Transporter-Driven Engineering of a Genetic Biosensor for the Detection and Production of Short-Branched Chain Fatty Acids in Saccharomyces cerevisiae
- 1NUS Synthetic Biology for Clinical and Technological Innovation (SynCTI), National University of Singapore, Singapore, Singapore
- 2Synthetic Biology Translational Research Programme, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore
- 3Department of Biochemistry, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore
- 4Science & Innovation Center, Mitsubishi Chemical Corporation, Yokohama, Japan
Biosensors can be used for real-time monitoring of metabolites and high-throughput screening of producer strains. Use of biosensors has facilitated strain engineering to efficiently produce value-added compounds. Following our recent work on the production of short branched-chain fatty acids (SBCFAs) in engineered Saccharomyces cerevisiae, here we harnessed a weak organic acid transporter Pdr12p, engineered a whole-cell biosensor to detect exogenous and intracellular SBCFAs and optimized the biosensor’s performance by varying PDR12 expression. We firstly constructed the biosensor and evaluated its response to a range of short-chain carboxylic acids. Next, we optimized its sensitivity and operational range by deletion and overexpression of PDR12. We found that the biosensor responded to exogenous SBCFAs including isovaleric acid, isobutyric acid and 2-methylbutanoic acid. PDR12 deletion enhanced the biosensor’s sensitivity to isovaleric acid at a low concentration and PDR12 overexpression shifted the operational range towards a higher concentration. Lastly, the deletion of PDR12 improved the biosensor’s sensitivity to the SBCFAs produced in our previously engineered SBCFA-overproducing strain. To our knowledge, our work represents the first study on employing an ATP-binding-cassette transporter to engineer a transcription-factor-based genetic biosensor for sensing SBCFAs in S. cerevisiae. Our findings provide useful insights into SBCFA detection by a genetic biosensor that will facilitate the screening of SBCFA-overproducing strains.
Introduction
Carboxylic acids are used as platform chemicals for manufacturing a variety of industrial products such as plastics, solvents, polymers and serve as “building blocks” for pharmaceuticals, fragrances, foods and other valuable products (Sauer et al., 2008). In the industry, carboxylic acids are produced from petrochemicals, which has brought concerns about unsustainability and environmental pollution. Therefore, there are strong interests in developing economical and environmentally friendly routes to produce carboxylic acids using microbial hosts (Chen and Nielsen, 2016). Isovaleric acid (IVA), isobutyric acid (IBA) and 2-methylbutanoic acid (2MBA) are representative short branched-chain fatty acids (SBCFAs) containing a methyl branch located on one or two carbon atoms (Yu et al., 2016). IVA derivatives are used as drugs, e.g., anticonvulsant and sedative and flavourings (Suerbaev et al., 2012; Wang et al., 2018). IBA derivatives are used as plastics, flavorings, fragrances, textile auxiliaries, surfactants and plasticizers (Zhang et al., 2011; Lang et al., 2014; Hammer et al., 2020). 2MBA is used for the synthesis of pharmaceuticals and flavoring (Kwon et al., 2000). Recently, several microorganisms, including Saccharomyces cerevisiae, Pseudomonas putida and Bacillus licheniformis, have been engineered to produce these SBCFAs (Lang et al., 2014; Yu et al., 2016; Shi et al., 2019). However, their production level in the engineered microbes is still low.
Genetic biosensors can be developed and used to facilitate strain engineering and optimization by biosensor-assisted high-throughput screening (HTS) of microbes for high SBCFA producers. Typically, a genetic biosensor consists of an allosterically regulated transcription factor (TF), which binds a specific ligand and associates with a target promoter to regulate the expression of a reporter gene (Teo and Chang, 2015; Taylor et al., 2016; Chen et al., 2018; Xia et al., 2019; D'Ambrosio et al., 2020; Hossain et al., 2020). For constructing an SBCFA biosensor, the weak organic acid-activated TF, War1p, from S. cerevisiae is a suitable candidate. War1p is constitutively expressed and binds to the weak organic acid response (War) element in the PDR12 promoter (pPDR12) (Kren et al., 2003; Gregori et al., 2008). When it binds to a weak organic acid, War1p is phosphorylated and conformationally changed to activate the expression of PDR12 (Mollapour and Piper, 2012). Based on this inducible system, Baumann et al. have developed a genetic biosensor which consists of War1p and pPDR12 coupled with green fluorescent protein (GFP) to detect short- and medium-chain fatty acids in S. cerevisiae, in which the fatty acid sensing was decoupled from the production using a two-cell sensor system (Baumann et al., 2018). Compared to the two-cell sensor system, a one-cell sensor system that produces and senses SBCFAs is favored for HTS of SBCFA-producing strains (Lim et al., 2018).
Optimizations are commonly needed to achieve desired properties of the genetic biosensors such as dynamic and operational ranges for the effective detection of target compounds (Hossain et al., 2020). The main strategies for such optimization include tuning of the TF’s receptor level, engineering of the DNA binding domain, the promoter’s upstream region, operator sequence or the 5′-untranslated region of the reporter gene and control of the target chemical level. For instance, Snoek et al. have reported that the evolution-guided engineering of the ligand binding domain of a bacterial TF (BenM) in S. cerevisiae led to a 15-fold increase in sensitivity and a 40-fold change in operational range (Snoek et al., 2020). Dabirian et al. have inserted the binding sites into various positions in the core promoter region of five native promoters in S. cerevisiae, which led to a higher sensitivity of malonyl-CoA-responsive TF (FapR) (Dabirian et al., 2019). Williams et al. have developed a pPDR12-based genetic biosensor to detect p-hydroxybenzoic acid and propionic acid through a positive-feedback activation of War1p by promoter engineering (Williams et al., 2017). Reduction of the intracellular level of target chemical by activating an exporter shifts the operational range of an allosteric TF-based biosensor in E. coli (Raman et al., 2014). In yeast, the ATP-binding-cassette (ABC) transporter Pdr12p plays roles in regulating the intracellular level of weak organic acids by export and its expression is selectively induced by weak organic acids (Piper et al., 1998; Hatzixanthis et al., 2003). Therefore, we hypothesized that tuning of PDR12 expression could be employed to optimize the properties of genetic biosensors for the detection of weak organic acids such as SBCFAs.
In this study, we aimed to develop a genetic biosensor to detect SBCFAs (exemplified by IVA, IBA and 2MBA) produced by our previously engineered S. cerevisiae strain (Yu et al., 2016). To this end, we firstly constructed a biosensor based on the War1p TF and pPDR12 and characterized its substrate spectrum. Furthermore, we optimized its sensitivity and operational range by deletion and overexpression of PDR12 in the engineered SBCFA-overproducing strain. This study provides useful insights into SBCFA detection by a one-cell biosensor system that can potentially facilitate the HTS of SBCFA-overproducing strains.
Materials and Methods
Plasmids and Strains
Plasmids and strains used in this study are shown in Table 1. Oligonucleotide primers (Supplementary Table S1) were synthesized by Integrated DNA Technologies (Singapore). Chemically competent Escherichia coli TOP10 was used for subcloning. The transformants were selected on LB agar plates with 100 μg/mL ampicillin.
