- 1Departments of Veterinary Microbiology and Preventive Medicine, College of Veterinary Medicine, Iowa State University, Ames, IA, United States
- 2National Risk Assessment Laboratory for Antimicrobial Resistance of Animal Original Bacteria, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 3Veterinary Diagnostic and Production Animal Medicine, College of Veterinary Medicine, Iowa State University, Ames, IA, United States
Campylobacter jejuni is a leading cause of foodborne illnesses worldwide. As a microaerobic pathogen, C. jejuni is subjected to DNA damages caused by various stresses such as reactive oxygen species (ROS) and UV radiations. The base excision repair (BER) system plays an important role in preventing mutations associated with oxidative DNA damage, but the system remains poorly characterized in Campylobacter. In this study, a BER homolog encoded by cj0595c (named nth) in C. jejuni was analyzed for endonuclease III activity and for its role in maintaining genomic stability. It was found that inactivation of nth resulted in elevated frequencies of spontaneous fluoroquinolone-resistant (FQR) and oxidative stress resistant (OXR) mutants, compared with the wild-type strain in C. jejuni. Sequencing analysis of the FQR and OXR mutants revealed that the elevated mutation rates were associated with C → T or G → A transition in gyrA (FQR mutants) or perR (for OXR mutants). In an in vitro assay, a purified recombinant C. jejuni Nth protein demonstrated endonuclease III activity that recognized and excised the thymine glycol (Tg) base from a double stranded DNA. These findings indicate that Nth functions as a BER repair enzyme in C. jejuni and is important for the repair of DNA damage, protecting the bacteria from stresses encountered within a host and in the environment.
Introduction
Campylobacter jejuni, a major bacterial foodborne pathogen, is among the most common causes of bacterial diarrhea in humans worldwide (Nachamkin et al., 2000). According to the United States Centers for Disease Control and Prevention (CDC), C. jejuni is estimated to cause 1.3 million infections and 120 deaths each year in the United States (Centers for Disease Control and Prevention [CDC], 2014). The main source of human Campylobacter infections is via contaminated food, water, or milk (Tauxe, 2002; Kassenborg et al., 2004). To survive in animal hosts and the environment, C. jejuni must be able to deal with various stresses such as reactive oxygen species (ROS) and UV radiations, which may lead to a variety of damages in the DNA structure, including single- or double-strand breaks, nucleotide modifications, abasic sites, bulky adducts, and mismatches (Eisen and Hanawalt, 1999). Bacteria have evolved sophisticated DNA repair mechanisms to defend against DNA damages and to retain their genetic integrity, including direct repair, excision repair and combinational repair, which have been well characterized in Escherichia coli and several other bacterial species (Eisen and Hanawalt, 1999; Friedberg, 2008). However, DNA repair mechanisms remain poorly defined in Campylobacter. It has been revealed through whole genome sequencing, that C. jejuni has an incomplete version of a DNA repair system compared to that found in E. coli (Parkhill et al., 2000; Gundogdu et al., 2007). It lacks important genes involved in DNA repair including lexA, which allows bacteria to mount a SOS response and survive sudden increases in DNA damage (Michel, 2005). MutH and MutL, which are methyl-directed mismatch repair (MMR) enzymes and repair replication errors that arise from mis-incorporations (mismatches) and strand slippage (frameshift errors) (Modrich, 1991), and MutM for base excision repair (BER), are also absent in C. jejuni.
In previous work, Gaasbeek et al. (2009) knocked out several putative DNA repair genes in C. jejuni including mutS, uvrB, ung, and recA and analyzed their effect on the emergence of spontaneous point mutations. None of the knocked-out mutants showed altered spontaneous point mutation frequency in C. jejuni. This is in contrast to findings in other bacteria where, mutations in the mutS loci have been associated with a hypermutability phenotype (Hsieh, 2001; Young and Ornston, 2001). Mutation of ung was also found to mediate increased mutation frequency in Helicobacter pylori (Huang et al., 2006), which is a close relative of C. jejuni. Therefore, the fact that inactivation of the known DNA repair genes did not change the spontaneous mutation rate in Campylobacter, suggests that there may be other mechanisms contributing to the repair of spontaneous point mutations in this organism. Indeed, our previous study revealed that MutY, an adenine glycosylase, which belongs to the BER system, plays an important role in the prevention of spontaneous point mutations, specifically the G → T or C → A transversion, in C. jejuni (Dai et al., 2015), indicating the BER system is functional in C. jejuni.
The BER system detects and repairs modified bases and plays an important role in preventing mutations associated with oxidative DNA damage such as 8-oxoguanine (David et al., 2007; Zharkov, 2008). The function of the BER system starts with the excision of the damaged bases by dedicated enzymes, namely DNA glycosylases (Zharkov, 2008). In E. coli, three major DNA glycosylases have been reported: the Uracil-DNA glycosylase superfamily, the Nth superfamily, and the Fpg/Nei superfamily (Zharkov, 2008). In H. pylori, strains lacking a functional endonuclease III (HpNth) showed slightly increased spontaneous mutation rates and were more susceptible than the parental strain to killing by exposure than to oxidative stress (O’Rourke et al., 2003). However, the function of the BER system has not been fully understood in Campylobacter. It remains unknown if BER enzymes other than MutY function in Campylobacter, affecting the spontaneous mutation rates and adaptating to endogenous or environmental stresses such as antimicrobials, oxidative stress and UV irradiation.
The goal of this study was to investigate the presence and functionality of BER homologs in C. jejuni. Using various methods, we found that the nth gene (cj0595c) encodes Endonuclease III of the BER system and inactivation of nth increased the spontaneous mutation frequencies with the C:G → T:A transition change, thus promoting the emergence of fluoroquinolone resistant (FQR) and oxidative stress resistant (OXR) mutants in C. jejuni. Furthermore, it was found that a purified C. jejuni Nth protein was able to recognize and excise an oxidative damaged base from a double stranded DNA in vitro. The work identifies a new functional BER protein in C. jejuni, providing new insights into the DNA repair and adaptation mechanisms in this organism.
Materials and Methods
Identification of Putative BER Homologs in C. jejuni
The genome sequence of C. jejuni NCTC 11168 was searched for homologs of major BER enzymes of E. coli (Krwawicz et al., 2007; Zharkov, 2008) using the Basic local alignment search tool (BLAST)1. The similarities between the C. jejuni BER homologs and their E. coli equivalents were evaluated according to the query coverage and maximum identity values provided by the BLAST programs after alignment.
Bacterial Strains and Growth Conditions
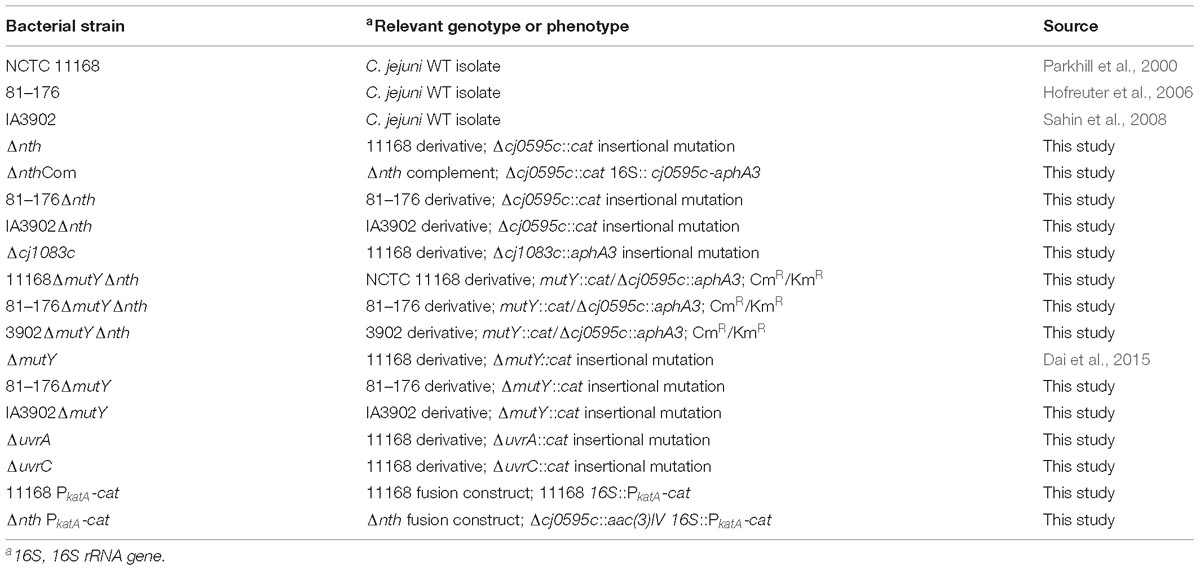
Bacterial strains used in this study are listed in Table 1. C. jejuni was cultured using Mueller-Hinton (MH) broth or agar (Difco) at 42°C under microaerobic conditions in jars filled with premixed gasses (5% O2, 10% CO2, and 85% N2). E. coli was grown on Luria-Bertani (LB) agar or in an LB broth at 37°C for 24 h, under aerobic conditions.
