activity 7 days after injury. Afterward, macroscopic images of Masson’s trichrome staining were applied to evaluate the deposition and maturation of collagen on day 7 (Figure 3F). Ki67 immunohistochemistry was used to investigate cell proliferation (Yang et al., 2020). Compared to the other four groups, the experimental groups showed stronger positive staining of Ki67 (Supplementary Figure S3), suggesting that both exosomes and EMs could facilitate cell proliferation during the healing period (Supplementary Figure S3), thereby accelerating granulation tissue formation and enhancing collagen deposition. The in vivo experiments showed that GelMA-loaded EMs could promote skin wound healing, and these results provide promising prospects for clinical application.
Proteomic Profiling Reveals a Distinct Protein Landscape Between Exosome Mimetics and Exosomes
The aforementioned data show that compared with hUMSC-derived exosomes, hUMSC-derived EMs have a similar efficiency in wound healing and even better efficiency under some circumstances. To determine the molecular difference between exosomes and hUMSC-derived EMs, we carried out label-free mass spectrometry-based proteomics. Proteomic profiling identified 2,469 proteins in exosomes and 3,427 proteins in EMs from a total of 3,673 proteins. A number of proteins (2,223) were commonly found in both samples (Figure 4A). The differentially enriched protein between samples was examined by the t-test and p values, indicating that 1,666 proteins were significantly differentially enriched (FC > 1.5, p < 0.05). A volcano plot was generated to visualize the differential enrichment between the two groups, as shown in Figure 4B. Next, the upregulated proteins in hUMSC-derived EMs were further analyzed based on the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (http://eggnog5.embl.de) (Huerta-Cepas et al., 2019). Subcellular analysis of upregulated proteins in hUMSC-derived EMs was performed (Figure 4C). Interestingly, in addition to the extracellular matrix, cytoskeleton, and cell membrane, many proteins were localized in the mitochondria. Furthermore, the results of enrichment analysis of the KEGG signaling pathway showed that EM proteins were enriched in oxidative phosphorylation- and tricarboxylic acid cycle (TCA cycle)-related pathways (Figure 4D). Subsequent analysis of the proteomic data revealed that EMs-upregulated proteins are mainly concentrated in the enzymes related to material metabolism in the TCA cycle and the respiratory chain complex (Figure 4E, red) (Fernie et al., 2004). Taken together, we showed that distinguishable protein profiles between EMs and exosomes are upregulated, especially in the mitochondria and oxidative phosphorylation. Protein profiling also revealed that EMs may promote wound healing by enhancing the oxidative phosphorylation of cells in the wound.
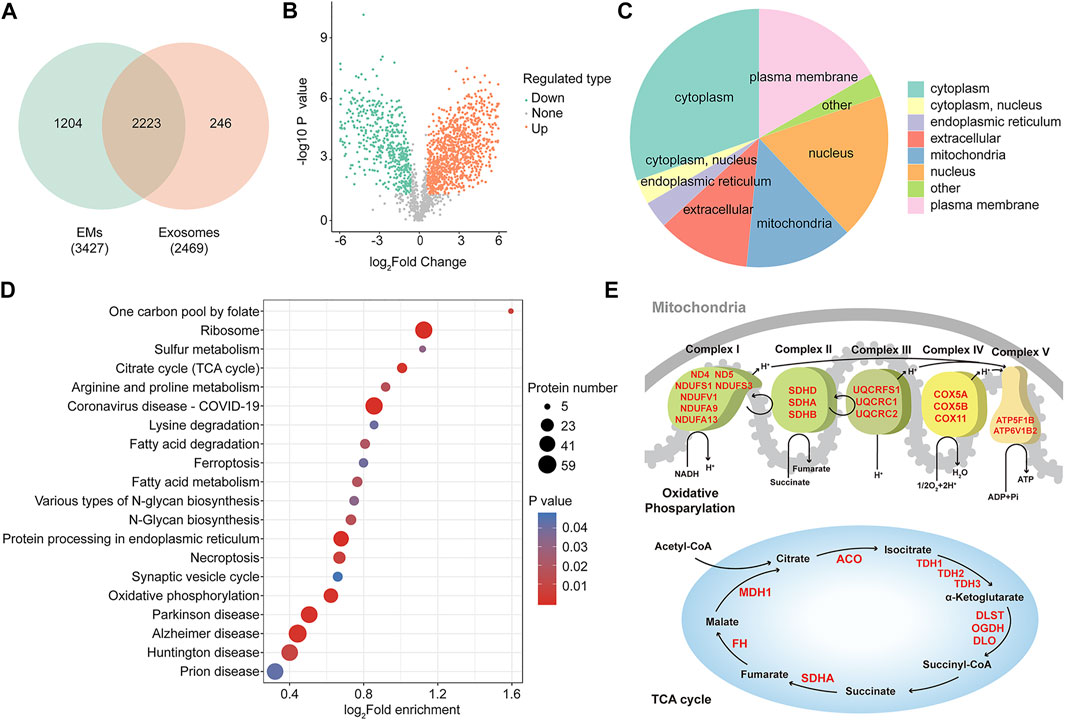
FIGURE 4. Proteomic profiling reveals a distinct protein landscape between EMs and exosomes. (A) Venn diagram showing the number of identified proteins of hUMSC-derived exosomes and EMs (n = 3). (B) Volcano plots are useful to visualize differentially enriched protein patterns among hUMSC-derived EMs and exosomes (the quantitative difference in enriched protein between EMs to exosomes using p-value > 0.05, FC ≥ 1.5). (C) Subcellular analysis of differentially enriched protein annotations between EMs and exosomes. (D) KEGG enrichment analysis of differentially enriched proteins annotations between EMs and exosomes. (E) Upregulated protein enriched in mitochondrial oxidative phosphorylation and the TCA cycle.
