Using Unnatural Protein Fusions to Engineer a Coenzyme Self-Sufficiency System for D-Phenyllactic Acid Biosynthesis in Escherichia coli
- 1School of Chemistry and Chemical Engineering, Chongqing University, Chongqing, China
- 2Department of Biological Systems Engineering, Washington State University, Pullman, WA, United States
The biosynthetic production of D-penyllactic acid (D-PLA) is often affected by insufficient supply and regeneration of cofactors, leading to high production cost, and difficulty in industrialization. In this study, a D-lactate dehydrogenase (D-LDH) and glycerol dehydrogenase (GlyDH) co-expression system was constructed to achieve coenzyme NADH self-sufficiency and sustainable production of D-PLA. Using glycerol and sodium phenylpyruvate (PPA) as co-substrate, the E. coli BL21 (DE3) harboring a plasmid to co-express LfD-LDH and BmGlyDH produced 3.95 g/L D-PLA with a yield of 0.78 g/g PPA, similar to previous studies. Then, flexible linkers were used to construct fusion proteins composing of D-LDH and GlyDH. Under the optimal conditions, 5.87 g/L D-PLA was produced by expressing LfD-LDH-l3-BmGlyDH with a yield of 0.97 g/g PPA, which was 59.3% increased compared to expression of LfD-LDH. In a scaled-up reaction, a productivity of 5.83 g/L/h was reached. In this study, improving the bio-catalytic efficiency by artificial redox self-equilibrium system with a bifunctional fusion protein could reduce the bio-production cost of D-PLA, making this bio-production of D-PLA a more promising industrial technology.
Introduction
Phenyllactic acid (PLA) widely exists in honey and fermented food and includes two enantiomers as D-phenyllactic acid (D-PLA) and L-phenyllactic acid (L-PLA) (Sorrentino et al., 2018; Luo et al., 2020a). Due to its safety, antimicrobial activity, low odor and good hydrophilicity, PLA has potential applications in the food, animal feed, pharmaceutical, and cosmetic industries (Valerio et al., 2016; Sun et al., 2019). Previous researchers found that the bactericidal effect of D-PLA was stronger than that of L-PLA (Dieuleveux et al., 1998) and D-PLA can be used as biological preservatives, antiviral compounds, hypoglycemic drugs, and protein inhibitors (Xu et al., 2015; Zhu et al., 2017; Liu et al., 2021). Moreover, because PLA can be polymerized into the aromatic polymers poly-PLA, it is a promising bio-based material (Wu et al., 2020). Although chemical methods of PLA production have been well studied, some limitations such as complex technology routes, harsh reaction conditions, excessive by-products and environmental pollution have hampered the chemical synthesis of PLA. (Wang et al., 2018a; Costa et al., 2020; Xu et al., 2020). Compared with chemical methods, biological methods have advantages of mild action conditions, energy savings, and environmental compatibility (Zhao et al., 2018). Therefore, the development of a eco-friendly biosynthesis method for PLA production is highly desirable.
PLA could be generally produced by a wide range of lactic acid bacteria or Escherichia coli with PPA, phenylalanine or glucose as starting materials in previous studies (Sorrentino et al., 2018; Wu et al., 2020). L. plantarum IMAU10124, P. pentosaceus, E. coli GK1, and L. crustorum NWAFU1078 produced 0.229 g/L (Zhang et al., 2014), 0.136 g/L (Yu et al., 2015a), 1.429 g/L (Kawaguchi et al., 2015), and 2.526 g/L (Xu et al., 2020) of PLA grown in MRS broth. Overall, since these methods produce various by-products and have relatively low yields, enzymatic/whole-cell cascade onecatalyst emerged as a green alternative for PLA production (Hou et al., 2019). Whole-cell catalytic conversion of PPA to PLA by strain overexpressing of NADH-dependent lactate dehydrogenase (LDH) is regarded as a more cost-efficient technology (Li et al., 2008; Mu et al., 2012; Luo et al., 2020b). D-LDH was overexpressed to produce D-PLA from PPA in Leuconostoc mesenteroides ATCC 8293, and the results showed that growing cells produced 35 mM D-PLA with a yield of 75.2–83.3% (Li et al., 2014). A novel NADH-dependent LDH gene, named lrldh, was cloned from Lactobacillus rossiae and heterologously expressed in recombinant E. coli/pET28a-lrldh. 20.5 g/L D-PLA was produced with a productivity of 49.2 g/L/d in a fed-batch biotransformation process (Luo et al., 2020a). Glucose and Phe need to undergo intermediate metabolism to produce PLA, which results in the increased fermentation time and decreased productivity. PPA is considered as a feasible precursor for the large-scale production of PLA, because it can biosynthesize PLA in one-step method and improve the catalytic efficiency (Li et al., 2014; Wang et al., 2018b).
However, the conversion from PPA to PLA by LDH requires the consumption of expensive coenzyme NADH. So the pathway involved in NADH regeneration is considered as an auxiliary pathway for PLA biosynthesis (Wu et al., 2020). Therefore, the whole-cell cascade catalysis using recombinant E. coli co-expression of glucose dehydrogenase (GDH)/formate dehydrogenase (FDH) and LDH has been widely used in the synthesis of PLA (Zhao et al., 2018; Rajanikar et al., 2021). Recombinant E. coli co-expressing LDH and GDH produced 17.25 g/L PLA from PPA with a productivity of 0.86 g/L/h (Zhu et al., 2017). Recombinant E. coli co-expressing D-LDH and FDH produced 10.02 g/L D-PLA from phenylalanine with a productivity of 1.67 g/L/h (Zheng et al., 2018). Unlike FDH or GDH, glycerol dehydrogenase (GlyDH) catalyzes the reduction of glycerol to dihydroxyacetone without the formation of toxic formic acid or gluconic acid (Xu et al., 2016), therefore, among these whole-cell reaction systems, co-expression of GlyDH and LDH might be a novel strategy for PLA synthesis. Furthermore, the co-substrate glucose was replaced with cost-effective glycerol to decrease the cost and improve the intracellular cofactor concentration.