Construction of C. jejuni Knockout Mutants
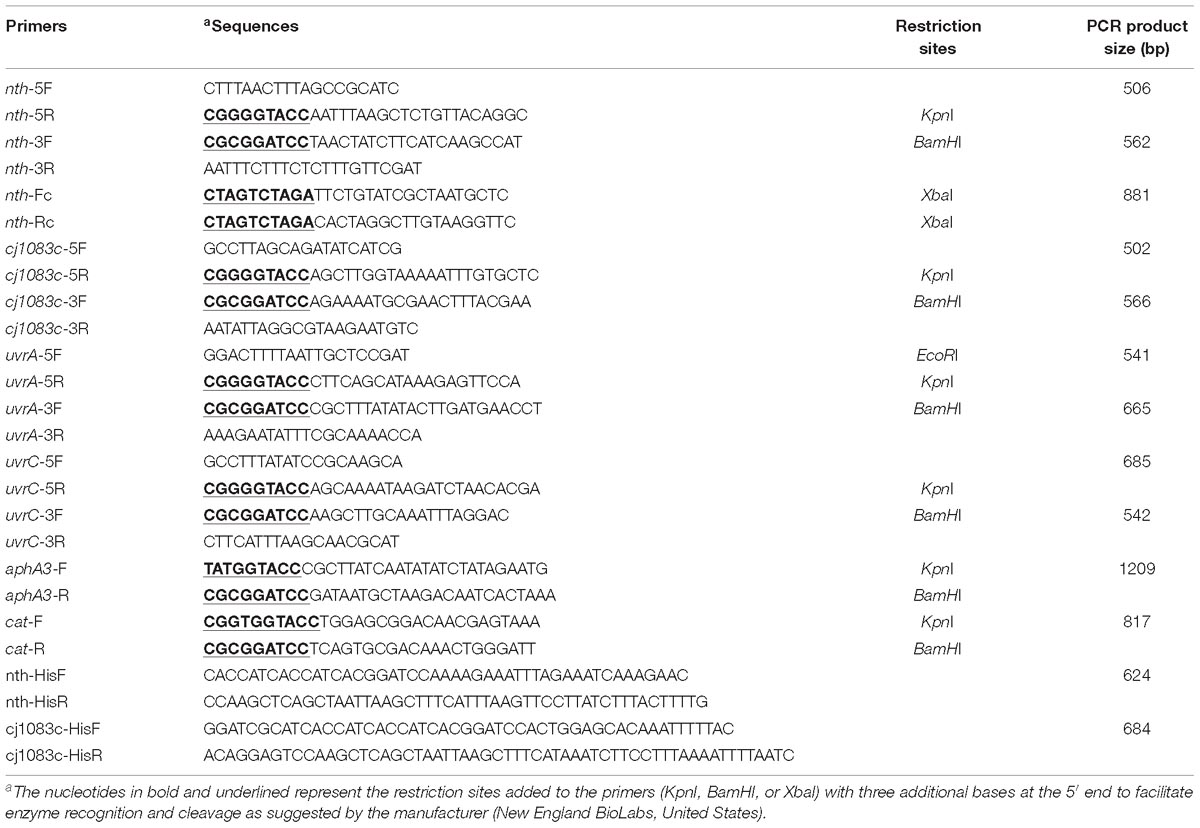
The putative BER genes (nth and cj1083c) and two Nucleotide Excision Repair (NER) genes [uvrA (cj0342c) and uvrC (cj1246c)] of C. jejuni NCTC 11168 were knocked out by insertional mutagenesis. The primers used for gene inactivation are listed in Table 2. To construct a Δnth strain, primers nth-5F and nth-5R were used to amplify a 506 bp fragment containing 5′ part of nth and its upstream region (nth-5′ fragment), while primers nth-3F and nth-3R were used to amplify a 562 bp fragment containing 3′ part of nth and its downstream region (nth-3′ fragment) (Supplementary Figure S1). Primer pairs aphA3-F/aphA3-R or cat-F/cat-R were used to amplify the aphA3 (1,209 bp) or cat gene (817 bp) from pMW10 or pRY112 encoding kanamycin or chloramphenicol resistance, respectively, using the Phusion High-Fidelity DNA Polymerase (New England Biolabs). After KpnI and BamHI digestion, the nth-5′, cat and nth-3′ PCR fragments were ligated by the T4 DNA ligase (New England Biolabs) and PCR amplified utilizing nth-5F and nth-3R primers, resulting in the construction of the nth-5′-cat-nth-3′ PCR product. The purified nth-5′-cat-nth-3′ product was then electroporated into C. jejuni NCTC 11168. Transformants were selected on MH agar plates containing 10 mg/L of chloramphenicol. To construct a ΔmutYΔnth double knockout strain, a similar nth-5′-aphA3-nth -3′ PCR product was constructed as described for Δnth above. The purified nth-5′-aphA3-nth-3′ product was then electroporated into the C. jejuni ΔmutY strain, which was constructed in our previous study (Dai et al., 2015). Transformants were selected on MH agar plates containing both 30 mg/L kanamycin and 10 mg/L of chloramphenicol. The insertion of the resistance marker into the nth and mutY genes was confirmed by PCR using primers nth-5F/nth-3R (Table 2) and mutY-5F/mutY-3R (Dai et al., 2015). A similar strategy was used to knockout mutants Δcj1083c, ΔuvrA, and ΔuvrC in C. jejuni strains NCTC 11168, 81–176 and IA3902.
Complementation of the C. jejuni Mutant Strains
The Δnth mutant was complemented by inserting a wild-type copy of the nth gene between the 16S and 23S rRNAs as described by Muraoka and Zhang (2011). The primers used are listed in Table 2. Briefly, primers nth-Fc and nth-Rc were used to amplify the intact nth gene including its promoter. The amplicon was digested with XbaI and cloned into the pRRK plasmid, which contains an aphA3 cassette in the opposite orientation to the ribosomal genes, to obtain plasmid construct pRRK-nth. The direction of the insertion was confirmed by primers nth-Fc and aphA3-F/aphA3-R. The construct with nth in the same transcriptional direction as the ribosomal genes was selected and used as the suicide vector to insert the nth gene into the chromosome of the Δnth strain. The complemented strain named ΔnthCom was selected on MH agar containing 30 μg/ml of kanamycin and was confirmed by PCR using primers nth-Fc and nth-Rc.
Assay of Spontaneous FQR Mutation Frequencies
To determine the spontaneous FQR mutation frequencies in Campylobacter, the methods of Bjorkholm et al. (2001) and Hanninen and Hannula (2007) were used with minor modifications. Briefly, bacterial strains (FQ-susceptible) were grown in 3 ml of MH broth for 24 h, after which 30 μl aliquots (∼105 CFU) of each strain were distributed into 20 tubes containing 3 ml of MH broth. Cultures were incubated with shaking for 24 h at 42°C. After incubation, colony counts (CFU) of evolved mutants in each tube were determined by spreading 1.5 ml on MH agar plates containing 1 μg/mL ciprofloxacin (10 × MIC). The number of total viable bacteria was determined from three tubes by dropping 10 μl of 10−4, 10−5, and 10−6 dilutions on non-selective MH agar plates. After 2–4 days of incubation in a microaerobic atmosphere at 42°C, colonies were counted. The frequency of resistant mutants was expressed as the median number of resistant colonies divided by the mean of the total number of viable cells. The mutation frequencies were calculated from the median of 20 cultures for the respective strain. Thus, the occasional jackpot cultures had no impact on the calculation and the obtained values (Bjorkholm et al., 2001). Several colonies growing on selective MH plates from the spontaneous FQR mutation frequency test were randomly picked to sequence the QRDR region in the gyrA gene. To ensure that the 30 μl of bacterial cultures used to inoculate the 3 ml broth samples were free of any preexisting resistant mutants, the same volume (30 μl) was also plated on a selective plate. The mutation frequency of each strain was measured only if no preexisting mutants were present in the 30 μl aliquots used as inoculum.
