- 1Graduate Program in Microbiology, College of Agriculture and Life Sciences, North Carolina State University, Raleigh, NC, United States
- 2Department of Food, Bioprocessing and Nutrition Sciences, North Carolina State University, Raleigh, NC, United States
- 3Department of Medicine, Division of Gastroenterology and Hepatology, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 4Center for Gastrointestinal Biology and Disease, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
Health-promoting aspects attributed to probiotic microorganisms, including adhesion to intestinal epithelia and modulation of the host mucosal immune system, are mediated by proteins found on the bacterial cell surface. Notably, certain probiotic and commensal bacteria contain a surface (S-) layer as the outermost stratum of the cell wall. S-layers are non-covalently bound semi-porous, crystalline arrays of self-assembling, proteinaceous subunits called S-layer proteins (SLPs). Recent evidence has shown that multiple proteins are non-covalently co-localized within the S-layer, designated S-layer associated proteins (SLAPs). In Lactobacillus acidophilus NCFM, SLP and SLAPs have been implicated in both mucosal immunomodulation and adhesion to the host intestinal epithelium. In this study, a S-layer associated serine protease homolog, PrtX (prtX, lba1578), was deleted from the chromosome of L. acidophilus NCFM. Compared to the parent strain, the PrtX-deficient strain (ΔprtX) demonstrated increased autoaggregation, an altered cellular morphology, and pleiotropic increases in adhesion to mucin and fibronectin, in vitro. Furthermore, ΔprtX demonstrated increased in vitro immune stimulation of IL-6, IL-12, and IL-10 compared to wild-type, when exposed to mouse dendritic cells. Finally, in vivo colonization of germ-free mice with ΔprtX led to an increase in epithelial barrier integrity. The absence of PrtX within the exoproteome of a ΔprtX strain caused morphological changes, resulting in a pleiotropic increase of the organisms’ immunomodulatory properties and interactions with some intestinal epithelial cell components.
Introduction
Lactic acid bacteria (LAB) are a clade of diverse Gram-positive, microaerophilic, and non-sporulating microbes which ferment hexoses primarily to lactic acid. Many of these bacteria, which include species from the genera Lactococcus, Enterococcus, Pediococcus, Oenococcus, Streptococcus, Leuconostoc, and Lactobacillus, have evolved through 1000s of years of fermentation in numerous food and drink substrates (Makarova et al., 2006; Douglas and Klaenhammer, 2010; Johnson and Klaenhammer, 2014). Predicated on this history of consumption in foods, many LAB are generally recognized as safe and serve vital industrial roles as starter and adjunct cultures for fermentation of dairy, vegetable, meat, and wine foodstuffs (Wood, 1998; Makarova et al., 2006).
In addition to their evolution to dairy environments, some LAB are niche-associated with the mucosal surfaces of animals, including the gastrointestinal and urogenital tracts (O’Flaherty and Klaenhammer, 2010a; Johnson and Klaenhammer, 2014). Many of these, most notably in the Lactobacillus genera, have been used as probiotics, which are defined by the FAO/WHO as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (FAO/WHO, 2002; Hill et al., 2014). One such organism is Lactobacillus acidophilus NCFM, an industrially significant probiotic strain, delivered commercially in various dairy and dietary supplement formulations for the past 35 years (Sanders and Klaenhammer, 2001). Furthermore, L. acidophilus NCFM is one of the most studied and well characterized probiotic bacteria (Sanders and Klaenhammer, 2001; Altermann et al., 2005; Klaenhammer et al., 2005, 2008).
Proteins and macromolecules at the cell surface of probiotic bacteria play a critical role in mediating strain-specific beneficial effects of probiotics toward the host (Lebeer et al., 2010; Bron et al., 2013). For L. acidophilus NCFM, the specificity of probiotic activity on the host has been characterized for numerous cell surface components, including lipoteichoic acid (Mohamadzadeh et al., 2011), sortase-dependent proteins (Call et al., 2015), an aggregation promoting factor (Goh and Klaenhammer, 2010), and a myosin-cross reactive protein (O’Flaherty and Klaenhammer, 2010b). Proteins at the Surface (S-) layer, which are non-covalently bound to the apical component of the cell wall peptidoglycan, are also of interest to the understanding of microbe-host interactions (Hynönen and Palva, 2013; Johnson and Klaenhammer, 2014).
Surface-layers are non-covalently bound, semi-porous, crystalline arrays comprised of self-assembling (glyco) protein subunits called S-layer proteins (SLPs; Sára and Sleytr, 2000). In L. acidophilus, the S-layer array is comprised of a dominant protein constituent, SlpA (46 kDa) with minor constituents SlpB (47 kDa) and SlpX (51 kDa; Goh et al., 2009). In vitro studies using intestinal epithelial cell lines suggest SLPs as a major factor in intestinal adhesion for L. acidophilus (Buck et al., 2005; Frece et al., 2005). Furthermore, SlpA of L. acidophilus NCFM has been shown to bind dendritic cell (DC) C-type lectin receptors (Konstantinov et al., 2008) and exert immunomodulatory signals which mitigate inflammatory disease states and promote maintenance of healthy intestinal barrier function (Lightfoot et al., 2015). Recent evidence has also shown that there are additional proteins which non-covalently co-localize to the outermost stratum of the cell surface with the S-layer, called S-layer associated proteins (SLAPs; Johnson et al., 2013, 2016). Due to the apparent importance of Lactobacillus cell surface proteins for probiotic roles, these SLAPs have been a prime target for investigation in recent years.
S-layer associated proteins were first characterized in L. acidophilus NCFM (Johnson et al., 2013), but have since been characterized in Lactobacillus helveticus, Lactobacillus crispatus, Lactobacillus amylovorus, and Lactobacillus gallinarum (Johnson et al., 2016). Notably, these SLAP-containing organisms are S-layer forming members of the L. acidophilus-Lactobacillus delbrueckii homology group. However, no SLAPs were proteomically identified within the non-covalent exoproteomes of the closely related, non-S-layer forming members of the homology group, including Lactobacillus gasseri, Lactobacillus johnsonii, as well as the taxonomic progenitor L. delbrueckii subsp. bulgaricus (Johnson et al., 2016). Preliminary functional analyses in L. acidophilus NCFM have revealed that SLAPs have a broad spectrum of both cellular and probiotic functionality, including cell division (Altermann et al., 2004; Johnson and Klaenhammer, 2016), autolysin activity (Johnson and Klaenhammer, 2016), immunomodulation (Johnson et al., 2013), and adhesion to extracellular matrices (Johnson and Klaenhammer, 2016; Hymes et al., 2016).
One of the most prevalent SLAPs identified in the exoproteome of L. acidophilus NCFM is a 72 kDa, uncharacterized serine protease encoded by the gene lba1578 (Johnson et al., 2013). In this study, the S-layer associated serine protease, designated PrtX, was selected for functional analysis.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. L. acidophilus strains were propagated in de Man Rogosa and Sharpe (MRS) broth (Difco) under ambient atmospheric conditions, statically or on MRS solid medium containing 1.5% (w/v) agar (Difco) under anaerobic conditions at 37°C, or at 42°C where indicated. Recombinant strains were selected in the presence of 2 μg/ml of erythromycin (Em; Sigma–Aldrich) and/or 2–5 μg/ml of chloramphenicol (Cm; Sigma). Escherichia coli strains were grown in brain heart infusion (Difco) medium at 37°C with aeration. E. coli EC101 was grown in the presence of 40 μg/ml kanamycin (Kn; Sigma–Aldrich) while NCK1911 and transformants were grown with 40 μg/ml Kn and 150 μg/ml Em. Counterselection of plasmid-free excision recombinants was performed using 5-fluorouracil-supplemented glucose semi-defined medium, as previously described (Goh et al., 2009).