Fusion proteins have been emerged recently as a new technology in biocatalysts, protein switches and therapeutics, aiming to channel substrates in sequential reactions, reduce effective reaction volume, facilitate cofactor regeneration and improve electron transfer (Yu et al., 2015b; Aalbers and Fraaije, 2019; Wu et al., 2021). Linkers have a critical role in the functionality and bioactivity of the destined fusion proteins, which can increase the stability/folding, expression, improve the biological activity, target to specific sites (Anami et al., 2018; He et al., 2019). Fan et al. constructed bifunctional and trifunctional fusions Mdh-Hps, Hps-Phi, and Mdh-Hps-Phi with flexible linkers (GGGGS)3 and (GGGGS)6, and the results showed that fusing Mdh with Hps or Hps-Phi enhanced methanol conversion to fructose-6-phosphate by 30% (Fan et al., 2018). Patgiri et al. engineered a fusion protein composed of lactate oxidase and catalase, which normalized the intracellular NADH:NAD + ratio by converting lactate and oxygen to pyruvate and water (Patgiri et al., 2020).
In this study, fusion protein engineering of D-LDH and GlyDH is conducted to aim for effective regeneration of coenzyme NADH for D-PLA biosynthesis. Firstly, a NADH-dependent D-LDH was overexpressed in recombinant E. coli CP101 to produce D-PLA from PPA. Then, plasmids for co-expression of D-LDH (two different genes) and GlyDH (two different genes) were transformed in E. coli (CP201, CP202, CP203, CP204) to achieve coenzyme self-sufficiency. To further improve the regeneration efficiency of coenzyme and to increase the yield of D-PLA, artificial fusion protein technology was used to construct D-LDH and GlyDH bifunctional fusion proteins as shown in Figure 1.
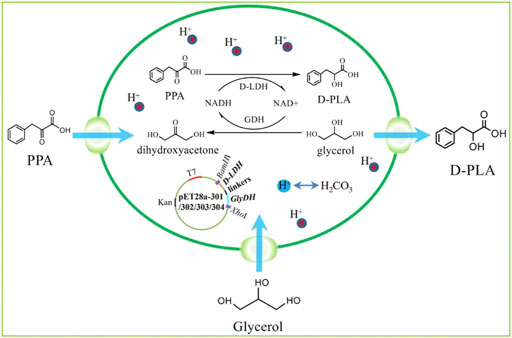
FIGURE 1. The program of redox self-balanced coenzyme regeneration and whole-cell synthesis of D-PLA.
Materials and Methods
Strains, Plasmids, and Chemicals
Enzymes (DNA polymerases, T4 DNA ligase, BamHI, and XhoI) and kits (DNA purification kit, plasmid isolation, DNA ligation kit, and competent cell preparation kit) were supplied by Takara (Dalian, China). With the exception of PPA, PLA, and NADH (Tsingke Biotechnology Co. Ltd., Beiing, China), all chemical reagents were purchased from China National Medicines Co. Ltd. (Beijing, China).
The Construction Process of the Plasmids
All bacterial strains and plasmids used in this study are listed in Supplementary Table S1. The D-LDH genes of Lactobacillus fermentum, Lactobacillus sp.SK007, and GlyDH genes of Bacillus megaterium, Exiguobacterium sibiricum were amplified via polymerase chain reaction (PCR) by using the relevant primer pairs listed in Supplementary Table S3. The PCR products were digested with BamHI and XhoI and ligated to pET28a to construct pET28a-LfD-LDH, pET28a-LsD-LDH, pET28a-BmGlyDH and pET28a-EsGlyDH, and then introduced into BL21 (DE3) individually to form the novel corresponding recombinant strains CP101, CP102, CP103, and CP104. The LfD-LDH and BmGlyDH were ligated by fusion PCR, and then the PCR product was digested with BamHI and XhoI and ligated to pET28a to construct pET28a-LfD-LDH-BmGlyDH. Using the same method as above, plasmids pET28a-LfD-LDH-EsGlyDH, pET28a-LsD-LDH-BmGlyDH, and pET28a-LsD-LDH-EsGlyDH could be constructed. Subsequently, plasmids above were introduced into BL21 (DE3) individually to form the novel corresponding recombinant strains CP201, CP202, CP203, and CP204. To construct fused D-LDH-GlyDH genes, fragments encoding flexible linkers were added to the C-terminal of D-LDH and N-terminal of GlyDH by overlap extension PCR using the primer pairs listed in Supplementary Table S3. Flexible linkers (GGGGS)1 (GGGGS)2, (GGGGS)3, and (GGGGS)6 listed in Supplementary Table S2 were added to the C-terminal of LfD-LDH and N-terminal of BmGlyDH and then ligated to pET28a to construct pET28a-LfD-LDH-l1-BmGlyDH, pET28a-LfD-LDH-l2-BmGlyDH, pET28a-LfD-LDH-l3-BmGlyDH, and pET28a-LfD-LDH-l6-BmGlyDH, and then introduced into BL21 (DE3) individually to construct recombinant strains CP301, CP302, CP303, and CP304. Successfully constructed recombinant plasmids were verified by DNA sequencing (Tsingke Biotechnology Co., Ltd., Beijing, China). CP100 carrying the backbone plasmid pET28a was constructed as the control strain.
Preparation of Whole-Cell Biocatalyst
Recombinant E. coli were inoculated into 40 ml LB medium (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) containing kanamycin (50 μg/ml) and grown in a rotary shaker (200 RPM) at 37°C overnight. 400 µL seed culture was inoculated into 40 ml LB medium containing 50 µg/ml kanamycin, and incubated at 37°C and 200 RPM to OD600 = 0.8. The recombinant E. coli was induced with 0.2 mM isopropyl-β-D-thiogalactoside (IPTG) at 25°C for 10 h. The cells were obtained via centrifugation at 4°C and 6,000 r/min for 10 min and washed three times with pH 7.0 PBS buffer. Cell catalyst (DCW) concentration was checked spectrophotometrically (722s, Shanghai Precision Scientific Instrument Co., Ltd., Shanghai, China) at an optical density of 600 (Yang et al., 2013). The concentrations of NADH, PPA, and PLA were measured as reported previously (Hou et al., 2017).
Optimization of D-LDH Induction and Enzymatic Catalysis Conditions
The recombinant E. coli was inoculated into 40 ml LB medium containing kanamycin (50 μg/ml) and grown overnight in a rotary shaker (200 RPM) at 37°C. 400 µL seed culture was inoculated into 50 ml LB medium for expression. The induction conditions, including temperature (20°C, 25°C, 30°C, 35°C, and 40°C), pH (6.0, 6.5, 7.0, 7.5, and 8.0), and concentration of IPTG (0.1, 0.2, 0.3, 0.4 and 0.5 mM), were investigated.
