Substrate-bound protein gradients to study haptotaxis
- 1McGill Program in Neuroengineering, Department of Neurology and Neurosurgery, Montreal Neurological Institute, McGill University, Montreal, QC, Canada
- 2Genome Quebec Innovation Centre, McGill University, Montréal, QC, Canada
- 3McGill Program in Neuroengineering, Department of Biomedical Engineering, McGill University, Montreal, QC, Canada
Cells navigate in response to inhomogeneous distributions of extracellular guidance cues. The cellular and molecular mechanisms underlying migration in response to gradients of chemical cues have been investigated for over a century. Following the introduction of micropipettes and more recently microfluidics for gradient generation, much attention and effort was devoted to study cellular chemotaxis, which is defined as guidance by gradients of chemical cues in solution. Haptotaxis, directional migration in response to gradients of substrate-bound cues, has received comparatively less attention; however, it is increasingly clear that in vivo many physiologically relevant guidance proteins – including many secreted cues – are bound to cellular surfaces or incorporated into extracellular matrix and likely function via a haptotactic mechanism. Here, we review the history of haptotaxis. We examine the importance of the reference surface, the surface in contact with the cell that is not covered by the cue, which forms a gradient opposing the gradient of the protein cue and must be considered in experimental designs and interpretation of results. We review and compare microfluidics, contact printing, light patterning, and 3D fabrication to pattern substrate-bound protein gradients in vitro. The range of methods to create substrate-bound gradients discussed herein makes possible systematic analyses of haptotactic mechanisms. Furthermore, understanding the fundamental mechanisms underlying cell motility will inform bioengineering approaches to program cell navigation and recover lost function.
Introduction
Migrating cells rely on extracellular cues to direct motility. Cues can influence cellular responses in a wide variety of ways ranging from differences in surface stiffness to direct protein–protein interactions (Lara Rodriguez and Schneider, 2013). Cytoskeletal rearrangement directed by extracellular cues dictates the navigation of cells through the environment. Cells may encounter gradients of cues, often proteins, where the local distribution of a guidance protein determines the response (Kim and Peyton, 2012), and can induce a directional change in migration. Protein gradients exist in two forms in vivo: diffusible, where proteins diffuse away from a source, or substrate-bound, where the cues are fastened either to cell surfaces or to the surrounding extracellular matrix (ECM). It is important to note that the same protein may act both as a diffusible and a surface-bound cue. For example, a secreted protein may diffuse a short distance, become bound, and then perhaps be released, travel further by diffusion, only to be bound again. Alternatively, a transmembrane protein that is initially firmly anchored to a cell membrane may be proteolyzed to release a soluble ectodomain that then becomes a diffusible cue.
Gradients of diffusible proteins have been investigated using a variety of methods (Wu et al., 2013). These have led to a better understanding of chemotropic cell responses and the downstream signal transduction cascades activated. However, some of these same guidance cues likely also function bound to a surface, which critically allows a cell to develop traction force by using the immobilized protein as an anchor in a process called mechanotransduction (Gillespie and Walker, 2001).
The investigation of chemotaxis, the guided navigation of cells in response to gradients of diffusible cues, has a long history, initially being described in the late 1800s (Engelmann, 1881; Pfeffer, 1884; Jennings, 1906). Particularly, well-studied examples include the migration of E. coli bacteria toward food sources (e.g., glucose) (Adler, 1966); the common slime mold dictyostelium, which responds to secreted cAMP gradients to direct growth (Gerisch et al., 1975); and lymphocytes, which respond to chemokine gradients to locate a site of immune response (Schall et al., 1990). Based on observations of lymphocyte navigation, Santiago Ramón y Cajal predicted in 1892 that extending axons navigate in response to gradients of secreted guidance cues (Sotelo, 2002).
The earliest studies of cells migrating in culture quickly provided evidence for the significance of the substrate. For example, classic studies by Ross Grainville Harrison used filaments derived from spider webs to show that cells require a solid substrate to migrate (Harrison, 1914). Extending these findings, Paul Weiss developed the principal of contact guidance, arguing that migrating cells and axons are directed by substrate topography during embryonic development (Weiss, 1934). However, it was not until 1965 that Carter (1965) defined the word haptotaxis, differentiating chemotactic guidance in response to a soluble cue from haptotactic migration on a substrate-bound cue. In this seminal study, Carter formed gradients of palladium by using a wire as a shadow mask to scatter a gradient of evaporated metal onto acetate-coated glass. Cells adhered more strongly to the palladium, and directed cell migration was observed from the acetate up the metal gradient. The mechanism was termed haptotaxis (from the Greek: “haptein” to fasten and “taxis” arrangement) to reflect that the cells were navigating in response to the relative strength of the adhesive contacts made with the substrate (Carter, 1965, 1967). Carter suggested that all cell movements within a tissue could be considered haptotactic, including movement traditionally considered to be chemotactic, speculating that the mechanism of action of chemotactic gradients arose from the adsorption of the cue to a surface to form a haptotactic gradient. Such claims, that haptotaxis might underlie all cell movements, not surprisingly, generated controversy, and drew counter arguments from those studying chemotaxis (Keller et al., 1979). Despite numerous studies over the last 35 years that have refined the terminology and included such terms as haptokinesis to differentiate between directed and random navigation (Schumann et al., 2010), this basic debate continues. In many cases, soluble cues may bind to surfaces and many studies of chemotaxis do not explicitly state or investigate possible contributions of haptotaxis. An additional source of confusion is the possible contribution of the substrate, the reference surface (RS), on which the protein cue is patterned. The formation of a protein gradient with gradually increasing surface coverage generally implies an opposing gradient of the RS, which is discussed in detail below.
Gradient geometry varies greatly in vivo, but exact characterization has been challenging and is essentially limited to findings from immunohistochemical analyses and assumptions derived from computational modeling. For instance, gradients are expected to have a dynamic range that spans three to four orders in magnitude (OM), and while overall they may increase from one extremity to the other, locally the complex 3D structure of the environment may result in non-monotonic profiles with local changes in gradient slope. This can be observed, for instance, in spinal cord cross-sections stained for the secreted chemotropic guidance protein netrin-1, where the fluorescence increases along the length of the gradient, but includes local fluctuations in fluorescence intensity due to uneven distribution of the protein around cells (Kennedy et al., 2006). Gradient methods need to be versatile to allow parameters such as dynamic range and gradient slope to be defined and manipulated.
In the late twentieth century, molecular biological insights identified multiple families of extracellular molecular cues underlying haptotaxis, clearly demonstrating that the patterns that instruct cell and axon migration during embryogenesis are not merely topographic. Early studies addressed the function of ECM components themselves, demonstrating critical influences on adhesion and migration (Letourneau et al., 1994a,b). Here, we review a wide range of methods to pattern substrate-bound protein gradients to investigate haptotaxis in vitro, as well as the experimental and conceptual framework that underpins these studies. Rather than providing a historical perspective into the development of these methods, we limit our discussion to techniques developed over the last three decades, describing their potential applicability to studies investigating mechanisms underlying haptotaxis.
The Opposing Reference Surface Gradient
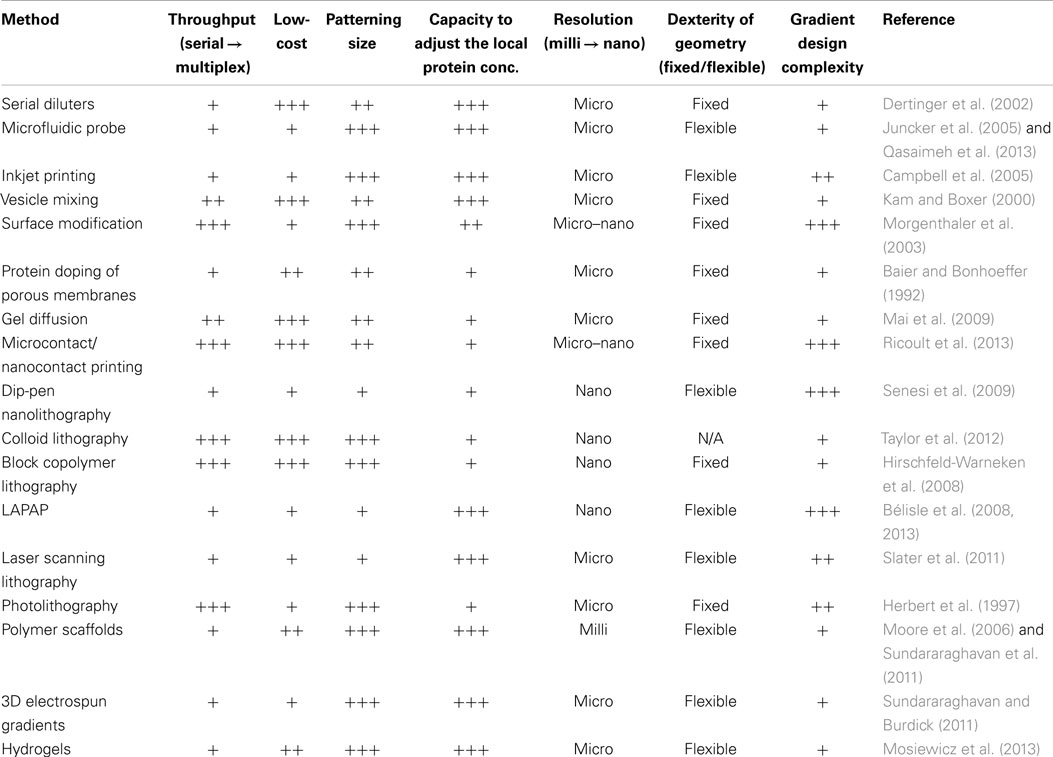
Surface-bound gradients of a guidance cue necessarily entail the presence of an opposite gradient of the underlying RS in the exposed areas of the surface devoid of the immobilized cue of interest (Figure 1).