Molecular Techniques and Statistical Analysis
Genomic DNA from L. acidophilus strains was isolated using a Zymo Research Fungal/Bacterial DNA MiniPrep kit (Zymo Research). Plasmid DNA from E. coli was isolated using a QIAprep Spin Miniprep kit (Qiagen). Restriction enzyme digestions and ligations were performed using Roche restriction enzymes (Roche Diagnostics) and T4 DNA ligase (New England Biolabs), respectively. PCR primers (Table 1) were designed based on the genomic sequence data and synthesized by Integrated DNA Technologies (Coralville, IA, United States). PCRs were performed in Bio-Rad MyCycler thermocyclers (Bio-Rad Laboratories) using Choice-Taq Blue DNA polymerase (Denville Scientific) for screening of recombinants and PfuUltra II fusion HS DNA polymerase (Agilent Technologies) for cloning. PCR amplicons were analyzed on 0.8% agarose gels and purified using QIAquick Gel Extraction kits (Qiagen).
Escherichia coli EC101 cells were made competent using a rubidium chloride competent cell protocol (Hanahan, 1983). L. acidophilus cells were prepared for electrotransformation using a modified penicillin treatment protocol (Wei et al., 1995; Walker et al., 1996; Goh et al., 2009).
All statistical analyses were performed using unpaired student’s t-test. P-values below 0.05 were considered significant.
Construction of a ΔprtX Strain of L. acidophilus NCFM
The upp-based counterselection gene replacement method (Goh et al., 2009) was used to create an internal deletion of 1966 bp in prtX (lba1578) of NCK1909, a upp-deficient background strain of L. acidophilus NCFM. Using splicing by overlap extension PCR (Horton et al., 1989), 439 and 548 bp regions flanking the deletion target were spliced together resulting in a 987 bp product with a BamHI restriction site added to the 5′ end and SacI restriction site to the 3′ end. This construct was digested with BamHI and SacI, then ligated into the polylinker of the similarly digested integration plasmid pTRK935 and transformed into competent E. coli EC101. The resulting recombinant plasmid, pTRK1073, was transformed into L. acidophilus NCK1909 harboring the helper plasmid pTRK669 (NCK1910). Single crossover integrants were screened as described previously (Goh et al., 2009). Colonies with the ΔprtX genotype were screened among the double recombinants recovered on glucose semi-defined medium agar plates containing 5-fluorouracil. Deletion was confirmed by PCR with primer pair 1578up and 1578dw (Table 1) and DNA sequencing. The resulting ΔprtX strain was designated NCK2282.
Lithium Chloride Extraction of Extracellular S-Layer Associated Proteins
Non-covalently bound cell surface proteins, including SLPs and SLAPs were extracted from NCK1909 and ΔprtX L. acidophilus NCFM strains using LiCl denaturing salt, as described previously (Johnson et al., 2013). Proteins were quantified via bicinchoninic acid assay kit (Thermo Scientific) and visualized via SDS–PAGE using precast 4–20% Precise Tris-HEPES protein gels (Thermo Scientific). Gels were stained using AcquaStain (Bulldog Bio) according to the instructions of the manufacturer.
Morphological Assessment and Electron Microscopy
Morphological assessment of L. acidophilus NCFM and ΔprtX was performed using a phase-contrast light microscope at 40× magnification (Nikon Eclipse E600). Cells were observed over a growth period of 24 h in MRS broth at 37°C. Pictures were taken using a QImaging MicroPublisher 5.0 RTV camera attachment at 1, 3, 5, 8, 10, and 13-h time points.
Sample processing for scanning electron microscopy (SEM) was performed by the Center for Electron Microscopy (CEM) at North Carolina State University. L. acidophilus NCFM strains were grown in 35 ml of MRS to logarithmic and stationary phases. Cells were pelleted by centrifugation at 3,166 × g for 15 min at room temperature. Pellets were resuspended in a fresh 1:1 (vol/vol) fixative mixture of 6% glutaraldehyde and 0.2 M sodium cacodylate (pH 5.5) and submitted to CEM for sample processing. SEM samples were viewed with a JEOL JSM 5900LV scanning electron microscope at 15 kV. For both light and electron microscopy analyses, images that were representative of the observed populations were used for assessment.
Adherence Assays
Mucin and extracellular matrices (ECM) binding assays were performed as described previously (Goh and Klaenhammer, 2010). Mucin (type III from porcine stomach, Sigma) was dissolved in PBS to a final concentration of 10 mg/ml. Fibronectin (from human plasma, Sigma), collagen (type IV from human cell culture, Sigma), and laminin (from Engelbreth-Holm-Swarm murine sarcoma/basement membrane; Sigma) were diluted in 50 mM carbonate-bicarbonate buffer (pH 9.6, Sigma) to a final concentration of 10 μg/ml. For each assay, a Nunc Maxisorp 96-well microplate (Sigma) was coated with 100 μl/well of substrate and incubated at 4°C overnight. The wells were then washed twice with PBS (pH 7.4) to remove excess substrate before blocking with 150 μl of 2% bovine serum albumin (BSA) solution (Sigma) for 2 h at 37°C. Excess BSA was removed by two washes with PBS.
Bacterial cells were grown in MRS broth to stationary phase (16 h) in preparation for the assay. Cultures were centrifuged (1,771 × g, 15 min, room temperature), washed once with PBS, and resuspended in PBS (pH 4.75). Cell density was adjusted to ∼1 × 108 CFU/mL based on previously calculated OD600/CFU ratios. Cell suspensions (100 μl) were added to each mucin or ECM-coated well. Initial cell counts of samples were enumerated on MRS plates. After incubation for 1 h at 37°C, the wells were gently washed five times with 200 μl/well of PBS. Adhered cells were recovered by adding 100 μl of 0.05% Triton X-100 solution (FisherBiotech, prepared in PBS) to each well and agitating on an orbital shaker (200 rpm) for 15 min. Cell suspensions were transferred into 900 μl of 0.1X MRS broth before being further diluted and plated in duplicate on MRS plates. Colonies were enumerated and calculated as a percent of relative adherence (mutant CFU/parent CFU), where parent (NCK1909) CFU were set at 100%.
Bacterial-Dendritic Cell Co-incubation Assay and Cytokine Measurement
In vitro bacterial-DC co-incubation assays were performed as described previously (Johnson et al., 2013; Call et al., 2015). Cytokine measurements for IL-6, IL-10, and IL-12 were quantified using Single-Analyte ELISArray kits (Qiagen), according to the manufacturer’s instructions. Following cytokine quantification, the cytokine expression data were statistically compared between the parent and mutant strains using student t-test.
Mono-Colonization of WT and ΔprtX L. acidophilus Strains in a Germ-Free Mouse Model
Germ-free 129S6/SvEv mice utilized for in vivo experiments were taken from breeding colonies maintained at the North Carolina State University Gnotobiotic Core of the Center for Gastrointestinal Biology and Disease, as described previously (Goh and Klaenhammer, 2014; Call et al., 2015). Mice were maintained in cages in germ-free flexible film isolators housed in a room with 12 h of light and darkness. They were provided access to a standard diet (Prolab RMH 3500, LabDiet) and water ad libitum. Germ-free status was evaluated at least once a month through culturing fecal samples in thioglycollate broth, blood agar, and Sabouraud agar. Prior to colonization with L. acidophilus strains, the mice were also verified germ-free by culturing fecal samples aerobically and anaerobically on plate count agar and MRS agar. Animal use protocols were approved by the Institutional Animal Care and Use Committee of North Carolina State University, Raleigh, NC, United States (Protocol 15-026-B). All mice handlers were certified by the American Association for Laboratory Animal Science. The Animal Welfare Assurance # is: A3331-01.