Chemicals and Reagents
The Q5 high-fidelity DNA polymerase, NEBuilder HiFi DNA Assembly Cloning Kit, Restriction enzymes, T4 DNA ligase and PCR reagents were purchased from New England Biolabs (Beverly, MA, United States). QIAquick Gel Extraction Kit, QIAprep Spin Miniprep Kit and RNeasy Mini Kit were purchased from Qiagen (Valencia, CA, United States). Peptone was purchased from Oxoid Ltd. (Basingstoke, Hampshire, UK). Dropout SC amino acid mixture formulation (without Uracil) for S. cerevisiae growth media was purchased from MP Biomedicals (Santa Ana, CA, United States). All other reagents were purchased from Sigma Aldrich (St. Louis, MO, United States) or Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan) unless otherwise stated.
Construction of the SBCFA Biosensor
To construct the SBCFA biosensor, a GFP reporter gene was cloned under the control of pPDR12 which is inducible by weak organic acids. Specifically, pPDR12 was amplified by PCR from the genomic DNA of S. cerevisiae BY4741 using primers PDR12p-f with a SacI site and PDR12p-r (Supplementary Table S1). The GFP gene was amplified from pKT127 harboring the yEGFP gene (Accession No. MK178572) (Sheff and Thorn, 2004) using primers yEGFP-f and yEGFP-r with an XhoI site. pESC-URA was digested with SacI and XhoI. The amplified pPDR12, yEGFP and the digested pESC-URA fragment were purified and assembled by NEBuilder to yield the SBCFA biosensor namely pPDR12-GFP. The sequence of the biosensor plasmid was confirmed by Sanger sequencing (1st Base, Singapore).
Deletion and Overexpression of PDR12 in S. cerevisiae
To tune the expression of PDR12 gene and study its effect on the biosensor’s behavior, PDR12 was deleted and over-expressed in S. cerevisiae. Briefly, the PDR12 gene deletion was carried out using the Cre-LoxP system (Gueldener et al., 2002). The plasmid pUG72 (carrying the loxP–URA3–loxP cassette) was used to generate the gene deletion cassette and its derivative plasmid namely pUG72-TEF1 (carrying the strong constitutive promoter pTEF1) was used to replace the pPDR12 with pTEF1 in the chromosome (Yu et al., 2016). The plasmid pSH69 (carrying pGAL1-CRE) which contains a hygromycin B resistance gene was used for marker rescue (Hegemann and Heick, 2011).
To delete the PDR12 gene, the URA3 selection marker flanked by loxP sites was amplified from pUG72 by PCR using primers PDR12-Del-f and PDR12-Del-r, which contained a 42 bp homology to the integration site of PDR12 gene (Supplementary Table S1). The obtained DNA fragments were purified and transformed into BY4741 and the SBCFA-overproducing strain 4G-ΔADH6 using a modified yeast transformation method (Gietz and Schiestl, 2007; Ling et al., 2015). The transformants were selected on solid defined medium lacking uracil (SCD-U) and confirmed by colony PCR using the primers PDR12-CheGD-f PDR12-CheGD-r (Supplementary Figure S1). The obtained strains were transformed with the CRE recombinase-expressing plasmid pSH69 and grown on YPD plates containing 200 μg/mL hygromycin B (YPDH) to remove the URA3 marker gene between the loxP sites. The obtained strains were evaluated by PCR to confirm removal of the URA3 gene and designated as TDL01 (derived from BY4741) and TDL02 (derived from 4G-ΔADH6).
The replacement of the native promoter of PDR12 gene (pPDR12) to TEF1 promoter (pTEF1) was carried out using the same procedure as described for PDR12 deletion. The pTEF1 was amplified by using primers PDR12-pTEF1-F and PDR12-pTEF1-R and pGU72-TEF1 as a template. The promoter replacement in the obtained transformants were confirmed by PCR with the primers TEFp-UP-f and PDR12-CheTEFp-r (Supplementary Figure S1). After marker rescue, the obtained strains were evaluated by PCR with primers PDR12-CheTEFp-f and PDR12-CheTEFp-r to confirm removal of the URA3 gene. The confirmed strain was used for transformation with the constructed SBCFA biosensor as described above.
Detection of Exogenous SBCFAs by the SBCFA Biosensor
Prior to the detection of exogenous SBCFAs, the strains harboring the SBCFA biosensor pPDR12-GFP were grown in SCD-U with 100 mM potassium phosphate buffer (KPB, pH6.5) overnight at 30°C with shaking at 200 rpm. The overnight culture was diluted to OD600 of 0.1 in 2 mL of fresh medium and grown until the OD600 reached 0.4–0.6. The cells were centrifuged and resuspended in fresh SCD-U with 100 mM KPB (pH6.5) and 100 μL of cells was transferred to each well in black 96-well plates with clear flat bottom. Subsequently, 90 μL of fresh SCD-U with KPB was mixed with 10 μL of SBCFAs (Supplementary Table S2) to final concentrations of 0.05–10 mM, where the SBCFA stocks were prepared at 1 M in 70% ethanol and pH neutralized with sodium hydroxide solution. The SBCFA stocks were diluted to 1–200 mM using sterile water before adding into the microplate wells.
To measure the fluorescence signal of the SBCFA biosensor, the samples described as above in the 96-well plate were incubated at 30°C with shaking at 600 rpm in a Synergy H1 plate reader (BioTek, VT, United States). The cell density (OD600) was measured at 600 nm and the fluorescence signal was measured with excitation at 470 nm and emission at 515 nm every 30 min. The fluorescence intensity was normalized to OD600 and the normalized fluorescence intensity was compared between the conditions with and without SBCFAs. The EC50 (50% effective concentration) values were calculated by nonlinear regression using GraphPad Prism version 9 software (Graph Pad Software, CA, United States).
Detection of Intracellularly Produced SBCFAs by the SBCFA Biosensor
Besides the detection of exogenous SBCFAs, the intracellular SBCFAs produced by our previous SBCFAs-overproducing strain 4G-ΔADH6 was detected. The strains were cultivated in SCD-U medium at 30°C with shaking at 220 rpm overnight. The overnight culture was re-inoculated to an initial OD600 of 0.05 in 25 mL fresh SCD-U with 100 mM KPB (pH6.5) and grown under the same conditions in 250 mL shake flasks. For time course experiments, culture samples were collected every 12 h to determine the SBCFA levels and cell density (OD600) was measured by a spectrophotometer BioPhotometer Plus (Eppendorf, Germany). The culture samples were diluted by SCD-U with KPB to adjust the cell concentration and the fluorescence intensity was measured on a Synergy H1 microplate reader and normalized to the cell density (Jung et al., 2021) as described above.