The enzymatic catalysis conditions of PPA to D-PLA were optimized. For D-PLA bioconversion, the catalytic system had 5 ml, which included 5 g/L PPA, 5.61 g/L glycerol, 9 g/L DCW, and 2% glucose. The bioconversion reactions were performed at 35°C on a 200 RPM shaker for 10 min for D-PLA production. The effects of temperature (25°C, 30°C, 35°C, 37°C, and 40°C), pH (6.0, 6.5, 7.0, 7.5, and 8.0), and PPA concentration (5, 6, 7, 8, and 9 g/L) on D-PLA production were determined.
Scale-Up of Bioconversions
Scale-up biotransformation is usually achieved by fed-batch cultures in a bioreactor (Abadli et al., 2021; Parizotto et al., 2021). The cell culture was concentrated and suspended (OD600 = 1.6) in 1-L PBS buffer (pH 7.0) with 60 g/L PPA and 66.5 g/L glycerol. Additional 40 g/L PPA and 16.78 g/L glycerol were added after 8 h bioconversion. The total concentration of PPA used in the system was 100 g/L. The scaled-up bioconversion was performed in a 5-L fermenter at 200 RPM, pH 7.0, 35°C.
Analytical Methods
The successful synthesis of PLA was confirmed by HPLC-MS (Agilent1260 series, Hewlett-Packard) with a C-18 5 µm column (4.6*25 mm) and the analysis was performed at 40°C with a mobile phase comprising 20% acetonitrile in water at a flow rate of 1 ml/min, wavelength 210 nm, 10 µL injection volume (Cheng et al., 2020) (Supplementary Figures S1, S2).
Results and Discussion
Construction of the D-PLA Biosynthetic Pathway in E. coli
Firstly, the synthesis pathway of D-PLA was constructed in single-enzyme expressing strains by overexpressing the D-LDH or GlyDH, so as to select suitable genes for the construction of co-expressing strains as shown in Figure 2A. CP101 with LfD-LDH and CP103 with BmGlyDH showed better productivity than CP102 with LsD-LDH and CP104 with EsGlyDH, with maximum conversion rates of 60.7 and 70.4% for PPA and glycerol, respectively, within 12 h. At the same time, their production capacity to the D-PLA was measured as shown in Figure 2B. CP101 could achieve the highest yield of D-PLA (5 g/L PPA to 3.07 g/L D-PLA) without co-expression of a cofactor regeneration system, while CP100, CP103 and CP104 lacking D-LDH gene could hardly produce PLA as reported (Jung et al., 2019). No D-PLA was detected in the control system, indicating that no enzyme could catalyze PPA to D-PLA in CP100. Therefore, one-step biosynthesis of D-PLA using PPA as raw material has been successfully realized.
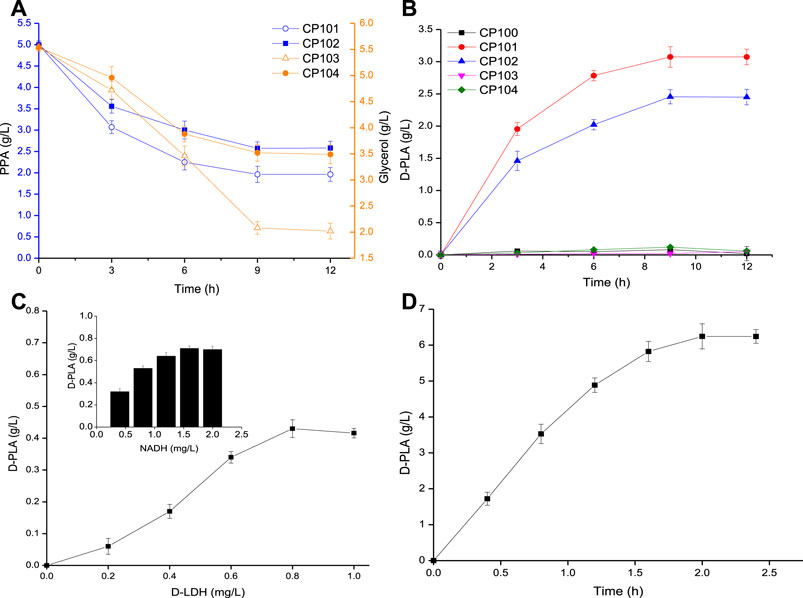
FIGURE 2. (A) Time course of the consumption of PPA (by CP101, CP102), and glycerol (by CP103, CP104); (B) Time course of the production of PLA by CP100, CP101, CP102, CP103, and CP104; (C) The effect of D-LDH and NADH concentration on D-PLA titer; (D) Time course of the production of D-PLA by enzyme catalysis. Data are means ± SD (n = 3).
D-LDH is a NADH-dependent enzyme, which has commonly used in the biosynthesis of D-PLA (Li et al., 2008; Mu et al., 2012). The effects of D-LDH and NADH concentration on D-PLA titer were determined. As it could be seen from Figure 2C, with the increase of enzyme dosage, D-PLA increased up to a maximum of 0.43 g/L at 0.8 mg/L D-LDH and 0.71 g/L at 1.6 mg/L NADH within 10 min, respectively. D-PLA cannot be produced without NADH addition indicating that NADH-dependent D-LDH must rely on NADH to catalyze the conversion of PPA to D-PLA. Similar studies have reported that the enzymatic production of PLA by P. pentosaceus improved by 30-folds on supplementation with NADH and NADH-regeneration catalyst (Yu et al., 2014). As it could be seen from Figure 2D, under the optimal conditions at pH 7 and 35°C, with PPA 7 g/L, 0.8 mg/L D-LDH, and NADH 1.6 mg/L in the fermentation broth, the D-PLA titer reached 6.24 g/L with a yield of 0.881 g/g PPA, respectively, within 2 h. Although it has been proved that the production of D-PLA by D-LDH enzyme catalysis can be achieved by adding coenzyme NADH, the high cost of NADH limits its industrial application.
The Self-Sufficient System of NADH With Coordinated Enzyme
In order to make up for the shortage of NADH in single-enzyme expressing strains and further increase yield of D-PLA in recombinant E. coli, a co-expression system was constructed for PLA biosynthesis. As it could be seen from Figure 3A, the co-expressing strains CP201, CP202, CP203, and CP204 catalyzed the synthesis of D-PLA, and obtained 3.95, 3.62, 3.73, and 3.50 g/L D-PLA from 5 g/L PPA in the biotransformation process for 12 h, respectively, while the control CP101 only obtained 3.07g/L. Therefore, for the single-enzyme expression of D-LDH, the co-expression of D-LDH and GlyDH can increase the yield of D-PLA by about 29.7%.