In preparation for mono-colonization of the germ-free mice, L. acidophilus NCK56 or the ΔprtX strain was propagated in MRS broth at 37°C overnight. Bacteria were harvested at stationary phase (16 h) via centrifugation (1735 × g, 10 min, room temperature). They were subsequently washed with and resuspended in PBS to an OD600 corresponding to 5 × 109 CFU/ml for each strain. Germ-free 129S6/SvEv mice were separated into two experimental groups: the first treated with NCK56 (n = 6, 3 males and 3 females, 17–18 weeks old) and the second treated with ΔprtX (n = 6, 3 males and 3 females, 17–18 weeks old). Mice were gavaged with ∼1 × 109 cells in 200 μl PBS per mouse. Fecal samples were collected from each mouse on the day of gavage (day 0), days 2, 5, and 7. Fecal samples were weighed, resuspended in 1 ml of PBS, diluted and plated on MRS agar for enumeration.
In Vivo FITC-Dextran Epithelial Barrier Integrity in a Germ-Free Mouse Model
Germ-free 129S6/SvEv mice were mono-colonized with either wild-type (WT) or ΔprtX strains of L. acidophilus NCFM, as described above. On the day of the assay, mice were denied access to food but allowed access to water ad libitum for 3 h, after which 150 μl of 3–5 kDa fluorescein isothiocyanate (FITC)-dextran at a concentration of 80 mg/ml was administered to each mouse using an oral gavage needle. Two hours after administration of the FITC-dextran, blood was collected using a 1 ml tuberculin syringe (BD) and stored in Microtainer serum separator tubes (BD). Blood was placed in the dark for 1 h and allowed to clot, after which the serum was separated via centrifugation (13,000 rpm, 4°C). Fluorescence in the serum was measured using a FLOUStar Optima Microtiter plate reader (BMG Technologies) with the excitation filter set for 490 nm, the emission filter set for 520 nm, and a 1250 gain setting for fluorescence intensity. FITC-dextran fluorescence was measured in the serum collected from NCK56 mono-colonized mice (n = 5, 2 males, 3 females, 17–18 weeks old) and ΔprtX mono-colonized mice (n = 5, 2 males, 3 females, 17–18 weeks old). Serum from a single germ-free control was used to measure background fluorescence. Using a standard of known concentrations of FITC-dextran in PBS, the concentration of FITC-dextran was calculated.
Results
PrtX, a S-Layer Associated Serine Protease of L. acidophilus
One of the most prevalent SLAPs in the non-covalently bound exoproteome of L. acidophilus NCFM is a 72 kDa uncharacterized serine protease encoded by the gene lba1578 (Johnson et al., 2013). The protein is 684 amino acids in length and contains a predicted signal peptide sequence located between amino acid 34 and 35 (VKA-AD). In silico analysis of LBA1578 revealed that there is discreet homology to serine proteases in Lactobacillus acetotolerans DSM 20749 (33% identity), Lactobacillus gigeriorum DSM 23908 (32%), Lactobacillus pasteurii DSM 23907 (31%), L. amylovorus DSM 20351 (31%), Lactobacillus kitasatonis DSM 16761 (31%), and Lactobacillus amylolyticus DSM 11664 (30%). Due to the uncharacterized nature of LBA1578 and the conserved annotation of serine protease among discreet homologs, this protein was designated PrtX.
Using RNA-seq transcriptional analysis from a previous study (Johnson et al., 2016), it was determined that lba1578 (prtX) mRNA is polycistronically expressed downstream of murC, which encodes a UDP-N-acetylmuramate-L-alanine ligase putatively involved in peptidoglycan biosynthesis (Figure 1A). Because PrtX is an uncharacterized serine protease which is prominently featured in the non-covalent exoproteome in L. acidophilus, it was selected for functional analysis. The upp-based counterselective gene replacement method (Goh et al., 2009) was used to create a markerless chromosomal deletion of 1966 base pairs in the coding region of prtX in L. acidophilus NCFM (Figure 1B). SDS–PAGE analysis of extracted SLAPs confirmed the presence of PrtX in the non-covalently bound exoproteome of the WT L. acidophilus NCFM and its absence from the ΔprtX strain (Figure 1C).
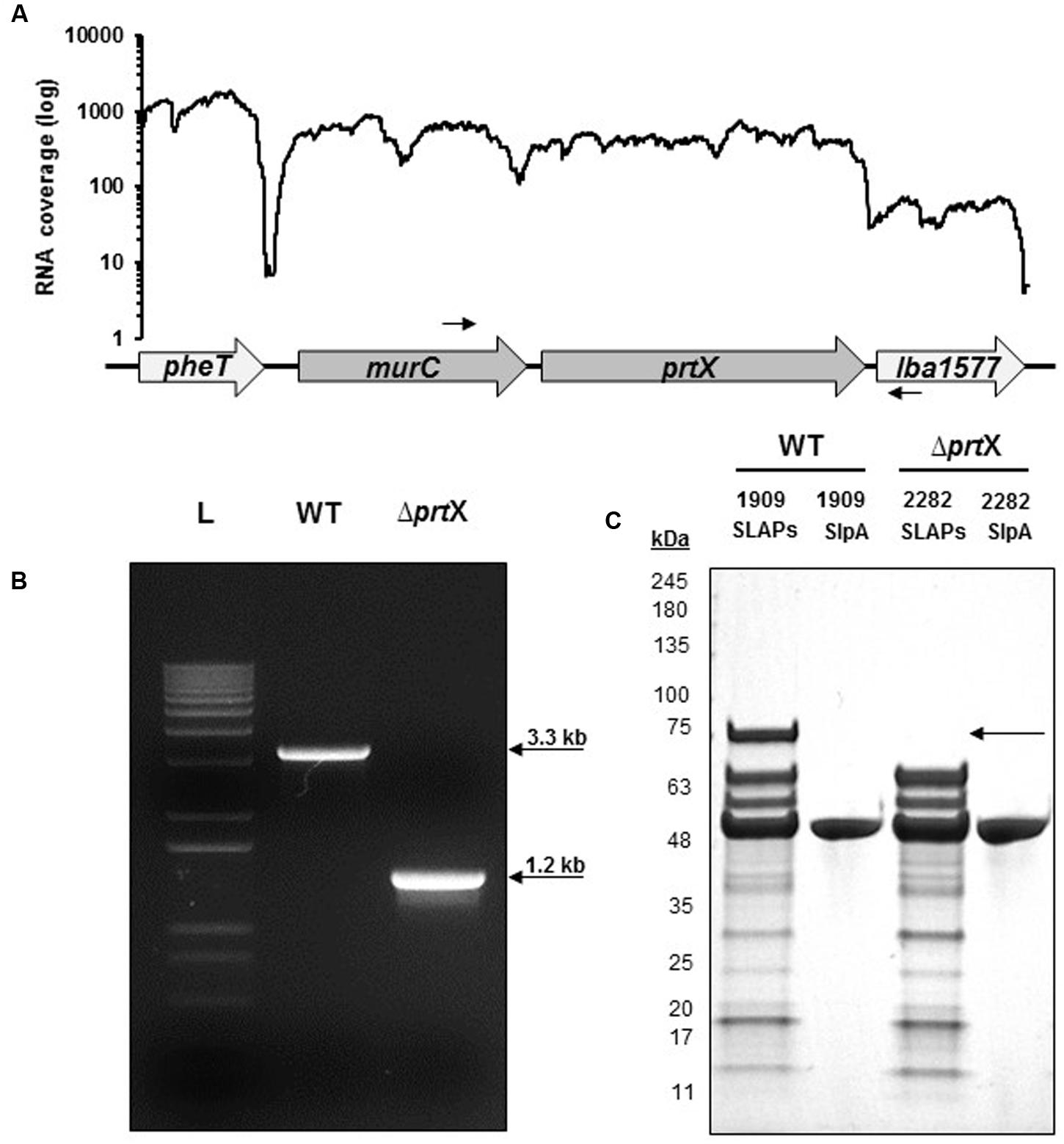
FIGURE 1. The gene encoding PrtX was deleted from the chromosome of Lactobacillus acidophilus NCFM. (A) RNA-seq analysis demonstrates that prtX is polycistronically expressed with murC, which encodes for a UDP-N-acetylmuramate-L-alanine ligase putatively involved in peptidoglycan biosynthesis. Black arrows indicate the forward and reverse primer pair used to confirm the deletion of prtX. (B) Gel electrophoresis of PCR products using the primers indicated in (A) for the parent strain (WT; 3.3 kb) compared to the PrtX-deficient strain (ΔprtX; 1.2 kb). The deletion was confirmed by sequencing. (C) SDS–PAGE of the non-covalently bound extracellular S-layer proteins (SLP) and S-layer associated proteins (SLAPs) isolated from both WT and ΔprtX. Absence of the 72 kDa band corresponding to PrtX within the SLAP extracts from ΔprtX confirmed the elimination of PrtX from the exoproteome of ΔprtX.
