- 1National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan, China
- 2Chinese Academy of Tropical Agricultural Sciences, Hainan, China
Bidirectional promoters, which show great application potential in genetic improvement of plants, have aroused great research interest recently. However, most bidirectional promoters were cloned individually in the studies of single genes. Here, we initiatively combined RNA-seq data and cDNA microarray data to discover the potential bidirectional promoters in rice genome. Based on the expression level and correlation of each adjacent and oppositely transcribed gene pair, we selected four candidate gene pairs. Then, the intergenic region between each pair was isolated and cloned into a dual reporter vector pDX2181 for functional identification. GUS and GFP assays of the transgenic plants indicated that all the intergenic regions showed bidirectional expression activity in various tissues. Through 5′ and 3′ deletion analysis on one of the above bidirectional promoters, we identified the enhancing region which sharply increased its bidirectional expression efficiency and the essential regions respectively responsible for its 5′ and 3′ basic expression activity. The bidirectional arrangement of the four gene pairs in six gramineous plants was also analyzed, showing the conserved characteristics of the four bidirectional promoters identified in our study. In addition, two novel cis-sequences conserved in the four bidirectional promoters were discovered by bioinformatic identification. Our study proposes a feasible method for selecting, cloning, and functionally identifying bidirectional promoters as well as for discovering their bidirectional regulatory regions and conserved sequences in rice.
Introduction
Plant architecture, development, and interaction with environment are controlled by the expression of a series of genes (Chen F. et al., 2010; Li C. et al., 2011; Zhu et al., 2011). As a critical regulator of gene expression, promoters are important in plant biotechnology and functional genomics research for their great application potential in genetic engineering and theoretical significance in the exploration of transcriptional regulation mechanism (Cai et al., 2007; Yi et al., 2011; Walcher and Nemhauser, 2012; Ye et al., 2012; Balasubramani et al., 2014). Many researches have been focused on the cloning and analysis of unidirectional promoters, such as constitutive promoters, spatiotemporal promoters, and inducible promoters (McElroy et al., 1990; Pan et al., 2015; Vijayan et al., 2015). Bidirectional promoters, which generally refer to the intergenic region between two adjacent genes transcribed in opposite directions, show better applicability than unidirectional promoters in genetic improvement (Trinklein et al., 2004; Mitra et al., 2009; Yang et al., 2013). That is because a bidirectional promoter can drive the expression of two genes simultaneously, and thus can be time-saving in constructing expression vectors and pyramiding of multiple genes (Kumar et al., 2015). Besides, it is very critical for transgenic breeding to enable the functionally related genes to express in the same pattern in the receptors (Ha et al., 2010; Ogo et al., 2013). However, the unidirectional promoters with the same specific expression patterns are only available in limited quantities, and repetitious use of the promoters may have a negative impact on the stability and expression of transgenes (Peremarti et al., 2010). According to previous reports, the expression patterns of bidirectional promoters in opposite directions are similar in many cases due to the co-expression of the adjacent genes (Huang et al., 2007; Wang et al., 2009; Chen W. et al., 2010; Didych et al., 2013). Therefore, bidirectional promoters could also compensate for the lack of unidirectional promoters with the same expression pattern.
Much work has been done to analyze the bidirectional promoters in mammalian genome with experimental and bioinformatic methods (Trinklein et al., 2004; Yang and Elnitski, 2008; Uwanogho et al., 2010). The results suggest that the divergent gene pairs regulated by bidirectional promoters exhibit the characteristics of conserved arrangement, coexpression, and functional association (Xu et al., 2012; Didych et al., 2013; Meersseman et al., 2014; Yang and Elnitski, 2014). Since the sequences of promoters are known to be variable (Müller et al., 2007), for discovering bidirectional promoters, it is particularly helpful to investigate the characteristics of the divergent gene pairs regulated by them.
Bidirectional promoters have become a research focus in plants in recent years. With the development of plant genome sequencing, bioinformatic analyses in plants like rice, Arabidopsis and Populus have revealed that the divergent gene pairs regulated by bidirectional promoters have similar characteristics, such as coexpression, functional association, and conserved arrangement (Krom and Ramakrishna, 2008; Dhadi et al., 2009; Wang et al., 2009; Chen W. et al., 2010). The structural characteristics of bidirectional promoters in plants are similar to those in mammals, such as higher GC content and less TATA boxes (Dhadi et al., 2009). Besides, bidirectional promoters have been cloned in many species. In Arabidopsis, the tissue-specific and light-inducible bidirectional promoter between cab1 and cab2, the tissue-specific bidirectional promoter between At5g06290 and At5g06280, and the tissue-specific and stress-inducible bidirectional promoter between At4g35985 and At4g35987 have been cloned successively (Bondino and Valle, 2009; Mitra et al., 2009; Banerjee et al., 2013). All of these promoters can be widely used in gene functional analysis in Arabidopsis. Several bidirectional promoters from other species such as melon and Capsicum annuum, have been also cloned gradually (Shin et al., 2003; Wang et al., 2008). In rice, a few promoters have been found to show bidirectional expression activities (Huang et al., 2007; Singh et al., 2009; Dhadi et al., 2013). So far, there has been no report about the cloning and identification of bidirectional promoters using two reporter genes simultaneously with stable transformation in rice.
Rice is one of the most important food crops in the world and a model plant for functional genomic researches in cereals (Zhang, 2007). Therefore, it is highly necessary to introduce multiple genes into rice for genetic improvement (Ha et al., 2010; Yang et al., 2011; Ogo et al., 2013). Accordingly, discovery of bidirectional promoters in rice is very critical. Besides, more complete genomic information (Goff et al., 2002; Yu et al., 2002; Pan et al., 2014) and more explicit gene expression information (Wang et al., 2010; Kawahara et al., 2013; Sakai et al., 2013) will greatly facilitate the development of a high-throughput method for discovering bidirectional promoters.
Most of the known bidirectional promoters were found during the researches of single genes. In this study, we reported a method of selection, cloning, functional identification, and deletion analysis of bidirectional promoters for their de novo discovery in rice. We first selected four adjacent and oppositely transcribed gene pairs based on their expression levels and correlations, which were derived from the RNA-seq data of the Michigan State University Rice Genome Annotation Project Database (MSU), the Rice Annotation Project Database (RAP), and the microarray data of the rice cDNA microarray database (CREP). Subsequently, the intergenic regions between the four gene pairs were cloned for functional identification. GUS and GFP assays of the transgenic plants indicated that all the intergenic regions showed bidirectional expression activity in various tissues. With 5′ and 3′ deletion analysis of one bidirectional promoter above, we found the regulatory region responsible for its bidirectional expression activity. Meanwhile, the bidirectional arrangement of the four gene pairs in six gramineous plants showed the conserved characteristics of the four bidirectional promoters identified in our study. Then, we discovered two cis-sequences conserved in the four bidirectional promoters with MEME. The two cis-sequences showed overrepresentation in the intergenic regions between divergent gene pairs in rice genome under the reference of random promoters. Our study proposes a feasible method for selecting, cloning, and functionally identifying bidirectional promoters as well as for the discovery of their bidirectional expression regulatory regions and conserved sequences in rice.