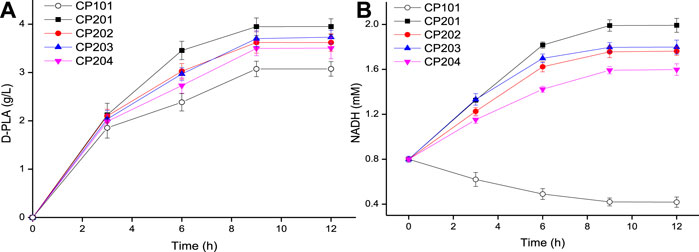
FIGURE 3. (A) Time course of the production of D-PLA by D-LDH and GlyDH co-expressed strains CP201, CP202, CP203, and CP204; (B) Time course of intracellular NADH concentration. Data are means ± SD (n = 3).
The above enzymatic catalysis results (Figure 2C) show that the concentration of NADH is an important limiting factor affecting the production of PLA by D-LDH catalysis. The cost of using NADH was expensive, so the intracellular NADH concentration was increased by using co-substrates (Hou et al., 2019). Therefore, the NADH concentration can be increased by using co-substrates (PPA and glycerol) in the co-expression system. The concentration of NADH in whole-cell E. coli was measured during the biosynthesis of D-PLA as shown in Figure 3B. In the control strain CP101 with PPA as the substrate of D-LDH single-enzyme expression, NADH decreased by 47.8% at 9 h. However, the intracellular NADH concentration of co-expressing strains (CP201, CP202, CP203, and CP204) with PPA and glycerol as co-substrates showed an increasing trend. This is because GlyDH in co-expressing strains oxidizes glycerol to dihydroxyacetone and produces coenzyme NADH, so more abundant NADH can be provided. A similar study reported whole-cell biocatalyst of co-expressing GDH and D-LDH was constructed to produce 262.8 g/L/d D-PLA without supplement of NADH (Luo et al., 2020b). Therefore, the co-substrate and co-expression system successfully constructed in this study can realize the regeneration of NADH to better produce D-PLA.
Improve the Efficiency of the Self-Sufficient System by Fusion Enzyme Engineering
In the area of enzyme engineering, early attempts were made to create two-protein fusions either to increase consecutive enzyme reaction rates or to generate bifunctional enzymes (Lindbladh et al., 1992; Beguin, 1999; Xiong et al., 2021). For example, a 77-fold improvement in the final product titer using a synthetic scaffold protein by recruiting three heterologous pathway enzymes in a designable manner (Dueber et al., 2009). For another example, a bifunctional enzyme coupling dihydroxyacetone kinase and fructose-1,6-bisphosphate aldolase was constructed to promote a 20-fold increase in the initial rate of the overall aldol reaction (Iturrate et al., 2010). According to previous studies, fusion enzyme engineering is a promising method for in situ recovery of co-enzyme NADH (Prachayasittikul et al., 2006).
Based on the results of the above researchers in the area of fusion enzyme engineering, flexible Linkers (GGGGS)1, (GGGGS)2, (GGGGS)3, and (GGGGS)6 were used to construct D-LDH and GlyDH bifunctional fusion proteins by over-lap PCR technique. As it could be seen from Figure 4, the four fusion protein strains CP301 (LfD-LDH-l1-BmGlyDH), CP302 (LfD-LDH-l2-BmGlyDH), CP303 (LfD-LDH-l3-BmGlyDH), and CP304 (LfD-LDH-l6-BmGlyDH) were used to catalyze the transformation of 5 g/L PPA into D-PLA, and 4.23, 4.35, 4.45, and 4.12 g/L PLA were obtained, respectively, within 12 h. The strain CP303 achieved 4.45 g/L titer D-PLA with a yield of 0.879 g/g PPA, which is 1.13 times that of the strain CP201. This benefits from the flexible peptide linker which was likely to bring enzyme moieties in close proximity for superior cofactor channeling, making D-LDH and GlyDH catalysis more efficient (Dueber et al., 2009). A similar study reported the application of peptide linker in the construction of bifunctional FDH and leucine dehydrogenase enzymatic complex for efficient cofactor regeneration, showing the production rate of fusion enzymatic complex with suitable flexible peptide linker was increased by 1.2 times compared with free enzyme mixture (Zhang et al., 2017). In addition, this linker might provide the proper space and flexibility to accommodate the different subunits of the LDH and GlyDH, as dehydrogenases are typically multimeric, and the fusion of dehydrogenases has been found to perturb proper oligomerization (Lerchner et al., 2016; Aalbers and Fraajie, 2017; Peters et al., 2017). Therefore, we successfully constructed D-LDH and GlyDH bifunctional fusion proteins to accelerate the cyclic regeneration of NADH and improve the efficiency of enzyme catalysis.
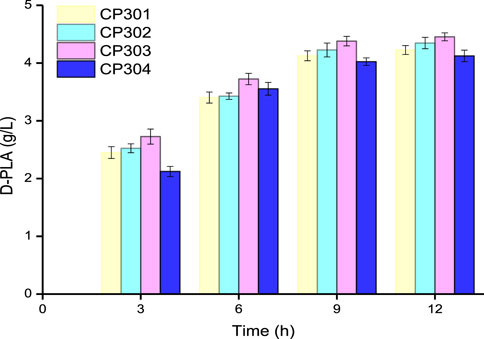
FIGURE 4. Time course of the production of D-PLA by fusion protein strains CP301, CP302, CP303, and CP304. Data are means ± SD (n = 3).
Optimization of Induction Conditions and Co-Substrates
In order to improve the ability and efficiency of the fusion proteins to catalyze the synthesis of D-PLA, the induction conditions of E. coli CP303 expressing D-LDH/GlyDH were optimized. Single factor experiments were used to optimize the induction conditions of recombinant E. coli in shake flasks, namely the influence of IPTG concentration, induction temperature, and pH on the catalytic synthesis of D-PLA by recombinant E. coli CP303. In addition, the ratio of substrate PPA, glycerol and cell catalyst were also optimized. Recombinant E. coli basic induction and co-substrate conditions: OD600 = 0.8, 0.1 mM IPTG, 5 g/L PPA, 5.61 g/L glycerol and induce expression at 25°C for 12 h.