Growth and Morphology of ΔprtX
The cellular morphology of the ΔprtX was examined using light microscopy over a 24-h period of growth in MRS broth (Figure 2A). Compared to the parent strain (WT), the ΔprtX strain demonstrated aberrant morphology and increased autoaggregation phenotypes (Figure 2A). Specifically, during lag phase (13 h; OD600 0.1–0.2) the ΔprtX cells appeared to have a longer chain length compared to WT cells. Logarithmic phase (5–10 h; OD600 0.5–1.0) cell chains of the ΔprtX mutant were only similarly longer than WT. Furthermore, the ΔprtX cells presented an increased autoaggregation phenotype which was not observed in the WT strain (Figure 2A). By stationary phase (13 h; OD600 1.4; Figure 2A) the autoaggregation phenotype was still observed in the ΔprtX cells.
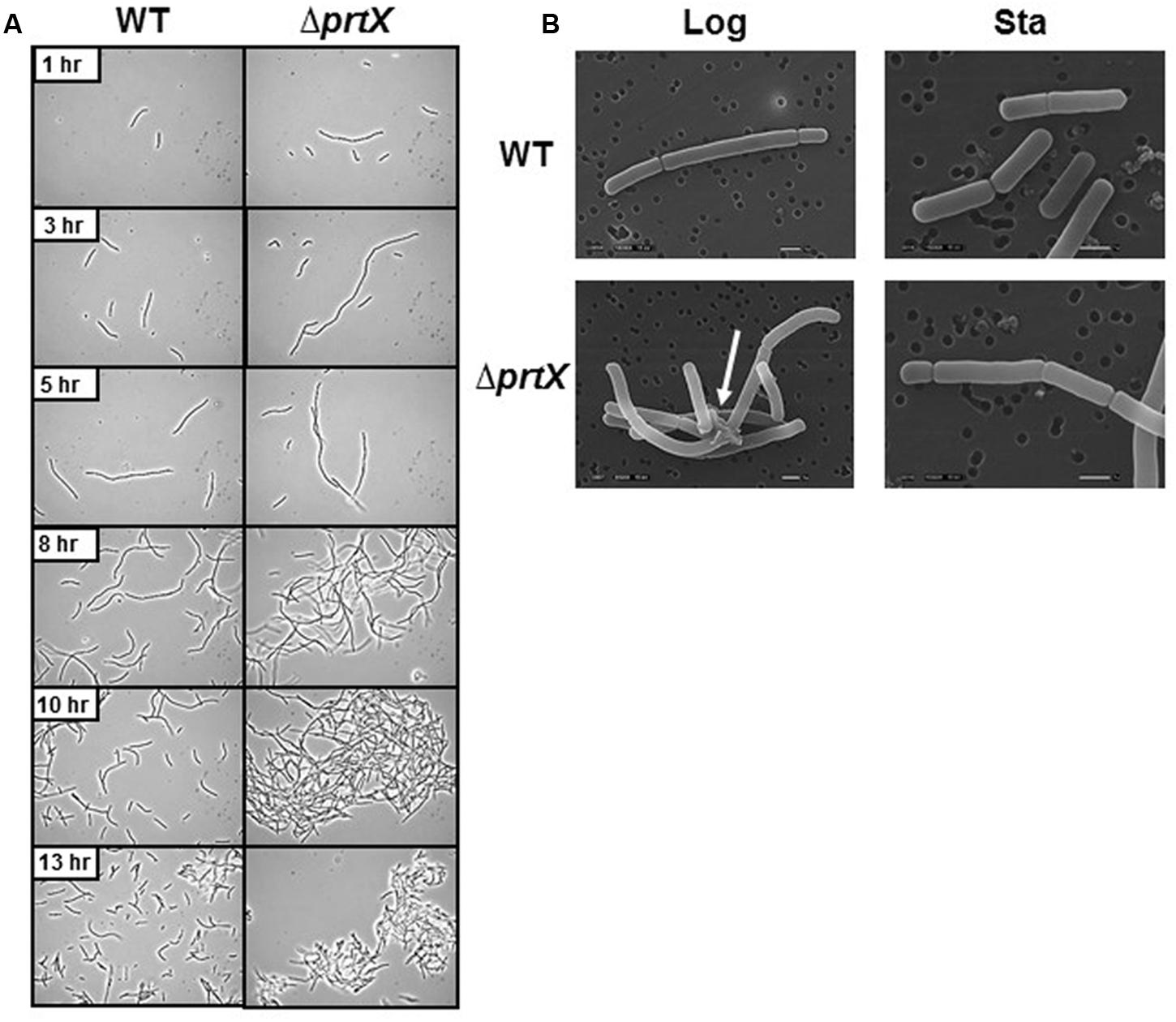
FIGURE 2. (A) The cellular morphology of the wild-type (WT) and mutant (ΔprtX) strains were assessed using phase-contrast light microscopy over a 13-h growth period. (B) Scanning electron micrographs were taken of WT and ΔprtX at logarithmic and stationary phases.
For a more detailed observation of these morphological phenotypes, WT and ΔprtX strains were examined using SEM at logarithmic phase (OD600 0.6) and stationary phase (OD600 1.9; Figure 2B). The SEM micrographs mirror much of the morphological assessment with the light microscope. Notably, protein complexes were repeatedly observed in the micrographs of log phase ΔprtX cells at 8,500× magnification, suggesting a build-up of proteins at the cell surface due to poor protein turnover (Figure 2B). Such complexes were not observed in the WT sample (Figure 2B). Furthermore, these complexes were not observed in the stationary phase samples of ΔprtX or WT cells. However, it is notable that compared to the WT stationary phase sample, much fewer cells were available for observation under the microscope. This is perhaps due to the difficulty of fixing large clusters of cells onto the platform for SEM. Collectively, these morphological data implicate that PrtX may be involved in protein turnover at the surface during log phase cell division.
Adherence of ΔprtX to Mucin and Extracellular Matrices
The ability of the prtX mutant to bind to mucin and the ECM fibronectin, collagen, and laminin was assessed and compared to WT, in vitro (Figure 3). The ΔprtX strain demonstrated a 40% increase in binding capacity to mucin, compared to the parent strain (Figure 3, p < 0.001). Similarly, ΔprtX showed a 20% increase in binding to fibronectin (Figure 3, p < 0.001). While ΔprtX appeared to have an increased binding capacity for laminin by 25%, this increase was not statistically significant. Finally, relative to WT, the ΔprtX strain did not show any increase in binding to collagen (Figure 3).