Discussion
Delayed wound healing remains an intractable affair for patients, families, and society. Recent studies of hUMSC-derived exosomes showed that, in addition to their cell communication function, they also play an important role in transferring therapeutic factors. The benefit of exosomes is that they have quantities of bioactive substances and lower immunological responses than the cells (Chen et al., 2018; Tao et al., 2018; Henriques-Antunes et al., 2019). The application of exosome-based cell-free therapy in wound healing and tissue regeneration attracts our attention. However, exosome-based therapy is still limited by their heterogeneity, therapeutic loading, large-scale manufacturing restrictions, and low yield for extraction and purification (Gurunathan et al., 2019). Recently, engineering nanoparticles are said to be obtained by polycarbonate filters with pores, microfluidic devices, supramolecular chemistry, and hybridization (Nasiri Kenari et al., 2020; Li et al., 2021). New studies have shown that EMs, as a kind of engineered nanoparticle, demonstrate reproducibility in the protocol, cost-effectiveness, higher yield production rates, and can simulate complex biological components (Ilahibaks et al., 2019; Li et al., 2021). Furthermore, Fan and his colleagues showed that hUMSC-derived EMs could promote bone regeneration (Fan et al., 2020). Therefore, we wondered whether the EMs could promote skin wound healing and tissue regeneration and whether there was a difference between extruded exosome mimetics and natural exosomes.
In this study, hUMSC-derived EMs were achieved by taking hUMSCs through polycarbonate filters with hUMSC-derived exosomes as their controls. As described in the results, we found that hUMSC-derived exosomes and EMs shared a similar appearance, indicating that EMs could potentially take advantage of nanoparticle properties such as biocompatibility, biological stability, and plasma protein interactions. Because EMs were eventually extruded by filters of 200 nm, compared to exosome particle sizes concentrated at approximately 100 nm, the particle sizes of EMs were concentrated between 100 and 200 nm. Moreover, besides the traditional cell membrane markers similar to exosomes, EMs also enriched some other proteins from their donor cells, which were shown in our Western blotting analysis. These proteins were also identified by mass spectrometry-based proteomics analysis. Biocompatible markers, such as CD55 and CD59, may help EMs escape from elimination and prolong the half-life of circulation.
GelMA hydrogels have been reported as promising biomaterials to deliver drugs/cells for wound healing and tissue regeneration because they have inherent advantages, such as absorbing wound fluid, preventing infection, and offering gaseous exchange (Pacelli et al., 2015). It could be easily achieved by conjugating lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) with GelMA by shielding it under UV rays (Yue et al., 2015). To increase the local drug distribution and allow exosomes and EMs to be continuously released, GelMA could be an ideal biomaterial. We successfully assembled exosomes and EMs on GelMA and they could be distributed evenly. Therefore, we proposed that bioactive hydrogels carrying hUMSC-derived EMs could be a highly efficient novel method for wound healing and tissue regeneration.