Determination of Spontaneous OXR Mutation Frequencies
It was not possible to determine the spontaneous OXR mutation frequency directly by using H2O2 as a selection agent, since this compound is not stable and tends to degrade rapidly in the culture medium. Therefore, a reporter system was developed by fusing the promoter of katA (an oxidative stress response gene) with the chloramphenicol resistance gene cat, generating PkatA-cat gene (Dai et al., 2017). This method was utilized in this study to compare spontaneous OXR mutation frequencies of the wild-type and the Δnth mutant strains. Since the fusion gene PkatA-cat plasmid construct harbors both aphA3 and cat resistance cassettes, an additional resistance cassette aac(3)IV, which contributes to apramycin resistance, was utilized to construct a new nth mutant in the 11168 isolate as described in a previous report (Cameron and Gaynor, 2014), generating Δnth::aac(3)IV. The PkatA-cat gene was then inserted into the genome of C. jejuni NCTC 11168 and Δnth::aac(3)IV isolates, generating 11168 PkatA-cat and Δnth PkatA-cat constructs. Similar to C. jejuni NCTC 11168, both constructs were susceptible to chloramphenicol (MIC = 1–2 μg/mL) as transcription of the PkatA-cat gene was inhibited by the oxidative stress regulator PerR. However, spontaneous perR mutations, which compromise the PerR function, would lead to the derepression of the katA promoter of PkatA-cat and consequently elevate the MIC of chloramphenicol (16–32 μg/mL) of the PerR mutants from both 11168 PkatA-cat and Δnth PkatA-cat (Dai et al., 2017). Therefore, MH agar plates containing 6 μg/mL of chloramphenicol were used in this study to measure spontaneous perR loss-of-function mutations in C. jejuni 11168 PkatA-cat and Δnth PkatA-cat. The spontaneous chloramphenicol resistance (CmR) mutation frequencies were determined as previously described for enumerating spontaneous ciprofloxacin and ampicillin resistance mutations in C. jejuni (Dai et al., 2015). Colonies growing on selective MH plates from the spontaneous CmR mutation frequency test were randomly picked to sequence the perR gene. In addition, these colonies were sub-cultured and tested for their susceptibility to H2O2 using disk diffusion assay as described below.
Oxidant Susceptibility Assay
A disk inhibition assay was utilized to compare the sensitivity to oxidants between C. jejuni WT and the OXR mutants as described previously (Palyada et al., 2009). Each C. jejuni strain was tested for susceptibility to 3% H2O2, 3% cumene hydroperoxide in DMSO, and 100 mM paraquat dichloride in H2O, respectively.
Assay of Susceptibility to UV Induced DNA Damage
Previous studies found that the disruption of the Nucleotide Excision Repair (NER) gene uvrB resulted in a significant reduction in viability after exposure to UV irradiation in C. jejuni, due to a lack of repair of bulky DNA damages induced by UV irradiation (Gaasbeek et al., 2009). To determine if C. jejuni BER genes were involved in the repair of bulky DNA damages, such as pyrimidine dimers induced by UV irradiation, C. jejuni wild-type 11168 and several DNA repair gene mutants were tested for susceptibility to UV irradiation. The methods of Gaasbeek et al. (2009) was utilized with minor changes. Overnight C. jejuni cultures grown on MH agar plates were suspended in MH broth to an OD600 of 0.2. Two hundred μl of each suspension was added into a 24-well cell culture plate (3 replicates/isolate), which was then placed in a UV cross-linker and exposed to UV irradiation at 312 nm. The irradiation was done with a density of 0.12 J cm−2 for 1 min. Then, serial dilutions (100–106) of UV-exposed and non-exposed cells were dropped on MH plates for enumeration of viable cells. After 24–48 h of incubation, the numbers of colonies were counted. All tests were performed in triplicates. The results were analyzed using a one-way ANOVA followed by a Dunnett’s test using the GraphPad Prism software.
Expression and Purification of rNth and rCj1083c From E. coli
The BER enzyme Nth in E. coli is known for its ability to remove damaged pyrimidines from double-stranded DNA, especially the Thymine glycol (Tg) (Saito et al., 1997), which is induced by oxidative damage and has been shown to be a lethal lesion in vitro and in vivo (Hariharan et al., 1977; Clark and Beardsley, 1986). To investigate if the Nth and Cj1083c proteins in C. jejuni repairs oxidative DNA damages, full-length histidine-tagged recombinant Nth (rNth) and Cj1083c (rCj1083c) of C. jejuni were produced in E. coli JM109 strain by using the pQE-30 vector (Qiagen). The complete coding sequences of nth and cj1083c gene in the isolates were amplified with primers nth-HisF/nth-HisR and cj1083c-HisF/cj1083c-HisR (Table 2). The amplified PCR products were ligated into the pQE-30 vector, which had been previously digested with BamHI and HindIII, utilizing the SLiCE cloning method (Zhang et al., 2012). The plasmid in the E. coli clones producing the Nth or Cj1083c proteins were sequenced, confirming the cloned constructs. E. coli harboring pQE-30-nth or pQE-30-cj1083c were grown in LB broth at 37°C with shaking at 250 rpm to an OD600 of 0.4–0.6. The expression of recombinant Nth and Cj1083c were induced by addition of 0.4 mM IPTG for 5 h at 28°C with shaking at 200 rpm. Purification of the rNth and Cj1083c proteins were performed following the procedures described previously (Zhang et al., 2000; Lin et al., 2002). The purified rNth and Cj1083c were desalted using PD-10 Desalting Columns (GE Healthcare Life Sciences, United States) and then stocked in 50% glycerol solution containing 20 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, and 1 mM 1,4-Dithiothreitol (DTT) at −20°C.
Oligonucleotide Substrate Preparation for Endonuclease III Activity Assay
A previously reported 30-mer oligonucleotide substrate oligo30-F-Tg, containing a Tg residue at position 13 (GATCCTCTAGAGTgCGACCTGCAGGCATGCA) (Marenstein et al., 2003), was synthesized and PAGE purified by Integrated DNA Technologies (United States). The 30-mer oligonucleotide oligo30-F containing a Thymine (T) of the identical sequence and the complementary oligonucleotide oligo30-R containing a mismatch base Guanine (G) against Tg or T were also synthesized. Nucleotides were annealed to the complementary strand according to the manufacturer’s instructions, which generated two double-strand DNA fragments nth-Tg-DNA and nth-control-DNA, containing the Tg:G or T:G pairs, respectively. These two DNA fragments were stocked at −20°C for subsequent endonuclease III activity assay.
Endonuclease III Activity Assay
Purified rNth and rCj1083c of C. jejuni were assayed for endonuclease III activity. A commercially available Nth from E. coli (New England Biolabs) was included as a positive control. An rPerR protein (Dai et al., 2017), which has no endonuclease III activity and was produced and purified from the same E. coli host with the same methods as for rNth and rCj1083c in this study, was used as a negative control. The endonuclease III activity assay was carried out following the manufacturer’s instructions. Briefly, 1 pmol of nth-Tg-DNA or nth-control-DNA was treated with either 1 μl (10 U) E. coli Nth, or C. jejuni rNth (2 pmol), rCj1083c (2 pmol) and rPerR (2 pmol) for overnight at 37°C in 1 X NEB reaction buffer containing 1 mM DTT, 20 mM Tris-HCl, and 1 mM EDTA (pH 8.0). The reaction mixture (10 μl) was then mixed with an equal volume of Gel loading Buffer II (Thermo Fisher Scientific, United States), followed by heating at 95°C for 5 min to denature any secondary DNA structure to generate single stranded DNA. Samples were then separated by 15% Denaturing urea polyacrylamide gel electrophoresis (Urea PAGE) in 7 M urea and 1 × Tris borate-EDTA (TBE) buffer at 200 V for 1 h. The Urea PAGE gel was stained using SYBR Gold Nucleic Acid Gel Stain (Thermo Fisher Scientific, United States). Gel images were taken with a digital imaging system under UV light at 254 nm.
Results
Identification of Putative BER Genes in C. jejuni
BLAST search of the C. jejuni NCTC11168 genome for BER genes did not identify any fpg or nei homologs in the genome. Searching for annotations of DNA glycosylases in C. jejuni revealed that cj0086c, nth, and cj1620c genes are likely to encode C. jejuni Uracil-DNA glycosylase (Ung), endonuclease III (Nth), and adenine DNA glycosylase (MutY), respectively (Table 3). Interestingly, the cj1083c gene was also predicted and annotated as an endonuclease III encoding gene in the NCTC 11168 genome, even though its protein sequence shares a relatively lower homology (30%) to Nth in E. coli. Further alignment of amino acid sequences of E. coli endonuclease III, C. jejuni Nth, and C. jejuni Cj1083c by PROMALS3D (Xia et al., 2009) revealed that all three proteins contain conserved alpha-helix secondary structures (Supplementary Figure S2). Deduced from the crystal structure of E. coli endonuclease III (Thayer et al., 1995), C. jejuni Nth and Cj1083c harbor a relatively conserved Helix-hairpin-helix (HhH) motif that may be involved in DNA binding (Supplementary Figure S2). However, amino acids that are predicted to be important for Nth functions in E. coli are less conserved in C. jejuni Cj1083c (Supplementary Figure S2). For example, Asp120 in E. coli Nth, which is important for the DNA lyase activity, was only found in C. jejuni Nth (Asp118) but not at the corresponding site in Cj1083c (Glu126). Gln41 in E. coli Nth, which may be important for DNA binding, is also conserved in C. jejuni Nth (Gln39) but not in Cj1083c (Asn42). Furthermore, the alignment showed that C. jejuni Cj1083c lacks an iron/sulfur cluster region, which was proposed to have a structural role for the DNA repair (Guan et al., 1998; Johnson et al., 2005). Among these genes, cj0086c (ung) and cj1620c (mutY) has been functionally characterized in Campylobacter (Gaasbeek et al., 2009; Dai et al., 2015). However, the role of nth and cj1083c in DNA repair has not been determined and is thus investigated in this study.