As it could be seen from Figure 5A, when the IPTG concentration increased from 0.1 to 0.5 mM, the D-PLA titer increased first and then decreased. Compared with 0.1 mM IPTG, the expression of D-LDH/GlyDH fusion increased by induction with 0.2 mM IPTG and achieved a highest titer (4.53 g/L) D-PLA. However, when the IPTG concentration increased to 0.3 mM, it began to inhibit the growth of E. coli CP303, especially when the IPTG concentration was 0.5 mM. Therefore, the optimal IPTG concentration was 0.2 mM, the cell growth was better and D-LDH/GlyDH enzyme activity was the highest at this concentration. pH can affect the spatial structure and activity of enzymes, so an optimal pH needs to be explored. The effect of pH on D-PLA production as shown in Figure 5B. When the reaction system pH was 7.0, the D-PLA titer catalyzed by E. coli CP303 was higher than other pH, indicating that the neutral environment can maintain the good catalytic activity of D-LDH and GlyDH at the same time, so the high yield of D-PLA can be achieved by maintaining the pH at 7.0. Temperature affects the growth rate, cell metabolic activity and protein production rate of E. coli CP303, so induction temperature has also been studied as shown in Figure 5C. When the induction temperature was 20°C, the slow metabolism of recombinant E. coli led to the slow expression rate of D-LDH/GlyDH fusion protein and the low yield of D-PLA. When the induction temperature was higher than 30°C, the yield of D-PLA began to decrease, mainly because the fusion proteins at high temperature began to exist in the inclusion form. Therefore, the optimal induction temperature is 25°C, the highest titer 4.68 g/L of D-PLA can be obtained, with a yield of 0.925 g/g PPA.
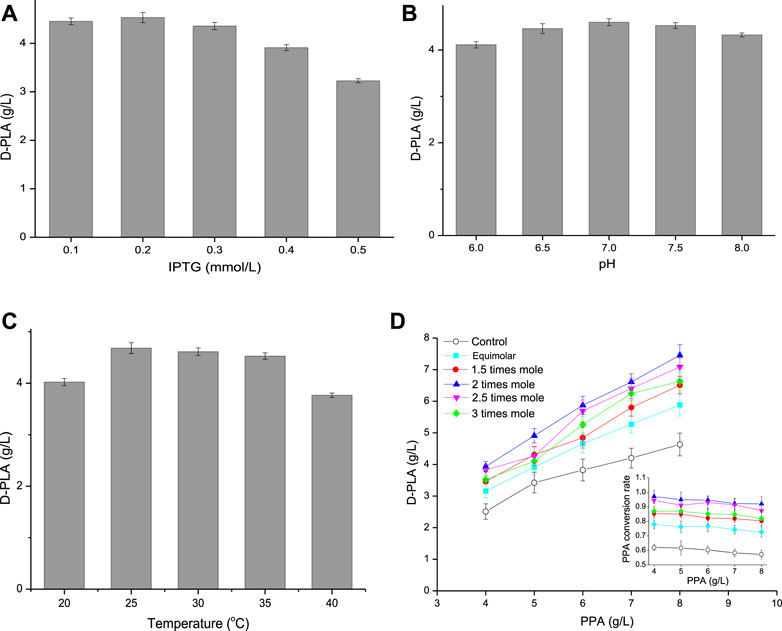
FIGURE 5. (A) Effect of IPTG concentration on D-PLA titer; (B) Effect of pH on D-PLA titer; (C) Effect of temperature on D-PLA titer; (D) Effect of substrate (glycerol, relative to PPA) on D-PLA titer. Data are means ± SD (n = 3).
The strategy of increasing intracellular NADH concentration by co-substrate has been demonstrated (Hou et al., 2019), and glycerol is also commonly used for dehydrogenation to produce lactic acid (Bharath et al., 2020). Therefore, in this study, intracellular NADH concentration was increased by using glycerol as the co-substrate of PPA. The influence of co-substrate on the yield of D-PLA was explored by adjusting the concentration and proportion of PPA and glycerol as shown in Figure 5D. D-PLA titer increased with the increase of PPA concentration, but the yield showed a trend of continuous decline, because the increase of PPA concentration enhanced the toxic effect on E. coli CP303. When the concentration of PPA increased to 7–8 g/L, the strong inhibitory effect made the conversion rate of PPA decreased greatly. Therefore, the optimal PPA concentration was 6 g/L. Furthermore, the addition amount of glycerol was optimized. When the glycerol concentration increased from equal molar concentration to 3 times molar concentration (relative to PPA), both the yield of D-PLA and PPA conversion showed a trend of first increasing and then decreasing at the same PPA concentration. This is because the low concentration of glycerol oxidizes itself to dihydroxyacetone through GlyDH and cannot provide sufficient coenzyme NADH, while the high concentration of glycerol has a certain inhibitory effect on the growth of E. coli and enzyme activity. Furthermore, without glycerol as the control, the maximum yield of D-PLA was only 4.63 g/L. Therefore, the optimal glycerol dosage was 2 times of the molar concentration of PPA. Under optimal conditions (6 g/L PPA, 6.73 g/L glycerol, 0.2 mM IPTG, pH 7, 25°C) of whole-cell catalytic production for 12 h, the maximum titer 5.87 g/L of D-PLA can be obtained with a yield of 0.967 g/g PPA.
Scale-Up of D-PLA Production
The whole-cell bioconversion was performed in a 5-L reactor using E. coli CP303 as biocatalyst as shown in Figure 6. When the fermentation executed for 8 h, and the concentration of substrate PPA and glycerol reduced from 60 g/L to 8.45 g/L and form 66.5 g/L to 37.6 g/L, respectively. Another PPA and glycerol addition into the bioreactor were conducted to reach a level of 48.45 g/L and 54.36 g/L, respectively. The whole-cell catalysis lasted for 16 h, and the final concentration of D-PLA was 93.3 g/L (561.53 mM), with a productivity of 5.83 g/L/h, and yield of 0.922 g/g PPA.
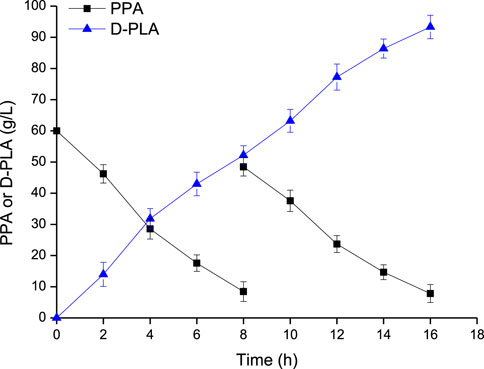
FIGURE 6. D-PLA production by whole-cell bioconversion using the CP303 strain in a 5 L bioreactor. Data are means ± SD (n = 3).