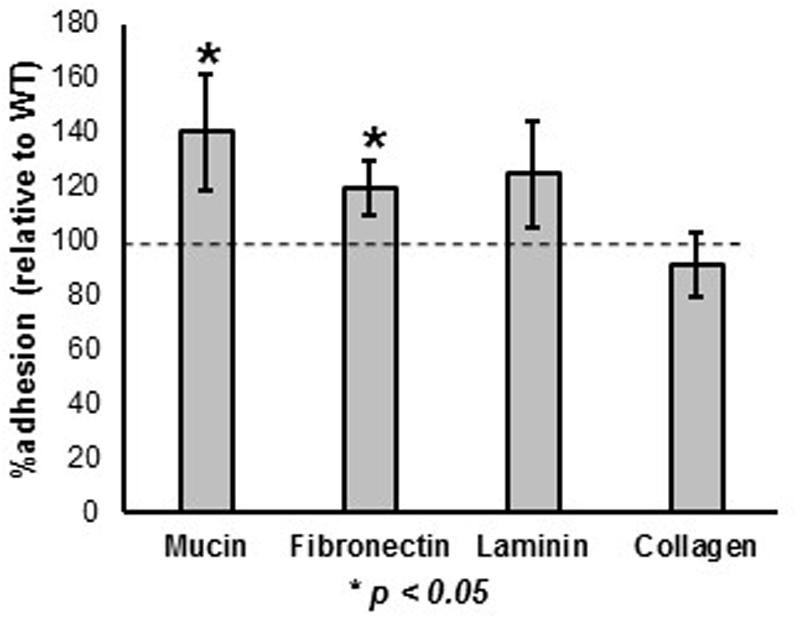
FIGURE 3. The ability of the ΔprtX mutant to bind to mucin and extracellular matrices (ECM) was assessed. Compared the WT reference (dotted line), ΔprtX showed a significant increase in binding to mucin and fibronectin. Asterisks indicate statistical significance (p < 0.001). Adherence assays were performed in triplicate; all error bars represent confidence intervals.
In Vitro Immunomodulation of Mouse DC Cells Exposed to ΔprtX
The immunomodulatory potential of the ΔprtX strain was assessed using an in vitro bacterial/murine DC co-incubation assay. Relative to the WT strain, ΔprtX demonstrated an overall increase in immunostimulation of the cytokines IL-6, IL-12, and IL-10 (Figure 4). Specifically, ΔprtX induced production of the pro-inflammatory cytokine IL-6 in DC at twice the level of that of the parent strain (Figure 4, IL-6). The pro-inflammatory cytokine IL-12 was also induced in ΔprtX compared to WT in a less pronounced, but statistically significant manner (Figure 4, IL-12). Similar to IL-6, in ΔprtX induced DC, the anti-inflammatory cytokine IL-10 was produced at twice the level of that produced by the WT-induced DC (Figure 4, IL-10). Although ΔprtX induced both the pro-inflammatory cytokine IL-12 and the anti-inflammatory cytokine IL-10, the IL-10/IL-12 ratio, which is a measure of the balance between pro-inflammatory and anti-inflammatory states (O’Garra and Murphy, 2009; Hamer et al., 2010), was higher in ΔprtX than in WT (Figure 4, IL-10/IL-12).
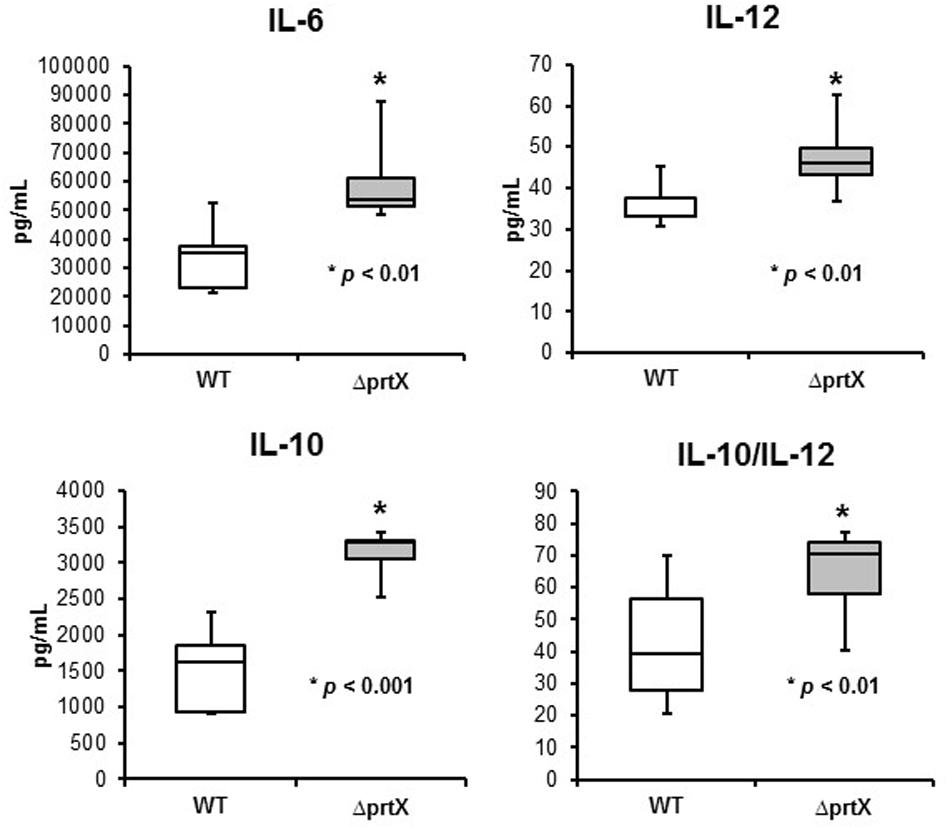
FIGURE 4. The immunomodulatory phenotype of ΔprtX (gray) compared to WT (white) was measured using a murine dendritic cell co-incubation assay. Cytokines IL-6, IL-12, and IL-10 were assessed using enzyme-linked immunosorbent assays. For all cytokines measured, ΔprtX demonstrated a significant increase in immunomodulation compared to WT (IL-6, p < 0.01; IL-12, p < 0.006; IL-10, p < 0.001). The IL-10/IL-12 ratio was also significantly increased in ΔprtX (p < 0.01). Co-incubation assays were performed in duplicate or triplicate; bars on the box-whisker plots represent the range.
In Vivo Mono-Colonization and Epithelial Barrier Integrity of Germ-Free Mice
Due to the increased adhesion and immunomodulatory properties of ΔprtX, in vitro, the biological relevance of the ΔprtX strain was explored in an in vivo germ-free mouse model. Germ-free mice were colonized with ∼1 × 109 CFU/g of either WT or ΔprtX strains; this mono-colonization was measured over 7 days (Figure 5A). Both WT and ΔprtX strains colonized the germ-free mice at similar rates and by day 7 the bacteria had colonized at 4.56 × 108 and 3.71 × 108 CFU/g fecal samples for WT and ΔprtX, respectively (Figure 5A). The gastrointestinal epithelial barrier integrity was examined in a second set of mice (Figure 5B). These mice were similarly colonized with WT or ΔprtX strains and fed FITC-dextran; the resulting FITC-dextran in the blood serum correlated to the gastrointestinal barrier integrity of the mouse. Compared to WT, serum from the ΔprtX colonized mice demonstrated a 21% reduction in FITC-dextran levels (Figure 5B, p < 0.05). These data indicate that the intestinal epithelial barrier integrity was improved in mice colonized with ΔprtX relative to mice colonized with WT.