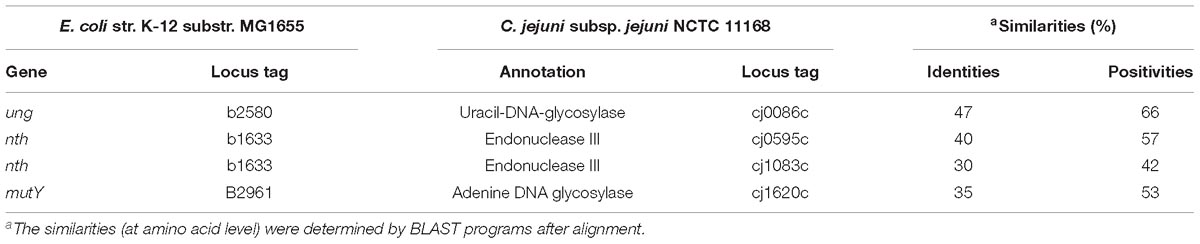
Table 3. BER gene orthologs identified in C. jejuni NCTC 11168 genome in comparison with those in E. coli K-12.
Elevated Spontaneous FQR Mutation Frequency in the Δnth Mutant but Not in the Δcj1083c Mutant
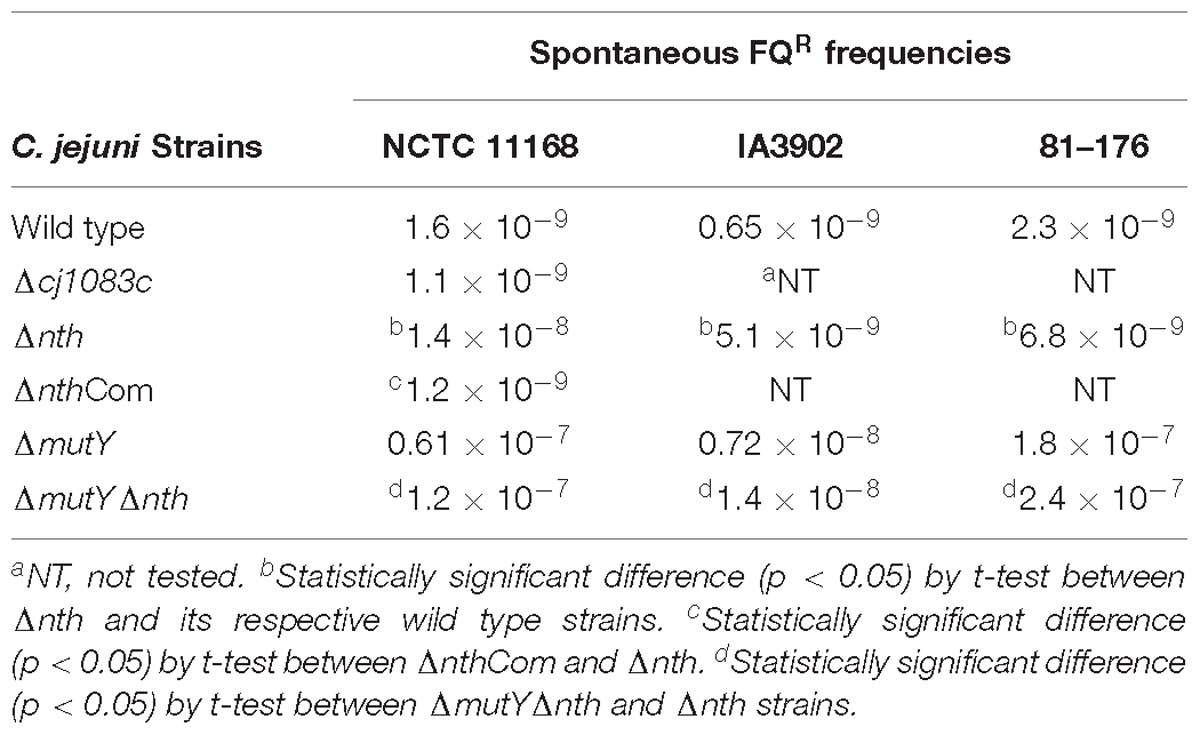
The nth and cj1083c genes were knocked out to determine their impact on frequencies of spontaneous ciprofloxacin resistant mutants (due to point mutations in the gyrA gene) (Table 4). Compared to the wild-type 11168, disruption of cj1083c did not cause any change in the FQR mutation frequency. However, inactivation of nth resulted in 3 to 10-fold (p < 0.05, t-test) increase in the frequencies of FQR mutants in three different C. jejuni strains including 11168, IA3902 and 81–176 (Table 4). Complementation of Δnth with an intact copy of the nth gene from the 11168 strain, restored the spontaneous FQR mutation frequencies to the wild type level (Table 4). Interestingly, the C. jejuni double knockout strain ΔmutYΔnth exhibited a further increase of the FQR mutation frequency compared with the Δnth strain (Table 4), but the increase was comparable to that from the MutY mutant alone, as reported in the previous study (Dai et al., 2015), suggesting that the mutY effect is dominant, which overshadows the impact of the nth mutation on the spontaneous FQR mutation frequencies in Campylobacter. Several FQR colonies, grown on selective plates, were picked from the Δnth background and subsequently sequenced for the QRDR region in the gyrA gene. The mutants harbored a C257 → T (Thr-86-Ile) substitution or a G268 → A (Asp-90-Asn) transition and had MIC values between 8 and 16 mg/L. Altogether, the results indicated that the Δnth mutation, but not Δcj1083c mutation, increased the spontaneous FQR mutation frequency in C. jejuni.
Elevated Spontaneous OXR Mutation Frequency in the Δnth Mutant
To further examine the role of nth in DNA repair, we determined its effect on the spontaneous OXR frequency, which is due to spontaneous loss-of-function mutations in the peroxide regulator perR in C. jejuni (Dai et al., 2017). Previously, we used a promoter-reporter fusion gene PkatA-cat to determine the spontaneous OXR mutation frequencies in C. jejuni (Dai et al., 2017). The fusion gene PkatA-cat was then inserted into the genome of C. jejuni 11168 and Δnth strains, generating 11168 PkatA-cat and Δnth PkatA-cat strains. The spontaneous chloramphenicol resistance (CmR) mutation frequencies were determined by plating on chloramphenicol-containing plates. The average CmR mutation frequency for the Δnth PkatA-cat isolate was 1.1 × 10−7, ∼6-fold higher than that of the C. jejuni 11168 PkatA-cat isolate (1.9 × 10−8) (P < 0.05; t-test). Colonies grown on chloramphenicol plates were randomly picked from both 11168 PkatA-cat or Δnth PkatA-cat and subsequently sequenced for the perR gene. All sequenced CmR colonies carried mutations in the perR gene (Table 5). For strain 11168 PkatA-cat, no unique mutation pattern was observed. However, all colonies sequenced from Δnth PkatA-cat carried a C → T or G → A transition in the PerR encoding sequence. These transition mutations led to amino acid (aa) changes or formation of a stop codon in PerR, which caused early termination of PerR translation. In addition, four CmR colonies from the Δnth PkatA-cat carried the same C250T mutation causing aa change at 84His in PerR, which is the metal binding site and essential for the regulatory function of PerR (Pohl et al., 2003; Lee and Helmann, 2007; Lucarelli et al., 2007). The result indicated that the CmR mutants generated from the Δnth strain had distinct perR mutation patterns, which are G:C → A:T transitions. This is consistent with the result found in E. coli, for which a previous study showed that mutation of Nth caused G:C → A:T transitions due to lack of repair of the pyrimidine oxidative damages in the E. coli mutants (Blaisdell et al., 1999). The spontaneous CmR mutants of both 11168 PkatA-cat and Δnth PkatA-cat strains were also assayed for susceptibility to different oxidants by a disk inhibition assay. As shown in Table 6, the CmR colonies were highly resistant to the oxidant treatments. Representative pictures of the disk inhibition results are shown in Supplementary Figure S3. This result is consistent with what we found in our previous work that the CmR phenotype corresponded with the OXR phenotype in C. jejuni strain carrying the fusion gene PkatA-cat (Dai et al., 2017). Altogether, these findings indicate that the Δnth mutation increased the loss-of-function mutations (C → T or G → A transition) in perR, leading to a PerR malfunction and a subsequent derepression of the PerR-controlled genes, which in turn led to the CmR and OXR phenotypes in C. jejuni.