Methods
Selection of Candidate Bidirectional Promoters
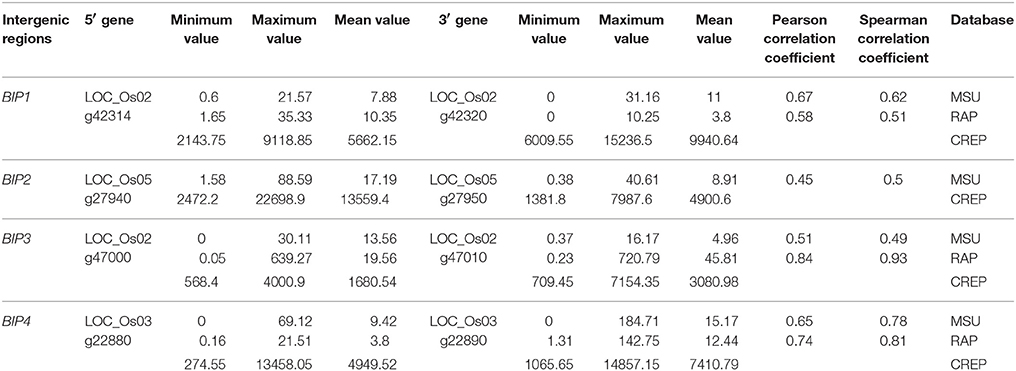
RNA-seq data were obtained from the Michigan State University Rice Genome Annotation Project Database (MSU) and the Rice Annotation Project Database (RAP; Kawahara et al., 2013; Sakai et al., 2013), and the microarray data were downloaded from the rice cDNA microarray database (CREP; Wang et al., 2010). Based on the expression characteristics, the criteria for candidate divergent gene pairs regulated by bidirectional promoters in our study were set as: the maximum expression value of the gene pair was simultaneously higher than 10 in RNA-seq data and higher than 5000 in microarray data, and the expression correlation coefficient between the gene pair from RNA-seq data of 95 samples was higher than 0.4. Because only 36 samples were available in microarray data, which were not sufficient for reliable correlation analysis, we did not consider the correlation coefficient of the data from microarray data. According to the criteria, we chose four divergent gene pairs (Table 1) and isolated their intergenic regions (here designated as BIP1, BIP2, BIP3, and BIP4, respectively) for functional identification.
Isolation and Vector Construction of BIP1, BIP2, BIP3, and BIP4
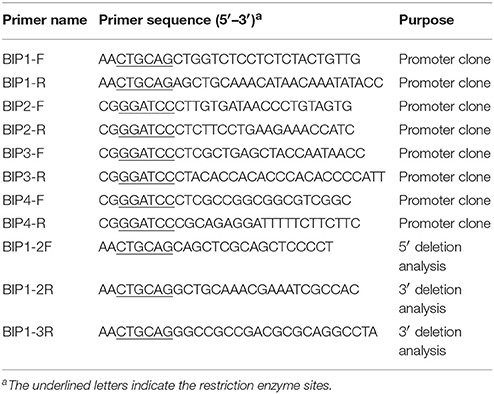
The genomic DNA of Minghui 63 (Oryza sativa L ssp. indica) was used as template to amplify BIP1, BIP2, BIP3, and BIP4 with specific primers (Table 2). The PCR-generated fragments were respectively inserted into T-vector (Promega) and confirmed by sequencing with primers SP6 and T7. The sequence-confirmed clone containing BIP1/BIP2/BIP3/BIP4 was digested by Pst I/BamH I/BamH I/BamH I and was respectively cloned into a dual reporter vector pDX2181 (Ye et al., 2012).
Agrobacterium-Mediated Rice Transformation
The sequence-confirmed clones were transformed into the Agrobacterium tumefaciens strain EHA105 by electroporation. Subsequently, all the constructs were introduced into Zhonghua11 (O. sativa L. ssp. japonica) by Agrobacterium-mediated transformation. The callus culture and transformation procedures were carried out as previously described (Hiei et al., 1994).
Histochemical and Fluorometric Analysis of GUS Activity
Histochemical staining of GUS activity in rice tissues was conducted essentially as previously described (Jefferson et al., 1987). Various tissues of T0 transgenic-positive transformants (root, leaf, sheath, panicle, stem, and mature seed) were incubated in GUS staining solution (50 mM sodium phosphate at pH 7.0, 10 mM Na2-EDTA, 0.1% Triton X-100, 1 mg/mL X-Gluc, 100 μg/ml chloramphenicol, 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide and 20% methanol) at 37°C for 2–10 h after 15-min vacuum filtration. After GUS staining, the samples were incubated in 70% ethanol to remove chlorophyll and photographs were taken under a dissecting microscope (Leica MZFLIII).
Quantitative analysis of GUS activity was conducted as previously described (Xu et al., 2010). The total protein concentration in the supernatant was quantified using the Bradford assay (Bradford, 1976). GUS protein in the supernatant was determined fluorometrically with an INFINITE 200 photometer (Tecan Austria Gmbh, Ltd, Grodig, Austria). GUS activity was determined fluorometrically by measuring the amount of 4-methylumbelliferone (Mu) produced under the catalysis of GUS in 1 mg of total protein per minute. Five biological replicates were assayed for each construct. Ten transgenic lines were randomly divided into five groups and two transgenic lines were considered as one biological replicate.
Histological and Quantitative Analysis of GFP
Histological analysis of GFP in rice tissues was detected and photographed under fluorescence microscope. Various tissues of T0 transgenic-positive transformants (root, leaf, sheath, panicle, stem, and mature seed) were sampled and observed under a fluorescence microscope (Leica MZ16F) using GFP filter sets and Leica Application Suite software.
The relative expression levels of GFP in rice tissues were detected by quantitative real-time PCR (qRT-PCR). Total RNAs of different rice tissues were extracted and reverse-transcribed as described previously (Wang et al., 2015a), and qRT-PCR was performed according to the same reference. The primers of GFP were GFP-F: 5′-ATCCGCCACAAC ATCGAGGA-3′ and GFP-R: 5′-TCGTCCATG CCGAGAGTGAT-3′, and the primers of GAPDH (the endogenous control) were GAPDH-F: 5′-CTGCAACTCAGA AGACCGTTG-3′ and GAPDH-R: 5′-CCTGTT GTCACCCTGGAAGTC-3′. Relative expression levels were determined using 2−ΔΔCT method (Livak and Schmittgen, 2001).
Melatonin Treatment
Melatonin (N-acetyl-5-methoxytryptamine), which is known as an indispensable hormone related to many physiological activities in animals, has also been identified as an important signaling molecule in response to many stresses in plants. In order to test the response of the four bidirectional promoters to melatonin, melatonin treatment was performed on the transgenic plants according to the procedure described by Shi and Chan (2014).