Gas Chromatography/Mass Spectrometry Analysis of SBCFAs
GC analysis of SBCFAs was performed using methods modified from previous studies (Teo et al., 2015; Yu et al., 2016; Ng et al., 2020). One milliliter of yeast culture was collected and centrifuged to obtain the supernatant. To quantify the SBCFAs in the supernatant, 0.4 mL of 10% hydrochloric acid–methanol (v/v) and heptadecanoic acid (C17:0) at a final concentration of 1 mM as an internal standard were added to 0.8 mL culture supernatant and the mixtures were vortexed for 2 min and incubated at 62°C for 3 h to methylate the SBCFAs. SBCFA standards (IVA, 2MBA, IBA) at 0.5 and 2.0 mM added to SCD-URA medium were extracted and methylated as describe above. To the reaction mixtures, 0.2 mL hexane was added, and the resulting fatty acid methyl esters (FAMEs) were extracted by vortexing for 2 min and phase separation by centrifugation. The organic extracts containing FAMEs were then subjected to GC/MS analysis using a 7890B GC system with an Agilent 5977A MSD and an HP-5MS column. The method for GC/MS analysis was as follows: oven temperature with initial hold at 45°C for 3 min, ramp to 50°C at 10°C/min and hold for 3 min, ramp to 280°C at 50°C/min and hold for 5 min. Helium was used as the carrier gas and set at a constant pressure of 13.8 psi. The injector was maintained at 250°C and the ion source temperature was set to 230°C. The injection volume was 1.0 μL in a splitless mode. FAMEs of the respective SBCFAs were identified by their retention time and the mass spectra of FAME standards. Data analysis was performed using the Agilent Enhanced Data Analysis software. Given that 2MBA and IVA are isomers that have the same mass spectrum and retention time, they were quantified as a mixture.
Results and Discussion
Construction and Characterization of the SBCFA Biosensor
We have previously engineered S. cerevisiae to produce SBCFAs that comprise IVA, IBA and 2MBA (Yu et al., 2016). Here, to construct a genetic biosensor to detect SBCFAs in S. cerevisiae, we designed a biosensor that consists of 1) War1p, a TF that phosphorylates upon binding to weak organic acids (e.g., hexanoic acid, heptanoic acid and octanoic acid), 2) pPDR12, which is activated by the phosphorylated War1p and 3) a yeast-enhanced GFP gene (yEGFP) under the control of pPDR12. To ensure a higher copy number of the target genes, we cloned the aforementioned genetic elements into a high-copy plasmid pESC-URA and introduced the resulting biosensor (namely SLCA) into S. cerevisiae strains (BY4741, TOE01, TDL01, TDL02, 4G-ΔADH6) to obtain the biosensor strains (BY4741-SLCA, TOE01-SLCA, TDL01-SLCA, TDL02-SLCA, 4G-ΔADH6-SLCA).
To determine a suitable condition for sensing SBCFAs, we measured and compared the fluorescence intensity of BY4741-SLCA in SCD-U medium under buffered pH (at pH6.5 and pH7.4) and unbuffered (at pH4.5) pH conditions added with IBA and IVA, respectively. Supplementary Figure S2 shows that SLCA with a starting pH4.5 and unbuffered had a higher background than that when the pH was buffered to 6.5 and 7.4 without SBCFAs, suggesting that the low pH effect can be mitigated by maintaining the pH at about 7.0. The background fluorescence without pH buffering increased with incubation, whereas it remained low and constant under the buffered pH condition. It was found that under the unbuffered condition, the pH dropped from pH4.5 to pH3.5 after overnight incubation and under the buffered condition the pH remained above pH6.0. The higher fluorescence background under the unbuffered condition (pH 4.5) was likely due to the higher concentration of proton than that at pH 6.0. The undissociated weak organic acids can pass through the plasma membrane by passive diffusion under an acidic condition and free weak organic acids can be taken up by active transport at a physiological condition near pH7 (Casal et al., 2008). The uptaking efficiency of the former is generally more efficient than the latter (Borrull et al., 2015). We hypothesized that the whole-cell biosensor BY4741-SLCA would give a higher response at pH6.5 and pH7.4 than at pH4.5. This was proven by a 7.3-fold and 2.3-fold higher fold-change (FC) of fluorescence intensity towards IBA under the buffered pH at 6.5 (FC 20.3) and 7.4 (FC 6.3) than that when unbuffered (FC 2.8) at about 3 h (Figure 1A). Supplementary Figure S2B also shows that the time-course responses (FC) to IVA at pH6.5 (FC 37.0) and 7.4 (FC 14.9) are significantly higher than at pH4.5 (FC 3.9) at about 3 h. Given higher fluorescence from at pH6.5 than pH7.4, the medium buffered at pH6.5 was chosen for biosensing SBCFAs in this study.
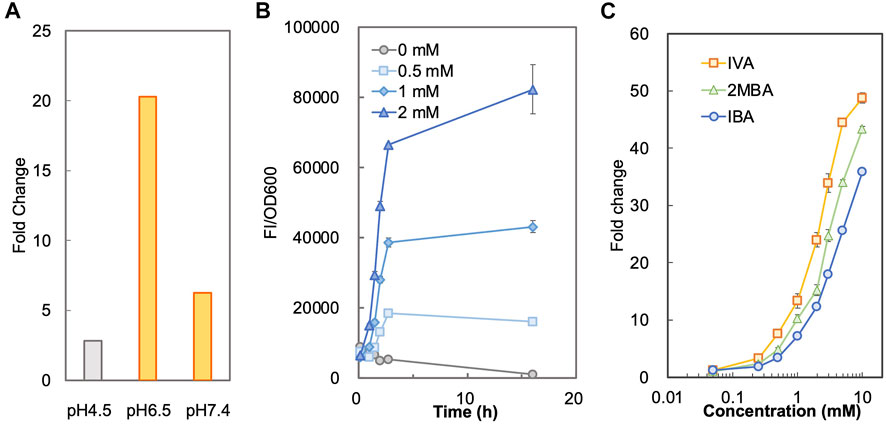
FIGURE 1. Characterization of biosensor BY4741-SLCA. (A) Responses of BY4741-SLCA cells (fold changes) to IBA under different pH conditions. BY4741-SLCA cells were grown in SCD-U medium under buffered (at pH6.5 and pH7.4) or unbuffered (at pH4.5) pH conditions. GFP fluorescence was measured after incubation with 1.0 mM IBA and normalized to cell density (OD600). The fluorescence of control without the IBA supply was normalized to its cell density and set to 1. (B) Time-course fluorescence intensity of BY4741-SLCA cells in response to IBA supplied into the medium. BY4741-SLCA cells were incubated in the buffered SCD medium (pH 6.5) with IBA supply (at 0.5, 1, and 2 mM). The fluorescence was measured and normalized to cell density (OD600). (C) Dose-dependent responses of BY4741-SLCA cells to respective SBCFA, i.e. IVA, 2MBA and IBA after a 3-h incubation. FI, fluorescence intensity. Values are means of three independent experiments. An error bar represents a standard deviation of three independent experiments.