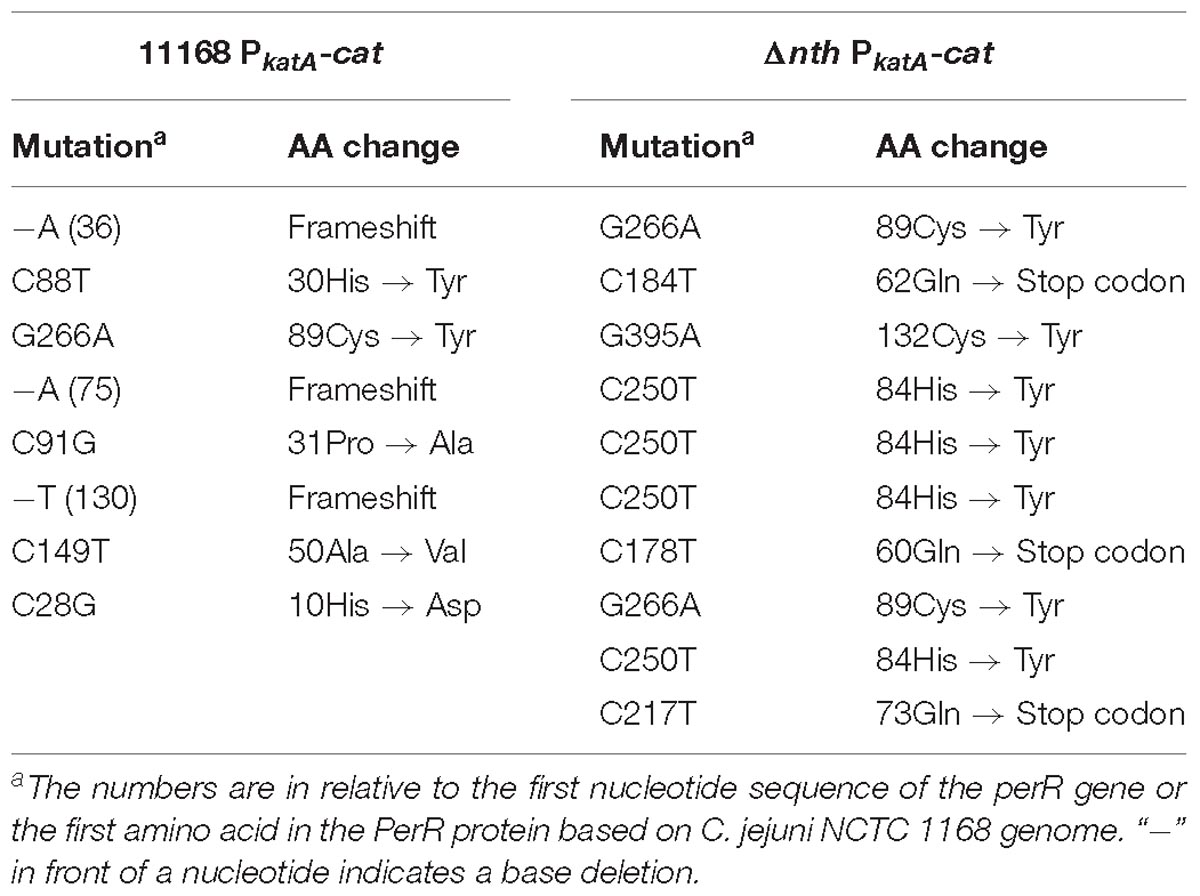
Table 5. perR mutations identified in the spontaneous OXR mutants from C. jejuni 11168 PkatA-cat and Δnth PkatA-cat isolates.
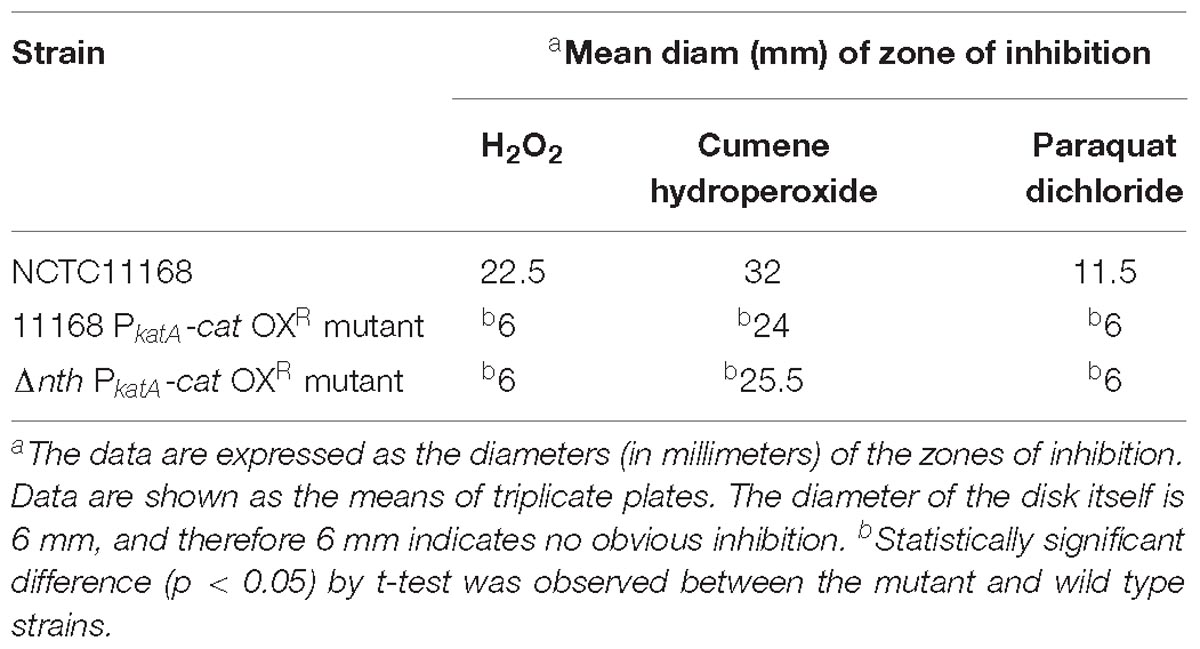
Table 6. Oxidative stress sensitivity of C. jejuni strains and constructs as measured by a disk diffusion assay.
C. jejuni Nth but Not Cj1083c Recognizes and Excises Thymine Glycol (Tg) Base From Double Stranded DNA
rNth and rCj1083c were successfully purified from E. coli with predicted molecular weights 25.03 and 27.79 kDa, respectively (Figure 1A). Figure 1B shows the activity of C. jejuni rPerR, rCj1083c, rNth, and E. coli Nth on two 30 bp double-strand DNA fragments: nth-Tg-DNA and nth-control-DNA, which contained the Tg:G or T:G pairs, respectively. As expected, E. coli Nth showed obvious endonuclease III activity on the nth-Tg-DNA fragment, which was shown by a cleaved fragment visible on the gel. Similarly, the C. jejuni rNth protein also exhibited endonuclease III activity on the nth-Tg-DNA fragment (Figure 1B). No cleaved product was observed for the nth-control-DNA fragment after treatment with either E. coli Nth or C. jejuni rNth. The negative control (rPerR) and rCj1083c did not yield a visible cleaved fragment on either of the nth-Tg-DNA or nth-control-DNA, suggesting that rCj1083c may not function as an endonuclease III in C. jejuni. The above results indicate that the C. jejuni Nth protein, instead of Cj1083c, recognizes and excises Tg base in the Tg:G pair instead of the T:G mismatch pair. The results also suggest that Nth is important for the repair of oxidative DNA damages in Campylobacter.