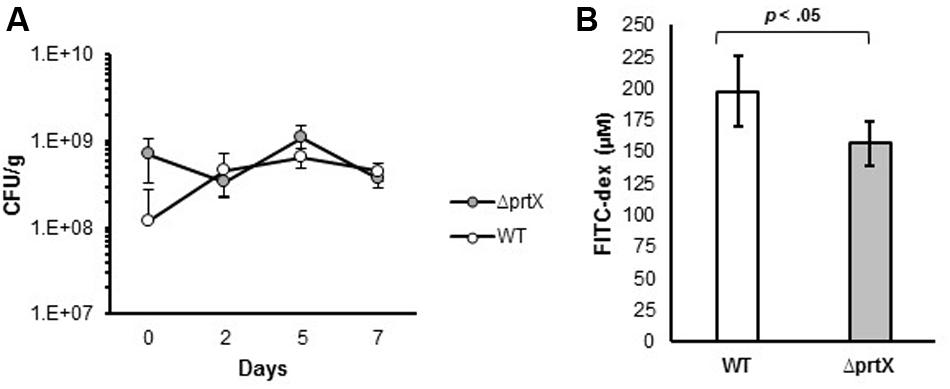
FIGURE 5. In vivo assessment of germ-free mice colonized with either WT or ΔprtX strains of L. acidophilus NCFM. (A) Mice were monocolonized with WT (white, n = 6) or ΔprtX (gray, n = 6) over 7 days and colonization levels were assessed by plating fecal samples on MRS agar. (B) Total gastrointestinal epithelial barrier integrity was assessed through the measurement of FITC-dextran in the serum of mice previously fed FITC-dex. Mice colonized with ΔprtX had a statistically significant reduction in FITC-dex in the serum, indicating an increase in epithelial barrier integrity (p < 0.05).
Discussion
In this study, the serine protease designated PrtX, was functionally and phenotypically characterized. Following the creation of a clean chromosomal deletion of prtX, the corresponding mutant strain (ΔprtX) was assessed for cellular morphology, in vitro mucin and ECM adhesion, in vitro immunomodulation, and in vivo mouse colonization and intestinal epithelial barrier integrity.
Morphological assessment of the ΔprtX isogenic mutant revealed a visible increase in autoaggregation, compared to the parent strain. This increased autoaggregation phenotype was further evaluated using SEM, in which logarithmic ΔprtX cells were observed to present aberrant cellular topology. These observations suggest that PrtX may be involved with protein turnover at the cell surface of L. acidophilus NCFM. Gene deletion studies of serine proteases in other Lactobacillus species, including PrtB in L. delbrueckii subsp. bulgaricus and PrtH in L. helveticus, have not reported a similar morphological or autoaggregation phenotypes (Gilbert et al., 1996; Pederson et al., 1999; Genay et al., 2009; Sadat-Mekmene et al., 2011). However, this difference in observed phenotypes may be due to the subcellular localization of the referenced serine proteases. PrtB and PrtH are cell-envelope serine proteases which are cell-membrane attached, whereas prtX is non-covalently attached to the cell wall peptidoglycan. Due to the lack of homologs for PrtX, it is difficult to contextualize the results of this study with similar studies in Lactobacillus species. However, a similar increased autoaggregation phenotype was reported during the functional analysis of S-layer associated autolysin, AcmB, in L. acidophilus NCFM (Johnson and Klaenhammer, 2016). Future proteomic analysis of the proteomic complexes observed in ΔprtX will undoubtedly aid in the description of PrtX and the proteins it may process.
Proteins localized at the cell surface of L. acidophilus NCFM are important mediators of adhesion to host intestinal epithelial mucus layer and ECM, in vitro (Buck et al., 2005; Azcarate-Peril et al., 2009; Goh and Klaenhammer, 2010; O’Flaherty and Klaenhammer, 2010b; Call et al., 2015; Hymes et al., 2016; Johnson and Klaenhammer, 2016). In this study, the ΔprtX strain demonstrated a significant increase in binding to mucin and fibronectin. These results are consistent with a recent analysis of the serine protease, PrtS in Streptococcus thermophilus, in which a PrtS-deficient strain bound to Caco-2 epithelial cells twice as effectively as the parent strain (Kebouchi et al., 2016). Furthermore, previous analysis of the S-layer associated fibronectin binding protein, FpbB, in L. acidophilus NCFM revealed that FpbB has specificity for binding fibronectin and mucin, in vitro (Hymes et al., 2016). Though the exact mechanism remains unclear, it is possible that the PrtX-deficient strain is deficient in protein turnover at the cell surface, including SLAPs such as FpbB, resulting in increased binding to mucin and fibronectin, specifically. It is possible that the generalized increase in autoaggregation resulted in an inflation of the CFU counts upon plating. However, this was not observed across all epithelial cell components (e.g., collagen), lending credence to the results’ representation of true adherence. More in-depth quantitative proteomic studies could elucidate this mechanism even further.
In addition to their role in adhesion, cell surface proteins of L. acidophilus NCFM, including SLP and SLAPs, have also been examined for roles in immunomodulation (Konstantinov et al., 2008; Mohamadzadeh et al., 2011; Johnson et al., 2013; Call et al., 2015; Lightfoot et al., 2015). Previous research regarding immunomodulation and CEPs, specifically, has focused on the production of bioactive compounds released during casein proteolysis in milk (Matar et al., 2001; Silva and Malcata, 2005; Korhonen and Pihlanto, 2006; Shah, 2007). For example, milk fermented with a non-proteolytic variant of L. helveticus was found to have a suppressed mucosal immune response compared to milk fermented by the WT strain of L. helveticus (Matar et al., 2001). Recent evidence, however, has pointed to a more specific immunomodulatory role of serine proteases in Lactobacillus. An extracellular lactocepin serine protease in Lactobacillus paracasei was found to exert anti-inflammatory effects by selectively degrading pro-inflammatory chemokines, such as CXCL10 (von Schillde et al., 2012). In the present study, we found that the absence of PrtX in the ΔprtX mutant resulted in a generalized increase in DC expression of cytokines IL-6, IL-12, and IL-10, compared to WT. One cytokine induced in DCs exposed to ΔprtX was the anti-inflammatory IL-10, which has been proposed as a biological therapy for chronic irritable bowel disease (IBD), including Crohn’s disease and colitis (Li and He, 2004). It is possible that PrtX may directly degrade certain cytokines, which may explain the observed accumulation of cytokines when DCs were exposed the PrtX-deficient strain, though this direct mechanism remains to be discovered.
Previous work has been performed in L. acidophilus NCFM using an in vivo germ-free mouse model. Through mutational analysis, sortase, an enzyme which covalently couples extracellular proteins containing an LPXTG motif to the cell surface, was found to contribute to gut retention of L. acidophilus NCFM (Call et al., 2015). Similarly, the glycogen biosynthesis pathway in L. acidophilus NCFM contributes to gut fitness and retention, in vivo (Goh and Klaenhammer, 2014). In the present study, the biological relevance of PrtX was examined in germ-free mice. Specifically, intestinal epithelial barrier integrity was assessed in germ-free mice colonized with either ΔprtX or the WT strains. Dysbiosis of the normal enteric gastrointestinal microbiome has been demonstrated as a key factor in the initiation and amplification of IBD (Tamboli et al., 2004). Recent evidence has implicated enteric and commensal proteases as one such mechanism for pathogenesis in IBD (Pruteanu et al., 2011; Carroll and Maharshak, 2013). In Enterococcus faecalis, a Gram-positive commensal bacterium of the gastrointestinal tract, extracellular proteases have been shown to mediate intestinal epithelial barrier disruption and contribute to intestinal inflammation (Steck et al., 2011; Maharshak et al., 2015). Here, we show that the ΔprtX mutant causes increased epithelial barrier integrity in the germ-free mice compared to WT colonized mice. PrtX may directly interact with various ECM components of intestinal epithelia eliciting a direct effect on intestinal epithelial integrity. It is also possible that the indirect immunomodulatory effects of ΔprtX in the germ-free mouse model resulted in the increased barrier integrity, as cytokines can modulate tight junction structure and function in intestinal epithelial cells (Walsh et al., 2000).
Conclusion
We have demonstrated that PrtX is a cell surface, S-layer associated serine protease homolog. Deletion of prtX from the chromosome of L. acidophilus NCFM revealed a distinct alteration in cell morphology, autoaggregation, and increased cell binding to mucin and fibronectin. Expression of IL-6, IL-12, and IL-10 was increased, in vitro, in mouse DCs. Furthermore, colonization of the ΔprtX strain in a germ-free mouse model improved gastrointestinal epithelial barrier integrity. Collectively, these data indicate that PrtX acts on the intestinal cell matrix and likely is involved in protein turnover during microbial cell division. PtrX, therefore, likely impacts how proteins are displayed within the microbial cell surface matrix and may alter the structure and properties of the epithelial intestinal cell matrix.