Conclusion
In this study, we reported for the first time the one-step synthesis of D-PLA using PPA as substrate by co-expression of D-LDH and GlyDH, achieving the regeneration of coenzyme NADH. At the same time, in order to further improve the regeneration efficiency of NADH and the yield of D-PLA, the D-LDH/GlyDH fusion protein strains were constructed with flexible glycine-rich linkers. The results showed that the fusion protein strain CP303 achieved a yield of 0.967 g/g PPA. In our study, cheap glycerol was used as co-substrate and coenzyme NADH was not needed to be added. A high-efficiency and low-cost pathway was constructed to sustainably produce high value-added product D-PLA, which is a technology with industrial potential.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
ZQ and RL performed the experiments, analyzed the data, and drafted the article. TL and PC analyzed the data. DW and XX conceived and coordinated the study. ZQ and DW finalized the article. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by the National Natural Science Foundation of China (21978027), the Fundamental Research Funds for the Central Universities (2020CDCGJ020, 2020CDCGHG068), Scientific Research Foundation of State Key Lab of Coal Mine Disaster Dynamics and Control (2011DA105287-ZR202002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.795885/full#supplementary-material
Abbreviations
DCW, cell catalyst; D-LDH, d-lactate dehydrogenase; D-PLA, d-phenyllactic acid; FDH, formate dehydrogenase; GDH, glucose dehydrogenase; GlyDH, glycerol dehydrogenase; HPLC-MS, high performance liquid chromatography-mass spectrometry; IPTG, isopropyl-β-d-thiogalactoside; l1, (GGGGS)1; l2, (GGGGS)2; l3, (GGGGS)3; l6, (GGGGS)6; PCR, polymerase chain reaction; PLA, phenyllactic acid; PPA, sodium phenylpyruvate.
References
Aalbers, F. S., and Fraaije, M. W. (2017). Coupled Reactions by Coupled Enzymes: Alcohol to Lactone cascade with Alcohol Dehydrogenase-Cyclohexanone Monooxygenase Fusions. Appl. Microbiol. Biotechnol. 101 (20), 7557–7565. doi:10.1007/s00253-017-8501-4
Aalbers, F. S., and Fraaije, M. W. (2019). Enzyme Fusions in Biocatalysis: Coupling Reactions by Pairing Enzymes. Chembiochem 20 (1), 20–28. doi:10.1002/cbic.201800394
Abadli, M., Dewasme, L., Tebbani, S., Dumur, D., and Vande Wouwer, A. (2021). An Experimental Assessment of Robust Control and Estimation of Acetate Concentration in Escherichia coli BL21(DE3) Fed-Batch Cultures. Biochem. Eng. J. 174, 108103. doi:10.1016/j.bej.2021.108103
Anami, Y., Yamazaki, C. M., Xiong, W., Gui, X., Zhang, N., An, Z., et al. (2018). Glutamic Acid-Valine-Citrulline Linkers Ensure Stability and Efficacy of Antibody-Drug Conjugates in Mice. Nat. Commun. 9 (1), 2512. doi:10.1038/s41467-018-04982-3
Béguin, P. (1999). Hybrid Enzymes. Curr. Opin. Biotechnol. 10 (4), 336–340. doi:10.1016/s0958-1669(99)80061-5
Bharath, G., Rambabu, K., Hai, A., Taher, H., and Banat, F. (2020). Development of Au and 1D Hydroxyapatite Nanohybrids Supported on 2D Boron Nitride Sheets as Highly Efficient Catalysts for Dehydrogenating Glycerol to Lactic Acid. ACS Sust. Chem. Eng. 8 (19), 7278–7289. doi:10.1021/acssuschemeng.9b06997
Cheng, Z., Ran, Q., Liu, J., Deng, X., Qiu, H., Jia, Z., et al. (2020). Rapid Determination for Benzoic Acid, Sorbic Acid, Phenyllactic Acid, Phenylalanine, and Saccharin Sodium in Vinegar by High-Performance Liquid Chromatography-UV. Food Anal. Methods 13 (8), 1673–1680. doi:10.1007/s12161-020-01784-6
Costa, J. R., Tonon, R. V., Cabral, L., Gottschalk, L., Pastrana, L., and Pintado, M. E. (2020). Valorization of Agricultural Lignocellulosic Plant Byproducts through Enzymatic and Enzyme-Assisted Extraction of High-Value-Added Compounds: A Review. ACS Sust. Chem. Eng. 8 (35), 13112–13125. doi:10.1021/acssuschemeng.0c02087
de Almeida Parizotto, L., Krebs Kleingesinds, E., Manfrinato Pedrotti da Rosa, L., Effer, B., Meira Lima, G., Herkenhoff, M. E., et al. (2021). Increased Glycosylated L-Asparaginase Production through Selection of Pichia pastoris Platform and Oxygen-Methanol Control in Fed-Batches. Biochem. Eng. J. 173, 108083. doi:10.1016/j.bej.2021.108083
Dieuleveux, V., Lemarinier, S., and Guéguen, M. (1998). Antimicrobial Spectrum and Target Site of D-3-Phenyllactic Acid. Int. J. Food Microbiol. 40 (3), 177–183. doi:10.1016/s0168-1605(98)00031-2
Dueber, J. E., Wu, G. C., Malmirchegini, G. R., Moon, T. S., Petzold, C. J., Ullal, A. V., et al. (2009). Synthetic Protein Scaffolds Provide Modular Control over Metabolic Flux. Nat. Biotechnol. 27 (8), 753–759. doi:10.1038/nbt.1557
Fan, L., Wang, Y., Tuyishime, P., Gao, N., Li, Q., Zheng, P., et al. (2018). Engineering Artificial Fusion Proteins for Enhanced Methanol Bioconversion. Chembiochem 19 (23), 2465–2471. doi:10.1002/cbic.201800424
He, R., Finan, B., Mayer, J. P., and DiMarchi, R. D. (2019). Peptide Conjugates with Small Molecules Designed to Enhance Efficacy and Safety. Molecules 24 (10), 1855. doi:10.3390/molecules24101855