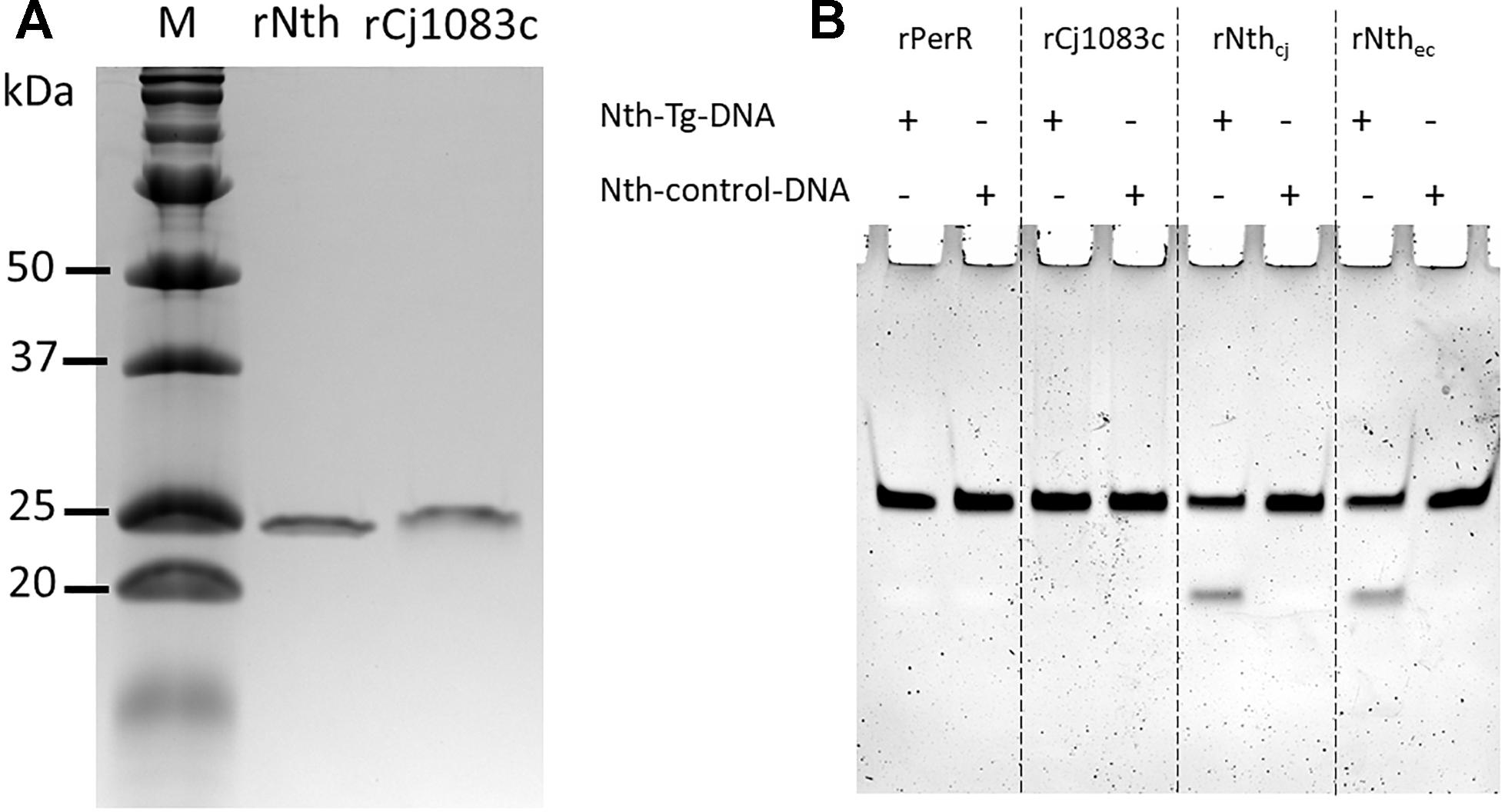
Figure 1. Production of the C. jejuni rNth and rCj1083c proteins in E. coli and evaluation of their Endonuclease III activities in vitro. (A) SDS-PAGE analysis of C. jejuni rNth and rCj1083c expressed in E. coli. Lane M contains molecular mass markers (Bio-Rad), while lane rNth or rCj1083c shows rNth or rCj1083c of C. jejuni NCTC 11168 purified by Ni-nitrilotriacetic acid affinity chromatography, respectively. (B) Urea PAGE analysis of endonuclease III activities of C. jejuni rPerR, rCj1083c, rNth (rNthcj) and E. coli Nth (rNthec), proteins. The ‘+’ and “–” signs indicate the presence and absence of the assayed DNA, respectively.
The BER Genes in C. jejuni Are Not Involved in the Bulky DNA Damage Repair
To evaluate if the BER genes are also involved in repairing bulky DNA damages, mutY and nth were assayed for their effect on susceptibility of C. jejuni to UV irradiation in this study. Two NER genes uvrA and uvrC, which encode a multienzyme complex UvrABC with uvrB gene, were included as positive controls in this study. As shown in Figure 2, knockout of either the uvrA or uvrC gene caused 104–106-fold reductions in viability compared to the wild type strain after UV irradiation (p < 0.05). However, the ΔmutY, Δnth mutant strains did not show a significant difference (p > 0.05) in viability compared to that of the C. jejuni 11168 wild type strain. These results indicate that in C. jejuni, the BER genes mutY and nth are not involved in repairing DNA damages caused by UV irradiation.
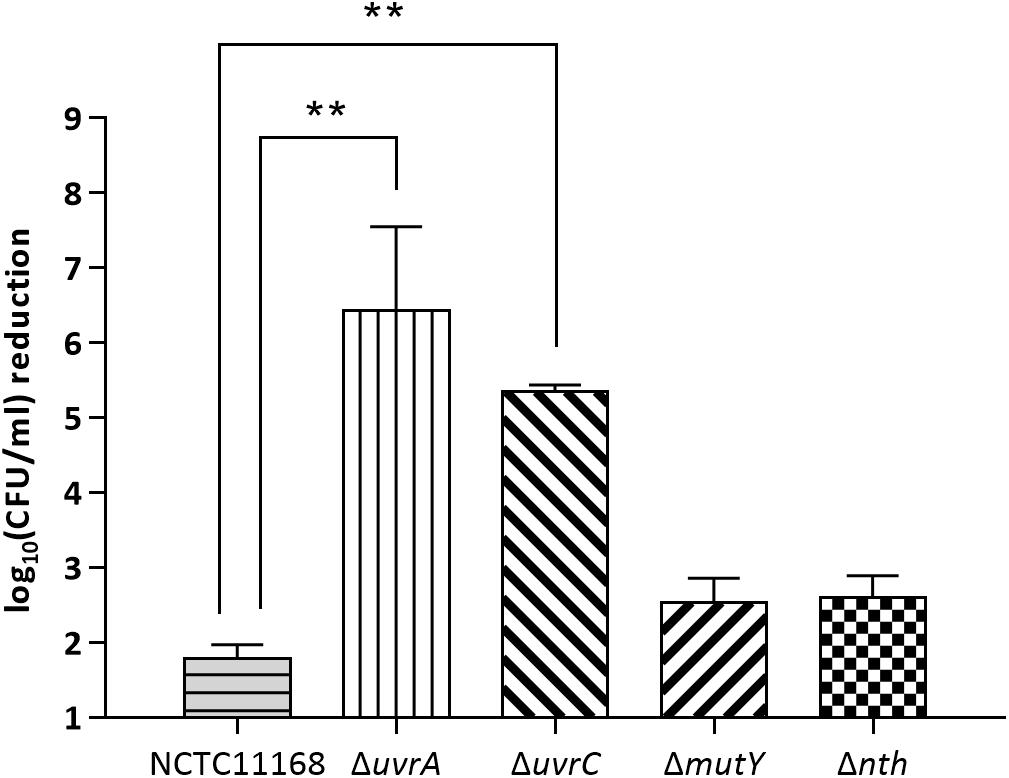
Figure 2. UV-mediated reduction of cell viability in C. jejuni NCTC 11168 and various mutant strains. The bars indicate standard errors of triplicate experiments. Statistically significant differences from the wild-type NCTC 11168 are marked by double asterisks (∗∗) (p < 0.01).
Discussion
Although the genome of C. jejuni harbors homologs of the excision repair systems, including NER and BER (Parkhill et al., 2000; Gaasbeek et al., 2009), most of them remain functionally uncharacterized. In this study two BER homologs, nth (cj0595c) and cj1083c, in C. jejuni were analyzed. It was revealed that inactivation of nth, but not cj1083c, increased FQR and OXR mutation frequencies, which were attributable to mutations in either the gyrA gene (related to the FQR phenotype) or the perR gene (related to the OXR phenotype). Notably, all of the spontaneous point mutations in the gyrA or perR genes of FQR or OXR mutants from the Δnth background are G:C → A:T transitions, which are regarded as a signature mutation pattern for Nth deficiency in E. coli (Blaisdell et al., 1999). Endonuclease III activity assay further revealed that the purified recombinant Nth protein recognized and excised Tg containing double-stranded DNA (Figure 1B). Altogether, these results indicate that Cj0595c functions as an Endonuclease III and is an Nth ortholog that is important for DNA damage repair in Campylobacter.