5′ and 3′ Deletion Analysis of BIP1
Among the four bidirectional promoters above, BIP1 showed the highest expression efficiency in both 5′ and 3′ orientations. Therefore, it was selected for 5′ and 3′ deletion analysis in order to find the regulatory regions responsible for bidirectional expression activity (Figure 1). The specific primers used for PCR amplification to generate different 5′ and 3′ truncated fragments are shown in Table 2. Vector construction, callus culture and transformation, histochemical and fluorometric analysis of GUS activity, histological and quantitative analysis of GFP were performed as described above.
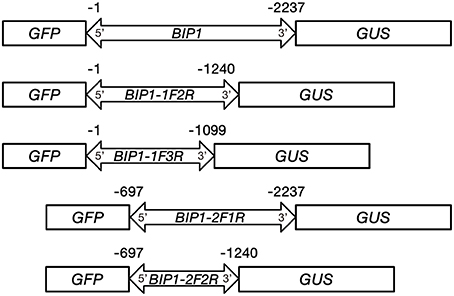
Figure 1. Schemes of constructs carrying BIP1 and different deleted versions fused with GFP and GUS reporter genes.
Conservation Analysis of the Four Bidirectional Promoters and Bioinformatic Identification of Their Conserved Sequences
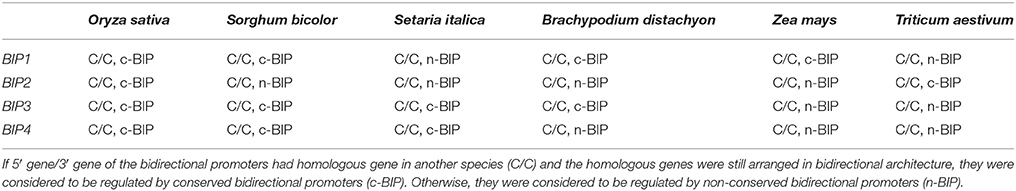
The conserved arrangements of the four gene pairs in six gramineous plants (O. sativa, Sorghum bicolor, Setaria italica, Brachypodium distachyon, Zea mays, and Triticum aestivum) were identified with the information from the Ensembl Plants database (http://plants.ensembl.org/index.html). The bidirectional genes whose homologous genes in other species were still arranged in a bidirectional architecture were considered to be regulated by conserved bidirectional promoters (c-BIP); otherwise, they were considered to be regulated by non-conserved bidirectional promoters (n-BIP).
The conserved sequences in the four bidirectional promoters were discovered by MEME (http://meme-suite.org//tools/meme) and their frequencies in the intergenic regions between divergent gene pairs in rice genome were identified by FIMO (p < 1E-8) using the reference of random promoters (http://meme-suite.org//tools/fimo).
Results
Selection of Four Novel Bidirectional Promoters in Rice Genome and Their Functional Characterization in Transgenic Plants
Based on RNA-seq and microarray data, we chose four divergent gene pairs (Table 1) according to the criteria in Section Methods and isolated their intergenic regions for functional identification. The four fragments were respectively cloned to a dual reporter vector pDX2181 and transformed into rice variety Zhonghua 11.
According to the results of GUS and GFP assays of the transgenic plants, all the intergenic regions showed bidirectional expression activity in various tissues. Histological GUS and GFP analysis of the transgenic plants showed that four novel bidirectional promoters (BIP1, BIP2, BIP3, and BIP4) were successfully identified in our work. Among them, BIP1, BIP2, and BIP3 showed bidirectional constitutive expression patterns and BIP4 showed bidirectional seed-specific expression pattern (Figure 2). Analysis of GUS fluorometric activities in various tissues of BIP1 transgenic plants (Figure 3) showed that the expression efficiency of BIP1 toward 3′ was the highest in the seed, which showed a GUS enzymatic activity of 17806 ± 2108 pmol 4-MU/min/mg protein, followed by in the root, which exhibited a GUS enzymatic activity of 14769 ± 1782 pmol 4-MU/min/mg protein, while the GUS enzymatic activities in the stem, sheath, panicle, and leaf were 9825 ± 1510, 8681 ± 834, 7380 ± 895, and 6092 ± 875 pmol 4-MU/min/mg protein, respectively. Analysis of GFP expression in BIP1 transgenic plants (Figure 3) revealed that the expression efficiency of BIP1 toward 5′ was the highest in the panicle, which was 2.4-fold higher than that in the leaf. While the expression levels of GFP in the root, sheath, stem, and seed were 1.8-, 1.5-, 0.4-, and 0.1-fold higher than that in the leaf, respectively. GUS assays of BIP2 transgenic plants (Figure 3) indicated that the expression efficiency of BIP2 toward 3′ was the highest in the leaf, which showed a GUS enzymatic activity of 3713 ± 445 pmol 4-MU/min/mg protein, and the GUS activities in the sheath, panicle, stem, and seed were 2548 ± 339, 1755 ± 220, 1063 ± 145, and 872 ± 173 pmol 4-MU/min/mg protein, respectively, while GUS activity was hardly detected in the root. Analysis of GFP expression in BIP2 transgenic plants (Figure 3) showed that the expression efficiency of BIP2 toward 5′ was the highest in the stem, which was 8.6-fold higher than that in the panicle, while the expression levels of GFP in the root, sheath, leaf, and seed were 3.9-, 2.6-, 2.3-, and 1.6-fold higher than that in the panicle, respectively. BIP3 transgenic plants showed GUS enzymatic activities of 1126 ± 128, 841 ± 117, 806 ± 85, and 734 ± 71 pmol 4-MU/min/mg protein in the sheath, seed, stem, and leaf, respectively, while GUS activity was hardly detected in the panicle and root. The expression efficiency of BIP3 toward 5′ was the highest in the panicle, which was 5.7-fold higher than that in the leaf, while the expression levels of GFP in the seed, stem, root, and sheath were 3.6-, 2.9-, 1.3-, and 0.9-fold higher than that in the leaf, respectively. BIP4 showed a bidirectional seed-specific expression pattern, as a GUS enzymatic activity of 1659 ± 211 pmol 4-MU/min/mg protein was detected in the seed while almost no GUS activity was detected in other tissues, and a 23-fold higher expression level of GFP was observed in the seed compared with in the leaf. In addition, these results indicate that all the bidirectional promoters identified here direct gene expression in an orientation-independent manner; namely, the expression patterns in opposite directions of these bidirectional promoters are similar, which is consistent with the co-expression characteristics of the adjacent genes.