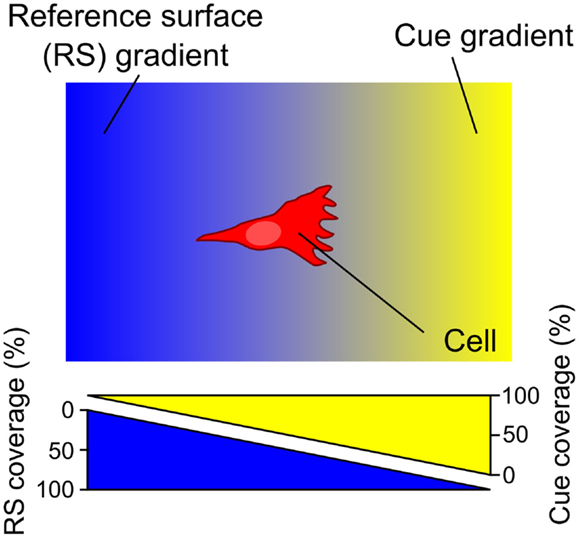
Figure 1. A surface gradient of a cue (yellow) entails an opposite gradient of the underlying reference surface (RS) (blue). The RS, acting relative to the guidance cue, either promotes or inhibits cell adhesion – in either case the RS gradient is expected to modulate the cell response. Consequently, haptotaxis of a cell on a surface-bound gradient is a response to both the gradient of the guidance cue of interest and the opposing RS gradient.
For all methods that form an immobilized gradient, it is critical to carefully consider the properties of the RS. To investigate its functional significance, the impact of changing the composition of the RS was studied using striped patterns of protein cues (Ricoult et al., 2014b). The gaps (also in the shape of stripes) were completely composed of the RS. A method was developed to tune the affinity of the RS by coating the surface with mixtures of high affinity [i.e., polylysine (PDL or PLL)] and low-affinity [i.e., polyethylene glycol (PEG)], and systematically change the ratio of PDL:PEG from 100:0 (high affinity) to 0:100 (low affinity). It was found that an RS composed of 100% high-affinity PDL, when adjacent to a protein known to promote cell adhesion, functioned to mask the influence of the printed protein, and significantly reduced the navigation speed of cells on the patterned surfaces. Conversely, an RS with 100% low-affinity PEG resulted in increased cell migration on non-guidance proteins, such as stripes of immunoglobulin (IgG).
In the context of a gradient, a high-affinity RS promotes migration in the direction opposite to the cue, while a low-affinity RS promotes migration in the same direction as the cue. Moreover, a low-affinity RS would inhibit adhesion of the cells at the low end of the gradient of the cue of interest and thus skew the distribution of cells on the gradient. In contrast, if the affinity of the RS is high, cells will strongly adhere, and motility will be reduced maximally at the low end of the gradient where a larger proportion of each cell contacts the RS. This reasoning holds for cases where high affinity is synonymous with strong adhesion, which is generally but not always the case. For example, cells may exhibit a high affinity for a laminin coated surface, but not adhere strongly to it (Calof and Lander, 1991). For cells at the low end of a linear protein gradient, the response to the cue will likely dominate as the relative change in protein density is large (e.g., a change from 1 to 2% cue density may likely be significant for a cell), while the relative change in the density of the RS is insignificant (e.g., changing from 99 to 98% coverage may be imperceptible to cells). Likewise, at the high end of the gradient only a small percentage of the RS will be exposed, and the relative change in RS will exceed the relative change in the cue, and hence may dominate cellular responses if the RS is not carefully chosen. While the relative change of exponential gradients is constant along the length of the gradient, the importance of the RS remains just as critical to cellular responses.
In many studies, it is assumed, often based on little if any evidence, that the RS only exerts a non-specific influence and does not trigger a specific biochemical response, yet this assumptions may not be accurate. For example, it is often taken for granted that PDL, which was used as the high-affinity component of our RS, is a biochemically neutral substrate; however, PDL-coated microspheres promote the differentiation of presynaptic specializations by axons (Lucido et al., 2009). Although the mechanism underlying this response remains unknown, its specificity suggests that PDL may activate downstream signaling pathways. More careful consideration of the RS and a better understanding of the molecular mechanism that may be triggered by cell–RS interactions will help refine the analysis of cell navigation. Depending on the experiment and cell type, it may be necessary to develop specific RSs that minimally confound cell navigation.
The response of a cell to a haptotactic cue in the presence of different RSs enables the creation of a cell-surface affinity curve that facilitates comparison between different cell types and surfaces (Ricoult et al., 2014b). The optimal RS will depend on the guidance cue being investigated, the cell type used, and may also depend on the physical properties of the underlying substrate (e.g., hydrogel vs. rigid surface). By choosing a RS of relatively extreme low or high affinity, it is possible to direct cells to migrate onto or off a guidance cue of interest, demonstrating tremendous potential for experimental artifact if the RS properties are not carefully considered. By carefully tuning the RS to a specific intermediate value, it is possible to evoke a biochemically appropriate response and study cell migration in response to a patterned guidance cue (Ricoult et al., 2014b). As such, the optimization of the RS is critical in studies of haptotaxis. Consistent with this, in studies using digital nanodot gradients described in detail in the Section “Protein Gradients Created by Microcontact and Nanocontact Printing,” directed cell migration was not detected using 100% PDL as the RS, but by optimizing the RS, appropriate cell migration then occurred (Ricoult et al., 2013). In summary, the presence of an RS is inherent in haptotaxis experiments. It is critical to carefully select and tune the RS, and to interpret cellular responses to a gradient of the protein cue of interest simultaneously in the context of the RS gradient.
Methods to Create Substrate-Bound Protein Gradients
An early method developed to create surface gradients of proteins employed vacuum application through membranes. Since then, a myriad of microfluidic devices, including open microfluidics have been developed. Methods of gradient formation from solution now include inkjet printing, chemical gradient formation to modulate adsorption of proteins, and diffusion from hydrogels. An alternative approach to manipulate the protein concentration is to change the density of microscopic “dots” or lines of proteins, which can be realized using techniques that include micro- and nanocontact printing, dip-pen nanolithography, and colloidal lithography. The use of light to form gradients has been explored using photolithography and direct laser writing. Haptotactic gradients can also be made in 3D by cross-linking guidance cues into hydrogels. Stationary microfluidic gradients can thus be “frozen” into 3D gradients using pH sensitive cues, or photopatterning, or by controlling the mixing of two hydrogels. It is also possible to make 3D gradients by making fiber scaffolds. This myriad collection of techniques developed to generate substrate-bound gradients over the last three decades is reviewed in detail in the following sections.
Protein Doping of Porous Membranes to Create Gradients
An early method to generate substrate-bound patterns, both stripes and gradients, employed a vacuum and a porous capillary filter (Figure 2A) (Baier and Bonhoeffer, 1992). Using this method, protein in solution is incubated on one side of the porous filter. A vacuum is then applied to pull the solution through the membrane, to which the biomolecules bind as they flow through the porous material. By placing silicon masks with slits on top of the porous filter, the protein flow is limited to the open portion of the mask resulting in patterns of protein adsorption on the surface matching the designs on the silicon mask, such as stripes. By placing a glass coverslip on top of a droplet of solution, the amount of solution available for passage through the membrane changes in a graded fashion. As the vacuum is applied, the solution first becomes depleted on the side of the coverslip with the biggest gap with the surface and move toward the smallest gap. This difference in the amount of protein that interacts with the matrix results in a substrate-bound protein gradient. Despite the efficacy of this approach, other methods with greater control over pattern geometry, a greater range of substrate materials (i.e., not reliant on a porous substrate), and a less complex experimental setup are now available and are discussed below.