In other bacteria, the endonuclease III (Nth) protein, together with endonuclease VIII (Nei), is responsible for repairing oxidized pyrimidines including Tg, which would cause G:C → A:T transitions in E. coli (Wang et al., 1998; Blaisdell et al., 1999). However, there are no endonuclease VIII homologs in the C. jejuni genome (Parkhill et al., 2000). Our results showed that inactivation of Nth elevated the mutation rate with the C → T or G → A transition in Campylobacter. This was evident from the observation that all the FQR mutants of the Nth mutant strain carried C257 → T or G268 → A transition in the QRDR region of gyrA and that all the spontaneous OXR mutants of the Nth mutant strain carried a C → T or G → A transition in the perR gene (Table 5). These results suggest that Nth functions effectively in Campylobacter despite the absence of a Nei homolog. Whether there are other DNA repair enzymes involved in the repair of oxidized pyrimidines in Campylobacter remains to be investigated in further studies.
We previously reported that the mutation of the BER protein MutY caused a >100 fold increase in spontaneous FQR and OXR mutation frequencies in C. jejuni (Dai et al., 2015, 2017), which is much higher than the increase of mutation frequencies caused by the Nth mutation (3–10-fold) as observed in the current study (Table 4). Similar results were also observed with E. coli and H. pylori, in which the Nth mutant only showed a mild mutator phenotype (Gifford and Wallace, 2000; O’Rourke et al., 2003). The difference between MutY and Nth in their impact on spontaneous mutation frequencies could be explained by the fact that they repair different types of mutations. Regardless, the successful complementation of the Δnth strain, with an intact nth gene to the wild-type level mutation frequency and the uniformed mutation patterns (G:C → A:T transitions) observed in the spontaneous FQR and OXR mutants of the Δnth mutant strains, clearly indicate that the increase in the spontaneous mutation frequencies is due to the Nth mutation in C. jejuni.
The mutations caused by MutY deficiency are predominantly G:C → T:A transversions in C. jejuni (Dai et al., 2015, 2017). This is due to the fact that mutY encodes an adenine glycosylase that specifically repairs the G:C → T:A transversions, which are caused by adenine/7,8-dihydro-8-oxoguanine (8-oxoG) mismatches (Cheng et al., 1992; Michaels and Miller, 1992). Tg and 8-oxoG are two of the most common DNA lesions that result from ROS or ionizing radiation (Basu et al., 1989; Kanvah et al., 2010). The ability of Nth and MutY to repair these DNA lesions, associated with oxidative damage, suggests that the BER system plays an important role in repairing oxidative DNA damages in Campylobacter, facilitating its adaptation in the host and environment.
Both the previous study (Dai et al., 2015) and the current work clearly demonstrates that Nth and MutY are responsible for fixing point mutations in Campylobacter. However, neither Nth nor MutY deficiency affected C. jejuni susceptibility to UV irradiation, which generates bulky DNA damage. On the contrary, inactivation of uvrA or uvrC significantly reduced the viability of C. jejuni after UV irradiation (104–106 fold) (Figure 2). This is consistent with a previous finding that uvrB mutation in C. jejuni drastically decreased the cell viability after a brief UV irradiation (Gaasbeek et al., 2009). These results suggest that C. jejuni requires the UvrABC system to repair UV induced bulky DNA damages. Thus, the BER and NER pathways in C. jejuni appear to have distinct and complementary functions, with the former functioning to repair single base changes and the latter fixing bulky DNA damages such as pyrimidine dimers that affect overall DNA structure.
Although Cj1083c is also annotated as endonuclease III in the genome of C. jejuni NCTC 11168 and shares 30% aa homology with the Nth in E. coli, mutation of this gene did not alter the spontaneous mutation frequencies in C. jejuni. Additionally, an in vitro enzymatic assay confirmed that rCj1083c lacks endonuclease III activity (Figure 1B). This is also consistent with the bioinformatics analysis (Supplementary Figure S2), which revealed that Cj1083c lacks an iron/sulfur cluster region important for DNA binding in E. coli Nth (Thayer et al., 1995). Although Cj1083c shares a general alpha-helix secondary structure with E. coli Nth and C. jejuni Nth, the key amino acids involved in DNA binding are less conserved. These observations suggest that Cj1083c may have a different function other than that of an endonuclease III. In H. pylori, the Cj1083c homolog, HP0602 (44% aa identify), was found to encode a novel 3-Methyladenine DNA Glycosylase (MagIII) within the Endonuclease III family (O’Rourke et al., 2000). MagIII was found to be able to recognize and release 3-methyladenine, instead of oxidized bases from modified DNA. Whether Cj1083c in C. jejuni has a similar function than a DNA repair enzyme in H. pylori requires further investigation.
In summary, the findings of the current study reveals a previously undefined Nth-like protein (encoded by cj0595c) involved in DNA repair in C. jejuni. This protein functions as Endonuclease III and repairs damaged pyrimidines in DNA. The presence of functional BER repair mechanisms, including MutY and Nth, protects C. jejuni cells from oxidative DNA damages encountered frequently within a host and in the environment.
Author Contributions
LD and QZ contributed conception and design of the study. LD wrote the first draft of the manuscript. LD, JX, OS, and QZ contributed to manuscript revision, read and approved the submitted version.
Funding
This work was supported by the Frank Ramsey Endowment Fund from Iowa State University. JX was supported by a fellowship from the Graduate Student Overseas Study Program of South China Agricultural University (Grant No. 2017LHPY030).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00698/full#supplementary-material
Footnotes
References
Basu, A. K., Loechler, E. L., Leadon, S. A., and Essigmann, J. M. (1989). Genetic effects of thymine glycol: site-specific mutagenesis and molecular modeling studies. Proc. Natl. Acad. Sci. U.S.A. 86, 7677–7681. doi: 10.1073/pnas.86.20.7677
Bjorkholm, B., Sjolund, M., Falk, P. G., Berg, O. G., Engstrand, L., and Andersson, D. I. (2001). Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. U.S.A. 98, 14607–14612. doi: 10.1073/pnas.241517298
Blaisdell, J. O., Hatahet, Z., and Wallace, S. S. (1999). A novel role for Escherichia coli endonuclease VIII in prevention of spontaneous G– > T transversions. J. Bacteriol. 181, 6396–6402.
Cameron, A., and Gaynor, E. C. (2014). Hygromycin B and apramycin antibiotic resistance cassettes for use in Campylobacter jejuni. PLoS One 9:e95084. doi: 10.1371/journal.pone.0095084
Centers for Disease Control and Prevention [CDC] (2014). Antibiotic Resistance Threats in the United States, 2013. Atlanta, GA: US Department of Health and Human Services, 61–62.
Cheng, K. C., Cahill, D. S., Kasai, H., Nishimura, S., and Loeb, L. A. (1992). 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G—-T and A—-C substitutions. J. Biol. Chem. 267, 166–172.