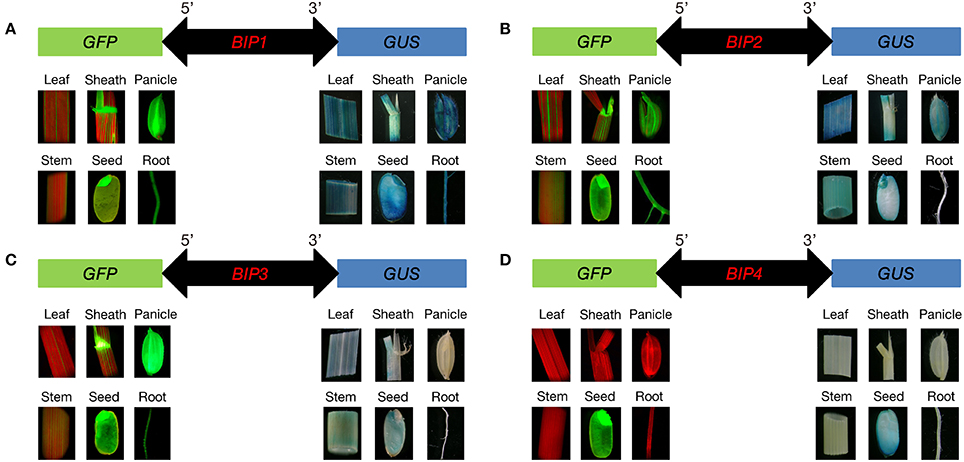
Figure 2. Histological analysis of GFP and GUS expression in various tissues of the transgenic plants containing different GFP/bidirectional promoter/GUS fusions. (A–D), plants containing GFP::BIP1-BIP4::GUS. Localization of GFP is shown at the left area; Localization of GUS is shown at the right area.
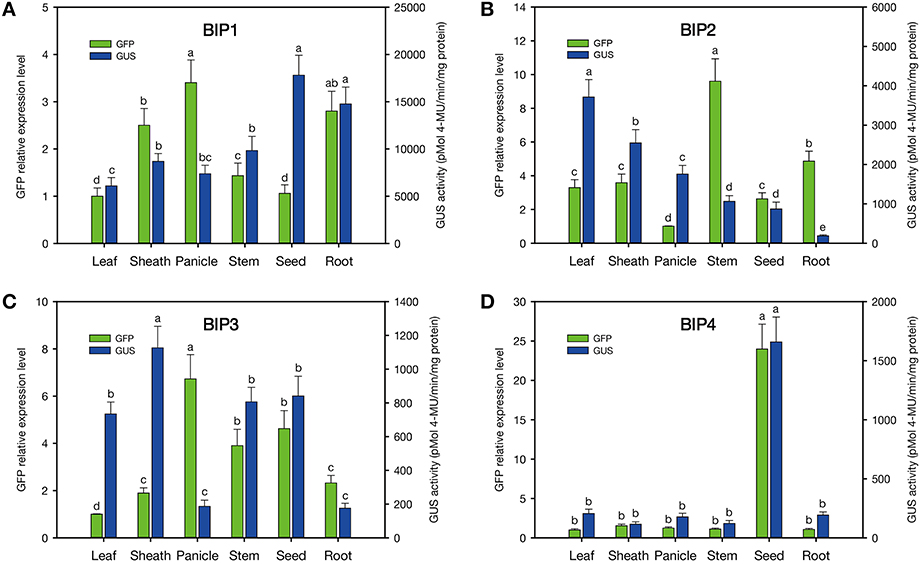
Figure 3. Quantitative analysis of GFP and GUS expression in various tissues of the transgenic plants containing different GFP/bidirectional promoter/GUS fusions. (A–D), plants containing GFP::BIP1-BIP4::GUS. a, b, c, d, e: significant difference (P < 0.05). Error bars indicate SE based on five independent biological replicates.
Melatonin is one of the most important hormones in plant and animal. In order to test the response of the four bidirectional promoters to melatonin, melatonin treatment was performed on the BIPs transgenic plants. The results of GUS and GFP assays indicated that the four bidirectional promoters were not induced by melatonin (Figure 4). Hence, it can be inferred that no melatonin-responsive cis-element was harbored in these promoters.
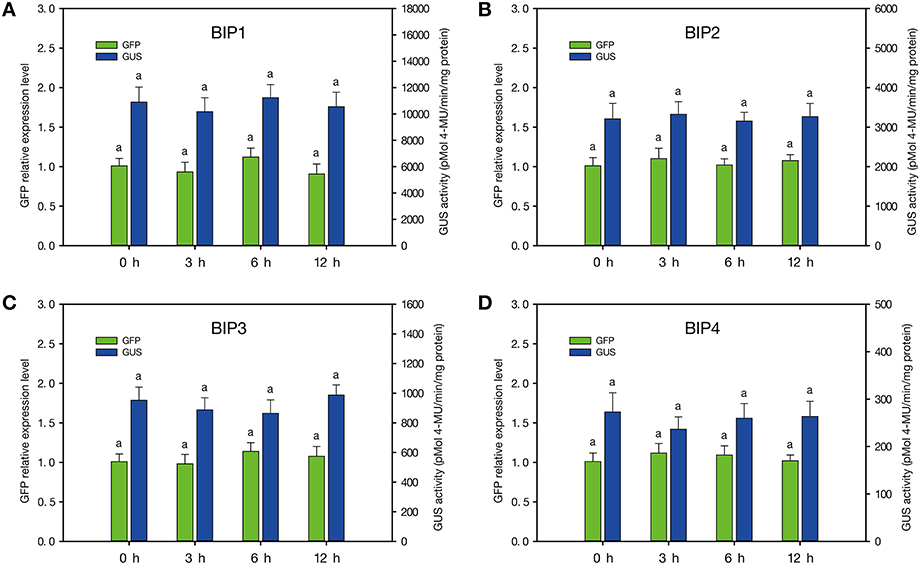
Figure 4. Quantitative analysis of GFP and GUS expression of the BIPs transgenic plants in response to melatonin. (A–D), plants containing GFP::BIP1-BIP4::GUS. a: no significant difference. Error bars indicate SE based on five independent biological replicates.
Identification of the Expression Regulatory Regions in BIP1
In order to identify the regulatory regions in BIP1, 5′ and 3′ deletion analysis of this promoter was performed. A series of truncated BIP1 were respectively cloned to pDX2181 and transformed into Zhonghua 11. Transgenic plants carrying BIP1-1F2R showed much lower GUS activity than BIP1 transgenic plants in various tissues, especially in the root, stem, seed and panicle, whose GUS activities were lower than 10% of that in the corresponding tissues of BIP1 transgenic plants (Figures 5, 6). It thus could be inferred that region 1 could greatly increase the transcriptional activity of BIP1 toward 3′. Meanwhile, BIP1-1F2R transgenic plants also showed an obvious decrease of GFP expression in various tissues compared with BIP1 transgenic plants (Figure 6). These results could be integrated to reveal that region 1 is a bidirectional transcription-enhancing region of BIP1. Further truncating in 3′ of BIP1 led to complete abolishment of GUS activity in BIP1-1F3R transgenic plants, while the GFP expression level was not obviously reduced in BIP1-1F3R transgenic plants compared with in BIP1-1F2R transgenic plants. These results suggest that region 2 is the essential region responsible for the basic expression activity of 3′ but not for that of 5′. Transgenic plants carrying BIP1-2F1R or BIP1-2F2R showed no expression of GFP, indicating that truncating 5′ of BIP1 will completely abolish 5′ expression activity of the promoter. GUS assays in BIP1-2F1R transgenic plants revealed that truncating 5′ of BIP1 caused slight changes of 3′ expression activity in most tissues except for the root, which showed obviously decreased GUS activity compared with that of BIP1 transgenic plants (Figure 6). These results suggest that region 3 is the essential region responsible for the basic expression activity of 5′ but not for that of 3′; however, it can positively regulate the expression activity of 3′ in the root.
