Cryptic Diversity and Database Errors Challenge Non-indigenous Species Surveys: An Illustration With Botrylloides spp. in the English Channel and Mediterranean Sea
- 1Laboratory AD2M (UMR7144), Station Biologique de Roscoff, Sorbonne Université, CNRS, Roscoff, France
- 2Department of Marine Ecology, Centre for Advanced Studies of Blanes (CEAB, CSIC), Blanes, Spain
- 3Citadel Hill Laboratory, Marine Biological Association of the United Kingdom, Plymouth, United Kingdom
Molecular tools have been extensively used in recent decades to examine biological invasion processes, and are increasingly being adopted as efficient tools to support non-indigenous species surveys, notably through barcoding approaches, i.e., the use of a reference sequence specific to a given species to validate its identification. The technique is easy to use but requires reliable reference sequences to be available in public databases. In addition, the increasing discovery of cryptic species in marine taxa may complicate taxonomic assignment. We illustrate these two issues in the ascidian genus Botrylloides, in which at least three global marine invaders have been recognized, including B. violaceus and B. diegensis. We obtained COI sequences from >750 colonies of Botrylloides spp. sampled in W Europe or provided by expert colleagues from other regions. Phylogenetic trees clearly distinguished our targeted taxa [i.e., B. violaceus, B. diegensis and B. leachii (native)]. They also revealed another discrete lineage apparently related to a recently described eastern Mediterranean species. By examining public databases, we found sequences of B. diegensis erroneously assigned to B. leachii. This observation has major implications as the introduced B. diegensis can be misidentified as a putatively native species. We also checked published sequences of the genus Botrylloides in the Mediterranean Sea, complemented with new samples. Based on our custom reference database, all published sequences of B. leachii corresponded to B. diegensis, although this NIS has hardly been reported at all in the Mediterranean region. Such database errors are unfortunate, as the barcoding approach is a powerful tool to identify the recognized Botrylloides species currently present in European seas. This is of particular importance because a trait often used during field assessment, i.e., single-color vs. two-color colonies, is misleading to distinguish B. violaceus and B. diegensis respectively: a substantial proportion of the single-color morph are actually B. diegensis in both the Mediterranean Sea and the English Channel. Altogether, this study exemplifies the advantages and disadvantages of molecular barcoding in NIS surveys and studies. The limitations that were identified are all easy to resolve once proper vouchers and collections are set up.
Introduction
In marine environments, biological introductions exert important pressure on natural ecosystems. Introduction of non-indigenous species (NIS) has occurred at an increasing rate since the 20th century, in pace with the increasing range and intensity of vectors (Nunes et al., 2014). Because successful eradications of invasive species in marine systems are very rare (Sambrook et al., 2014; Ojaveer et al., 2015), both pre-border prevention measures (e.g., limiting the introduction of NIS by ballast waters) and early detection focused on introduction hotspots (e.g., marinas and harbors) have been recommended (Ojaveer et al., 2014). Accurate identification is a pre-requisite for the implementation of policies and regulations such as the Marine Framework Strategy Directive (MSFD), in which Descriptor 2 is dedicated to NIS, with one criterion involving the detection and identification of new NIS, and another concerning the changing spatial extent of already-reported NIS.
However, identifying new or recently introduced NIS can be challenging when based only on traditional methods (Darling et al., 2017), such as rapid assessment surveys (RAS), which rely heavily on field recognition of species and have been commonly used to survey NIS in marinas and harbors (e.g., Cohen et al., 2005; Arenas et al., 2006; Campbell et al., 2007; Bishop et al., 2015a; Lehtiniemi et al., 2015). Many marine animal NIS in introduction hotspots belong to taxonomic groups, such as bryozoans, hydrozoans and tunicates, that require substantial taxonomic expertise. In addition, the development of molecular studies in the last three decades has revealed a large number of cryptic species (i.e., species that are not distinguishable based on morphological traits) in these taxonomic groups (Knowlton, 2000; Appeltans et al., 2012). Consequently, an increasing number of studies have recommended the use of molecular tools to complement the traditional methods, and thus achieve reliable taxonomic identification of marine NIS (Comtet et al., 2015; Darling et al., 2017; Dias et al., 2017).