As shown in the results, EMs promoted fibroblast proliferation and migration in vitro. Surprisingly, EMs have even better biological activities than exosomes under certain conditions. At the same time, the results in vivo showed a similar result that the EMs could promote wound healing as effectively as exosomes, and the GelMA-EMs group presented the best wound healing efficiency even over the GelMA-Exos group. The skin wound injury treatment outcomes were especially diverse regarding wound area shrinkage, tissue regeneration, and epidermal stretching. The manufacturing of engineered exosome mimetics, an alternative candidate of exosomes to mimic exosomes by size, morphology, and function, has been proposed in this article. The administration of exosomes and EMs by GelMA has better efficiency than mere subcutaneous administration. The obtained results suggested that EMs combined with wound dressing scaffolds could provide a novel cell-free method for wound healing and tissue regeneration.
Although the benefits of hUMSC-derived EMs address the issues in wound healing, the proteome cargo of EMs has not been comprehensively characterized and may potentially have an effect on recipient tissues and cells (Nasiri Kenari et al., 2019). We investigated the proteome cargo of EMs and compared it to that of natural exosomes secreted from the same cell line. A subset of proteins is highly abundant in hUMSC-derived EMs compared to well-known exosomal cargo proteins. We further applied GO and KEGG pathway analyses. There was a noteworthy enrichment of the mitochondrial-related proteins in EMs compared to exosomes. Therefore, we evaluated the content of mitochondrial-related proteins in EMs, and the results showed that there were abundant mitochondrial-related proteins in EMs (Supplementary Figure S4) (Driesen et al., 2009). The aforementioned results indicate that the functional mitochondrial components in hUMSCs may be packaged into EMs. As it is well known, the mitochondrion is an energy reservoir for the functional ability and individual activity. Hayakawa and his colleagues reported that astrocytic mitochondria could provide entry to adjacent neurons and amplify cell survival signals (Hayakawa et al., 2016). Many researchers have reported that mitochondrial transfer from the bone marrow-derived stromal cells could protect against acute lung injury (Islam et al., 2012), and it is envisaged that EMs may take effect in the means of MSC mitochondrial transfer to regulate wound healing and tissue regeneration processes.
Conclusion
In this study, the efficiency of EMs in repairing wounds was verified in vivo and in vitro. The obtained results showed that compared with hUMSC-derived natural exosomes, hUMSC-derived exosome mimetics could similarly promote wound healing by promoting the proliferation and migration of dermal fibroblasts. Further proteomics data revealed that EMs carry mitochondrial-related proteins derived from hUMSCs. Quantities of TCA cycle-related proteins and respiratory chain complexes are significantly enriched in EMs. The obtained results suggest that treatment with GelMA-carrying EMs for wound healing and tissue regeneration could have promising prospects for future clinical applications. EMs are promising candidates for natural cell-derived exosomes.
Data Availability Statement
The Proteomic raw data are available in iProX (www.iprox.org) under the accession number IPX0004319000.
Ethics Statement
The animal study was reviewed and approved by the Committee on Ethics of Biomedicine, Naval Medical University.