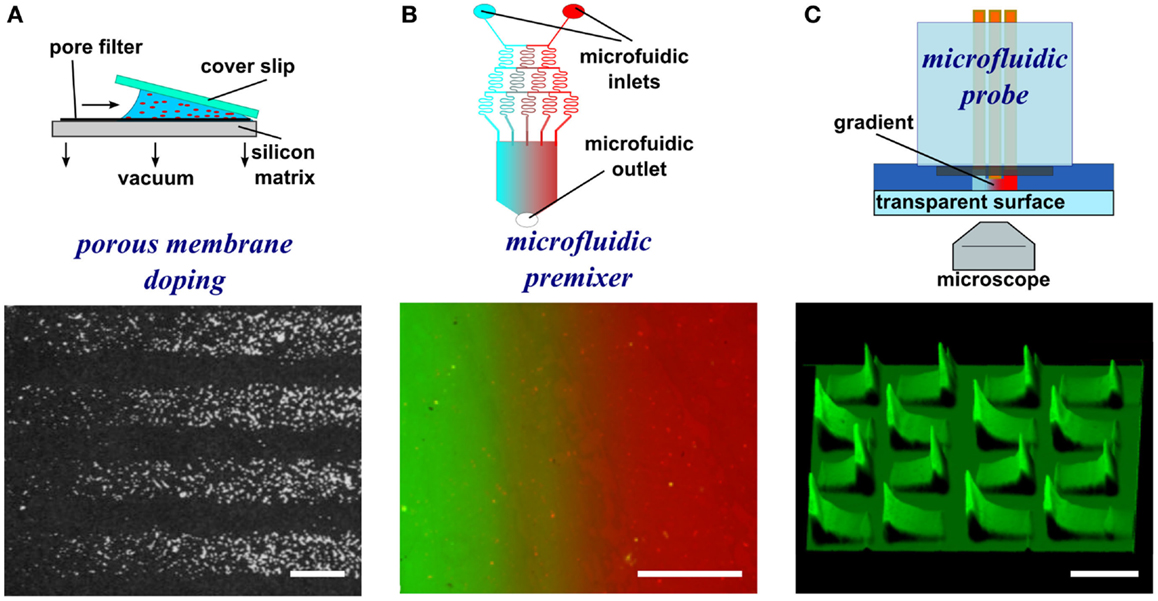
Figure 2. Schematic representations and experimental results of porous membrane doping and microfluidic methods to form substrate-bound protein gradients. (A) By applying a vacuum, protein solutions can be aspirated through a silicon matrix where the proteins bind. By placing an angled coverslip on top of the solution, a protein gradient can be adsorbed to the matrix. Reprinted with permission from AAAS (Baier and Bonhoeffer, 1992). (B) Micrograph image of a microfluidic serial dilutor forming a dilution series that is flowed into a wide chamber forming a gradient across it. With the appropriate surface chemistry, proteins adsorb to the surface, thereby capturing the diffusible gradient on the substrate. (C) A microfluidic probe with injection and aspiration apertures serves to create confined microfluidic flows that can pattern gradients (here fluorescence intensity is shown as topography) by continuously changing the writing speed while patterning. Scale bars are (A,B) 100 μm and (C) 500 μm.
Microfluidic Patterning of Surface-Bound Gradients
Microfluidics manipulates minute volumes of liquids at scales ranging from micrometers to millimeters. Microfluidic conditions suppress turbulence and impose laminar flows where mixing occurs solely through diffusion (Whitesides, 2006). Based on the advantages of microfluidics, an assortment of designs that follow closed-channel or open-channel configurations have been developed to create diffusible gradients suitable for chemotaxis studies. Microfluidic gradient generators in general create diffusible gradients that can be imprinted onto a surface by protein adsorption (Jiang et al., 2005). Operation of microfluidic devices may be compromised by the presence of minute obstructions, such as air bubbles that disturb flow and disrupt gradient geometry. It is important to note that the surface concentration profile may not reflect the profile in solution, and as flow continues, the surface may become saturated, or multilayers of proteins may form (Squires et al., 2008). Furthermore, accurate characterization of the immobilized gradients, beyond quantifying fluorescence intensity, which poorly predicts the concentration of immobilized protein, has not been achieved.
Microfluidic gradient generators
The simplest microfluidic design is a T-junction where two solutions merge into a perpendicular channel and a gradient is formed by the diffusion of a biomolecule across the laminar flow boundary (Walker et al., 2004). The simplicity of the design limits the size of the gradient, but the implementation of more complicated designs could yield gradients of more complex geometries as well as of greater width. One such example is a microfluidic serial diluter where multiple inlet solutions are repetitively split and merged to create microchannels and the dilution series released in a chamber where it rapidly forms a smooth gradient (Figure 2B) (Jeon et al., 2000). For example, substrate-bound protein gradients of the ECM protein laminin were generated on pre-coated poly-l-lysine substrates using this method. These have been used to investigate the response of rat hippocampal neurons to monotonic surface-bound gradients of laminin (Dertinger et al., 2002), and also the response of Xenopus spinal neurons to non-monotonic gradients of laminin in the presence of a diffusible BDNF gradient, the first reported application of a double gradient (Wang et al., 2008). Alternative approaches have been pursued that relatively easily create diffusible gradients (Lee et al., 2009). When adsorbing proteins onto a surface, the high surface-to-volume ratio of microfluidics often leads to depletion of molecules in solution, which can result in the spontaneous formation of substrate-bound gradients. An early demonstration took advantage of this phenomenon and formed gradients along the length of a channel, spanning up to 1.2 OM (Caelen et al., 2000). A more recent example developed channels with a triangular cross-section, thus making use of the higher resistance, reduced flow, and greater depletion at the edges to form gradients (Park et al., 2010). These few examples of microfluidic devices and a great majority of microfluidic devices that yield diffusible gradients in general could be used to pattern substrate-bound gradients by flowing the gradient on the appropriate surface for extended periods. Even though substrate-bound gradients are relatively easy to produce using microfluidic gradient generators, the design of microfluidic chips requires knowledge of microfluidic flow and microfabrication, and as mentioned above, their characterization is inexact.
Open microfluidic gradient generators
The closed-channel configuration of most microfluidic devices limits their use to patterning a single continuous gradient, even though non-monotonic gradients can be generated (Figure 2C). To overcome this limitation, microfluidic probes were developed (Juncker et al., 2005) that combine the attributes of microfluidic systems and scanning probes. The probe can be moved over a surface by either controlling the position of the microfluidic probe with micromanipulators or by moving the substrate with a motorized stage. Once an appropriate location is identified, flow can be turned on, and by controlling the flow rates and the lag rate of the probe on the surface, substrate-bound protein gradients can be patterned, such as graded distributions of fluorescently labeled IgG (Juncker et al., 2005; Qasaimeh et al., 2013). By exploiting the attributes of the microfluidic probe, the first gradient array, composed of 16 gradients 500 μm in length, were patterned onto a surface within minutes. Although versatile, one limitation of this method is that operation of such microfluidic probes requires a dedicated microscope as well as a motorized stage to move the MFP.
Inkjet Printed Gradients
Inkjet printing is a common non-contact surface patterning approach whereby ink droplets are propelled onto a surface through a nozzle at a desired location. The minimal dot size is dictated by the diameter of the nozzle, which defines the diameter of the droplet. Patterned dots from conventional bio-inkjet printers are ~100 μm in diameter, resulting in a resolution of ~300 μm (Tan et al., 2010). Using a bio-inkjet developed to print protein, a substrate-bound continuous concentration gradient of 1.75 mm in length was generated by altering the number of printed rounds of a fixed concentration between adjacent printed dots of ~75 μm diameter on a glass surface. The gradient was formed by inkjeting between 1 and 20 droplets at the same location; however, a difference in fluorescence intensity of only 0.2 OM was achieved (Campbell et al., 2005), illustrating an important limitation of this method.
Surface Chemistry Modifications to Control Protein Adsorption
Rather than directly patterning the proteins as a gradient, it is also possible to first form a gradient in the chemical composition of the surface with different affinities, or “stickiness,” for proteins. For example, a chemical surface gradient may be formed by dip coating a gold-coated surface at a controlled rate in a solution of alkanethiols to yield a gradient of thiol coverage (Morgenthaler et al., 2003). Next, protein solutions are applied to the entire surface, and because proteins will preferentially adsorb to the hydrophobic alkane chains, the chemical gradient will be replicated as a gradient of the protein of interest. Even though the ease and low cost of this approach make it appealing, and variations of the slopes can be produced by controlling the dipping and retraction speeds, complex gradient geometries are challenging to produce. Furthermore, the chemical assembly relies on the presence of an underlying gold surface, which imposes restrictions on the substrates used and complicates cell imaging.
Gel Diffusion to Create Substrate-Bound Gradients
To generate gradients using gel diffusion, a protein solution is flowed through embedded capillaries in a hydrogel, the proteins diffuse through the gel from the source and adsorb as gradients on the surface (Figure 3A). For instance, agarose stamps with open channels on one of the surfaces are contacted with PLL-coated epoxy coverslips to create closed channels. In one example, netrin-1 or BDNF was then injected into microfluidic channels, allowed to diffuse through the agarose gel, and bound to the surface (Mai et al., 2009). Proteins bound to the surface are more abundant close to the source and became increasingly sparse further away. Gradients are fairly simple to form, but depend on the capacity to bind the diffusible protein to the hydrogel, and control over gradient geometry is very limited.