Hou, Y., Gao, B., Cui, J., Tan, Z., Qiao, C., and Jia, S. (2019). Combination of Multi-Enzyme Expression fine-tuning and Co-substrates Addition Improves Phenyllactic Acid Production with an Escherichia coli Whole-Cell Biocatalyst. Bioresour. Tech. 287, 121423. doi:10.1016/j.biortech.2019.121423
Hou, Y., Hossain, G. S., Li, J., Shin, H.-D., Du, G., Chen, J., et al. (2017). Metabolic Engineering of Cofactor Flavin Adenine Dinucleotide (FAD) Synthesis and Regeneration inEscherichia Colifor Production of α-keto Acids. Biotechnol. Bioeng. 114 (9), 1928–1936. doi:10.1002/bit.26336
Iturrate, L., Sánchez-Moreno, I., Oroz-Guinea, I., Pérez-Gil, J., and García-Junceda, E. (2010). Preparation and Characterization of a Bifunctional Aldolase/Kinase Enzyme: A More Efficient Biocatalyst for C-C Bond Formation. Chem. Eur. J. 16 (13), 4018–4030. doi:10.1002/chem.200903096
Jung, S., Hwang, H., and Lee, J.-H. (2019). Effect of Lactic Acid Bacteria on Phenyllactic Acid Production in Kimchi. Food Control 106, 106701. doi:10.1016/j.foodcont.2019.06.027
Kawaguchi, H., Teramura, H., Uematsu, K., Hara, K. Y., Hasunuma, T., Hirano, K., et al. (2015). Phenyllactic Acid Production by Simultaneous Saccharification and Fermentation of Pretreated Sorghum Bagasse. Bioresour. Tech. 182, 169–178. doi:10.1016/j.biortech.2015.01.097
Lerchner, A., Daake, M., Jarasch, A., and Skerra, A. (2016). Fusion of an Alcohol Dehydrogenase with an Aminotransferase Using a PAS Linker to Improve Coupled Enzymatic Alcohol-To-Amine Conversion. Protein Eng. Des. Selection 29 (12), 557–562. doi:10.1093/protein/gzw039
Li, L., Shin, S.-Y., Lee, K. W., and Han, N. S. (2014). Production of Natural Antimicrobial Compound D -phenyllactic Acid Using Leuconostoc Mesenteroides ATCC 8293 Whole Cells Involving Highly Active D -lactate Dehydrogenase. Lett. Appl. Microbiol. 59 (4), 404–411. doi:10.1111/lam.12293
Li, X., Jiang, B., Pan, B., Mu, W., and Zhang, T. (2008). Purification and Partial Characterization of Lactobacillus Species SK007 Lactate Dehydrogenase (LDH) Catalyzing Phenylpyruvic Acid (PPA) Conversion into Phenyllactic Acid (PLA). J. Agric. Food Chem. 56 (7), 2392–2399. doi:10.1021/jf0731503
Lindbladh, C., Persson, M., Bulow, L., and Mosbach, K. (1992). Characterization of a Recombinant Bifunctional Enzyme, Galactose Dehydrogenase/bacterial Luciferase, Displaying an Improved Bioluminescence in a Three-Enzyme System. Eur. J. Biochem. 204 (1), 241–247. doi:10.1111/j.1432-1033.1992.tb16630.x
Liu, J., Huang, R., Song, Q., Xiong, H., Ma, J., Xia, R., et al. (2021). Combinational Antibacterial Activity of Nisin and 3-Phenyllactic Acid and Their Co-production by Engineered Lactococcus Lactis. Front. Bioeng. Biotechnol. 9, 612105. doi:10.3389/fbioe.2021.612105
Luo, X., Zhang, Y., Yin, F., Hu, G., Jia, Q., Yao, C., et al. (2020a). Enzymological Characterization of a Novel D-Lactate Dehydrogenase from Lactobacillus Rossiae and its Application in D-Phenyllactic Acid Synthesis. 3 Biotech. 10 (3), 101. doi:10.1007/s13205-020-2098-5
Luo, X., Zhang, Y., Yin, L., Zheng, W., and Fu, Y. (2020b). Efficient Synthesis of D-Phenyllactic Acid by a Whole-Cell Biocatalyst Co-expressing Glucose Dehydrogenase and a Novel D-Lactate Dehydrogenase from Lactobacillus Rossiae. 3 Biotech. 10 (1), 14. doi:10.1007/s13205-019-2003-2
Mu, W., Yu, S., Jiang, B., and Li, X. (2012). Characterization of D-Lactate Dehydrogenase from Pediococcus Acidilactici that Converts Phenylpyruvic Acid into Phenyllactic Acid. Biotechnol. Lett. 34 (5), 907–911. doi:10.1007/s10529-012-0847-1
Patgiri, A., Skinner, O. S., Miyazaki, Y., Schleifer, G., Marutani, E., Shah, H., et al. (2020). An Engineered Enzyme that Targets Circulating Lactate to Alleviate Intracellular NADH:NAD+ Imbalance. Nat. Biotechnol. 38 (3), 309–313. doi:10.1038/s41587-019-0377-7
Peters, C., Rudroff, F., Mihovilovic, M. D., and T. Bornscheuer, U. (2017). Fusion Proteins of an Enoate Reductase and a Baeyer-Villiger Monooxygenase Facilitate the Synthesis of Chiral Lactones. Biol. Chem. 398 (1), 31–37. doi:10.1515/hsz-2016-0150
Prachayasittikul, V., Ljung, S., Isarankura-Na-Ayudhya, C., and Bülow, L. (2006). NAD(H) Recycling Activity of an Engineered Bifunctional Enzyme Galactose Dehydrogenase/lactate Dehydrogenase. Int. J. Biol. Sci. 2 (1), 10–16. doi:10.7150/ijbs.2.10
Rajanikar, R. V., Nataraj, B. H., Naithani, H., Ali, S. A., Panjagari, N. R., and Behare, P. V. (2021). Phenyllactic Acid: A green Compound for Food Biopreservation. Food Control 128, 108184. doi:10.1016/j.foodcont.2021.108184
Sorrentino, E., Tremonte, P., Succi, M., Iorizzo, M., Pannella, G., Lombardi, S. J., et al. (2018). Detection of Antilisterial Activity of 3-Phenyllactic Acid Using Listeria Innocua as a Model. Front. Microbiol. 9, 1373. doi:10.3389/fmicb.2018.01373
Sun, H.-L., Chuai, J., Wei, H., Zhang, X., and Yu, H. (2019). Multi-functional Organic Gelator Derived from Phenyllactic Acid for Phenol Removal and Oil Recovery. J. Hazard. Mater. 366, 46–53. doi:10.1016/j.jhazmat.2018.11.095