Author Contributions
YW, HJ, and LZ contributed to the supervision and design of the study. JZ, ZL, and LW carried out the experiments and organized methods. QJ, AD, and ND performed partly experiments and performed analysis. YZ performed partly experiments and provided approval for the work. JZ wrote the first draft of the manuscript. ZL and YW wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
The authors acknowledge the support of the China National Key Research and Development Program Stem Cell and Translational Research Key Projects (2018YFA0108300), the Shanghai Key Laboratory of Cell Engineering (14DZ2272300), and Naval Medical University and the University of Shanghai for Science and Technology Joint Projects (2020-RZ04).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.866505/full#supplementary-material
References
Bernardini, F. P., Gennai, A., Izzo, L., Zambelli, A., Repaci, E., Baldelli, I., et al. (2015). Superficial Enhanced Fluid Fat Injection (SEFFI) to Correct Volume Defects and Skin Aging of the Face and Periocular Region. Aesthet. Surg. J. 35 (5), 504–515. doi:10.1093/asj/sjv001
Biazar, E., and Keshel, S. H. (2013). The Healing Effect of Stem Cells Loaded in Nanofibrous Scaffolds on Full Thickness Skin Defects. J. Biomed. Nanotechnol. 9 (9), 1471–1482. doi:10.1166/jbn.2013.1639
Brown, B. C., McKenna, S. P., Siddhi, K., McGrouther, D. A., and Bayat, A. (2008). The Hidden Cost of Skin Scars: Quality of Life after Skin Scarring. J. Plast. Reconstr. Aesthet. Surg. 61 (9), 1049–1058. doi:10.1016/j.bjps.2008.03.020
Chen, C.-Y., Rao, S.-S., Ren, L., Hu, X.-K., Tan, Y.-J., Hu, Y., et al. (2018). Exosomal DMBT1 from Human Urine-Derived Stem Cells Facilitates Diabetic Wound Repair by Promoting Angiogenesis. Theranostics 8 (6), 1607–1623. doi:10.7150/thno.22958
Chen, P., Ruan, A., Zhou, J., Huang, L., Zhang, X., Ma, Y., et al. (2020). Extraction and Identification of Synovial Tissue-Derived Exosomes by Different Separation Techniques. J. Orthop. Surg. Res. 15 (1), 97. doi:10.1186/s13018-020-01604-x
Ding, D.-C., Shyu, W.-C., and Lin, S.-Z. (2011). Mesenchymal Stem Cells. Cel. Transpl. 20 (1), 5–14. doi:10.3727/096368910x
Driesen, R. B., Verheyen, F. K., Schaart, G., de Mazière, A., Viebahn, C., Prinzen, F. W., et al. (2009). Cardiotin Localization in Mitochondria of Cardiomyocytes In Vivo and In Vitro and its Down-Regulation during Dedifferentiation. Cardiovasc. Pathol. 18 (1), 19–27. doi:10.1016/j.carpath.2007.12.008
Driskell, R. R., Lichtenberger, B. M., Hoste, E., Kretzschmar, K., Simons, B. D., Charalambous, M., et al. (2013). Distinct Fibroblast Lineages Determine Dermal Architecture in Skin Development and Repair. Nature 504 (7479), 277–281. doi:10.1038/nature12783
Fan, J., Lee, C.-S., Kim, S., Chen, C., Aghaloo, T., and Lee, M. (2020). Generation of Small RNA-Modulated Exosome Mimetics for Bone Regeneration. ACS Nano 14 (9), 11973–11984. doi:10.1021/acsnano.0c05122
Fernie, A. R., Carrari, F., and Sweetlove, L. J. (2004). Respiratory Metabolism: Glycolysis, the TCA Cycle and Mitochondrial Electron Transport. Curr. Opin. Plant Biol. 7 (3), 254–261. doi:10.1016/j.pbi.2004.03.007
Gurtner, G. C., Werner, S., Barrandon, Y., and Longaker, M. T. (2008). Wound Repair and Regeneration. Nature 453 (7193), 314–321. doi:10.1038/nature07039
Gurunathan, S., Kang, M.-H., Jeyaraj, M., Qasim, M., and Kim, J.-H. (2019). Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 8 (4), 307. doi:10.3390/cells8040307
Han, G., and Ceilley, R. (2017). Chronic Wound Healing: A Review of Current Management and Treatments. Adv. Ther. 34 (3), 599–610. doi:10.1007/s12325-017-0478-y