The usefulness of the molecular barcoding approach is well established. The benefits of the approach have been demonstrated by numerous studies (for a review, see Comtet et al., 2015). Such an approach can be used to ascertain the presence of a new NIS (e.g., Asterocarpa humilis: Bishop et al., 2013), to reveal false morphology-based NIS identification (e.g., Crepidula spp.: McGlashan et al., 2008) or to determine the taxonomic status of previously unrecognized NIS (Ordóñez et al., 2016). However, the robustness of the molecular barcoding approach relies on some important pre-requisites, notably the availability of reliable reference sequence data for the targeted species. Thus, the prompt updating of databases is essential following taxonomic revision and discovery of unrecognized cryptic species. The sequences delivered in public databases such as GenBank (Benson et al., 2013) or BOLD [Barcode of Life Data System; (Ratnasingham and Hebert, 2007)] should thus be error-free and validated by taxonomic experts. BOLD is expected to provide such a framework. However, the public portal also retrieves non-curated sequences from Genbank that can propagate errors. There is so far a limited number of initiatives specifically aimed at delivering reference data for marine NIS. Although rare, such initiatives should be strongly encouraged, notably to support international regulations and policies such as the MSFD in Europe (Darling et al., 2017). Such an initiative has recently been undertaken in Western Australia, where a reference collection (vouchers, DNA and sequences for one marker (COI)) was assembled for 75 species (out of 79) of the “Western Australian Prevention List for Introduced Marine Pests” (Dias et al., 2017). Similarly, facing the issue of cryptic diversity, integrative taxonomy coupling molecular phylogenetics with morphology and other species attributes (e.g., ecology, life-history traits) is needed (Pante et al., 2015).
We here illustrate the issues described above using the colonial tunicate genus Botrylloides as a particularly relevant case study. At least two Botrylloides species, B. violaceus Oka, 1927 and B. diegensis Ritter and Forsyth, 1927, with the recent addition of B. giganteus (Pérés, 1949) (Rocha et al., 2019) are globally invasive. B. violaceus and B. diegensis are both native to the N Pacific. While the former is native to the NW Pacific, there is more uncertainty regarding the native range of the latter: although B. diegensis was originally described from the NE Pacific (southern California), it is likely that the species had been introduced from the western or southern Pacific (Carlton, 2009). The two species are important members of the fouling community colonizing artificial substrates on the Pacific coast of the United States, for instance in harbors and marinas (e.g., Nydam and Stachowicz, 2007). The same two species have also been introduced in Europe, notably in the English Channel (EC), where they are well established in marinas of the United Kingdom and N France (Bishop et al., 2015a, b). In Europe, one putatively native species, B. leachii (Savigny, 1816), is also recognized, often showing coloration somewhat similar to the two-tone color pattern seen in B. diegensis (Supplementary Figure S3, panel 6). The original description (Savigny, 1816) of B. leachii was based on material “Communiqué par M[onsieur]. Leach, directeur du Musée britannique” (i.e., William Elford Leach, then working in the Department of Natural History of the British Museum) and the specimen was thought by Savigny to have originated probably from the English coast (“Habite les côtes de l’Angleterre?”). Conversely to the introduced species, the native taxon seems uncommon in artificial habitats in some parts of the EC (J. Bishop, L. Lévêque, and F. Viard, pers. obs.).
The first reports of non-indigenous Botrylloides spp. in the English Channel were made after RAS in 2004 (Arenas et al., 2006). A single species, B. violaceus, was reported, recognized by its single-colored colonies and large and morphologically distinctive brooded larvae. Colonies of B. violaceus could occur in a range of single colors, including violet, cream, yellow (Supplementary Figure S3, panel 4), brick-red and (commonly) orange. A second species was encountered during the 2004 surveys, but was not reported by Arenas et al. (2006) since its identity was uncertain (Bishop et al., 2015a, b). This second species showed a strong and distinctive two-colored pattern featuring a broad ring of solid pigmentation (commonly orange or cream) surrounding each buccal orifice, against a contrasting darker background (Supplementary Figure S3, panel 2), and was subsequently identified as B. diegensis. This distinction, based on color morphs, was used by Lambert and Lambert (2003) to distinguish B. violaceus and B. diegensis in southern California, United States. In 2011, the present authors and colleagues collected extensive samples on both sides of the English Channel for an intended population-genetic study of B. violaceus; single-color colonies were thus collected, with or without confirmation of the distinctive but seasonally brooded larvae of that species, under the assumption that these would all be the target species. Preliminary molecular analyses revealed that these single-color colonies included specimens attributable to B. diegensis, so that B. diegensis had been misidentified in the field as B. violaceus on the criterion of possessing single-colored, rather than two-colored, colonies. B. diegensis in the English Channel could thus occur in a single-color morphotype (e.g., Supplementary Figure S3, panel 1) easily confused with B. violaceus, even by those supposedly familiar with the taxa. In addition, during our surveys some color morphs believed to belong to B. leachii were encountered that had a two-color pattern similar to the two-color pattern of B. diegensis, although differing in detail (Supplementary Figure S3, panel 6).