Author Contributions
BJ designed the study, conducted the research, interpreted the results, and wrote the paper. SOF, YG, and IC conducted the research, interpreted the results, and contributed/edited the paper. RB interpreted results and contributed/edited the paper. TK designed the study, interpreted results, and contributed/edited the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was funded, in part, by the North Carolina Agricultural Foundation and DuPont Nutrition & Health. The authors would like to thank Dr. Sue Tonkonogy and Rosemary Sanozky-Dawes for advice, technical assistance, and support, as well as Dr. Anthony Blikslager for the FITC-Dextran serum protocol. We also thank Karen Flores of the Gnotobiotic Core, College of Veterinary Medicine, North Carolina State University for technical assistance with experiments using germ-free and gnotobiotic mice. We acknowledge Valerie Lapham at the North Carolina State University Center for Electron Microscopy for assistance with scanning electron microscopy imaging. The Gnotobiotic Core is a facility of the Center for Gastrointestinal Biology and Disease, funded by NIH grant P30 DK034987.
References
Altermann, E., Buck, L. B., Cano, R., and Klaenhammer, T. R. (2004). Identification and phenotypic characterization of the cell-division protein CdpA. Gene 342, 189–197. doi: 10.1016/j.gene.2004.08.004
Altermann, E., Russell, W. M., Azcarate-Peril, M. A., Barrangou, R., Buck, B. L., McAuliffe, O., et al. (2005). Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. U.S.A. 102, 3906–3912. doi: 10.1073/pnas.0409188102
Azcarate-Peril, M. A., Tallon, R., and Klaenhammer, T. R. (2009). Temporal gene expression and probiotic attributes of Lactobacillus acidophilus during growth in milk. J. Dairy Sci. 92, 870–886. doi: 10.3168/jds.2008-1457
Bron, P. A., Tomita, S., Mercenier, A., and Kleerebezem, M. (2013). Cell surface-associated compounds of probiotic lactobacilli sustain the strain-specificity dogma. Curr. Opin. Microbiol. 16, 262–269. doi: 10.1016/j.mib.2013.06.001
Buck, B. L., Altermann, E., Svingerud, T., and Klaenhammer, T. R. (2005). Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71, 8344–8351. doi: 10.1128/AEM.71.12.8344-8351.2005
Call, E. K., Goh, Y. J., Selle, K., Klaenhammer, T. R., and O’Flaherty, S. (2015). Sortase-deficient lactobacilli: effect on immunomodulation and gut retention. Microbiology 161, 311–321. doi: 10.1099/mic.0.000007
Carroll, I. M., and Maharshak, N. (2013). Enteric bacterial proteases in inflammatory bowel disease-pathophysiology and clinical implications. Wolrd J. Gastroenterol. 19, 7531–7543. doi: 10.3748/wjg.v19.i43.7531
Douglas, G. L., and Klaenhammer, T. R. (2010). Genomic evolution of domesticated microorganisms. Annu. Rev. Food Sci. Technol. 1, 397–414. doi: 10.1146/annurev.food.102308.124134
Frece, J., Kos, B., Svetec, I. K., Zgaga, Z., Mrsa, V., and Suskoviæ, J. (2005). Importance of S-layer proteins in probiotic activity of Lactobacillus acidophilus M92. J. Appl. Microbiol. 98, 285–292. doi: 10.1111/j.1365-2672.2004.02473.x
Genay, M., Sadat, L., Gagnaire, V., and Lortal, S. (2009). prtH2, not prtH, is the ubiquitous cell wall proteinase gene in Lactobacillus helveticus. Appl. Environ. Microbiol. 75, 3238–3249. doi: 10.1128/AEM.02395-08
Gilbert, C., Atlan, D., Blanc, B., Portailer, R., Germond, J. E., Lapierre, L., et al. (1996). A new cell surface proteinase: sequencing and analysis of the prtB gene from Lactobacillus delbrueckii subsp. bulgaricus. J. Bacteriol. 178, 3059–3065. doi: 10.1128/jb.178.11.3059-3065.1996
Gilliland, S. E., Speck, M. L., and Morgan, C. G. (1975). Detection of Lactobacillus acidophilus in feces of humans, pigs, and chickens. Appl. Microbiol. 30, 541–545.
Goh, Y. J., Azcarate-Peril, M. A., O’Flaherty, S., Durmaz, E., Valence, F., Jardin, J., et al. (2009). Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 75, 3093–3105. doi: 10.1128/AEM.02502-08
Goh, Y. J., and Klaenhammer, T. R. (2010). Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 76, 5005–5012. doi: 10.1128/AEM.00030-10
Goh, Y. J., and Klaenhammer, T. R. (2014). Insights into glycogen metabolism in Lactobacillus acidophilus: impact on carbohydrate metabolism, stress tolerance and gut retention. Microb. Cell Fact. 13:94. doi: 10.1186/s12934-014-0094-3
Hamer, H. M., Jonkers, D. M. A. E., Vanhoutvin, S. A. W. L., Troost, F. J., Rijkers, G., de Bruine, A., et al. (2010). Effect of butyrate enemas on inflammation and antioxidant status in the colonic mucosa of patients with ulcerative colitis in remission. Clin. Nutr. 29, 738–744. doi: 10.1016/j.clnu.2010.04.002
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/S0022-2836(83)80284-8
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Horton, R. M., Hunt, H. D., Ho, S. N., Pullen, J. K., and Pease, L. R. (1989). Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68. doi: 10.1016/0378-1119(89)90359-4
Hymes, J., Johnson, B. R., Barrangou, R., and Klaenhammer, T. R. (2016). Functional analysis of an S-layer-associated fibronectin-binding protein in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 82, 2676–2685. doi: 10.1128/AEM.00024-16
Hynönen, U., and Palva, A. (2013). Lactobacillus surface layer proteins: structure, function and applications. Appl. Microbiol. Biotechnol. 97, 5225–5243. doi: 10.1007/s00253-013-4962-2
Johnson, B., Selle, K., O’Flaherty, S., Goh, Y. J., and Klaenhammer, T. (2013). Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiology 159, 2269–2282. doi: 10.1099/mic.0.070755-0
Johnson, B. R., Hymes, J., Sanozky-Dawes, R., DeCrescenzo Henriksen, E., Barrangou, R., and Klaenhammer, T. R. (2016). Conserved S-layer-associated proteins revealed by exoproteomic survey of S-layer-forming lactobacilli. Appl. Environ. Microbiol. 82, 134–145. doi: 10.1128/AEM.01968-15
Johnson, B. R., and Klaenhammer, T. R. (2014). Impact of genomics on the field of probiotic research: historical perspectives to modern paradigms. Antonie Van Leeuwenhoek 106, 141–156. doi: 10.1007/s10482-014-0171-y
Johnson, B. R., and Klaenhammer, T. R. (2016). AcmB is an S-layer-associated β-N-acetylglucosaminidase and functional autolysin in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 82, 5687–5697. doi: 10.1128/AEM.02025-16
Kebouchi, M., Galia, W., Genay, M., Soligot, C., Lecomte, X., Awussi, A. A., et al. (2016). Implication of sortase-dependent proteins of Streptococcus thermophilus in adhesion to human intestinal epithelial cell lines and bile salt tolerance. Appl. Microbiol. Biotechnol. 100, 3669–3679. doi: 10.1007/s00253-016-7322-1
Klaenhammer, T. R., Altermann, E., Pfeiler, E. A., Buck, B. L., Goh, Y. J., O’Flaherty, S., et al. (2008). Functional genomics of probiotic Lactobacilli. J. Clin. Gastroenterol. 42, 160–162. doi: 10.1097/MCG.0b013e31817da140