Han, J., and Sidhu, K. (2011). Embryonic Stem Cell Extracts: Use in Differentiation and Reprogramming. Regenerative Med. 6 (2), 215–227. doi:10.2217/rme.11.8
Hayakawa, K., Esposito, E., Wang, X., Terasaki, Y., Liu, Y., Xing, C., et al. (2016). Transfer of Mitochondria from Astrocytes to Neurons after Stroke. Nature 535 (7613), 551–555. doi:10.1038/nature18928
Henriques-Antunes, H., Cardoso, R. M. S., Zonari, A., Correia, J., Leal, E. C., Jiménez-Balsa, A., et al. (2019). The Kinetics of Small Extracellular Vesicle Delivery Impacts Skin Tissue Regeneration. ACS Nano 13 (8), 8694–8707. doi:10.1021/acsnano.9b00376
Huang, Y.-Z., Gou, M., Da, L.-C., Zhang, W.-Q., and Xie, H.-Q. (2020). Mesenchymal Stem Cells for Chronic Wound Healing: Current Status of Preclinical and Clinical Studies. Tissue Eng. B: Rev. 26, 555–570. doi:10.1089/ten.TEB.2019.0351
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 47 (D1), D309–d314. doi:10.1093/nar/gky1085
Ilahibaks, N. F., Lei, Z., Mol, E. A., Deshantri, A. K., Jiang, L., Schiffelers, R. M., et al. (2019). Biofabrication of Cell-Derived Nanovesicles: A Potential Alternative to Extracellular Vesicles for Regenerative Medicine. Cells 8 (12), 1509. doi:10.3390/cells8121509
Islam, M. N., Das, S. R., Emin, M. T., Wei, M., Sun, L., Westphalen, K., et al. (2012). Mitochondrial Transfer from Bone-Marrow-Derived Stromal Cells to Pulmonary Alveoli Protects Against Acute Lung Injury. Nat. Med. 18 (5), 759–765. doi:10.1038/nm.2736
Kalluri, R., and LeBleu, V. S. (2020). The Biology, Function, and Biomedical Applications of Exosomes. Science 367 (6478), eaau6977. doi:10.1126/science.aau6977
Kim, M., Rhee, J.-K., Choi, H., Kwon, A., Kim, J., Lee, G. D., et al. (2017). Passage-dependent Accumulation of Somatic Mutations in Mesenchymal Stromal Cells during In Vitro Culture Revealed by Whole Genome Sequencing. Sci. Rep. 7 (1), 14508. doi:10.1038/s41598-017-15155-5
Klotz, B. J., Gawlitta, D., Rosenberg, A. J. W. P., Malda, J., and Melchels, F. P. W. (2016). Gelatin-Methacryloyl Hydrogels: Towards Biofabrication-Based Tissue Repair. Trends Biotechnol. 34 (5), 394–407. doi:10.1016/j.tibtech.2016.01.002
Li, P., Kaslan, M., Lee, S. H., Yao, J., and Gao, Z. (2017). Progress in Exosome Isolation Techniques. Theranostics 7 (3), 789–804. doi:10.7150/thno.18133
Li, Y.-J., Wu, J.-Y., Liu, J., Xu, W., Qiu, X., Huang, S., et al. (2021). Artificial Exosomes for Translational Nanomedicine. J. Nanobiotechnol. 19 (1), 242. doi:10.1186/s12951-021-00986-2
Liu, F., Feng, X. X., Zhu, S. L., Huang, H. Y., Chen, Y. D., Pan, Y. F., et al. (2018). Sonic Hedgehog Signaling Pathway Mediates Proliferation and Migration of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis via MAPK/ERK Signaling Pathway. Front. Immunol. 9, 2847. doi:10.3389/fimmu.2018.02847
Livshits, M. A., Khomyakova, E., Evtushenko, E. G., Lazarev, V. N., Kulemin, N. A., Semina, S. E., et al. (2015). Isolation of Exosomes by Differential Centrifugation: Theoretical Analysis of a Commonly Used Protocol. Sci. Rep. 5, 17319. doi:10.1038/srep17319
Lu, M., and Huang, Y. (2020). Bioinspired Exosome-Like Therapeutics and Delivery Nanoplatforms. Biomaterials 242, 119925. doi:10.1016/j.biomaterials.2020.119925
Nakamura, Y., Ishikawa, H., Kawai, K., Tabata, Y., and Suzuki, S. (2013). Enhanced Wound Healing by Topical Administration of Mesenchymal Stem Cells Transfected with Stromal Cell-Derived Factor-1. Biomaterials 34 (37), 9393–9400. doi:10.1016/j.biomaterials.2013.08.053
Nasiri Kenari, A., Kastaniegaard, K., Greening, D. W., Shambrook, M., Stensballe, A., Cheng, L., et al. (2019). Proteomic and Post‐Translational Modification Profiling of Exosome‐Mimetic Nanovesicles Compared to Exosomes. Proteomics 19 (8), 1800161. doi:10.1002/pmic.201800161