Valerio, F., Di Biase, M., Lattanzio, V. M. T., and Lavermicocca, P. (2016). Improvement of the Antifungal Activity of Lactic Acid Bacteria by Addition to the Growth Medium of Phenylpyruvic Acid, a Precursor of Phenyllactic Acid. Int. J. Food Microbiol. 222, 1–7. doi:10.1016/j.ijfoodmicro.2016.01.011
Wang, X., Hou, Y., Liu, L., Li, J., Du, G., Chen, J., et al. (2018a). A New Approach for Efficient Synthesis of Phenyllactic Acid from L-Phenylalanine: Pathway Design and Cofactor Engineering. J. Food Biochem. 42 (5), e12584. doi:10.1111/jfbc.12584
Wang, Y., Ren, H., and Zhao, H. (2018b). Expanding the Boundary of Biocatalysis: Design and Optimization of In Vitro Tandem Catalytic Reactions for Biochemical Production. Crit. Rev. Biochem. Mol. Biol. 53 (2), 115–129. doi:10.1080/10409238.2018.1431201
Wu, W., Deng, G., Liu, C., Gong, X., Ma, G., Yuan, Q., et al. (2020). Optimization and Multiomic Basis of Phenyllactic Acid Overproduction by Lactobacillus Plantarum. J. Agric. Food Chem. 68 (6), 1741–1749. doi:10.1021/acs.jafc.9b07136
Wu, X., Zhang, C., Xing, X. H., Yun, Z., Zhao, L., and Wu, Q. (2021). Construction and Characterization of Novel Bifunctional Fusion Proteins Composed of Alcohol Dehydrogenase and NADH Oxidase with Efficient Oxidized Cofactor Regeneration. Biotechnol. Appl. Biochem doi:10.1002/bab.2225
Xiong, W., Liu, B., Shen, Y., Jing, K., and Savage, T. R. (2021). Protein Engineering Design from Directed Evolution to De Novo Synthesis. Biochem. Eng. J. 174, 108096. doi:10.1016/j.bej.2021.108096
Xu, G.-C., Zhang, L.-L., and Ni, Y. (2016). Enzymatic Preparation of D-Phenyllactic Acid at High Space-Time Yield with a Novel Phenylpyruvate Reductase Identified from Lactobacillus Sp. CGMCC 9967. J. Biotechnol. 222, 29–37. doi:10.1016/j.jbiotec.2015.12.011
Xu, J. J., Fu, L. J., Si, K. L., Yue, T. L., and Guo, C. F. (2020). 3‐phenyllactic Acid Production by Free‐whole‐cells of Lactobacillus Crustorum in Batch and Continuous Fermentation Systems. J. Appl. Microbiol. 129 (2), 335–344. doi:10.1111/jam.14599
Xu, J., Xu, X., Wang, Q., and Fan, X. (2015). Chiral Separation of Phenyllactic Acid by Helical Structure from spring Dextrin. J. Incl Phenom Macrocycl Chem. 82 (3-4), 515–521. doi:10.1007/s10847-015-0487-x
Yang, H., Liu, L., Shin, H.-d., Chen, R. R., Li, J., Du, G., et al. (2013). Comparative Analysis of Heterologous Expression, Biochemical Characterization Optimal Production of an Alkaline α-amylase from alkaliphilicAlkalimonas amylolyticainEscherichia coliandPichia Pastoris. Biotechnol. Prog. 29 (1), 39–47. doi:10.1002/btpr.1657
Yu, K., Liu, C., Kim, B.-G., and Lee, D.-Y. (2015a). Synthetic Fusion Protein Design and Applications. Biotechnol. Adv. 33 (1), 155–164. doi:10.1016/j.biotechadv.2014.11.005
Yu, S., Zhou, C., Zhang, T., Jiang, B., and Mu, W. (2015b). 3-Phenyllactic Acid Production in Milk by SK25 during Laboratory Fermentation Process. J. Dairy Sci. 98 (2), 813–817. doi:10.3168/jds.2014-8645
Yu, S., Zhu, L., Zhou, C., An, T., Jiang, B., and Mu, W. (2014). Enzymatic Production of D-3-Phenyllactic Acid by Pediococcus Pentosaceus D-Lactate Dehydrogenase with NADH Regeneration by Ogataea Parapolymorpha Formate Dehydrogenase. Biotechnol. Lett. 36 (3), 627–631. doi:10.1007/s10529-013-1404-2
Zhang, X. X., Abd Elazim, A. M., Ruiz, G., and Yu, R. C. (2014). Fracture Behaviour of Steel Fibre-Reinforced concrete at a Wide Range of Loading Rates. Int. J. Impact Eng. 71, 89–96. doi:10.1016/j.ijimpeng.2014.04.009
Zhang, Y., Wang, Y., Wang, S., and Fang, B. (2017). Engineering Bi-functional Enzyme Complex of Formate Dehydrogenase and Leucine Dehydrogenase by Peptide Linker Mediated Fusion for Accelerating Cofactor Regeneration. Eng. Life Sci. 17 (9), 989–996. doi:10.1002/elsc.201600232
Zhao, W., Ding, H., Lv, C., Hu, S., Huang, J., Zheng, X., et al. (2018). Two-step Biocatalytic Reaction Using Recombinant Escherichia coli Cells for Efficient Production of Phenyllactic Acid from L-Phenylalanine. Process Biochem. 64, 31–37. doi:10.1016/j.procbio.2017.09.019
Zheng, Z., Xia, M., Fang, X., Jiang, T., and Ouyang, J. (2018). Enhanced Biosynthesis of Chiral Phenyllactic Acid from L-Phenylalanine through a New Whole-Cell Biocatalyst. Bioproc. Biosyst Eng 41 (8), 1205–1212. doi:10.1007/s00449-018-1949-5
Keywords: coenzyme self-sufficiency, d-Lactate dehydrogenase, phenyllactic acid, D-PLA, glycerol dehydrogenase, fusion protein
Citation: Qin Z, Wang D, Luo R, Li T, Xiong X and Chen P (2021) Using Unnatural Protein Fusions to Engineer a Coenzyme Self-Sufficiency System for D-Phenyllactic Acid Biosynthesis in Escherichia coli. Front. Bioeng. Biotechnol. 9:795885. doi: 10.3389/fbioe.2021.795885
Received: 15 October 2021; Accepted: 01 December 2021;
Published: 17 December 2021.
Edited by:
Jingyu Wang, Westlake Institute for Advanced Study (WIAS), ChinaReviewed by:
Min Jiang, Nanjing Tech University, ChinaFriso Aalbers, University of Groningen, Netherlands
Copyright © 2021 Qin, Wang, Luo, Li, Xiong and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan Wang, dwang@cqu.edu.cn
 Zhao Qin
Zhao Qin Dan Wang
Dan Wang Ruoshi Luo1
Ruoshi Luo1  Peng Chen
Peng Chen