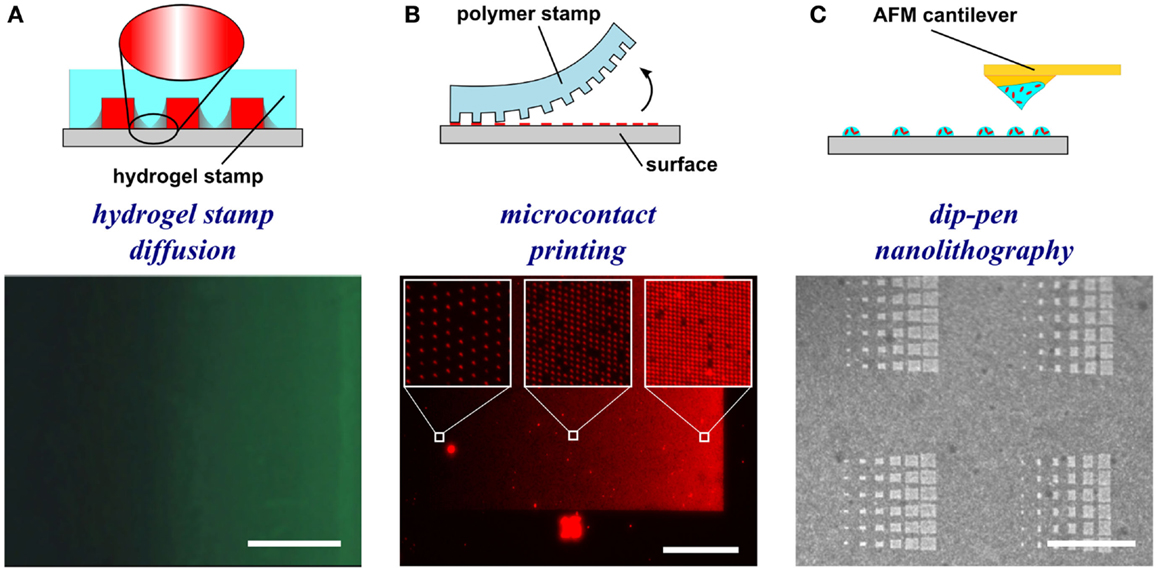
Figure 3. Schematic representations and experimental results of hydrogel-based diffusion, microcontact printing, and dip-pen nanolithography methods to form substrate-bound protein gradients. (A) Hydrogel stamps in contact with the substrate create closed channels that can be filled with a protein solution. The proteins then diffuse through the hydrogel and adsorb to the surface in a graded distribution. Republished with permission of the Journal of Neuroscience (Mai et al., 2009); permission conveyed through Copyright Clearance Center, Inc. (B) In microcontact printing, polymer stamps with topography are inked with a protein solution and used to stamp protein in a gradient geometry by limiting the regions of contact between the stamp and the surface. (C) Dip-pen nanolithography employs an AFM cantilever inked with a protein solution to transfer localized nanovolumes of protein solution onto the surface in any desired geometry (e.g., dot gradient). Reprinted with permission from AAAS (Huo et al., 2008). Scale bars are (A) 100 μm, (B) 1 mm, and (C) 30 μm.
Protein Gradients Created by Microcontact and Nanocontact Printing
Microcontact printing employs a polymer stamp with relief to create protein patterns of specific geometry (Figure 3B). The stamp is inked with a protein solution, incubated to allow the protein to adsorb, and subsequently contacted to the surface to transfer the protein only in the contacting areas. A number of different designs have been reported to date that can either be continuous or digital (Baier and Bonhoeffer, 1992; Lang et al., 2008). Continuous gradients can be formed by adsorption of proteins from solution onto a stamp using the methods described in the preceding paragraphs, and then printing the stamp on a substrate (Figure 4A). In continuous gradients, the density of proteins on the surface changes smoothly. Digital gradients on the other hand are formed by patterning proteins as discrete surface-bound “patches” and forming a discontinuous gradient by changing the surface coverage density of the patches. Conventionally, the concentration of proteins in the patches is constant, in many cases saturated with a “monolayer” of proteins, while the patches can be shaped as dots, dashes, or lines. Note that continuous gradients are in fact also digital since, ultimately, individual proteins adsorbed on the surface form discrete “patches,” but the term digital gradient is reserved for gradients that were designed to be discontinuous at a much larger scale ranging from tens of nanometers to millimeters.
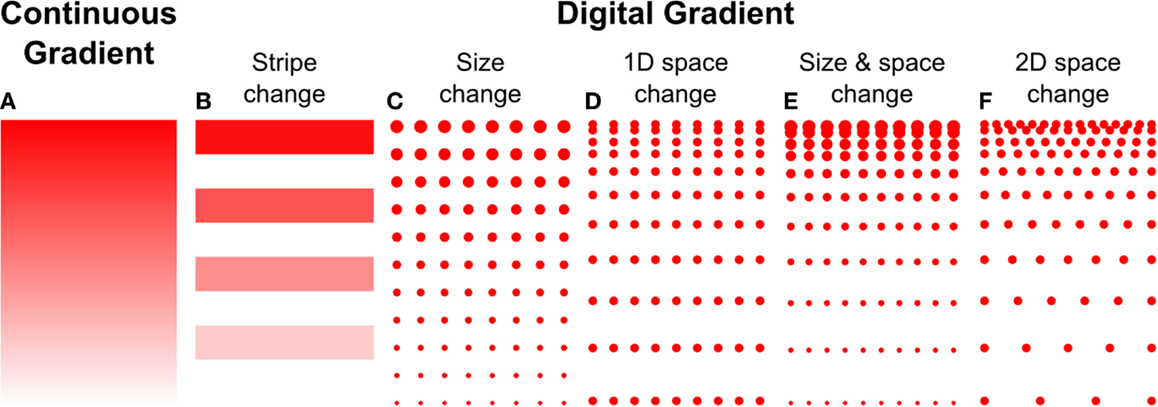
Figure 4. Haptotaxis gradients can either be continuous or digital. (A) Continuous gradients have constant changes in protein concentration, and usually result from the adsorption of biomolecules from diffusible gradients. Digital gradients can be formed in a number of ways, (B) by patterning biomolecules as lines or dots and (C) changing their size, (D) spacing in 1D, or (E) both. (F) The spacing can also be changed in 2D.
A great variety of digital, discontinuous gradients have been made. One conceptually simple approach is to form lines with increasing concentrations by flowing different concentrations of the same molecule through a series of parallel channels on a flat PDMS stamp, resulting in protein adsorption to the stamp. Subsequently, the stamp was used for microcontact printing to transfer the lines onto a flat substrate (Figure 4B) (Lang et al., 2008). A more common approach to form digital gradients is to form patches – a dot or a line – and continuously change the spacing or the size, or both, in the direction of the gradient. Digital gradients with constant spacing and a variable patch size (Figure 4C) were formed by a dashed line with a centrally located node where the length of the dashes is changed based on the distance from the central node (Fricke et al., 2011). The two gradients described above are straightforward to make, but exhibit relatively large surfaces without guidance cue and feature a limited dynamic range. Digital gradients with the spacing of features changed in one direction (Figure 4D) were demonstrated with dots and lines at the microscale (von Philipsborn et al., 2006) and at the nanoscale (Coyer et al., 2007). Coyer et al. also demonstrated gradients with changes in both spacing and feature size (Figure 4E); however, the dynamic range of these gradients was limited to 1.4 OM, and the overall fabrication process remained costly for the nanoscale gradients. Digital nanodot gradients were introduced with the spacing of 200 nm dots changed along two directions (Figure 4F) along with a low-cost patterning method that reduced the cost of individual gradients to a few cents (Ricoult et al., 2013). An advantage of digital gradients is that the local concentration can be read from the dot density, which unambiguously allows defining the density over a large dynamic range. It will be of interest to determine how the size of the nanodots and the protein concentration modulate cellular responses. Whereas prior studies indicated that a feature size of 200 nm was needed to elicit a response (Geiger et al., 2009), no consensus has been reached. The influence of the surface concentration of the protein that makes up the nanodots has not been studied, and a better understanding of the functional significance of these two parameters, in combination with the RS, and how they relate to continuous gradients, will help refine the design of gradient studies in general.