Following from these preliminary observations and molecular results, in this study we examined a large number of Botrylloides colonies displaying a single-color pattern, using sequencing data to make an in-depth assessment of the association between color-morph and species. We chose to analyze a fragment of the mitochondrial gene region cytochrome oxidase I (COI) which has been previously investigated to study B. violaceus in its N American introduced range (Bock et al., 2011; Lejeusne et al., 2011), besides being a marker commonly used in barcoding studies of marine invertebrates (Comtet et al., 2015) and a standard for BOLD (Hebert et al., 2016). This dataset was completed with data from specimens kindly provided by taxonomic experts (see acknowledgment section); this allowed us to get reliable reference sequences for the three Botrylloides species, B. violaceus, B. diegensis and B. leachii, putatively present in the EC. We further analyzed if similar confusion existed in the Mediterranean, where six Botrylloides species have been reported: B. leachii (native), B. violaceus (introduced), B. pizoni Brunetti and Mastrototaro, 2012 (likely a synonym of the invasive B. giganteus; Reem et al., 2018), B. niger Herdman, 1886 (likely a Lessepsian migrant), B. anceps (Herdman, 1891) (likely a Lessepsian migrant), and B. israeliense Brunetti (2009) (uncertain status). To this end, we used all existing Mediterranean sequences of the genus in GenBank, complemented with new samples, for comparison with our reference sequences. The objectives of our work were four-fold: (i) ascertaining the distribution of the two NIS in the study