Next, under the condition with the pH buffered to 6.5, we characterized the operational range of BY4741-SLCA against individual SBCFAs (IVA, 2MBA, IBA) fed at various concentrations. We firstly determined the minimal incubation time required for the SLCA to respond to the SBCFAs till the signal remained stable. Using IBA as an example, we found that there was a time-dependent exponential increase in the fluorescence signal against 0.5–2.0 mM of IBA till about 3 h. Afterwards, there was a slight decrease of signal from control (without IBA) (Figure 1B), which showed a 10-fold increase in cell density and a 2-fold increase in fluorescence (Supplementary Figure S3). This result suggests that a 3-h incubation is required prior to measuring the biosensor’s signal.
To determine the correlation of SBCFA doses to the signal, we exposed the BY4741-SLCA to IVA, 2MBA and IBA fed at various concentrations (0.05–10 mM) and measured the fluorescence signal. In Figure 1C, with a 3-h incubation, BY4741-SLCA demonstrated a dose-dependent increase of the response (FC) against the respective SBCFAs ranging from 0.05 mM (∼FC 2) to 10 mM (>FC 36). It is noted that, starting from 0.25 mM till 10 mM, the response from IVA was the highest (up to FC 49) amongst the three tested SBCFAs, suggesting that IVA is a preferred sensing target (EC50 2.2) over 2MBA (up to FC 43, EC50 3.6) and IBA (up to FC 36, EC50 5.0). Our results show that our biosensor is suitable for screening SBCFA-overproducing strains that produce SBCFAs at 0.25–10 mM, including the engineered S. cerevisiae strain producing 4 mM SBCFAs (Yu et al., 2016).
Structurally, the branched chain of IBA (C4) and 2MBA (C5) is located at the α position, whereas IVA’s (C5) branched chain is at the β position further from the carboxylic group. Given the differences in the branched-chain positions, the binding affinity of War1p to these SBCFAs will differ, depending on the varying binding tendency towards the structurally dissimilar carbon chain. It is known that War1p prefers binding to lipophilic weak acids over hydrophilic weak acids (Kim et al., 2019). Therefore, the differences in the binding efficacy of War1p to the SBCFAs could cause different responses of the biosensor. In addition, intracellular concentration of these SBCFAs could vary due to different import/export efficiency, hence causing variable responses of the biosensor.
Substrate Spectrum of the SBCFA Biosensor
Besides SBCFAs, we investigated the substrate spectrum of SLCA against other representative industry-relevant short-chain weak organic acids, including short linear chain fatty acids and lipophilic carboxylic acid derivatives such as phenolic acids, methacrylic acid, hydroxy carboxylic acids, keto acid and branched-chain amino acid (Castano-Cerezo et al., 2019).
To determine if shorter linear chain fatty acids and the lipophilic carboxylic acids with various chemical groups (vinyl, methyl, oxo, phenyl, hydroxyl and amino) were substrates of the biosensor BY4741-SLCA, we investigated its response to acetic (C2), propionic (C3) and octanoic (C8) acids and several lipophilic carboxylic acid derivatives (Figure 2). We found that SLCA was responsive to propionic acid (FC 3.8) and octanoic acid (FC 34.0), but not acetic acid. This result suggests that the pPDR12-based genetic biosensor respond to linear chain fatty acids with 3 carbons (C3) and longer. In addition, the biosensor SLCA showed a strong response (FC 40.0), consistent with a previous study (Baumann et al., 2018).
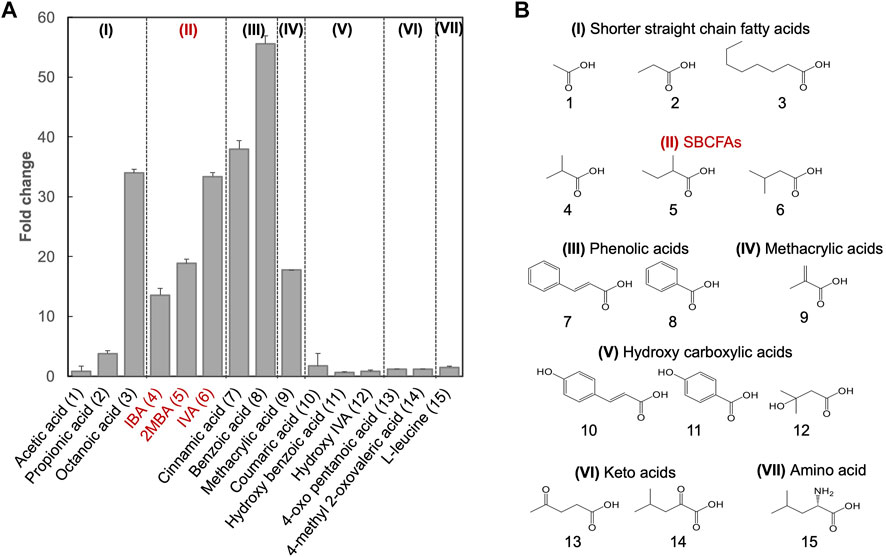
FIGURE 2. Substrate spectral analysis of biosensor BY4741-SLCA. (A) Responses of BY4741-SLCA cells (fold change) to weak organic acids categorized based on the similarity of their chemical structures. GFP fluorescence was measured after overnight incubation at 30°C and normalized to cell density (OD600) and further to control without weak organic acid supply. Values are the means of three independent experiments, and an error bar represents a standard deviation of three independent experiments. (B) Molecular structures of the tested weak acids, comprising of shorter straight chain fatty acids (I), and lipophilic carboxylic acid derivatives including SBCFAs (II), phenolic acids (III), methacrylic acid (IV), hydroxy carboxylic acid (V), keto acids (VI) and amino acid Leucine (VII). Their numbers are indicated, and SBCFAs are highlighted in red.
For methacrylic acid, a lipophilic carboxylic acid derivative with a vinyl group, a significant response was observed from the biosensor (FC 17.7). In line with previous studies (Hatzixanthis et al., 2003; Kren et al., 2003), the biosensor showed an FC 55.6 in response to benzoic acid (with a phenyl group) and an FC 38.0 in response to cinnamic acid (with a phenyl and a vinyl group). To our knowledge, this is the first evidence on sensing cinnamic acid using the pPDR12-based system in S. cerevisiae. We did not observe any responses (FC < 2.0) to the SBCFA’s precursors including L-leucine (with an amino group) and 4-methyl 2-oxovalerate (with a methyl and an oxo group), as well as hydroxylated derivatives including hydroxy isovaleric acid, hydroxy benzoic acid and coumaric acid. Supplementary Table S2 shows that some carboxylic acids (e.g., hydroxy isovaleric acid and 4-oxo pentanoic acid) have a relatively low hydrophobic constant (logP), i.e., a relatively low solubility in the medium. Interestingly, coumaric acid and hydroxy benzoic acid have high logP values. The lack of response to these compounds might be due to their low intracellular concentration caused either by low uptake, high efflux, or cellular metabolism (Barnhart-Dailey et al., 2019).