Using these new designs, the gradients reached an unprecedented dynamic range of 3.85 OM (Ongo et al., 2014), which could more accurately represent the expected dynamic range of gradients in vivo. More complex algorithms were also developed to introduce noise into the gradients at the nanoscale by pseudo-randomly distributing dots within a row of constant density and compensating for dot overlap based on the probability of overlap at the given density. Furthermore, noise was also introduced at the microscale by developing gradients where local dot density was fit to non-monotonic functions (Figure 5A). The power of these designs, the relative ease and high throughput of the technique result in a very flexible method. As the resolution and availability of prototyping methods continue to increase (e.g., direct writers and 3D printers) the time and cost associated with the electron beam lithography production of the digital nanodot gradient masters will almost certainly be further minimized. Furthermore, new advances in microcontact printing, such as humidified microcontact printing (Ricoult et al., 2014a), have facilitated the printing of nanopatterns composed of multiple proteins and could lead to the patterning of multiple gradients of different cues.
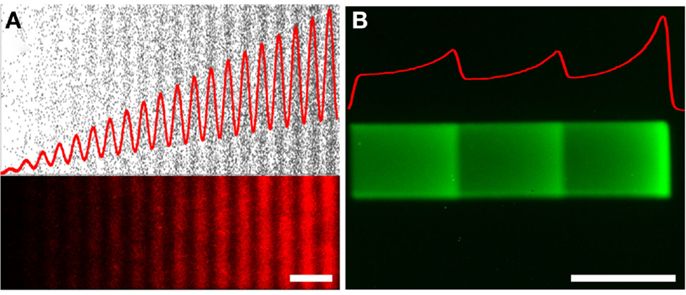
Figure 5. Non-monotonic haptotaxis gradients. Non-monotonic protein gradients achieved with (A) low-cost lift-off nanocontact printing showing (top) the dots of the digital design (black), and (bottom) the fluorescence image of a red fluorescently labeled IgG of the same non-monotonic DNG. (B) Non-monotonic gradients can also be achieved through laser-assisted protein patterning (LAPAP), here illustrated by patterning a fluorescently labeled IgG. The red curves illustrate the density functions of the patterned gradients. Scale bars are (A,B) 50 μm.
Dip-Pen Nanolithography
In dip-pen nanolithography, cantilevers are employed as reservoirs to pattern surfaces (Figure 3C). By using an agarose solution embedded with protein, biologically active protein can be patterned on high energy, activated surfaces (Senesi et al., 2009). By changing the contact time of the tip with the surface, the humidity level of the environment, and viscosity of the agarose, the dot size can be reduced to 50 nm in diameter. By changing these parameters from dot to dot, an array of protein dots with increasing size can be produced, thereby creating a gradient. Recently, polymer pyramid arrays have been employed in essentially the same way, based on regulating the force applied to control the portion of the pyramid that contacts the surface and therefore the size of the feature (Huo et al., 2008). Furthermore, embedded heaters have been added to polymer pyramid arrays to control polymer expansion under each pyramid to individually manipulate dot size (Brown et al., 2013). Dip-pen lithography provides one of the highest resolutions among the various patterning techniques currently available and recent developments such as low-cost cantilever arrays or individually computer controlled cantilevers have made this method very promising; however, in its current state, the method remains relatively slow and impractical for routine use in most biology laboratories.
Colloid Lithography
In colloid lithography, polystyrene spheres in solution are deposited on a surface, and as the solvent evaporates, the spheres form a regular, packed monolayer (Taylor et al., 2012) (Figure 6A). Spheres are then deformed by exposing them to a temperature gradient, which melts the spheres differentially based on the heat applied. By having a heat source to create a temperature gradient on the surface, the extent of sphere melting goes from high, close to the source, to absent, at the most distant position. A PEG silane is then evaporated on the surface and self assembles where the beads do not contact and protect the surface. Spheres are then detached and proteins incubated to fill the gaps previously protected by the beads. The colloid lithography approach yields very large patterned surfaces; however, the limited control of dot placement results in frequent defects. Furthermore, this approach is currently limited to generating linear gradients.
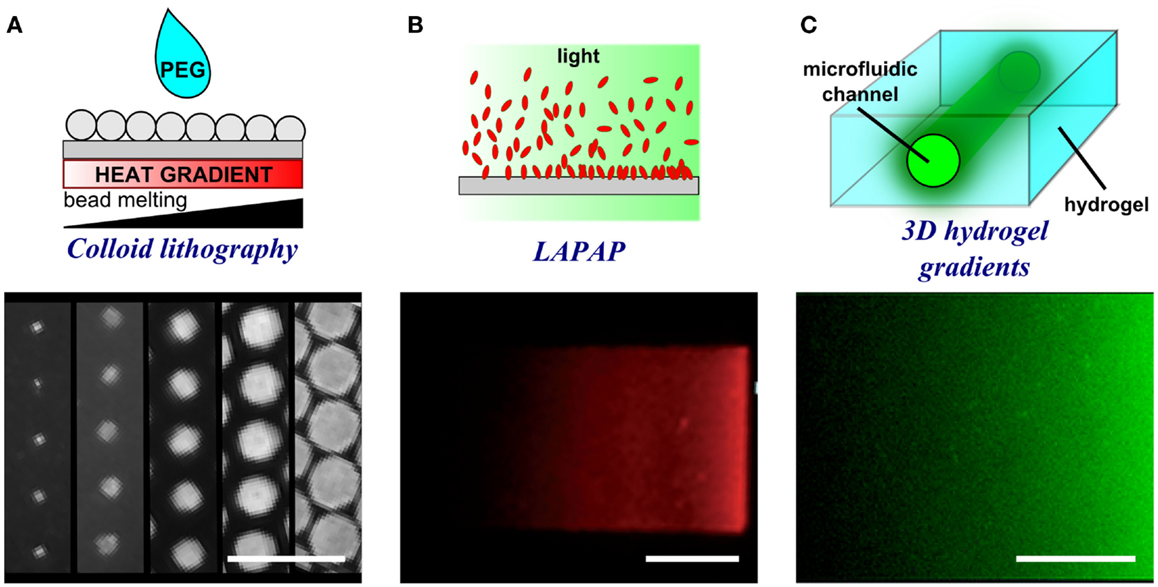
Figure 6. Schematic representations and experimental results of porous membrane doping and microfluidic methods to form substrate-bound protein gradients. (A) By applying a heat gradient under the surface, microspheres melt and interact with the surface to different extents thereby creating a gradient of dots of different size. Adapted with permission from Taylor et al. (2012). Copyright 2012 American Chemical Society. (B) Controlled photobleaching of fluorescently labeled proteins can also be used to increase the reactivity of the proteins with the surface in specific locations and by controlling light intensity or exposure, gradients can be formed. Reproduced in part from Bélisle et al. (2008) with permission of The Royal Society of Chemistry. (C) The presence of a microfluidic channel within a hydrogel enables the filling of a point source solution within a 3D environment. The proteins diffuse from the source and create a gradient in a similar fashion to hydrogel stamp diffusion. Reproduced in part from Lienemann et al. (2015) with permission of The Royal Society of Chemistry. Scale bars are (A) 20 μm, (B) 25 μm, and (C) 250 μm.
Block Copolymer Lithography
In block copolymer nanolithography, polymeric micelles with a polar core and containing metal nanoparticles self-assemble on a surface in an arrangement defined by the architecture of the micelles (Spatz et al., 1999). The polymer portion of the micelles can then be eliminated from the surface by plasma exposure to leave behind patterns of nanoparticles. A dipping machine is then used to change the time of exposure of the surface to the solution. By changing the 2D dot spacing from 55 to 85 nm over 3 mm, it was possible to produce a gradient of ~0.5 OM (Hirschfeld-Warneken et al., 2008). The metal particles were then selectively decorated with thiol-linked peptides, based on the high affinity of thiols for gold surfaces. Block copolymer lithography is an appealing method for nanopatterning, given the ease of the procedure and the commercial availability of a range of micelles; however, the dynamic ranges of the gradients generated remain limited and the capacity of the method to create complex gradients remains unclear.
Patterning with Light
Photolithography
An early technique to pattern with light is photolithography, a relatively harsh microfabrication method that uses strong UV light projected through a metal mask to expose specific regions on a surface. The surface may be coated with a photopolymer, or photoresist, that upon exposure to UV light alters the level of polymer crosslinking. Alternatively, selective UV light exposure photolithography has been employed to eradicate specific portions of self-assembled monolayers (SAMs) of thiols terminated with PEG end groups. The eradication of such groups then opens space on the surface for proteins to bind (Hynes and Maurer, 2012). Peptides can also be selectively adsorbed to surfaces coated with thiol SAMs when exposed to light in a process called photoimmobilization. By selectively exposing the surface to light of graded intensity with a photomask, surface-bound peptide gradients can be patterned (Herbert et al., 1997). Photolithography provides a means to pattern very large surfaces in one step; however, the method typically requires a costly clean-room environment. Furthermore, its dependence on light limits resolution to the microscale.