EC area, (ii) providing further molecular reference data, specific to European seas, (iii) assessing if field identification, based on the typical single-color pattern of B. violaceus, is truly reliable, and (iv) updating Mediterranean records of Botrylloides spp. backed by sequence data.
Materials and Methods
Specimen Collection
Colonies (N = 627) displaying a single color, as typically described for B. violaceus, were randomly sampled along floating pontoons, from April to September 2011, in 20 marinas along the southern coastline of England from Plymouth to Brighton (9 marinas) and along the coastlines of Brittany from Quiberon to Saint-Malo (11 marinas) (Table 1 and Figure 1). RAS done in these same locations have shown the presence of non-indigenous Botrylloides spp. (Bishop et al., 2015a; FV and L. Lévêque, unpublished data). All the sampling was undertaken by SCUBA diving or from the surface by removing colonies from the pilings and pontoons in marinas. Samples were preserved in 96% ethanol prior to genetic analysis.
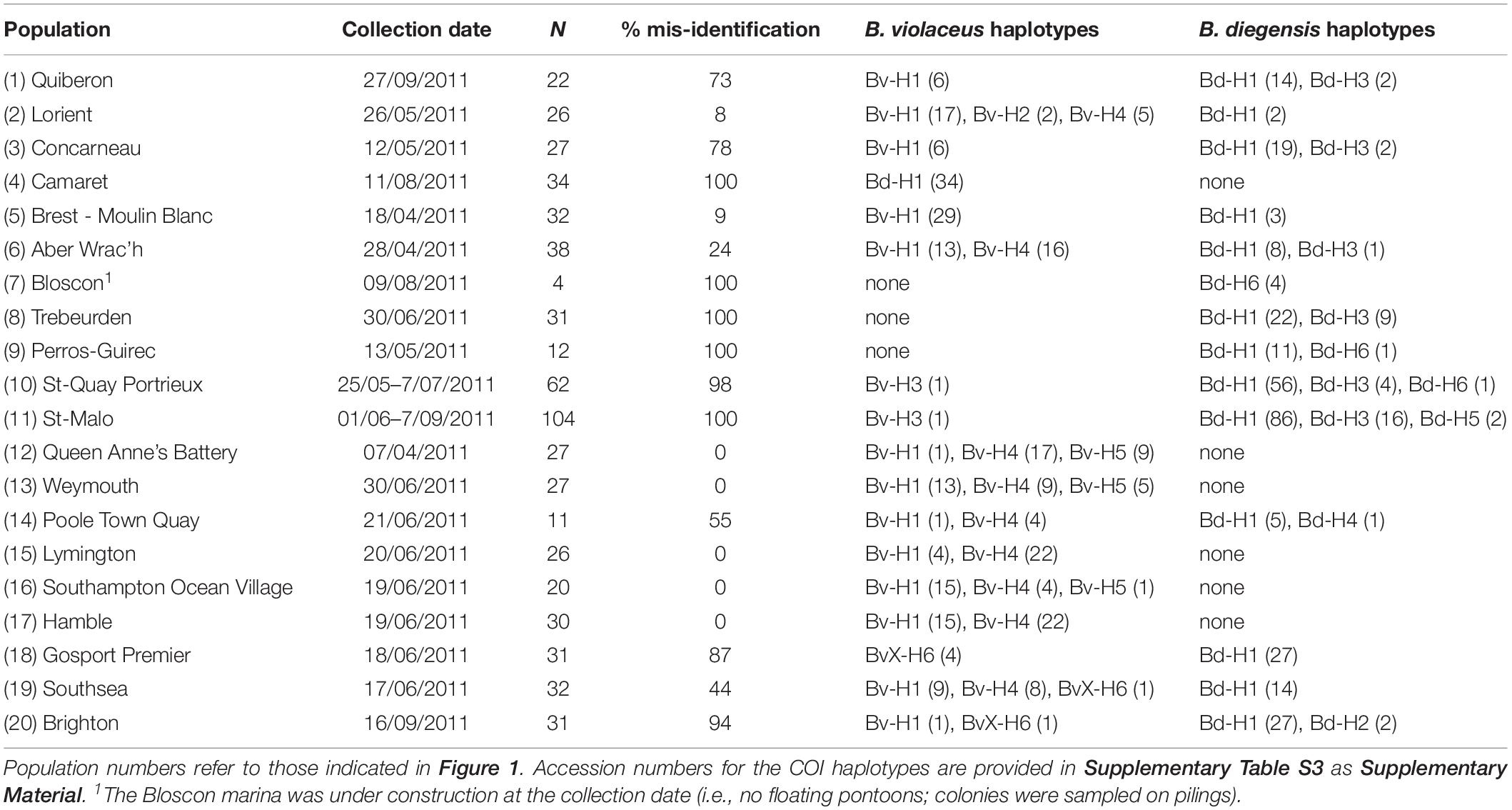
Table 1. Population sampling, proportion of mis-identification (based on COI sequencing) and haplotype counts.
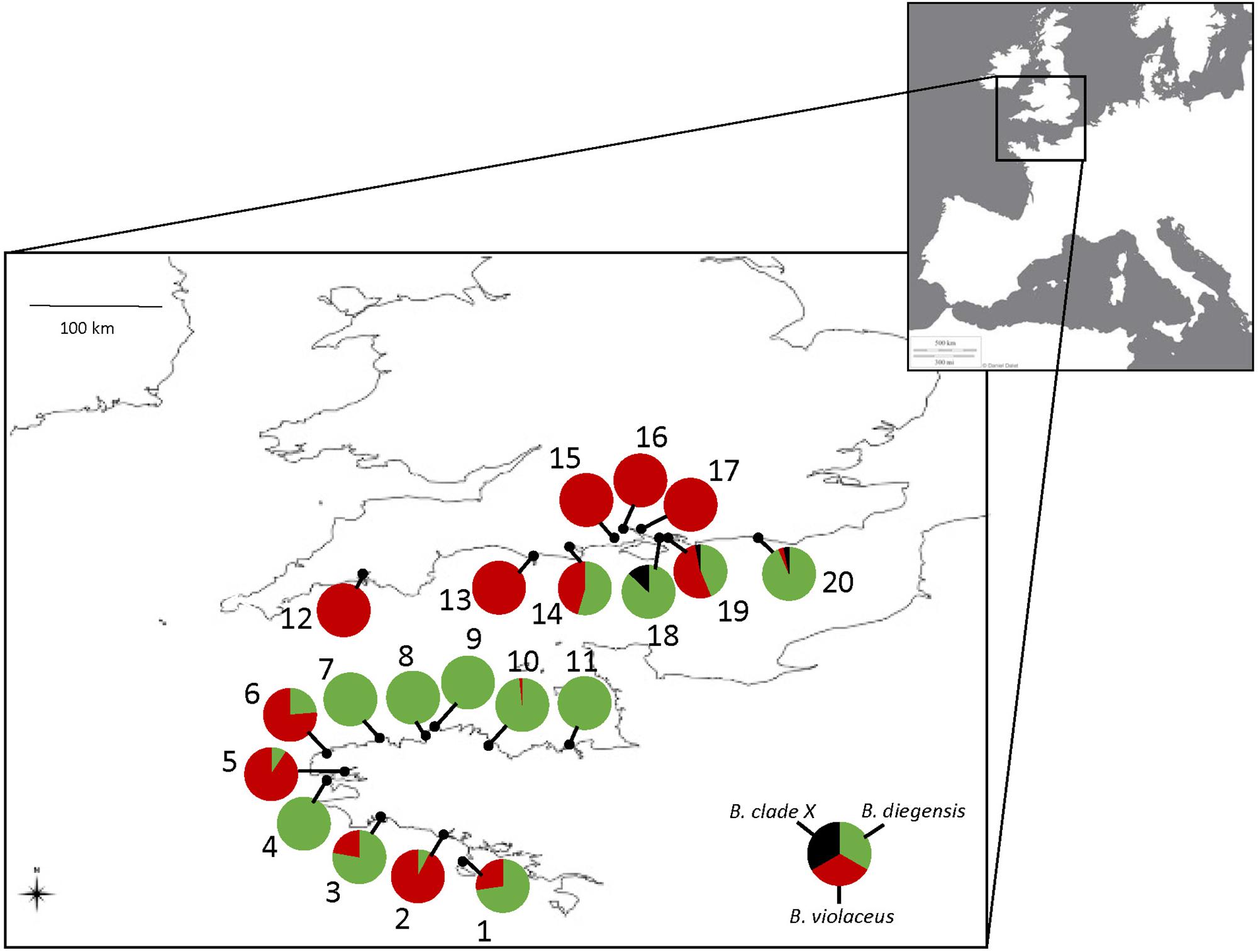
Figure 1. Study area and sampling localities of the 627 single-color colonies sampled in the English Channel and morphologically assigned to B. violaceus, with pie charts indicating the percentage of the three Botrylloides taxa, after being identified with molecular barcoding (see Table 1). Numbers refer to population name as indicated in Table 1. The source for the European map in the inset is ©Terre Ouverte.