Optimization of the SBCFA Biosensor by PDR12 Deletion and Overexpression
Given the role of transporters in regulating the intracellular level of SBCFAs (Piper et al., 1998; Nygard et al., 2014), we examined the effect of deleting and overexpressing an efflux pump gene PDR12 on the SLCA’s behaviors (sensitivity and operational range). We hypothesized that the deletion of PDR12 would result in a higher intracellular level of SBCFAs than that in the wild-type cells and vice versa for PDR12 overexpression.
Based on the above hypothesis, we sought to enhance the biosensor’s sensitivity through PDR12 deletion and to shift the operational range through PDR12 overexpression (Figure 4A). Firstly, we deleted PDR12 gene (Supplementary Figure S1) and examined its effect on cell growth in the presence of 15 mM IVA. We found that the PDR12-deficient strain (TDL01) had a slight growth retardation after 12 h in the absence of IVA. In the presence of IVA, TDL01 showed a significant growth retardation till the log-phase when it had a cell density comparable to its parental strain BY4741 (Supplementary Figure S4). The significant growth retardation suggests a growth inhibition caused by the higher intracellular IVA accumulated in TDL01. Secondly, we introduced the biosensor plasmid pPDR12-GFP into TDL01 and measured the FC of the resulting strain TDL-SLCA. TDL-SLCA was found to have a response of FC 107.9 to 2 mM IVA, which is over 3-fold higher than BY4741 (FC 33.4). The increase in response to IVA in TDL-SLCA confirms the significant improvement in sensitivity. In addition, there was an increase in response by about three times to 2 mM of benzoic acid and a slight increase against cinnamic acid. We observed no change in the response toward other carboxylic acids (i.e., hydroxyl isovaleric acid, hydroxyl benzoic acid and coumaric acid) (Figure 3). These results show that the PDR12 deletion improves the sensitivity of the pPDR12-GFP biosensor to its target sensing substrates. Thirdly, to evaluate the operational range of the biosensor, we investigated its response to IVA at various doses in the PDR12-deficient (TDL01) and -overexpressing (TOE01) strains (Figure 4B and Supplementary Figure S1). Given a higher expression level of PDR12 upon the replacement of pPDR12 with the TEF1 promoter (pTEF1) in S. cerevisiae (Yu et al., 2016), in the absence of exogenous IVA supply, the biosensor TOE01-SLCA showed the lowest background fluorescence in SCD medium (Supplementary Figure S5). TOE01-SLCA showed a shift of the operational range towards higher concentrations to exogenous IVA (2–25 mM) compared to BY4741-SLCA (Control). Given that the response was not saturated at 25 mM IVA, there would be a continuing increase of the response to IVA at >25 mM. Thus, we speculated an operational range that is wider than 2–25 mM. TDL01-SLCA showed a higher sensitivity (up to FC18.9) toward exogenous IVA (0.2–10 mM). EC50 of TDL01-SLCA (1.3 mM) was 7.7-fold lower than that of BY4741-SLCA (10 mM).
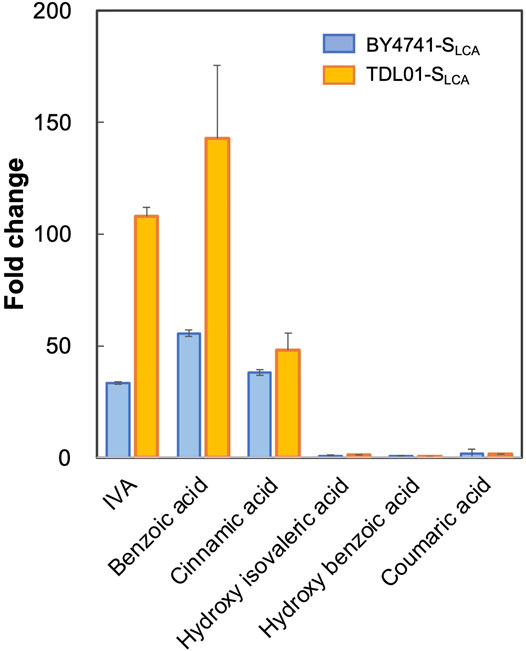
FIGURE 3. Responses of biosensor TDL01-SLCA with PDR12 gene deletion to representative weak organic acid including IVA, benzoic acid, cinnamic acid, hydroxy isovaleric acid, hydroxy benzoic acid and coumaric acid. BY4741-SLCA and TDL01-SLCA cells were grown until the exponential phase. After a 16-h incubation in a buffered SCD-U medium (at pH6.5) with the supply of weak organic acids (2 mM each), GFP fluorescence was measured, and fold changes were calculated. Values are means of three independent experiments, and an error bar represents standard deviation of three independent experiments.
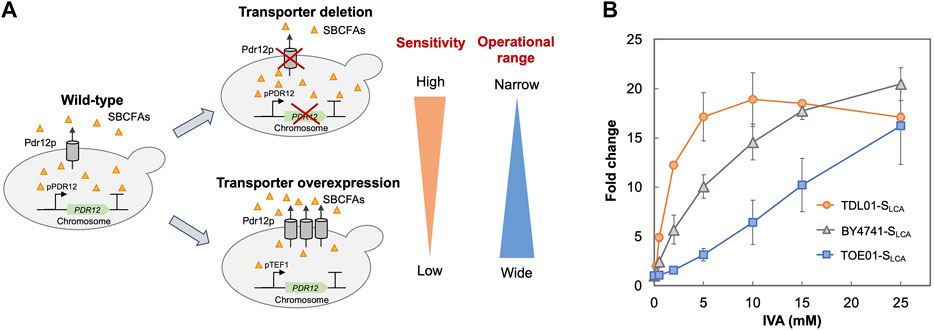
FIGURE 4. Optimization of biosensor’s sensitivity and operational range. (A) Schematic presentation of strategies employed to optimize the biosensor. The ABC transporter Pdr12p is known to regulate the intracellular level of weak organic acids by export. To improve the biosensor’s sensitivity, an ABC transporter gene PDR12 was deleted to increase the intracellular SBCFA level (TDL-SLCA), which will increase the fluorescence. To extend the biosensor’s operational range, PDR12 was over-expressed under the control of a strong constitutive promoter pTEF1 to reduce the intracellular SBCFA level (TOE-SLCA) through SBCFA export, which will enhance the SBCFA saturation level. (B) Dose-dependent responses of biosensors against IVA fed at 0.5, 2, 5, 10, and 25 mM. Their responses (fold change) were compared to control BY4741-SLCA. Fluorescence was measured after a 5-h incubation with IVA. Values are the means of three independent experiments, and an error bar represents a standard deviation of three independent experiments.
The higher sensitivity (TDL01-SLCA) and wider operational range (TOE01-SLCA) could be due to changes in export efficiency caused by PDR12 overexpression and deletion. Our results suggest that maintenance of the appropriate intracellular level of target biochemical can be used to enhance the operational range or widen the sensitivity of a whole-cell biosensor. Whole-cell biosensors with various operational ranges or sensitivities are desired for pathway engineering and strain optimization. For instance, at the early stage of pathway engineering when the production level of target biochemical is low, a biosensor with a higher sensitivity like TDL01-SLCA is favored for screening strain screening. On the other hand, at a later stage, a biosensor with a wider operational range like TOE01-SLCA is suitable for optimizing the production to achieve a higher production level.