Laser-based lithography
In this approach, the fluorescence light source of a microscope is used to either selectively bind or detach biomolecules from a surface. In LAPAP, fluorescently labeled biotin is photobleached, rendering it reactive and causing it to covalently bind to an underlying BSA-coated surface (Figure 6B) (Bélisle et al., 2008, 2013). Streptavidin is then bound to the immobilized biotin and serves as a site to link navigational guidance cues. In contrast, in laser scanning lithography light is used to detach bound molecules from the surface (Slater et al., 2011), for example, to replace detached non-fouling thiols with peptide-binding thiols on a gold surface.
Any desired pattern of high complexity can be designed computationally and sent to a microscope to automatically create the surface-bound patterns (Figure 5B). For instance, by altering the laser intensity and the displacement velocity of the fluorescence focal spot, gradients can be produced (Bélisle et al., 2008, 2013). Notably, this method combines a high level of flexibility with regard to the pattern produced and the capacity to locally coat a substrate with different concentrations of protein. Furthermore, by conducting multiple light exposure and incubation steps, multi-protein patterns can be readily achieved, even generating protein gradients running in opposite directions (Bélisle et al., 2009). Laser-based patterning methods are powerful techniques where intricate designs spanning up to 3.0 OM can be patterned, but the chemistry required to link the protein to the surface is not universal, placing some limits on the proteins that can be used for this type of assembly.
3D Gradients
Three dimensional (3D) substrates have grown in popularity due to their potential to generate patterns of even greater relevance to mechanisms that occur in vivo; however, with the implementation of 3D systems, greater complexity arises and data extraction and analysis becomes more complicated. Nevertheless, a number of methods have been developed to generate gradients in 3D.
Hydrogel-based protein gradients
The creation of 3D biomaterials for tissue engineering applications also commonly employs hydrogels (Figure 6C) (Lutolf and Hubbell, 2005). A main attribute of hydrogels is that they are relatively easy to use and their chemistry can be readily designed to include protein binding sites, for example, to capture tagged proteins from solution and integrate them into a PEG-based gel (Lienemann et al., 2015). By forming a soluble gradient in the gel using either a point source diffusion (Lienemann et al., 2015), or a microfluidic gradient generator (Allazetta et al., 2011), proteins can be captured in distributions that reflect the geometry of the soluble gradient. Microfluidics can also be used to create gradients of cross-linked peptides in collagen gels (Sundararaghavan et al., 2011). Alternatively, by electrically manipulating pH within a PEG hydrogel, it is possible to create gradients of gel polymerization based on the distance from the pH electrode (Milleret et al., 2014). Motifs have also been patterned within a hydrogel by integrating photolabile caged biomolecules and using light to selectively release the biomolecules and initiate enzymatic patterning of a PEG gel (Mosiewicz et al., 2013). Gradients presented in hydrogels better mimic the in vivo system; however, control over gradient geometry is currently limited, but could be further improved by adapting some of the methods for high resolution 2D patterning to 3D patterning techniques, such as nanocontact printing or dip-pen nanolithography.
Spin coating fibrous scaffolds of graded protein content
A popular method to create 3D scaffolds is to spin coat fibers onto a surface. One such material that can be spin coated is hyaluronic acid (HA), an ECM component widespread in mammalian brain (Bignami et al., 1993). Sundararaghavan et al. (2011) altered the conventional method of spin coating by adding a T-channel to the spinneret of the machine, which allowed the two solutions to be mixed at different rates over time. By changing the mixing ratio as the fibers were deposited on the surface, a gradient along the z axis could be created within the 3D fibrous scaffold. By mixing either two HA solutions of different modulus or two HA solutions containing different proteins, mechanical and chemical gradients were created that were then used to investigate durotaxis and haptotaxis, respectively (Sundararaghavan and Burdick, 2011). This method of layering spin coated fibers provides a means to achieve more complex 3D gradients; however, the size of the gradients achieved through this method currently greatly exceed that of typical biological gradients and it is unclear whether cells have the capacity to respond to gradients patterned at this scale.
Conclusions and Outlook
There is substantial incentive to better understand cell navigation, providing further insight into the mechanisms underlying embryonic development and enabling the production of effective biomaterials for regenerative medicine. Multiple proteins known to direct cell and axon migration during development are bound to cell surfaces and the ECM, suggesting that haptotactic mechanisms play a critical role. To better understand haptotaxis, many methods have been developed to create substrate-bound gradients in vitro, which are summarized above and in Table 1.
For all of these assays, it is important to carefully consider the RS. For example, a method was developed to adjust the RS in cell-surface affinity screens that were conducted on stripe patterns (Ricoult et al., 2014b), and to select the condition deemed optimal for nanodot gradient studies (Ricoult et al., 2013). In future studies, the RS should be carefully engineered, tuned, and characterized to allow evaluation of the contribution of the opposing RS gradient to the cell response. Appropriate consideration of the RS will facilitate comparisons with other studies and accelerate understanding of the mechanisms underlying motility.
Gradients patterned using the methods described above can be divided into two classes: continuous and digital, where both have advantages and disadvantages. For instance, continuous gradients can easily be achieved through the diffusion of a protein from a source, but the accurate quantitative characterization of such gradients is challenging and often inexact. Digital gradients are also easily produced, but they are deterministic, and as a result the exact local concentration can be calculated. However, the size of the patterned dots of protein aggregates formed typically remain much larger than the size of individual proteins forming such aggregates in natural gradients, which may influence cellular responses.
The numerous technologies that are now available to form substrate-bound protein gradients provide multiple options to study the mechanisms underlying haptotaxis. Patterning techniques such as LAPAP or dip-pen nanolithography offer the capacity for designs with high resolution, whereas substrate-bound gradients achieved through microfluidic gradients or microcontact printing can rapidly pattern large surfaces at a low cost without need for particularly specialized equipment. Furthermore, using digital nanodot gradients, large arrays of distinct gradients can be patterned onto a same substrate by lift-off nanocontact printing, enabling the evaluation of large numbers of parameters in a single experiment, quantifying their effects, and providing insight into the mechanisms underlying haptotaxis.
Current methods such as LAPAP and low-cost lift-off nanocontact printing enable technically straight-forward patterning of substrate-bound protein gradients with high dynamic ranges (i.e., 3.85 OM), non-monotonic complexity, high resolution, and a composition of multiple proteins. Further developments aim to recreate gradients of high biological relevance in environments that better match those found in vivo. By combining protein immobilization with the above-mentioned methods, 3D multi-protein patterns have been achieved, but their resolution remains to be improved to achieve the high resolution possible using methods such as dip-pen nanolithography. Further improvements are focused on developing gradients that will change geometry as the biomaterial ages, for instance, to reflect variations in the gradient geometry that occur as development takes place (Kennedy et al., 2006). Lastly, ongoing studies aim to develop methods to ensure that the conformation of the patterned protein is minimally impacted by the stresses induced by the patterning technique and the surface chemistry. For example, in a landmark study, it was shown that fibronectin adsorbed onto surfaces of distinct wettabilities elicited different ligand–receptor interactions and cell differentiation (García et al., 1999), which was attributed to conformational changes in the fibronectin. The study by García et al., however, did not consider that changing the RS might also have an effect on the cells (Ricoult et al., 2014b). Using the recently developed humidified μCP method (Ricoult et al., 2014a), it is possible to pattern proteins onto a range of RSs with different hydrophobicities and evaluate the cell response. Indeed, it was observed that cells, when in the presence of an appropriate RS, respond to printed proteins through predictable biological pathways, seemingly indistinguishable from the response observed on passively adsorbed protein (Ricoult et al., 2014b). Taken together, these studies highlight that there are many open technical and biological questions related to haptotaxis that remain to be explored. The continued development of methods for forming immobilized gradients on a variety of RS in 2D and 3D will provide better models to study cell biology and benefit many other areas such as tissue engineering.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Greta Thompson-Steckel for comments on the manuscript. We also thank Grant Ongo, Loïc Binan and Santiago Costantino for providing the schematics and images for the non-monotonic gradient figure. We acknowledge support from CIHR (Regenerative and Nanomedicine grant), the Canadian Foundation for Innovation, and the McGill Nanotools Microfabrication Laboratory (funded by CFI and McGill University). SGR was supported by the CREATE Integrated Sensor Systems and Neuroengineering programs funded both by NSERC and The Molson Foundation. TEK was supported by a Scholarship from the Killam Trust and by a Chercheur Nationaux Award from the Fonds de la Recherche en Santé du Québec. DJ holds a Canada Research Chair.