A reference collection of 128 specimens was gathered for analysis (Supplementary Table S1). It included a total of 84 specimens of B. diegensis, of which 39 were kindly provided by expert colleagues from the W coast of United States, Italy and New Zealand. The remainder were sampled in four marinas in Brittany. In every case, these colonies displayed the typical two-color pattern of B. diegensis. We also included 17 reference specimens for B. leachii from United Kingdom, Ireland and France. These latter specimens mostly displayed the color pattern of Botrylloides radiata Alder and Hancock, 1848, as illustrated by Alder and Hancock (1912), pl. 64, Figures 10 and 11), a form synonymized with B. leachii by Berrill (1950) and Millar (1970) (Supplementary Figure S3, panels 5 and 6). These specimens will be hereafter referred to as the ‘radiata’ morph of B. leachii. Nine typical violet B. violaceus colonies, also displaying the distinctive large larvae of this species, were obtained in two French localities. Finally, to be used as an outgroup, 18 specimens of Botryllus schlosseri were collected in United Kingdom and France. All these samples were preserved in 96% ethanol after their collection.
In order to update Mediterranean records, a third dataset was constructed, using samples obtained from the NE of the Iberian Peninsula (oyster cultures in the Ebro Delta, 40°46′27.43′′N, 00°44′27.11′′E), the S of France (marina of Sète, 43°23′44.2″N, 3°41′51.0′′E), the SW of Italy (Miseno Lagoon, Naples, 40°47′34.19′′N, 14°04′39.37′′E) and the Adriatic (Venetian Lagoon, 45°25′50.54′′N, 12°22′07.77′′E), as well as a compilation of all Mediterranean Botrylloides COI sequences available in GenBank (Supplementary Table S2).
Mitochondrial DNA Amplifications and Sequencing
Genomic DNA was extracted from a single zooid (dissected under the microscope), using the Nucleospin 96 Tissue Kit (Macherey-Nagel, Düren, Germany) following the manufacturer’s protocol and using a final elution volume of 80 μl.
A fragment of the cytochrome c oxidase subunit I (COI) gene was amplified using the LCO1490 and HCO2198 primers (Folmer et al., 1994). Amplifications were carried out in a 30 μl reaction volume with 4 μl of genomic DNA, 1X PCR buffer (Thermoprime AbGene), 2 mM of MgCl2 (Thermoprime AbGene), 0.05 mM of each dNTP, 0.4 μM of each primer and 0.33 U of Taq DNA polymerase (Thermoprime AbGene). PCR was performed following the protocol of Lejeusne et al. (2011): initial denaturing step at 94°C for 3 min, followed by 5 amplification cycles (94°C for 50 s, 45°C for 50 s, 72°C for 60 s), 30 further cycles (94°C for 50 s, 50°C for 50 s, 72°C for 60 s), and a final elongation step at 72°C for 5 min. Sequencing reactions were performed at the LGC Genomics platform (Berlin, Germany) on purified (ExoSAP®-it) PCR products using the reverse primer (HCO2198). Mediterranean amplicons were obtained with the same primers and protocols, and sequenced at Macrogen Company (Korea).