Sensing of SBCFAs in the SBCFAs-Overproducing Strain
After we studied SLCA’s responses to exogenously supplied SBCFAs, we used it to sense SBCFAs in an SBCFA-overproducing strain. We previously engineered S. cerevisiae BY4741 (4G-ΔADH6) to overproduce SBCFAs at up to 3.7 mM SBCFAs (Yu et al., 2016). Given that the intracellular level of SBCFAs lies within the identified operational range of TDL01-SLCA (0.2–10 mM), we employed the strategy of PDR12 deletion for the biosensing. To this end, we deleted PDR12 in 4G-ΔADH6, resulting in TDL02. Next, we introduced the pPDR12-GFP plasmid into 4G-ΔADH6, TDL02 and analyzed the fluorescence signals of the resulting strains 4G-ΔADH6-SLCA and TDL02-SLCA.
Figure 5A shows that the fluorescence signal of 4G-ΔADH6-SLCA intensified with the increase in SBCFA production. The fluorescence signal of TDL02-SLCA also showed an increase along with the SBCFA production, but up to 76.7% higher than 4G-ΔADH6-SLCA. The fluorescence signal of both 4G-ΔADH6-SLCA and TDL02-SLCA showed a linear correlation to the produced SBCFAs (R2 > 0.93) (Figure 5B). Given that the SBCFA production level is comparable in 4G-ΔADH6 and TDL02, the fluorescence signal suggests the biosensor SLCA can monitor the SBCFA production real-time and the PDR12 deletion enhances the sensitivity of the biosensor in the SBCFA-overproducing strain. The sensitivity enhancement is consistent with BY4741 and TDL01 with SBCFA supplementation. At 48 h, the signal of TDL02-SLCA is slightly lower than 4G-ΔADH6-SLCA, likely due to a slightly lower SBCFA production in TDL02-SLCA (0.34 mM) than 4G-ΔADH6-SLCA (0.44 mM) (Supplementary Figure S6).
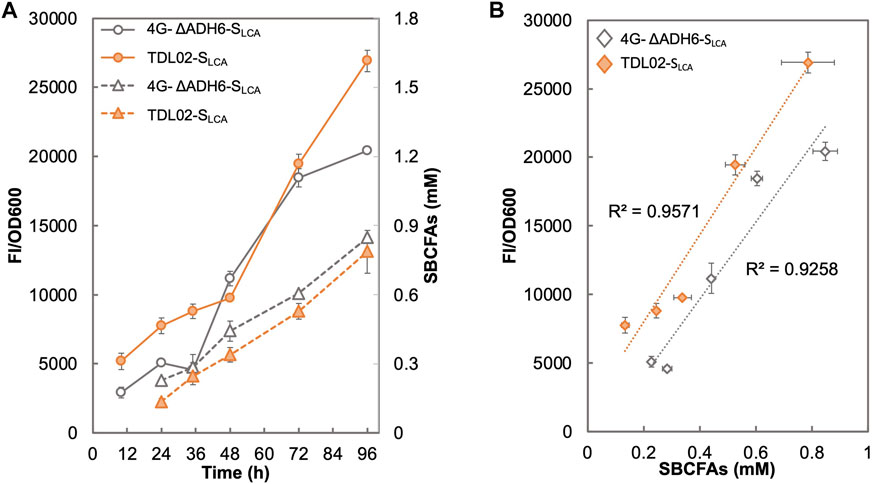
FIGURE 5. Responses of SBCFA biosensor in a S. cerevisiae strain overproducing SBCFAs. PDR12 was deleted in a previously engineered SBCFA-overproducing strain 4G-ΔADH6, resulting in strain TDL02. The biosensor SLCA was introduced into TDL02 and 4G-ΔADH6 respectively. (A) The fluorescence (solid lines) and the over-produced SBCFAs (dotted lines) were measured after fermentation in buffered SCD-U (at pH6.5) medium for up to 96 h. (B) Correlation between GFP fluorescence with SBCFAs produced in 4G-ΔADH6-SLCA and TDL02-SLCA. Trendlines with respective R2 value are shown in orange (4G-ΔADH6-SLCA) and grey (TDL02-SLCA). Values are the means of three independent experiments, and an error bar represents a standard deviation of three independent experiments.
In terms of the biosensing substrates, TDL02-SLCA and 4G-ΔADH6-SLCA sensed the mixture of SBCFAs, where the fluorescence signal attributes to the responses from such a mixture of SBCFAs. However, the contribution of each SBCFA in the mixture to the fluorescence is still unclear. In addition, we observed a 3-fold lower SBCFA titer in both 4G-ΔADH6 and TDL02 and a lower proportion of IVA/2MBA than those reported in our previous study (Yu et al., 2016). Such changes in the production titer and SBCFA profiles could be due to the use of a defined medium SCD that allows the SBCFA production from glucose, whereas in our previous study, the SBCFAs were likely produced from glucose and branched-chain amino acids present in YPD medium. Nevertheless, the SBCFA levels obtained using SCD were shown to lie within the operational range of our biosensor TDL01-SLCA which allows real-time monitoring of the SBCFA production in the engineered strains.
Conclusion
In this study, we engineered a genetic biosensor based on War1p and pPDR12. The biosensor detected SBCFAs (e.g., IVA, IBA and 2MBA) that were exogenously fed and intracellularly produced by engineered S. cerevisiae strains. The biosensor’s properties including sensitivity and operational range were optimized by varying the expression level of PDR12 encoding an ABC transporter responsible for SBCFA export. Besides SBCFAs, the biosensor can also sense short linear chain fatty acids and lipophilic carboxylic acid derivatives. Our findings suggest that the engineered biosensor is potentially a useful tool for monitoring the production of biochemicals, such as IVA, IBA, 2MBA, benzoic acid, cinnamic acid, propionic acid and octanoic acid, and for screening and discovering high producer strains.
Given the low sensitivity observed when the SBCFAs were at low concentrations (<0.25 mM), future efforts could be focused on improving the sensitivity. One strategy could be to introduce a positive-feedback loop on War1p to enhance the sensitivity. Williams et al. reported that the promoter replacement of the WAR1 promoter with pPDR12 in the genome, where War1p regulates its own expression in response to propionic acid (Williams et al., 2017), resulting in a dramatic increase in the sensitivity (from FC 3.6 to FC 10). In our current study, we confirmed an operational range of up to ∼0.8 mM of the intracellularly produced SBCFA mixture. To effectively detect SBCFAs produced by a higher producer, the operational range could be further evaluated and improved through optimization of the binding affinity between SBCFAs and the receptor domain of War1p, DNA binding domain of War1p or WARE sequence of pPDR12 (Gregori et al., 2008).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
RM designed and performed the study. RM and HL wrote the manuscript. HL, JF, NF, and MC analyzed the data and edited the manuscript. MC supervised the study. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Mitsubishi Chemical Corporation (2019-0449), the NUS Medicine Synthetic Biology Translational Research Programme (NUHSRO/2020/077/MSC/02/SB), the Summit Research Program of the National University Health System (NUHSRO/2016/053/SRP/05), the Synthetic Biology Initiative of the National University of Singapore (DPRT/943/09/14), the Synthetic Biology R&D Programme (SBP-P2, SBP-P7, SBP-P9), the U.S. Army (ARO/2019/74459LS) and the Ministry of Education, Singapore (NUHSRO/2020/046/T1/3). We thank Ms. Sofia Sigal-Passeck for assisting in strain characterization and fluorescence measurements.