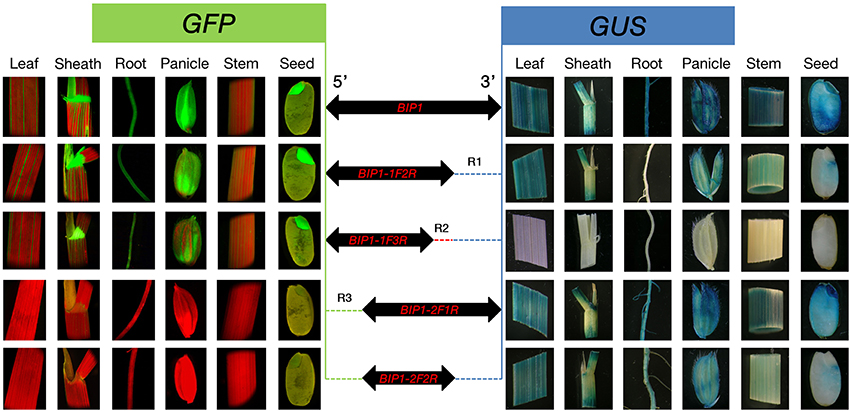
Figure 5. Histological analysis of GFP and GUS expression in various tissues of the transgenic plants containing different GFP/BIP1 deleted version/GUS fusions. Localization of GFP is shown at the left area; Localization of GUS is shown at the right area. R1 (blue dashed line), region 1; R2 (red dashed line), region 2; R3 (green dashed line), region 3.
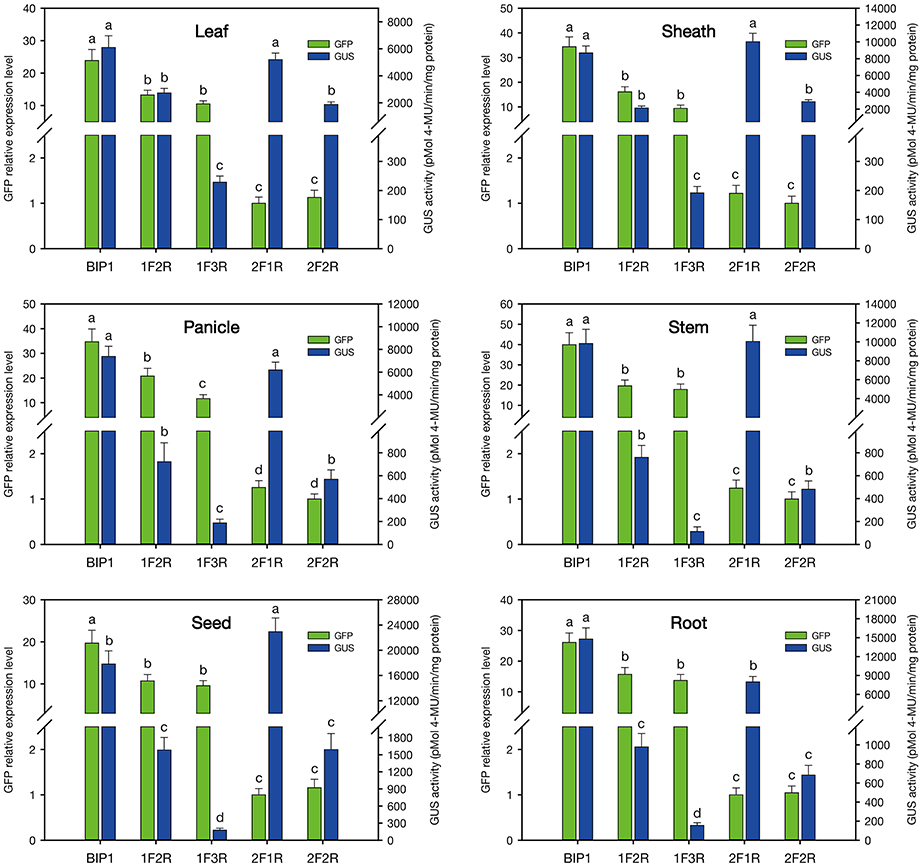
Figure 6. Quantitative analysis of GFP and GUS expression in various tissues of the transgenic plants containing different GFP/BIP1 deleted version/GUS fusions. 1F2R, plants containing GFP::BIP1-1F2R::GUS; 1F3R, 2F1R and 2F2R follow the same pattern. a, b, c, d: significant difference (P < 0.05). Error bars indicate SE based on five independent biological replicates.
Conserved Arrangement of the Four Gene Pairs Regulated by Bidirectional Promoters
The sequences of promoters are known to be variable (Müller et al., 2007). Therefore, in order to analyze the conservation of the four bidirectional promoters in different species, we investigated the conservation of the four gene pairs regulated by these promoters.
The conserved arrangement of the four gene pairs in six gramineous plants was identified with information from the Ensembl Plants database. The bidirectional genes whose homologous genes in other species were still arranged in a bidirectional architecture were considered to be regulated by c-BIP; otherwise, they were considered to be regulated by n-BIP (Table 3). It was found that BIP1 and BIP3 were the most c-BIP in the six gramineous plants. BIP1 was conserved in O. sativa, S. bicolor, B. distachyon, and Z. mays; BIP3 was conserved in O. sativa, S. bicolor, S. italic, and B. distachyon; BIP4 was conserved in O. sativa, S. bicolor, and S. italica; while BIP2 was only conserved in O. sativa and T. aestivum.
Potential cis-Sequences Involved in Bidirectional Expression
By MEME, two conserved cis-sequences in the four bidirectional promoters were identified (Figure 7). Cis-sequence 1 was a G/C-rich sequence, while cis-sequence 2 was an A/T-rich sequence. Subsequently, the frequencies of these cis-sequences in the intergenic regions between divergent gene pairs in rice genome were analyzed by FIMO using the reference of random promoters. Consistent with the expectation, the two cis-sequences conserved in the four bidirectional promoters both showed overrepresentation in potential bidirectional promoters in rice genome compared with random promoters. This result further reveals that the two novel cis-sequences are probably involved in bidirectional expression.