Mitochondrial DNA Analyses
The sequences were edited using CodonCode Aligner v. 3.7.1 (CodonCode Corporation, Dedham, MA). They were aligned using BioEdit v.7.1.981 33 (Hall, 1999). After alignment, a 455 base-pair fragment was retained, further reduced to 435 base pairs for the EC dataset and to 395 base pairs for the Mediterranean dataset, for comparison with sequences obtained for our reference samples or available in public databases. DnaSP version 5 (Rozas et al., 2003) was used to compute the number and frequency of haplotypes (i.e., unique sequences) per taxon or locality and estimate polymorphism. The number of base substitutions per site across sequences was computed with MEGA7 (Tamura et al., 2011), using the Maximum Composite Likelihood model (Tamura et al., 2004) with positions with less than 95% site coverage eliminated.
Phylogenetic trees were built with MEGA7 (Tamura et al., 2011) using 32 unique sequences for the analyses of the EC dataset and 54 sequences with the Mediterranean dataset (details in Results). Goodness of fit with various evolution models was first tested: based on both BIC criteria and AICc value, the best model explaining the data was the Hasegawa-Kishino-Yano model with 54–61% of the sites evolutionarily invariable, according to the trees. Rooted phylogenetic trees were constructed using a maximum likelihood method with heuristic search (Subtree-Pruning-Regrafting method). To assess the reliability of the inferred trees, bootstrap tests were carried out (1000 bootstraps). Note that similar topologies were obtained using other tree construction methods, in particular a simple distance-based method (i.e., Neighbor-joining).
To start investigating the status of a new reported lineage (see results), GMYC (Pons et al., 2006) and mPTP (Kapli et al., 2017) species delimitation analyses were carried out on the EC and Mediterranean unique sequences of Botrylloides spp. These two approaches differ in their properties (e.g., GYMC requires ultrametric trees), and one may be more confident in the inferred species delimitation when similar results are obtained with both. GMYC analysis was carried out with the R library splits, using an ultrametric tree built using the Beast2 software (Bouckaert et al., 2019), with a Yule tree prior and site model as obtained with MEGA analysis (see above). The mPTP analysis was carried out with the web-service available at http://mptp.h-its.org, with the tree produced with MEGA (note that the same results were obtained with the tree produced by the Beast2 Bayesian phylogenetic inference).
Results
Haplotypic Diversity and Comparison to Reference Samples
Altogether, over the 627 colonies sampled in 20 marinas in 2011, we obtained 12 mitochondrial haplotypes (Table 1) over 455 base pairs, with no missing data. The polymorphism was very high with 122 sites showing substitutions, for a total number of 140 mutations. In addition, although these colonies were all assigned to B. violaceus based on their color, evolutionary divergence over all sequence pairs was substantial, with a number of base substitutions per site of 0.175 over the 12 sequence pairs. Marked variations were observed among pairwise sequence divergence estimates, ranging from 0.002 to 0.301. On the other hand, the analysis of the 127 reference samples including the three Botrylloides spp. putatively present and Botryllus schlosseri (outgroup) yielded 20 mitochondrial haplotypes (Supplementary Table S1).
A phylogenetic tree was built to map the 12 haplotypes obtained over the 627 colonies with the 20 sequences obtained for the 127 reference samples (Figure 2). This tree displayed three monophyletic groups associated with 1) B. diegensis, 2) B. leachii (‘radiata’ morph) and 3) B. violaceus reference sequences. It showed that 11 haplotypes obtained from the colonies sampled actually belonged to two groups, namely those clustering with B. violaceus and B. diegensis references. As these colonies were all originally thought to be B. violaceus, based on the single-color pattern, this result thus clearly indicates mis-identification during collection in the field. No sequence clustered with our B. leachii references. Among the 11 haplotypes, seven were 100% identical to one of the 20 reference sequences: four (out of six assigned to the B. diegensis clade) were identical to B. diegensis references (Bd-H1.Ref, Bd-H2.Ref, Bd-H3.Ref, and Bd-H6.Ref in Figure 2) and three (out of five) to B. violaceus references (Bv-H1.Ref, Bv-H3.Ref, Bv-H4.Ref).