Conflict of Interest
Authors RM and NF were employed by company Mitsubishi Chemical Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.838732/full#supplementary-material
References
Barnhart-Dailey, M. C., Ye, D., Hayes, D. C., Maes, D., Simoes, C. T., Appelhans, L., et al. (2019). Internalization and Accumulation of Model Lignin Breakdown Products in Bacteria and Fungi. Biotechnol. Biofuels 12, 175. doi:10.1186/s13068-019-1494-8
Baumann, L., Rajkumar, A. S., Morrissey, J. P., Boles, E., and Oreb, M. (2018). A Yeast-Based Biosensor for Screening of Short- and Medium-Chain Fatty Acid Production. ACS Synth. Biol. 7, 2640–2646. doi:10.1021/acssynbio.8b00309
Borrull, A., López-Martínez, G., Poblet, M., Cordero-Otero, R., and Rozès, N. (2015). New Insights into the Toxicity Mechanism of Octanoic and Decanoic Acids onSaccharomyces Cerevisiae. Yeast 32, 451–460. doi:10.1002/yea.3071
Casal, M., Paiva, S., Queirós, O., and Soares-Silva, I. (2008). Transport of Carboxylic Acids in Yeasts. FEMS Microbiol. Rev. 32, 974–994. doi:10.1111/j.1574-6976.2008.00128.x
Castaño-Cerezo, S., Fournié, M., Urban, P., Faulon, J. L., and Truan, G. (2019). Development of a Biosensor for Detection of Benzoic Acid Derivatives in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 7, 372. doi:10.3389/fbioe.2019.00372
Chen, B., Lee, H. L., Heng, Y. C., Chua, N., Teo, W. S., Choi, W. J., et al. (2018). Synthetic Biology Toolkits and Applications in Saccharomyces cerevisiae. Biotechnol. Adv. 36, 1870–1881. doi:10.1016/j.biotechadv.2018.07.005
Chen, Y., and Nielsen, J. (2016). Biobased Organic Acids Production by Metabolically Engineered Microorganisms. Curr. Opin. Biotechnol. 37, 165–172. doi:10.1016/j.copbio.2015.11.004
D'Ambrosio, V., Pramanik, S., Goroncy, K., Jakočiūnas, T., Schönauer, D., Davari, M. D., et al. (2020). Directed Evolution of VanR Biosensor Specificity in Yeast. Biotechnol. Notes 1, 9–15. doi:10.1016/j.biotno.2020.01.002
Dabirian, Y., Li, X., Chen, Y., David, F., Nielsen, J., and Siewers, V. (2019). Expanding the Dynamic Range of a Transcription Factor-Based Biosensor in Saccharomyces cerevisiae. ACS Synth. Biol. 8, 1968–1975. doi:10.1021/acssynbio.9b00144
Gietz, R. D., and Schiestl, R. H. (2007). High-efficiency Yeast Transformation Using the LiAc/SS Carrier DNA/PEG Method. Nat. Protoc. 2, 31–34. doi:10.1038/nprot.2007.13
Gregori, C., Schüller, C., Frohner, I. E., Ammerer, G., and Kuchler, K. (2008). Weak Organic Acids Trigger Conformational Changes of the Yeast Transcription Factor War1 In Vivo to Elicit Stress Adaptation. J. Biol. Chem. 283, 25752–25764. doi:10.1074/jbc.m803095200
Gueldener, U., Heinisch, J., Koehler, G. J., Voss, D., and Hegemann, J. H. (2002). A Second Set of loxP Marker Cassettes for Cre-Mediated Multiple Gene Knockouts in Budding Yeast. Nucleic Acids Res. 30, e23. doi:10.1093/nar/30.6.e23
Hammer, S. K., Zhang, Y., and Avalos, J. L. (2020). Mitochondrial Compartmentalization Confers Specificity to the 2-ketoacid Recursive Pathway: Increasing Isopentanol Production in Saccharomyces cerevisiae. ACS Synth. Biol. 9, 546–555. doi:10.1021/acssynbio.9b00420
Hatzixanthis, K., Mollapour, M., Seymour, I., Bauer, B. E., Krapf, G., Schüller, C., et al. (2003). Moderately Lipophilic Carboxylate Compounds Are the Selective Inducers of theSaccharomyces cerevisiaePdr12p ATP-Binding Cassette Transporter. Yeast 20, 575–585. doi:10.1002/yea.981
Hegemann, J. H., and Heick, S. B. (2011). Delete and Repeat: a Comprehensive Toolkit for Sequential Gene Knockout in the Budding Yeast Saccharomyces cerevisiae. Methods Mol. Biol. 765, 189–206. doi:10.1007/978-1-61779-197-0_12
Hossain, G. S., Saini, M., Miyake, R., Ling, H., and Chang, M. W. (2020). Genetic Biosensor Design for Natural Product Biosynthesis in Microorganisms. Trends Biotechnol. 38, 797–810. doi:10.1016/j.tibtech.2020.03.013
Jung, H., Ling, H., Tan, Y. Q., Chua, N. H., Yew, W. S., and Chang, M. W. (2021). Heterologous Expression of Cyanobacterial Gas Vesicle Proteins in Saccharomyces cerevisiae. Biotechnol. J. 16, e2100059. doi:10.1002/biot.202100059
Kim, M. S., Cho, K. H., Park, K. H., Jang, J., and Hahn, J.-S. (2019). Activation of Haa1 and War1 Transcription Factors by Differential Binding of Weak Acid Anions inSaccharomyces Cerevisiae. Nucleic Acids Res. 47, 1211–1224. doi:10.1093/nar/gky1188
Kren, A., Mamnun, Y. M., Bauer, B. E., Schu¨ller, C., Wolfger, H., Hatzixanthis, K., et al. (2003). War1p, a Novel Transcription Factor Controlling Weak Acid Stress Response in Yeast. Mol. Cell Biol 23, 1775–1785. doi:10.1128/mcb.23.5.1775-1785.2003
Kwon, D. Y., Hong, Y.-J., and Yoon, S. H. (2000). Enantiomeric Synthesis of (S)-2-methylbutanoic Acid Methyl Ester, Apple Flavor, Using Lipases in Organic Solvent. J. Agric. Food Chem. 48, 524–530. doi:10.1021/jf990871w
Lang, K., Zierow, J., Buehler, K., and Schmid, A. (2014). Metabolic Engineering of Pseudomonas Sp. Strain VLB120 as Platform Biocatalyst for the Production of Isobutyric Acid and Other Secondary Metabolites. Microb. Cell Fact 13, 2. doi:10.1186/1475-2859-13-2