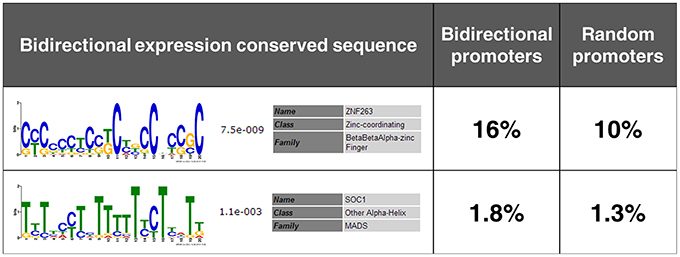
Figure 7. Two novel cis-sequences conserved in the four identified bidirectional promoters. The frequencies of the two cis-sequences in the intergenic regions between divergent gene pairs in rice genome and in the random promoters used as a reference set are shown on the second column and the third column, respectively.
Discussion
In this study, we initiatively combined RNA-seq data and cDNA microarray data to discover potential bidirectional promoters in rice. Four adjacent and oppositely transcribed gene pairs were selected based on their expression levels and correlations. The intergenic regions between the four gene pairs were successfully cloned, and all of them were identified to be bidirectional promoters, confirming the feasibility of our method for discovering bidirectional promoters. This is the first study to clone and identify bidirectional promoters using two reporter genes simultaneously with stable transformation in rice. Among the four identified bidirectional promoters, BIP1 shows high expression efficiency in various tissues, and thus has a high application potential in genetic engineering, such as driving two resistant genes simultaneously in transgenic breeding against pest/disease stress, which can confer more strong, broad, and durable resistance in rice (Du et al., 2009; Shah et al., 2009; Yang et al., 2011; Wang et al., 2015a). Rice is one of the most important food crops in the world, and its seed is the edible part consumed by human. Therefore, it is highly necessary to improve the nutrient quality of the seed (Ha et al., 2010; Li Y. et al., 2011, 2014; Ogo et al., 2013). Efficient and specific expression of multiple target genes for seed improvement in rice could hardly be realized without seed-specific promoter. In this work, BIP4 shows a bidirectional seed-specific expression pattern, indicating its high application potential in the improvement of seed quality by specifically driving multiple genes. The results of 5′ and 3′ deletion analysis reveal that region 1 is the bidirectional transcription-enhancing region of BIP1; region 2 is the essential region specifically responsible for the basic expression activity of 3′; region 3 is the essential region responsible for the basic expression activity of 5′ but not for that of 3′, while it can positively regulate the 3′ expression activity in the root.
Conservation analysis of the four bidirectional promoters in gramineous plants reveals the possible co-evolution of adjacent genes regulated by these promoters. The bidirectional arrangement of LOC_Os02g42314 and LOC_Os02g42320, which are regulated by BIP1, is conserved in four species of six gramineous plants, suggesting that they are relatively conserved during co-evolution. The functional annotations of these two genes, which are “ubiquitin-conjugating enzyme” and “peptidase,” show that both of them are structural genes conserved during evolution. Moreover, the functional relationship between the two genes further supports their co-evolutionary conservation. LOC_Os02g47000 and LOC_Os02g47010, which are regulated by BIP3, also show conserved arrangement in four species of six gramineous plants. Although the functional annotation of LOC_Os02g47000 is unclear, considering that LOC_Os02g47010 is annotated to encode “secretory carrier-associated membrane protein,” we speculate that LOC_Os02g47000 may encode a structural protein related to secretory pathway.
So far, many cis-regulatory sequences have been identified, which are involved in inducible expression (Liu et al., 2010, 2014; Yuan et al., 2011; Koschmann et al., 2012; Walcher and Nemhauser, 2012) and tissue-specific expression (Hartmann et al., 2005; Cai et al., 2007; Ye et al., 2012; Wang et al., 2015b). A previous report also has suggested that several known cis-sequences might be related to bidirectional expression (Dhadi et al., 2009). Here, we used the experimentally verified bidirectional promoters to predict two cis-sequences related to bidirectional expression which had not been identified in rice genome. Subsequently, overrepresentation of the two novel cis-sequences in the intergenic regions between divergent gene pairs further reveals their involvement in bidirectional expression. Interestingly, cis-sequence 1 is a G/C-rich sequence, which is consistent with the characteristics of higher GC content in bidirectional promoters; however, cis-sequence 2 is an A/T-rich sequence, which might be a new finding in the sequence characteristics of bidirectional promoters. Overall, the novel bidirectional promoters identified using two reporter genes simultaneously with stable transformation in rice are expected to have high applicability in genetic engineering. Our study proposes a feasible method for selecting, cloning, and functionally identifying bidirectional promoters as well as for discovering their bidirectional regulatory regions and conserved sequences in rice.
Author Contributions
YL and RW conceived and designed the experiments. RW, YY, MZ, and MY performed the experiments. RW and YY performed the data analysis. RW and YL wrote the paper. FZ and HC revised the paper. YL secured the funds to support this research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the National Program of Transgenic Variety Development of China (2014ZX08001-001), the National High Technology Research and Development Program of China (863 Program) and the National Natural Science Foundation of China. And we thank Prof. Zuoxiong Liu (Huazhong Agricultural University) for language improvement.