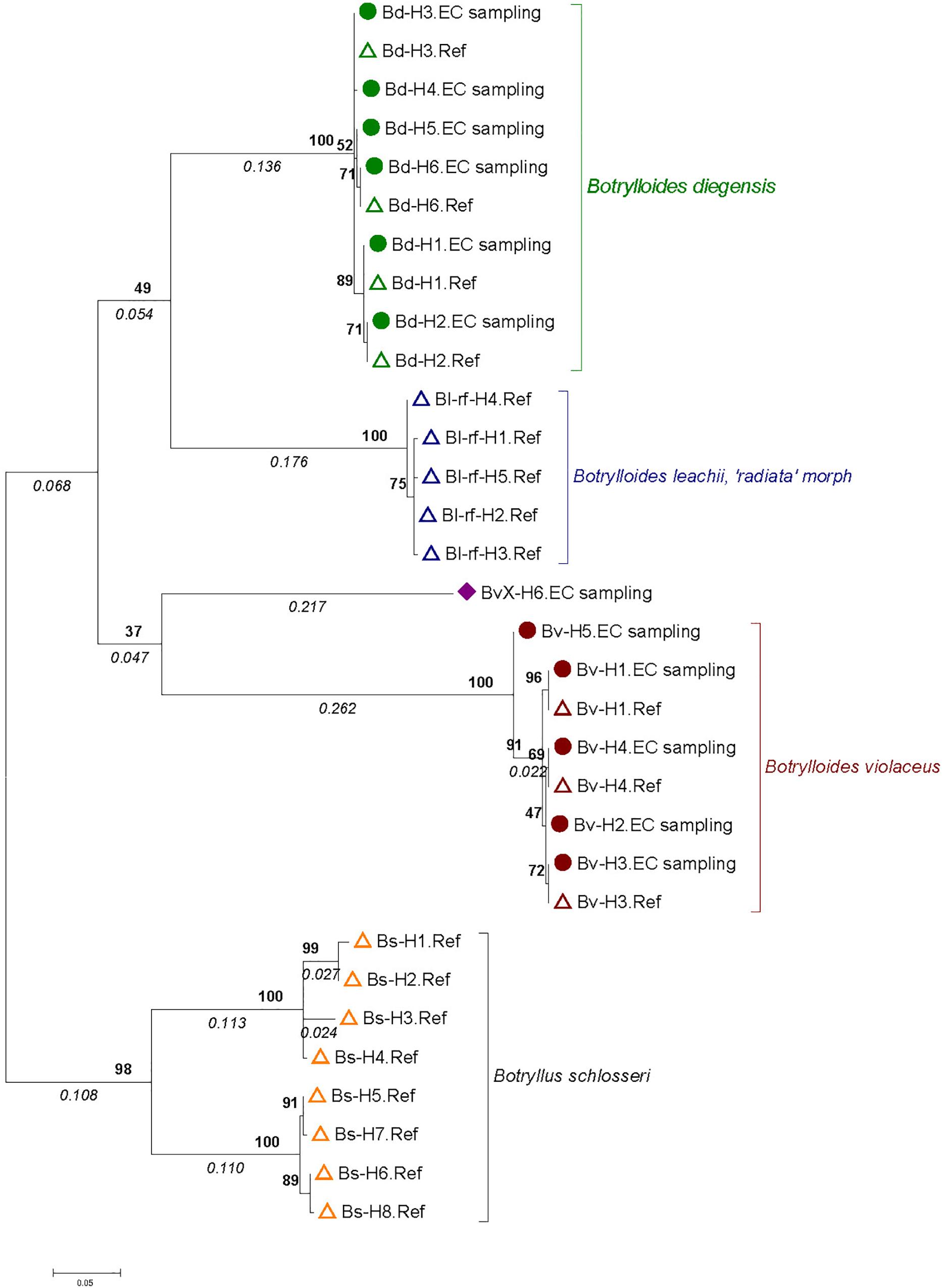
Figure 2. Molecular phylogenetic tree based on 30 Botrylloides spp. and Botryllus schlosseri haplotypes. The tree was built by the Maximum Likelihood Method based on the Hasegawa-Kishino-Yano model with 61% of sites evolutionarily invariable. It is made of 12 haplotypes obtained from 627 single-color Botrylloides spp. colonies sampled in the English Channel (solid circles and diamond, name ending with “EC sampling”) and 20 haplotypes from reference samples (open triangles; name ending with ‘Ref’) from Botrylloides spp. and Botryllus schlosseri colonies (see text and Table 1 and Supplementary Table S1 for details). A total of 435 positions were analyzed. The tree is drawn to scale, with branch lengths measured as number of substitutions per site indicated in italics below the branch. Numbers in bold on nodes indicate percent bootstrap support values (1000 bootstraps).
One haplotype (‘BvX-H6.EC sampling’ in Figure 2) clustered with the five B. violaceus haplotypes but the topology was poorly supported by bootstrap values. In addition, the evolutionary divergence of this haplotype (hereafter named BvX-H6) from the other haplotypes of B. violaceus was particularly high (19.6%) (Table 2). This strong divergence is clearly reflected in the mismatch distribution shown in Supplementary Figure S1 provided as Supplementary Material. When removing this haplotype from the analyses, the three targeted species (i.e., B. diegensis, B. violaceus and B. leachii) were characterized by a low level of within-species divergence (0.5 to 1.29%) and much higher level (15.5–19.6%) of divergence among them (Table 2), highlighting that the COI marker used is a robust barcoding marker to distinguish them, in particular B. diegensis from B. violaceus, and that BvX-H6 corresponds to an evolutionarily divergent lineage as compared to the three targeted species. Colonies with the BvX-H6 haplotype displayed a single-color pattern (Supplementary Figure S3, panels 7 and 8) broadly similar to B. violaceus.