Nasiri Kenari, A., Cheng, L., and Hill, A. F. (2020). Methods for Loading Therapeutics into Extracellular Vesicles and Generating Extracellular Vesicles Mimetic-Nanovesicles. Methods 177, 103–113. doi:10.1016/j.ymeth.2020.01.001
Pacelli, S., Paolicelli, P., Dreesen, I., Kobayashi, S., Vitalone, A., and Casadei, M. A. (2015). Injectable and Photocross-Linkable Gels Based on Gellan Gum Methacrylate: A New Tool for Biomedical Application. Int. J. Biol. Macromol. 72, 1335–1342. doi:10.1016/j.ijbiomac.2014.10.046
Rodrigues, M., Kosaric, N., Bonham, C. A., and Gurtner, G. C. (2019). Wound Healing: A Cellular Perspective. Physiol. Rev. 99 (1), 665–706. doi:10.1152/physrev.00067.2017
Singer, A. J., and Clark, R. A. F. (1999). Cutaneous Wound Healing. N. Engl. J. Med. 341 (10), 738–746. doi:10.1056/nejm199909023411006
Takeo, M., Lee, W., and Ito, M. (2015). Wound Healing and Skin Regeneration. Cold Spring Harbor Perspect. Med. 5 (1), a023267. doi:10.1101/cshperspect.a023267
Tao, S.-C., Rui, B.-Y., Wang, Q.-Y., Zhou, D., Zhang, Y., and Guo, S.-C. (2018). Extracellular Vesicle-Mimetic Nanovesicles Transport LncRNA-H19 as Competing Endogenous RNA for the Treatment of Diabetic Wounds. Drug Deliv. 25 (1), 241–255. doi:10.1080/10717544.2018.1425774
Tonnard, P. L., Verpaele, A. M., and Zeltzer, A. A. (2013). Augmentation Blepharoplasty: A Review of 500 Consecutive Patients. Aesthet. Surg. J. 33 (3), 341–352. doi:10.1177/1090820x13478966
Wang, M., Wang, C., Chen, M., Xi, Y., Cheng, W., Mao, C., et al. (2019). Efficient Angiogenesis-Based Diabetic Wound Healing/Skin Reconstruction through Bioactive Antibacterial Adhesive Ultraviolet Shielding Nanodressing with Exosome Release. ACS Nano 13 (9), 10279–10293. doi:10.1021/acsnano.9b03656
Yang, J., Chen, Z., Pan, D., Li, H., and Shen, J. (2020). Umbilical Cord-Derived Mesenchymal Stem Cell-Derived Exosomes Combined Pluronic F127 Hydrogel Promote Chronic Diabetic Wound Healing and Complete Skin Regeneration. Ijn 15, 5911–5926. doi:10.2147/ijn.S249129
Yue, K., Trujillo-de Santiago, G., Alvarez, M. M., Tamayol, A., Annabi, N., and Khademhosseini, A. (2015). Synthesis, Properties, and Biomedical Applications of Gelatin Methacryloyl (GelMA) Hydrogels. Biomaterials 73, 254–271. doi:10.1016/j.biomaterials.2015.08.045
Zhang, E., Gao, B., Yang, L., Wu, X., and Wang, Z. (2016). Notoginsenoside Ft1 Promotes Fibroblast Proliferation via PI3K/Akt/mTOR Signaling Pathway and Benefits Wound Healing in Genetically Diabetic Mice. J. Pharmacol. Exp. Ther. 356 (2), 324–332. doi:10.1124/jpet.115.229369
Keywords: exosome mimetics, hydrogel, wound healing, oxidative phoshorylation, exosome
Citation: Zhu J, Liu Z, Wang L, Jin Q, Zhao Y, Du A, Ding N, Wang Y, Jiang H and Zhu L (2022) Exosome Mimetics-Loaded Hydrogel Accelerates Wound Repair by Transferring Functional Mitochondrial Proteins. Front. Bioeng. Biotechnol. 10:866505. doi: 10.3389/fbioe.2022.866505
Received: 31 January 2022; Accepted: 31 March 2022;
Published: 20 May 2022.
Edited by:
Wolfgang Holnthoner, Ludwig Boltzmann Institute for Experimental and Clinical Traumatology, AustriaReviewed by:
Joaquim Miguel Oliveira, University of Minho, PortugalSeverin Mühleder, Centro Nacional de Investigaciones Cardiovasculares (CNIC), Spain
Copyright © 2022 Zhu, Liu, Wang, Jin, Zhao, Du, Ding, Wang, Jiang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue Wang, wangyuesmmu@163.com; Hua Jiang, dosjh@126.com; Lie Zhu, zhulie19811@smmu.edu.cn
†These authors have contributed equally to this work and share first authorship
 Jie Zhu1†
Jie Zhu1†  Ling Wang
Ling Wang Yue Wang
Yue Wang Lie Zhu
Lie Zhu