References
Balasubramani, A., Winstead, C. J., Turner, H., Janowski, K. M., Harbour, S. N., Shibata, Y., et al. (2014). Deletion of a conserved cis-element in the Ifng locus highlights the role of acute histone acetylation in modulating inducible gene transcription. PLoS Genet. 10:e1003969. doi: 10.1371/journal.pgen.1003969
Banerjee, J., Sahoo, D. K., Dey, N., Houtz, R. L., and Maiti, I. B. (2013). An intergenic region shared by At4g35985 and At4g35987 in Arabidopsis thaliana is a tissue specific and stress inducible bidirectional promoter analyzed in transgenic Arabidopsis and tobacco plants. PLoS ONE 8:e79622. doi: 10.1371/journal.pone.0079622
Bondino, H. G., and Valle, E. M. (2009). A small intergenic region drives exclusive tissue-specific expression of the adjacent genes in Arabidopsis thaliana. BMC Mol. Biol. 10:95. doi: 10.1186/1471-2199-10-95
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cai, M., Wei, J., Li, X., Xu, C., and Wang, S. (2007). A rice promoter containing both novel positive and negative cis-elements for regulation of green tissue-specific gene expression in transgenic plants. Plant Biotechnol. J. 5, 664–674. doi: 10.1111/j.1467-7652.2007.00271.x
Chen, F., Gao, M., Miao, Y., Yuan, Y., Wang, M., Li, Q., et al. (2010). Plasma membrane localization and potential endocytosis of constitutively expressed XA21 proteins in transgenic rice. Mol. Plant 3, 917–926. doi: 10.1093/mp/ssq038
Chen, W., Meaux, J. D., and Lercher, M. J. (2010). Co-expression of neighbouring genes in Arabidopsis: separating chromatin effects from direct interactions. BMC genomics 11:178. doi: 10.1186/1471-2164-11-178
Dhadi, S. R., Deshpande, A., Driscoll, K., and Ramakrishna, W. (2013). Major cis-regulatory elements for rice bidirectional promoter activity reside in the 5′-untranslated regions. Gene 526, 400–410. doi: 10.1016/j.gene.2013.05.060
Dhadi, S. R., Krom, N., and Ramakrishna, W. (2009). Genome-wide comparative analysis of putative bidirectional promoters from rice, Arabidopsis and Populus. Gene 429, 65–73. doi: 10.1016/j.gene.2008.09.034
Didych, D., Shamsutdinov, M., Smirnov, N., Akopov, S., Monastyrskaya, G., Uspenskaya, N., et al. (2013). Human PSENEN and U2AF1L4 genes are concertedly regulated by a genuine bidirectional promoter. Gene 515, 34–41. doi: 10.1016/j.gene.2012.11.058
Du, B., Zhang, W., Liu, B., Hu, J., Wei, Z., Shi, Z., et al. (2009). Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. U.S.A. 106, 22163–22168. doi: 10.1073/pnas.0912139106
Goff, S. A., Ricke, D., Lan, T. H., Presting, G., Wang, R., Dunn, M., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. Japonica). Science 296, 92–100. doi: 10.1126/science.1068275
Ha, S. H., Liang, Y. S., Jung, H., Ahn, M. J., Suh, S. C., Kweon, S. J., et al. (2010). Application of two bicistronic systems involving 2A and IRES sequences to the biosynthesis of carotenoids in rice endosperm. Plant Biotechnol. J. 8, 928–938. doi: 10.1111/j.1467-7652.2010.00543.x
Hartmann, U., Sagasser, M., Mehrtens, F., Stracke, R., and Weisshaar, B. (2005). Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 57, 155–171. doi: 10.1007/s11103-004-6910-0
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Huang, Y., Xiao, B., and Xiong, L. (2007). Characterization of a stress responsive proteinase inhibitor gene with positive effect in improving drought resistance in rice. Planta 226, 73–85. doi: 10.1007/s00425-006-0469-8
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. doi: 10.1186/1939-8433-6-4
Koschmann, J., MacHens, F., Becker, M., Niemeyer, J., Schulze, J., Bülow, L., et al. (2012). Integration of bioinformatics and synthetic promoters leads to the discovery of novel elicitor-responsive cis-regulatory sequences in Arabidopsis. Plant Physiol. 160, 178–191. doi: 10.1104/pp.112.198259
Krom, N., and Ramakrishna, W. (2008). Comparative analysis of divergent and convergent gene pairs and their expression patterns in rice, Arabidopsis, and Populus. Plant Physiol. 147, 1763–1773. doi: 10.1104/pp.108.122416
Kumar, S., AlAbed, D., Whitteck, J. T., Chen, W., Bennett, S., Asberry, A., et al. (2015). A combinatorial bidirectional and bicistronic approach for coordinated multi-gene expression in corn. Plant Mol. Biol. 87, 341–353. doi: 10.1007/s11103-015-0281-6
Li, C., Wang, Y., Liu, L., Hu, Y., Zhang, F., Mergen, S., et al. (2011). A rice plastidial nucleotide sugar epimerase is involved in galactolipid biosynthesis and improves photosynthetic efficiency. PLoS Genet. 7:e1002196. doi: 10.1371/journal.pgen.1002196
Li, Y., Fan, C., Xing, Y., Jiang, Y., Luo, L., Sun, L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. doi: 10.1038/ng.977
Li, Y., Fan, C., Xing, Y., Yun, P., Luo, L., Yan, B., et al. (2014). Chalk5 encodes a vacuolar H+-translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 46, 398–404. doi: 10.1038/ng.2923
Liu, L., Zhou, Y., Szczerba, M. W., Li, X., and Lin, Y. (2010). Identification and application of a rice senescence-associated promoter. Plant Physiol. 153, 1239–1249. doi: 10.1104/pp.110.157123
Liu, W., Mazarei, M., Peng, Y., Fethe, M. H., Rudis, M. R., Lin, J., et al. (2014). Computational discovery of soybean promoter cis-regulatory elements for the construction of soybean cyst nematode-inducible synthetic promoters. Plant Biotechnol. J. 12, 1015–1026. doi: 10.1111/pbi.12206
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
McElroy, D., Zhang, W., Cao, J., and Wu, R. (1990). Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2, 163–171. doi: 10.1105/tpc.2.2.163
Meersseman, C., Léjard, V., Rebours, E., Boussaha, M., Maftah, A., Petit, D., et al. (2014). Bovine TWINKLE and mitochondrial ribosomal protein L43 genes are regulated by an evolutionary conserved bidirectional promoter. Gene 537, 154–163. doi: 10.1016/j.gene.2013.11.088
Mitra, A., Han, J., Zhang, Z. J., and Mitra, A. (2009). The intergenic region of Arabidopsis thaliana cab1 and cab2 divergent genes functions as a bidirectional promoter. Planta 229, 1015–1022. doi: 10.1007/s00425-008-0859-1
Müller, F., Demény, M. A., and Tora, L. (2007). New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J. Biol. Chem. 282, 14685–14689. doi: 10.1074/jbc.R700012200
Ogo, Y., Ozawa, K., Ishimaru, T., Murayama, T., and Takaiwa, F. (2013). Transgenic rice seed synthesizing diverse flavonoids at high levels: a new platform for flavonoid production with associated health benefits. Plant Biotechnol. J. 11, 734–746. doi: 10.1111/pbi.12064
Pan, Y., Deng, Y., Lin, H., Kudrna, D. A., Wing, R. A., Li, L., et al. (2014). Comparative BAC-based physical mapping of Oryza sativa ssp. indica var. 93-11 and evaluation of the two rice reference sequence assemblies. Plant J. 77, 795–805. doi: 10.1111/tpj.12412