References
Allazetta, S., Cosson, S., and Lutolf, M. P. (2011). Programmable microfluidic patterning of protein gradients on hydrogels. Chem. Commun. 47, 191–193. doi:10.1039/c0cc02377a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baier, H., and Bonhoeffer, F. (1992). Axon guidance by gradients of a target-derived component. Science 255, 472–475. doi:10.1126/science.1734526
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bélisle, J., Mazzaferri, J., and Costantino, S. (2013). Laser-assisted adsorption by photobleaching. Methods Cell Biol. 119, 125–140. doi:10.1016/B978-0-12-416742-1.00007-X
Bélisle, J. M., Correia, J. P., Wiseman, P. W., Kennedy, T. E., and Costantino, S. (2008). Patterning protein concentration using laser-assisted adsorption by photobleaching, LAPAP. Lab. Chip 8, 2164–2167. doi:10.1039/b813897d
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bélisle, J. M., Kunik, D., and Costantino, S. (2009). Rapid multicomponent optical protein patterning. Lab. Chip 9, 3580–3585. doi:10.1039/b911967a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bignami, A., Hosley, M., and Dahl, D. (1993). Hyaluronic-acid and hyaluronic acid-binding proteins in brain extracellular-matrix. Anat. Embryol. 188, 419–433. doi:10.1007/BF00190136
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brown, K. A., Eichelsdoerfer, D. J., Shim, W., Rasin, B., Radha, B., Liao, X., et al. (2013). A cantilever-free approach to dot-matrix nanoprinting. Proc. Natl. Acad. Sci. U.S.A. 110, 12921–12924. doi:10.1073/pnas.1311994110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Caelen, I., Bernard, A., Juncker, D., Michel, B., Heinzelmann, H., and Delamarche, E. (2000). Formation of gradients of proteins on surfaces with microfluidic networks. Langmuir 16, 9125–9130. doi:10.1021/la000851k
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Calof, A. L., and Lander, A. D. (1991). Relationship between neuronal migration and cell-substratum adhesion – laminin and merosin promote olfactory neuronal migration but are antiadhesive. J. Cell Biol. 115, 779–794. doi:10.1083/jcb.115.3.779
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Campbell, P. G., Miller, E. D., Fisher, G. W., Walker, L. M., and Weiss, L. E. (2005). Engineered spatial patterns of FGF-2 immobilized on fibrin direct cell organization. Biomaterials 26, 6762–6770. doi:10.1016/j.biomaterials.2005.04.032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carter, S. B. (1965). Principles of cell motility: the direction of cell movement and cancer invasion. Nature 208, 1183–1187. doi:10.1038/2081183a0
Carter, S. B. (1967). Haptotaxis and the mechanism of cell motility. Nature 213, 256–260. doi:10.1038/213256a0
Coyer, S. R., García, A. J., and Delamarche, E. (2007). Facile preparation of complex protein architectures with sub-100-nm resolution on surfaces. Angew. Chem. Int. Ed. 46, 6837–6840. doi:10.1002/anie.200790177
Dertinger, S. K. W., Jiang, X., Li, Z., Murthy, V. N., and Whitesides, G. M. (2002). Gradients of substrate-bound laminin orient axonal specification of neurons. Proc. Natl. Acad. Sci. U.S.A. 99, 12542–12547. doi:10.1073/pnas.192457199
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Engelmann, T. W. (1881). Neue methode zur untersuchung der sauerstoffausscheidung pflanzlicher und thierischer organismen. Pflügers Archiv. Eur. J. Physiol. 25, 285–292. doi:10.1007/BF01661982
Fricke, R., Zentis, P. D., Rajappa, L. T., Hofmann, B., Banzet, M., Offenhäusser, A., et al. (2011). Axon guidance of rat cortical neurons by microcontact printed gradients. Biomaterials 32, 2070–2076. doi:10.1016/j.biomaterials.2010.11.036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
García, A. J., Vega, M. A. D., and Boettiger, D. (1999). Modulation of cell proliferation and differentiation through substrate-dependent changes in fibronectin conformation. Mol. Biol. Cell 10, 785–798. doi:10.1091/mbc.10.3.785
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geiger, B., Spatz, J. P., and Bershadsky, A. D. (2009). Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10, 21–33. doi:10.1038/nrm2593
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gerisch, G., Hulser, D., Malchow, D., and Wick, U. (1975). Cell communication by periodic cyclic-AMP pulses. Philos. Trans. R. Soc. Lond. B Biol. Sci 272, 181–192. doi:10.1098/rstb.1975.0080
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gillespie, P. G., and Walker, R. G. (2001). Molecular basis of mechanosensory transduction. Nature 413, 194–202. doi:10.1038/35093011
Harrison, R. G. (1914). The reaction of embryonic cells to solid structures. J. Exp. Zool. 17, 521–544. doi:10.1002/jez.1400170403
Herbert, C. B., Mclernon, T. L., Hypolite, C. L., Adams, D. N., Pikus, L., Huang, C. C., et al. (1997). Micropatterning gradients and controlling surface densities of photoactivatable biomolecules on self-assembled monolayers of oligo(ethylene glycol) alkanethiolates. Chem. Biol. 4, 731–737. doi:10.1016/S1074-5521(97)90311-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hirschfeld-Warneken, V. C., Arnold, M., Cavalcanti-Adam, A., López-García, M., Kessler, H., and Spatz, J. P. (2008). Cell adhesion and polarisation on molecularly defined spacing gradient surfaces of cyclic RGDfK peptide patches. Eur. J. Cell Biol. 87, 743–750. doi:10.1016/j.ejcb.2008.03.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huo, F. W., Zheng, Z. J., Zheng, G. F., Giam, L. R., Zhang, H., and Mirkin, C. A. (2008). Polymer pen lithography. Science 321, 1658–1660. doi:10.1126/science.1162193
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hynes, M. J., and Maurer, J. A. (2012). Photoinduced monolayer patterning for the creation of complex protein patterns. Langmuir 28, 16237–16242. doi:10.1021/la303429a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jennings, H. S. (1906). Behavior of the Lower Organisms. New York: Columbia University Press, The Macmillan Company, agents.
Jeon, N. L., Dertinger, S. K. W., Chiu, D. T., Choi, I. S., Stroock, A. D., and Whitesides, G. M. (2000). Generation of solution and surface gradients using microfluidic systems. Langmuir 16, 8311–8316. doi:10.1021/la000600b
Jiang, X. Y., Xu, Q. B., Dertinger, S. K. W., Stroock, A. D., Fu, T. M., and Whitesides, G. M. (2005). A general method for patterning gradients of biomolecules on surfaces using microfluidic networks. Anal. Chem. 77, 2338–2347. doi:10.1021/ac048440m
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Juncker, D., Schmid, H., and Delamarche, E. (2005). Multipurpose microfluidic probe. Nat. Mater. 4, 622–628. doi:10.1038/nmat1435
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kam, L., and Boxer, S. G. (2000). Formation of supported lipid bilayer composition arrays by controlled mixing and surface capture. J. Am. Chem. Soc. 122, 12901–12902. doi:10.1021/ja0034038
Keller, H. U., Wissler, J. H., and Ploem, J. (1979). Chemotaxis is not a special case of haptotaxis. Experientia 35, 1669–1671. doi:10.1007/BF01953261
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kennedy, T. E., Wang, H., Marshall, W., and Tessier-Lavigne, M. (2006). Axon guidance by diffusible chemoattractants: a gradient of netrin protein in the developing spinal cord. J. Neurosci. 26, 8866–8874. doi:10.1523/JNEUROSCI.5191-05.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, H.-D., and Peyton, S. R. (2012). Bio-inspired materials for parsing matrix physicochemical control of cell migration: a review. Integr. Biol. (Camb) 4, 37–52. doi:10.1039/c1ib00069a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lang, S., Philipsborn, A., Bernard, A., Bonhoeffer, F., and Bastmeyer, M. (2008). Growth cone response to ephrin gradients produced by microfluidic networks. Anal. Bioanal. Chem. 390, 809–816. doi:10.1007/s00216-007-1363-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lara Rodriguez, L., and Schneider, I. C. (2013). Directed cell migration in multi-cue environments. Integr. Biol. (Camb) 5, 1306–1323. doi:10.1039/c3ib40137e
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, K., Kim, C., Ahn, B., Panchapakesan, R., Full, A. R., Nordee, L., et al. (2009). Generalized serial dilution module for monotonic and arbitrary microfluidic gradient generators. Lab. Chip 9, 709–717. doi:10.1039/b813582g
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Letourneau, P. C., Condic, M. L., and Snow, D. M. (1994a). Interactions of developing neurons with the extracellular matrix. J. Neurosci. 14, 915–928.