Lim, H. G., Jang, S., Jang, S., Seo, S. W., and Jung, G. Y. (2018). Design and Optimization of Genetically Encoded Biosensors for High-Throughput Screening of Chemicals. Curr. Opin. Biotechnol. 54, 18–25. doi:10.1016/j.copbio.2018.01.011
Ling, H., Pratomo Juwono, N. K., Teo, W. S., Liu, R., Leong, S. S. J., and Chang, M. W. (2015). Engineering Transcription Factors to Improve Tolerance against Alkane Biofuels in Saccharomyces cerevisiae. Biotechnol. Biofuels 8, 231. doi:10.1186/s13068-015-0411-z
Mollapour, M., and Piper, P. W. (2012). Activity of the Yeast Zinc-finger Transcription Factor War1 Is Lost with Alanine Mutation of Two Putative Phosphorylation Sites in the Activation Domain. Yeast 29, 39–44. doi:10.1002/yea.1915
Ng, T.-K., Yu, A.-Q., Ling, H., Pratomo Juwono, N. K., Choi, W. J., Leong, S. S. J., et al. (2020). Engineering Yarrowia Lipolytica towards Food Waste Bioremediation: Production of Fatty Acid Ethyl Esters from Vegetable Cooking Oil. J. Biosci. Bioeng. 129, 31–40. doi:10.1016/j.jbiosc.2019.06.009
Nygård, Y., Mojzita, D., Toivari, M., Penttilä, M., Wiebe, M. G., and Ruohonen, L. (2014). The Diverse Role of Pdr12 in Resistance to Weak Organic Acids. Yeast 31, 219–232. doi:10.1002/yea.3011
Piper, P., Mahe, Y., Thompson, S., Pandjaitan, R., Holyoak, C., Egner, R., et al. (1998). The Pdr12 ABC Transporter Is Required for the Development of Weak Organic Acid Resistance in Yeast. EMBO J. 17, 4257–4265. doi:10.1093/emboj/17.15.4257
Raman, S., Rogers, J. K., Taylor, N. D., and Church, G. M. (2014). Evolution-guided Optimization of Biosynthetic Pathways. Proc. Natl. Acad. Sci. USA 111, 17803–17808. doi:10.1073/pnas.1409523111
Sauer, M., Porro, D., Mattanovich, D., and Branduardi, P. (2008). Microbial Production of Organic Acids: Expanding the Markets. Trends Biotechnol. 26, 100–108. doi:10.1016/j.tibtech.2007.11.006
Sheff, M. A., and Thorn, K. S. (2004). Optimized Cassettes for Fluorescent Protein Tagging inSaccharomyces Cerevisiae. Yeast 21, 661–670. doi:10.1002/yea.1130
Shi, J., Zhan, Y., Zhou, M., He, M., Wang, Q., Li, X., et al. (2019). High-level Production of Short Branched-Chain Fatty Acids from Waste Materials by Genetically Modified Bacillus Licheniformis. Bioresour. Technol. 271, 325–331. doi:10.1016/j.biortech.2018.08.134
Snoek, T., Chaberski, E. K., Ambri, F., Kol, S., Bjørn, S. P., Pang, B., et al. (2020). Evolution-guided Engineering of Small-Molecule Biosensors. Nucleic Acids Res. 48, e3. doi:10.1093/nar/gkz954
Suerbaev, K. A., Chepaikin, E. G., and Zhaksylykova, G. Z. (2012). Hydromenthoxycarbonylation of Isobutylene in the Presence of Tetrakis(triphenylphosphine)palladium-Based Catalyst Systems. Pet. Chem. 52, 422–425. doi:10.1134/s0965544112060126
Taylor, N. D., Garruss, A. S., Moretti, R., Chan, S., Arbing, M. A., Cascio, D., et al. (2016). Engineering an Allosteric Transcription Factor to Respond to New Ligands. Nat. Methods 13, 177–183. doi:10.1038/nmeth.3696
Teo, W. S., and Chang, M. W. (2015). Bacterial XylRs and Synthetic Promoters Function as Genetically Encoded Xylose Biosensors inSaccharomyces Cerevisiae. Biotechnol. J. 10, 315–322. doi:10.1002/biot.201400159
Teo, W. S., Ling, H., Yu, A.-Q., and Chang, M. W. (2015). Metabolic Engineering of Saccharomyces cerevisiae for Production of Fatty Acid Short- and Branched-Chain Alkyl Esters Biodiesel. Biotechnol. Biofuels 8, 177. doi:10.1186/s13068-015-0361-5
Wang, Y., He, L., Pan, Q., Duan, C., and Wang, J. (2018). Effects of Basal Defoliation on Wine aromas: A Meta-Analysis. Molecules 23, 779. doi:10.3390/molecules23040779
Williams, T. C., Xu, X., Ostrowski, M., Pretorius, I. S., and Paulsen, I. T. (2017). Positive-feedback, Ratiometric Biosensor Expression Improves High-Throughput Metabolite-Producer Screening Efficiency in Yeast. Synth. Biol. (Oxf) 2, ysw002. doi:10.1093/synbio/ysw002
Xia, P.-F., Ling, H., Foo, J. L., and Chang, M. W. (2019). Synthetic Genetic Circuits for Programmable Biological Functionalities. Biotechnol. Adv. 37, 107393. doi:10.1016/j.biotechadv.2019.04.015
Yu, A.-Q., Pratomo Juwono, N. K., Foo, J. L., Leong, S. S. J., and Chang, M. W. (2016). Metabolic Engineering of Saccharomyces cerevisiae for the Overproduction of Short Branched-Chain Fatty Acids. Metab. Eng. 34, 36–43. doi:10.1016/j.ymben.2015.12.005
Keywords: short branched-chain fatty acids, weak organic acids, transporter, genetic biosensor, promoter, Saccharomyces cerevisiae
Citation: Miyake R, Ling H, Foo JL, Fugono N and Chang MW (2022) Transporter-Driven Engineering of a Genetic Biosensor for the Detection and Production of Short-Branched Chain Fatty Acids in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 10:838732. doi: 10.3389/fbioe.2022.838732
Received: 18 December 2021; Accepted: 28 February 2022;
Published: 18 March 2022.
Edited by:
Shihui Yang, Hubei University, ChinaReviewed by:
Xinqing Zhao, Shanghai Jiao Tong University, ChinaJifeng Yuan, Xiamen University, China
Copyright © 2022 Miyake, Ling, Foo, Fugono and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthew Wook Chang, bchcmw@nus.edu.sg
 Ryoma Miyake
Ryoma Miyake Hua Ling
Hua Ling Jee Loon Foo
Jee Loon Foo Nobutake Fugono4
Nobutake Fugono4  Matthew Wook Chang
Matthew Wook Chang