Pan, Y., Ma, X., Liang, H., Zhao, Q., Zhu, D., and Yu, J. (2015). Spatial and temporal activity of the foxtail millet (Setaria italica) seed-specific promoter pF128. Planta 241, 57–67. doi: 10.1007/s00425-014-2164-5
Peremarti, A., Twyman, R. M., Gómez-Galera, S., Naqvi, S., Farré, G., Sabalza, M., et al. (2010). Promoter diversity in multigene transformation. Plant Mol. Biol. 73, 363–378. doi: 10.1007/s11103-010-9628-1
Sakai, H., Lee, S. S., Tanaka, T., Numa, H., Kim, J., Kawahara, Y., et al. (2013). Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 54, e6. doi: 10.1093/pcp/pcs183
Shah, J. M., Raghupathy, V., and Veluthambi, K. (2009). Enhanced sheath blight resistance in transgenic rice expressing an endochitinase gene from Trichoderma virens. Biotechnol. Lett. 31, 239–244. doi: 10.1007/s10529-008-9856-5
Shi, H., and Chan, Z. (2014). The cysteine2/histidine2-type transcription factor zinc finger of Arabidopsis thaliana 6-activated c-repeat-binding factor pathway is essential for melatonin-mediated freezing stress resistance in Arabidopsis. J. Pineal Res. 57, 185–191. doi: 10.1111/jpi.12155
Shin, R., Kim, M. J., and Paek, K.-H. (2003). The CaTin1 (Capsicum annuum TMV-induced clone 1) and CaTin1-2 genes are linked head-to-head and share a bidirectional promoter. Plant Cell Physiol. 44, 549–554. doi: 10.1093/pcp/pcg069
Singh, A., Sahi, C., and Grover, A. (2009). Chymotrypsin protease inhibitor gene family in rice: genomic organization and evidence for the presence of a bidirectional promoter shared between two chymotrypsin protease inhibitor genes. Gene 428, 9–19. doi: 10.1016/j.gene.2008.09.028
Trinklein, N. D., Aldred, S. F., Hartman, S. J., Schroeder, D. I., Otillar, R. P., and Myers, R. M. (2004). An abundance of bidirectional promoters in the human genome. Genome Res. 14, 62–66. doi: 10.1101/gr.1982804
Uwanogho, D., Yasin, S. A., Starling, B., and Price, J. (2010). The intergenic region between the Mouse Recql4 and Lrrc14 genes functions as an evolutionary conserved bidirectional promoter. Gene 449, 103–117. doi: 10.1016/j.gene.2009.08.011
Vijayan, J., Devanna, B. N., Singh, N. K., and Sharma, T. R. (2015). Cloning and functional validation of early inducible Magnaporthe oryzae responsive CYP76M7 promoter from rice. Front. Plant Sci. 6:371. doi: 10.3389/fpls.2015.00371
Walcher, C. L., and Nemhauser, J. L. (2012). Bipartite promoter element required for auxin response. Plant Physiol. 158, 273–282. doi: 10.1104/pp.111.187559
Wang, C., Ding, D., Yan, R., Yu, X., Li, W., and Li, M. (2008). A novel bi-directional promoter cloned from melon and its activity in cucumber and tobacco. J. Plant Biol. 51, 108–115. doi: 10.1007/BF03030719
Wang, L., Xie, W., Chen, Y., Tang, W., Yang, J., Ye, R., et al. (2010). A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 61, 752–766. doi: 10.1111/j.1365-313X.2009.04100.x
Wang, Q., Wan, L., Li, D., Zhu, L., Qian, M., and Deng, M. (2009). Searching for bidirectional promoters in Arabidopsis thaliana. BMC Bioinformatics 10:S29. doi: 10.1186/1471-2105-10-S1-S29
Wang, R., Lu, L., Pan, X., Hu, Z., Ling, F., Yan, Y., et al. (2015a). Functional analysis of OsPGIP1 in rice sheath blight resistance. Plant Mol. Biol. 87, 181–191. doi: 10.1007/s11103-014-0269-7
Wang, R., Zhu, M., Ye, R., Liu, Z., Zhou, F., Chen, H., et al. (2015b). Novel green tissue-specific synthetic promoters and cis-regulatory elements in rice. Sci. Rep. 5:18256. doi: 10.1038/srep18256
Xu, C., Chen, J., and Shen, B. (2012). The preservation of bidirectional promoter architecture in eukaryotes: what is the driving force? BMC Syst. Biol. 6:S21. doi: 10.1186/1752-0509-6-s1-s21
Xu, L., Ye, R., Zheng, Y., Wang, Z., Zhou, P., Lin, Y., et al. (2010). Isolation of the endosperm-specific LPAAT gene promoter from coconut (Cocos nucifera L.) and its functional analysis in transgenic rice plants. Plant Cell Rep. 29, 1061–1068. doi: 10.1007/s00299-010-0892-y
Yang, M. Q., and Elnitski, L. (2014). Orthology-driven mapping of bidirectional promoters in human and mouse genomes. BMC Bioinformatics 15:S1. doi: 10.1186/1471-2105-15-S17-S1
Yang, M. Q., and Elnitski, L. L. (2008). Prediction-based approaches to characterize bidirectional promoters in the mammalian genome. BMC Genomics 9:S2. doi: 10.1186/1471-2164-9-S1-S2
Yang, S., Sleight, S. C., and Sauro, H. M. (2013). Rationally designed bidirectional promoter improves the evolutionary stability of synthetic genetic circuits. Nucleic Acids Res. 41, e33. doi: 10.1093/nar/gks972
Yang, Z., Chen, H., Tang, W., Hua, H., and Lin, Y. (2011). Development and characterisation of transgenic rice expressing two Bacillus thuringiensis genes. Pest Manag. Sci. 67, 414–422. doi: 10.1002/ps.2079
Ye, R., Zhou, F., and Lin, Y. (2012). Two novel positive cis-regulatory elements involved in green tissue-specific promoter activity in rice (Oryza sativa L. ssp.). Plant Cell Rep. 31, 1159–1172. doi: 10.1007/s00299-012-1238-8
Yi, N., Oh, S. J., Kim, Y. S., Jang, H. J., Park, S. H., Jeong, J. S., et al. (2011). Analysis of the Wsi18, a stress-inducible promoter that is active in the whole grain of transgenic rice. Transgenic Res. 20, 153–163. doi: 10.1007/s11248-010-9400-y
Yu, J., Hu, S., Wang, J., Wong, G. K., Li, S., Liu, B., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. doi: 10.1126/science.1068037
Yuan, T., Li, X., Xiao, J., and Wang, S. (2011). Characterization of Xanthomonas oryzae-responsive cis-acting element in the promoter of rice race-specific susceptibility gene Xa13. Mol. Plant 4, 300–309. doi: 10.1093/mp/ssq076
Zhang, Q. (2007). Strategies for developing green super rice. Proc. Natl. Acad. Sci. U.S.A. 104, 16402–16409. doi: 10.1073/pnas.0708013104
Keywords: rice, bidirectional promoter, stable transformation, GUS assay, GFP assay, deletion analysis, conservation analysis
Citation: Wang R, Yan Y, Zhu M, Yang M, Zhou F, Chen H and Lin Y (2016) Isolation and Functional Characterization of Bidirectional Promoters in Rice. Front. Plant Sci. 7:766. doi: 10.3389/fpls.2016.00766
Received: 13 March 2016; Accepted: 17 May 2016;
Published: 31 May 2016.
Edited by:
Haitao Shi, Hainan University, ChinaReviewed by:
Jiang Zhang, Hubei University, ChinaShiyi Zhou, Hubei University of Education, China
Zhijuan Feng, Zhejiang Academy of Agriculture, China
Copyright © 2016 Wang, Yan, Zhu, Yang, Zhou, Chen and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongjun Lin, yongjunlin@mail.hzau.edu.cn
 Rui Wang
Rui Wang Yan Yan2
Yan Yan2 Menglin Zhu
Menglin Zhu Mei Yang
Mei Yang Fei Zhou
Fei Zhou Yongjun Lin
Yongjun Lin