Letourneau, P. C., Snow, D. M., and Gomez, T. M. (1994b). Growth cone motility: substratum-bound molecules, cytoplasmic [Ca2+] and Ca(2+)-regulated proteins. Prog. Brain Res. 102, 35–48. doi:10.1016/S0079-6123(08)60530-0
Lienemann, P. S., Devaud, Y. R., Reuten, R., Simona, B. R., Karlsson, M., Weber, W., et al. (2015). Locally controlling mesenchymal stem cell morphogenesis by 3D PDGF-BB gradients towards the establishment of an in vitro perivascular niche. Integr. Biol. (Camb) 7, 101–111. doi:10.1039/c4ib00152d
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lucido, A. L., Suarez Sanchez, F., Thostrup, P., Kwiatkowski, A. V., Leal-Ortiz, S., Gopalakrishnan, G., et al. (2009). Rapid assembly of functional presynaptic boutons triggered by adhesive contacts. J. Neurosci. 29, 12449–12466. doi:10.1523/JNEUROSCI.1381-09.2009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lutolf, M. P., and Hubbell, J. A. (2005). Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 23, 47–55. doi:10.1038/nbt1055
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mai, J., Fok, L., Gao, H., Zhang, X., and Poo, M.-M. (2009). Axon initiation and growth cone turning on bound protein gradients. J. Neurosci. 29, 7450–7458. doi:10.1523/JNEUROSCI.1121-09.2009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Milleret, V., Simona, B. R., Lienemann, P. S., Vörös, J., and Ehrbar, M. (2014). Electrochemical control of the enzymatic polymerization of PEG hydrogels: formation of spatially controlled biological microenvironments. Adv. Healthc. Mater. 3, 508–514. doi:10.1002/adhm.201300479
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moore, K., Macsween, M., and Shoichet, M. (2006). Immobilized concentration gradients of neurotrophic factors guide neurite outgrowth of primary neurons in macroporous scaffolds. Tissue Eng. 12, 267–278. doi:10.1089/ten.2006.12.267
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morgenthaler, S., Lee, S. W., Zurcher, S., and Spencer, N. D. (2003). A simple, reproducible approach to the preparation of surface-chemical gradients. Langmuir 19, 10459–10462. doi:10.1021/la034707l
Mosiewicz, K. A., Kolb, L., Van Der Vlies, A. J., Martino, M. M., Lienemann, P. S., Hubbell, J. A., et al. (2013). In situ cell manipulation through enzymatic hydrogel photopatterning. Nat. Mater. 12, 1072–1078. doi:10.1038/nmat3766
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ongo, G., Ricoult, S. G., Kennedy, T. E., and Juncker, D. (2014). Ordered, random, monotonic and non-monotonic digital nanodot gradients. PLoS ONE 9:e106541. doi:10.1371/journal.pone.0106541
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Park, J., Kim, D. H., Kim, G., Kim, Y., Choi, E., and Levchenko, A. (2010). Simple haptotactic gradient generation within a triangular microfluidic channel. Lab. Chip 10, 2130–2138. doi:10.1039/b924222h
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pfeffer, W. F. P. (1884). Locomotorische Richtungsbewegungen Durch Chemische Reize. Untersuchungen aus dem botanischen Institut zu Tübingen Bd I, Vol. 3, pp. 363–482.
Qasaimeh, M. A., Ricoult, S. G., and Juncker, D. (2013). Microfluidic probes for use in life sciences and medicine. Lab. Chip 13, 40–50. doi:10.1039/c2lc40898h
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ricoult, S. G., Pla-Roca, M., Safavieh, R., Lopez-Ayon, G. M., Grütter, P., Kennedy, T. E., et al. (2013). Large dynamic range digital nanodot gradients of biomolecules made by low-cost nanocontact printing for cell haptotaxis. Small 9, 3308–3313. doi:10.1002/smll.201202915
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ricoult, S. G., Sanati Nezhad, A., Knapp-Mohammady, M., Kennedy, T. E., and Juncker, D. (2014a). Humidified microcontact printing of proteins: universal patterning of proteins on both low and high energy surfaces. Langmuir 30, 12002–12010. doi:10.1021/la502742r
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ricoult, S. G., Thompson-Steckel, G., Correia, J. P., Kennedy, T. E., and Juncker, D. (2014b). Tuning cell-surface affinity to direct cell specific responses to patterned proteins. Biomaterials 35, 727–736. doi:10.1016/j.biomaterials.2013.10.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schall, T. J., Bacon, K., Toy, K. J., and Goeddel, D. V. (1990). Selective attraction of monocytes and lymphocytes-T of the memory phenotype by cytokine rantes. Nature 347, 669–671. doi:10.1038/347669a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schumann, K., Lämmermann, T., Bruckner, M., Legler, D. F., Polleux, J., Spatz, J. P., et al. (2010). Immobilized chemokine fields and soluble chemokine gradients cooperatively shape migration patterns of dendritic cells. Immunity 32, 703–713. doi:10.1016/j.immuni.2010.04.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Senesi, A. J., Rozkiewicz, D. I., Reinhoudt, D. N., and Mirkin, C. A. (2009). Agarose-assisted dip-pen nanolithography of oligonucleotides and proteins. ACS Nano 3, 2394–2402. doi:10.1021/nn9005945
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Slater, J. H., Miller, J. S., Yu, S. S., and West, J. L. (2011). Fabrication of multifaceted micropatterned surfaces with laser scanning lithography. Adv. Funct. Mater. 21, 2876–2888. doi:10.1016/B978-0-12-416742-1.00011-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sotelo, C. (2002). The chemotactic hypothesis of Cajal: a century behind. Prog. Brain Res. 136, 11–20. doi:10.1016/S0079-6123(02)36004-7
Spatz, J. P., Mössmer, S., Hartmann, C., Möller, M., Herzog, T., Krieger, M., et al. (1999). Ordered deposition of inorganic clusters from micellar block copolymer films. Langmuir 16, 407–415. doi:10.1021/la990070n
Squires, T. M., Messinger, R. J., and Manalis, S. R. (2008). Making it stick: convection, reaction and diffusion in surface-based biosensors. Nat. Biotechnol. 26, 417–426. doi:10.1038/nbt1388
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sundararaghavan, H. G., and Burdick, J. A. (2011). Gradients with depth in electrospun fibrous scaffolds for directed cell behavior. Biomacromolecules 12, 2344–2350. doi:10.1021/bm200415g
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sundararaghavan, H. G., Masand, S. N., and Shreiber, D. I. (2011). Microfluidic generation of haptotactic gradients through 3D collagen gels for enhanced neurite growth. J. Neurotrauma 28, 2377–2387. doi:10.1089/neu.2010.1606
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tan, C. P., Cipriany, B. R., Lin, D. M., and Craighead, H. G. (2010). Nanoscale resolution, multicomponent biomolecular arrays generated by aligned printing with parylene peel-off. Nano Lett. 10, 719–725. doi:10.1021/nl903968s
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Taylor, Z. R., Keay, J. C., Sanchez, E. S., Johnson, M. B., and Schmidtke, D. W. (2012). Independently controlling protein dot size and spacing in particle lithography. Langmuir 28, 9656–9663. doi:10.1021/la300806m
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
von Philipsborn, A. C., Lang, S., Loeschinger, J., Bernard, A., David, C., Lehnert, D., et al. (2006). Growth cone navigation in substrate-bound ephrin gradients. Development 133, 2487–2495. doi:10.1242/dev.02412
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Walker, G. M., Ozers, M. S., and Beebe, D. J. (2004). Cell infection within a microfluidic device using virus gradients. Sens. Actuators B Chem. 98, 347–355. doi:10.1039/b816986a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, J. C., Li, X., Lin, B., Shim, S., Ming, G.-L., and Levchenko, A. (2008). A microfluidics-based turning assay reveals complex growth cone responses to integrated gradients of substrate-bound ECM molecules and diffusible guidance cues. Lab. Chip 8, 227–237. doi:10.1039/b713945d
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weiss, P. (1934). In vitro experiments on the factors determining the course of the outgrowing nerve fiber. J. Exp. Zool. 68, 393–448. doi:10.1002/jez.1400680304
Whitesides, G. M. (2006). The origins and the future of microfluidics. Nature 442, 368–373. doi:10.1038/nature05058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, J., Wu, X., and Lin, F. (2013). Recent developments in microfluidics-based chemotaxis studies. Lab. Chip 13, 2484–2499. doi:10.1039/c3lc50415h
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: haptotaxis, substrate-bound gradient, immobilized gradient, digital gradient, reference surface
Citation: Ricoult SG, Kennedy TE and Juncker D (2015) Substrate-bound protein gradients to study haptotaxis. Front. Bioeng. Biotechnol. 3:40. doi: 10.3389/fbioe.2015.00040
Received: 31 January 2015; Accepted: 13 March 2015;
Published online: 30 March 2015.
Edited by:
Anna Lagunas, CIBER-BBN, Spain; Instituto de Salud Carlos III, SpainReviewed by:
Tamal Das, Max Planck Institute for Intelligent Systems, GermanyClaudio G. Rolli, Weizmann Institute of Science, Israel
Copyright: © 2015 Ricoult, Kennedy and Juncker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Juncker, Department of Biomedical Engineering, Génome Québec Innovation Centre, McGill University, 740 Drive Penfield Avenue, Office #6206, Montreal, QC H3A 0G1, Canada e-mail: david.juncker@mcgill.ca
 Sébastien G. Ricoult
Sébastien G. Ricoult Timothy E. Kennedy
Timothy E. Kennedy David Juncker
David Juncker