Klaenhammer, T. R., Barrangou, R., Buck, B. L., Azcarate-Peril, M. A., and Altermann, E. (2005). Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol. Rev. 29, 393–409. doi: 10.1016/j.fmrre.2005.04.007
Konstantinov, S. R., Smidt, H., de Vos, W. M., Bruijns, S. C. M., Singh, S. K., Valence, F., et al. (2008). S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. U.S.A. 105, 19474–19479. doi: 10.1073/pnas.0810305105
Korhonen, H., and Pihlanto, A. (2006). Bioactive peptides: production and functionality. Int. Dairy J. 16, 945–960. doi: 10.1016/j.idairyj.2005.10.012
Law, J., Buist, G., Haandrikman, A., Kok, J., Venema, G., and Leenhouts, K. (1995). A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177, 7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995
Lebeer, S., Vanderleyden, J., and De Keersmaeker, S. C. J. (2010). Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol. 8, 171–184. doi: 10.1038/nrmicro2297
Li, M. C., and He, S. H. (2004). IL-10 and its related cytokines for treatment of inflammatory bowel disease. World J. Gastroenterol. 10, 620–625. doi: 10.3748/wjg.v10.i5.620
Lightfoot, Y. L., Selle, K., Yang, T., Goh, Y. J., Sahay, B., Zadeh, M., et al. (2015). SIGNR3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J. 34, 881–895. doi: 10.15252/embj.201490296
Maharshak, N., Young Huh, E., Paiboonrungruang, C., Shanahan, M., Thurlow, L., Herzog, J., et al. (2015). Enterococcus faecalis gelatinase mediates intestinal permeability via protease-activated receptor 2. Infect. Immun. 83, 2762–2770. doi: 10.1128/IAI.00425-15
Makarova, K., Slesarev, A., Wolf, Y., Sorokin, A., Mirkin, B., Koonin, E., et al. (2006). Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 15611–15616. doi: 10.1073/pnas.0607117103
Matar, C., Valdez, J. C., Medina, M., Rachid, M., and Perdigon, G. (2001). Immunomodulating effects of milks fermented by Lactobacillus helveticus and its non-proteolytic variant. J. Dairy Res. 68, 601–609. doi: 10.1017/S0022029901005143
Mohamadzadeh, M., Pfeiler, E. A., Brown, J. B., Zadeh, M., Gramarossa, M., Managlia, E., et al. (2011). Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc. Natl. Acad. Sci. U.S.A. 108, 4623–4630. doi: 10.1073/pnas.1005066107
O’Flaherty, S., and Klaenhammer, T. R. (2010a). The role and potential of probiotic bacteria in the gut, and the communication between gut microflora and gut/host. Int. Dairy J. 20, 262–268.
O’Flaherty, S. J., and Klaenhammer, T. R. (2010b). Functional and phenotypic characterization of a protein from Lactobacillus acidophilus involved in cell morphology, stress tolerance and adherence to intestinal cells. Microbiology 156, 3360–3367. doi: 10.1099/mic.0.043158-0
O’Garra, A., and Murphy, K. M. (2009). From IL-10 to IL-12: how pathogens and their products stimulate APCs to induce TH1 development. Nat. Immunol. 10, 929–932. doi: 10.1038/ni0909-929
Pederson, J. A., Mileski, G. J., Weimer, B. C., and Steele, J. L. (1999). Genetic characterization of a cell envelope-associated proteinase from Lactobacillus helveticus CNRZ32. J. Bacteriol. 181, 4592–4597.
Pruteanu, M., Hyland, N. P., Clarke, D. J., Kiely, B., and Shanahan, F. (2011). Degradation of the extracellular matrix components by bacterial-derived metalloproteases: implications for inflammatory bowel diseases. Inflamm. Bowel Dis. 17, 1189–1200. doi: 10.1002/ibd.21475
Russell, W. M., and Klaenhammer, T. R. (2001). Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl. Environ. Microbiol. 67, 4361–4364. doi: 10.1128/AEM.67.9.4361-4364.2001
Sadat-Mekmene, L., Jardin, J., Corre, C., Molle, D., Richoux, R., Delage, M. M., et al. (2011). Simultaneous presence of PrtH and PrtH2 proteinases in Lactobacillus helveticus strains improves breakdown of the pure αs1-casein. Appl. Environ. Microbiol. 77, 179–186. doi: 10.1128/AEM.01466-10
Sanders, M. E., and Klaenhammer, T. R. (2001). Invited review: the scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J. Dairy Sci. 84, 319–331. doi: 10.3168/jds.S0022-0302(01)74481-5
Sára, M., and Sleytr, U. B. (2000). S-layer proteins. J. Bacteriol. 182, 859–868. doi: 10.1128/JB.182.4.859-868.2000
Shah, N. P. (2007). Functional cultures and health benefits. Int. Dairy J. 17, 1262–1277. doi: 10.1016/j.idairyj.2007.01.014
Silva, S. V., and Malcata, F. X. (2005). Caseins as source of bioactive peptides. Int. Dairy J. 15, 1–15. doi: 10.1016/j.idairyj.2004.04.009
Steck, N., Hoffmann, M., Sava, I. G., Kim, S. C., Hahne, H., Tonkonogy, S. L., et al. (2011). Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology 141, 959–971. doi: 10.1053/j.gastro.2011.05.035
Tamboli, C. P., Neut, C., Desreumaux, P., and Colombel, J. F. (2004). Dysbiosis in inflammatory bowel disease. Gut 53, 1–4. doi: 10.1136/gut.53.1.1
von Schillde, M. A., Hormannsperger, G., Weiher, M., Alpert, C. A., Hahne, H., Bauerl, C., et al. (2012). Lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Cell Host Microbe 11, 387–396. doi: 10.1016/j.chom.2012.02.006
Walker, D. C., Aoyama, K., and Klaenhammer, T. R. (1996). Electrotransformation of Lactobacillus acidophilus group A1. FEMS Microbiol. Lett. 138, 233–237. doi: 10.1111/j.1574-6968.1996.tb08163.x
Walsh, S. V., Hopkins, A. M., and Nusrat, A. (2000). Modulation of tight junction structure and function by cytokines. Adv. Drug Deliv. Rev. 41, 303–313. doi: 10.1016/S0169-409X(00)00048-X
Wei, M. Q., Rush, C. M., Norman, J. M., Hafner, L. M., Epping, R. J., and Timms, P. (1995). An improved method for the transformation of Lactobacillus strains using electroporation. J. Microbiol. Methods 21, 97–109. doi: 10.1016/0167-7012(94)00038-9
Keywords: serine protease, S-layer, S-layer associated proteins, Lactobacillus, probiotic, intestinal barrier integrity, mucin, fibronectin
Citation: Johnson BR, O’Flaherty S, Goh YJ, Carroll I, Barrangou R and Klaenhammer TR (2017) The S-layer Associated Serine Protease Homolog PrtX Impacts Cell Surface-Mediated Microbe-Host Interactions of Lactobacillus acidophilus NCFM. Front. Microbiol. 8:1185. doi: 10.3389/fmicb.2017.01185
Received: 02 April 2017; Accepted: 12 June 2017;
Published: 30 June 2017.
Edited by:
Michael Gänzle, University of Alberta, CanadaReviewed by:
Francesca Turroni, University College Cork, IrelandEric Altermann, AgResearch, New Zealand
Copyright © 2017 Johnson, O’Flaherty, Goh, Carroll, Barrangou and Klaenhammer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Todd R. Klaenhammer, klaenhammer@ncsu.edu
 Brant R. Johnson
Brant R. Johnson Sarah O’Flaherty
Sarah O’Flaherty Yong Jun Goh
Yong Jun Goh Ian Carroll3,4
Ian Carroll3,4 Rodolphe Barrangou
Rodolphe Barrangou Todd R. Klaenhammer
Todd R. Klaenhammer