Clark, J. M., and Beardsley, G. P. (1986). Thymine glycol lesions terminate chain elongation by DNA polymerase I in vitro. Nucleic Acids Res. 14, 737–749. doi: 10.1093/nar/14.2.737
Dai, L., Muraoka, W. T., Wu, Z., Sahin, O., and Zhang, Q. (2015). A single nucleotide change in mutY increases the emergence of antibiotic-resistant Campylobacter jejuni mutants. J. Antimicrob. Chemother. 70, 2739–2748. doi: 10.1093/jac/dkv190
Dai, L., Sahin, O., Tang, Y., and Zhang, Q. (2017). A mutator phenotype promoting the emergence of spontaneous oxidative stress-resistant mutants in Campylobacter jejuni. Appl. Environ. Microbiol. 83:e01685-17. doi: 10.1128/AEM.01685-17
David, S. S., O’Shea, V. L., and Kundu, S. (2007). Base-excision repair of oxidative DNA damage. Nature 447, 941–950. doi: 10.1038/nature05978
Eisen, J. A., and Hanawalt, P. C. (1999). A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 435, 171–213. doi: 10.1016/S0921-8777(99)00050-6
Friedberg, E. C. (2008). A brief history of the DNA repair field. Cell Res. 18, 3–7. doi: 10.1038/cr.2007.113
Gaasbeek, E. J., van der Wal, F. J., van Putten, J. P., de Boer, P., van der Graaf-van Bloois, L., de Boer, A. G., et al. (2009). Functional characterization of excision repair and RecA-dependent recombinational DNA repair in Campylobacter jejuni. J. Bacteriol. 191, 3785–3793. doi: 10.1128/JB.01817-08
Gifford, C. M., and Wallace, S. S. (2000). The genes encoding endonuclease VIII and endonuclease III in Escherichia coli are transcribed as the terminal genes in operons. Nucleic Acids Res. 28, 762–769. doi: 10.1093/nar/28.3.762
Guan, Y., Manuel, R. C., Arvai, A. S., Parikh, S. S., Mol, C. D., Miller, J. H., et al. (1998). MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat. Struct. Biol. 5, 1058–1064. doi: 10.1038/4168
Gundogdu, O., Bentley, S. D., Holden, M. T., Parkhill, J., Dorrell, N., and Wren, B. W. (2007). Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics 8:162. doi: 10.1186/1471-2164-8-162
Hanninen, M. L., and Hannula, M. (2007). Spontaneous mutation frequency and emergence of ciprofloxacin resistance in Campylobacter jejuni and Campylobacter coli. J. Antimicrob. Chemother. 60, 1251–1257. doi: 10.1093/jac/dkm345
Hariharan, P. V., Achey, P. M., and Cerutti, P. A. (1977). Biological effect of thymine ring saturation in coliphage phiX174-DNA. Radiat. Res. 69, 375–378. doi: 10.2307/3574445
Hofreuter, D., Tsai, J., Watson, R. O., Novik, V., Altman, B., Benitez, M., et al. (2006). Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74, 4694–4707. doi: 10.1128/IAI.00210-06
Hsieh, P. (2001). Molecular mechanisms of DNA mismatch repair. Mutat. Res. 486, 71–87. doi: 10.1016/S0921-8777(01)00088-X
Huang, S., Kang, J., and Blaser, M. J. (2006). Antimutator role of the DNA glycosylase mutY gene in Helicobacter pylori. J. Bacteriol. 188, 6224–6234. doi: 10.1128/JB.00477-06
Johnson, D. C., Dean, D. R., Smith, A. D., and Johnson, M. K. (2005). Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281. doi: 10.1146/annurev.biochem.74.082803.133518
Kanvah, S., Joseph, J., Schuster, G. B., Barnett, R. N., Cleveland, C. L., and Landman, U. (2010). Oxidation of DNA: damage to nucleobases. Acc. Chem. Res. 43, 280–287. doi: 10.1021/ar900175a
Kassenborg, H. D., Smith, K. E., Vugia, D. J., Rabatsky-Ehr, T., Bates, M. R., Carter, M. A., et al. (2004). Fluoroquinolone-resistant Campylobacter infections: eating poultry outside of the home and foreign travel are risk factors. Clin. Infect. Dis. 38(Suppl. 3), S279–S284. doi: 10.1086/381597
Krwawicz, J., Arczewska, K. D., Speina, E., Maciejewska, A., and Grzesiuk, E. (2007). Bacterial DNA repair genes and their eukaryotic homologues: 1. Mutations in genes involved in base excision repair (BER) and DNA-end processors and their implication in mutagenesis and human disease. Acta Biochim. Pol. 54, 413–434.
Lee, J. W., and Helmann, J. D. (2007). Functional specialization within the Fur family of metalloregulators. Biometals 20, 485–499. doi: 10.1007/s10534-006-9070-7
Lin, J., Michel, L. O., and Zhang, Q. (2002). CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46, 2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002
Lucarelli, D., Russo, S., Garman, E., Milano, A., Meyer-Klaucke, W., and Pohl, E. (2007). Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J. Biol. Chem. 282, 9914–9922. doi: 10.1074/jbc.M609974200
Marenstein, D. R., Chan, M. K., Altamirano, A., Basu, A. K., Boorstein, R. J., Cunningham, R. P., et al. (2003). Substrate specificity of human endonuclease III (hNTH1). Effect of human APE1 on hNTH1 activity. J. Biol. Chem. 278, 9005–9012. doi: 10.1074/jbc.M212168200
Michaels, M. L., and Miller, J. H. (1992). The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174, 6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992
Michel, B. (2005). After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol. 3:e255. doi: 10.1371/journal.pbio.0030255
Modrich, P. (1991). Mechanisms and biological effects of mismatch repair. Annu. Rev. Genet. 25, 229–253. doi: 10.1146/annurev.ge.25.120191.001305
Muraoka, W. T., and Zhang, Q. (2011). Phenotypic and genotypic evidence for L-fucose utilization by Campylobacter jejuni. J. Bacteriol. 193, 1065–1075. doi: 10.1128/JB.01252-10
Nachamkin, I., Engberg, I., and Aarestrup, F. M. (2000). “Diagnosis and antimicrobial susceptibility of Campylobacter spp,” in Campylobacter, 2nd Edn, eds I. Nachamkin and M. J. Blaser (Washington, DC: ASM Press),45–66.
O’Rourke, E. J., Chevalier, C., Boiteux, S., Labigne, A., Ielpi, L., and Radicella, J. P. (2000). A novel 3-methyladenine DNA glycosylase from Helicobacter pylori defines a new class within the endonuclease III family of base excision repair glycosylases. J. Biol. Chem. 275, 20077–20083. doi: 10.1074/jbc.M001071200
O’Rourke, E. J., Chevalier, C., Pinto, A. V., Thiberge, J. M., Ielpi, L., Labigne, A., et al. (2003). Pathogen DNA as target for host-generated oxidative stress: role for repair of bacterial DNA damage in Helicobacter pylori colonization. Proc. Natl. Acad. Sci. U.S.A. 100, 2789–2794. doi: 10.1073/pnas.0337641100
Palyada, K., Sun, Y. Q., Flint, A., Butcher, J., Naikare, H., and Stintzi, A. (2009). Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10:481. doi: 10.1186/1471-2164-10-481
Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. doi: 10.1038/35001088
Pohl, E., Haller, J. C., Mijovilovich, A., Meyer-Klaucke, W., Garman, E., and Vasil, M. L. (2003). Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47, 903–915. doi: 10.1046/j.1365-2958.2003.03337.x
Sahin, O., Plummer, P. J., Jordan, D. M., Sulaj, K., Pereira, S., Robbe-Austerman, S., et al. (2008). Emergence of a tetracycline-resistant Campylobacter jejuni clone associated with outbreaks of ovine abortion in the United States. J. Clin. Microbiol. 46, 1663–1671. doi: 10.1128/JCM.00031-08
Saito, Y., Uraki, F., Nakajima, S., Asaeda, A., Ono, K., Kubo, K., et al. (1997). Characterization of endonuclease III (nth) and endonuclease VIII (nei) mutants of Escherichia coli K-12. J. Bacteriol. 179, 3783–3785. doi: 10.1128/jb.179.11.3783-3785.1997
Tauxe, R. V. (2002). Emerging foodborne pathogens. Int. J. Food Microbiol. 78, 31–41. doi: 10.1016/S0168-1605(02)00232-5
Thayer, M. M., Ahern, H., Xing, D., Cunningham, R. P., and Tainer, J. A. (1995). Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 14, 4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x
Wang, D., Kreutzer, D. A., and Essigmann, J. M. (1998). Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat. Res. 400, 99–115. doi: 10.1016/S0027-5107(98)00066-9
Xia, X., Zhang, S., Su, Y., and Sun, Z. (2009). MICAlign: a sequence-to-structure alignment tool integrating multiple sources of information in conditional random fields. Bioinformatics 25, 1433–1434. doi: 10.1093/bioinformatics/btp251
Young, D. M., and Ornston, L. N. (2001). Functions of the mismatch repair gene mutS from Acinetobacter sp. strain ADP1. J. Bacteriol. 183, 6822–6831. doi: 10.1128/JB.183.23.6822-6831.2001
Zhang, Q., Meitzler, J. C., Huang, S., and Morishita, T. (2000). Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter major outer membrane protein. Infect. Immun. 68, 5679–5689. doi: 10.1128/IAI.68.10.5679-5689.2000
Zhang, Y., Werling, U., and Edelmann, W. (2012). SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 40:e55. doi: 10.1093/nar/gkr1288
Keywords: Campylobacter, DNA repair, oxidative stress, antibiotic resistance, foodborne pathogens, endonuclease III
Citation: Dai L, Xia J, Sahin O and Zhang Q (2019) Identification of a nth-Like Gene Encoding an Endonuclease III in Campylobacter jejuni. Front. Microbiol. 10:698. doi: 10.3389/fmicb.2019.00698
Received: 29 November 2018; Accepted: 20 March 2019;
Published: 10 April 2019.
Edited by:
Abd El-Latif Hesham, Assiut University, EgyptReviewed by:
Abdi Elmi, London School of Hygiene & Tropical Medicine (LSHTM), United KingdomBanaz Star-Shirko, London School of Hygiene & Tropical Medicine (LSHTM), United Kingdom
Bhavna Gordhan, National Health Laboratory Service, South Africa
Copyright © 2019 Dai, Xia, Sahin and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qijing Zhang, zhang123@iastate.edu
 Lei Dai
Lei Dai Jing Xia1,2
Jing Xia1,2 Orhan Sahin
Orhan Sahin Qijing Zhang
Qijing Zhang