Table 2. Estimates of average molecular divergence over haplotype pairs within and between taxonomic groups (i.e., the three accepted species, examining separately the haplotype BvX-H6).
Mis-Identification Rates and Haplotype Distribution Across Populations
Based on the results of the phylogenetic analysis, each of the 627 colonies could be assigned to either B. diegensis, B. violaceus or the divergent BvX-H6 lineage. Altogether 373 (59%) of the 627 single-color colonies sampled in the United Kingdom and France as B. violaceus in 2011 were unambiguously assigned to B. diegensis on molecular criteria, as shown above. We observed large variations across regions and populations (Figure 1), with mis-identification ranging from 0 to 100%, and a mean mis-identification rate of 53% per population. The mis-identification was not associated with a particular haplotype, with six haplotypes found among these B. diegensis single-color colonies. The number of reference specimens is not large enough to encompass the whole diversity of B. diegensis but it is noteworthy that haplotypes Bd-H1, Bd-H2, Bd-H3 and Bd-H6 characterizing our B. diegensis two-color reference samples (Supplementary Table S1), were also found in single-color colonies of B. diegensis mis-identified as B. violaceus (Table 1).
Comparison With Sequences in Public Databases
We completed our analysis with a species identification search on the BOLD portal, using as a query each of the distinct haplotypes obtained in this study. The five B. violaceus haplotypes were found to match with reference sequences in the BOLD system (Supplementary Table S3): they were assigned with high similarity (97.59–97.62%) to ‘Botrylloides violaceum’ (note that this neuter form of the specific name is no longer accepted according to WoRMS, being replaced by the masculine form used here; see Ryland, 2015). These haplotypes (Bv-H1 to Bv-H5) matched with four haplotypes (named as Bv2, Bv8, Bv9, and Bv11) obtained as part of two population genetics studies of B. violaceus in N. America (Bock et al., 2011; Lejeusne et al., 2011).
The haplotype BvX-H6 did not match any sequences in BOLD, and the best match obtained using a BLAST search on the NCBI portal was with Botrylloides israeliense (Reem et al., 2018) but with only 93.27% similarity (for a 91% query cover over 461 bp), which indicates that the assignment is not robust.
None of the B. diegensis haplotypes matched with any sequences registered under this name in BOLD (Supplementary Table S3): they were all assigned at 99.76–100% similarity to references registered under the name B. leachii (or the incorrect variant spelling B. leachi) with different published (Griggio et al., 2014; López-Legentil et al., 2015) or unpublished sources. It is noteworthy that one of these sources (Griggio et al., 2014) is a full mitochondrial genome sequence.
The five haplotypes of the ‘radiata’ morph of B. leachii did not match with any references registered in BOLD (Supplementary Table S4). When searching in the NCBI portal, the best match was with a sequence (acc. no. KY235402) provided under the name B. leachii, but with an extremely low level of similarity (85.15–85.68%), thus well below the threshold used for taxonomic assignment. Interestingly this sequence matches, with high similarities (99.76–100%), our B. diegensis haplotypes in BOLD (not shown). Thus, we can conclude that there are no reference sequences corresponding to our specimens of the ‘radiata’ morph of B. leachii either in BOLD or NCBI.
Comparison With Mediterranean Sequences
A total of 22 sequences were retrieved from GenBank of Mediterranean Botrylloides (six of them from whole mitochondrial mtDNA data), and 17 new sequences were obtained from the samples collected in Spain, France and Italy (Supplementary Table S2). These samples were mapped onto a phylogenetic tree with the 25 haplotypes obtained with the EC dataset (EC sampling and reference sequences; Supplementary Tables S3, S4).
The results (Figure 3) showed that all sequences in GenBank previously assigned to B. leachii clustered unambiguously with B. diegensis. The only previous B. violaceus sequence (i.e., complete mitochondrial genome HF548552; Griggio et al., 2014) grouped with our reference sequences of B. violaceus. B. pizoni branched within the [B. diegensis – B. leachii] clade, with a higher similarity to our B. leachii references, whereas B. niger branched at the root of this clade. In both cases, very low bootstrap values indicated that the topology was poorly supported. The tree, however, indicates that they are distinct from the B. violaceus clade.
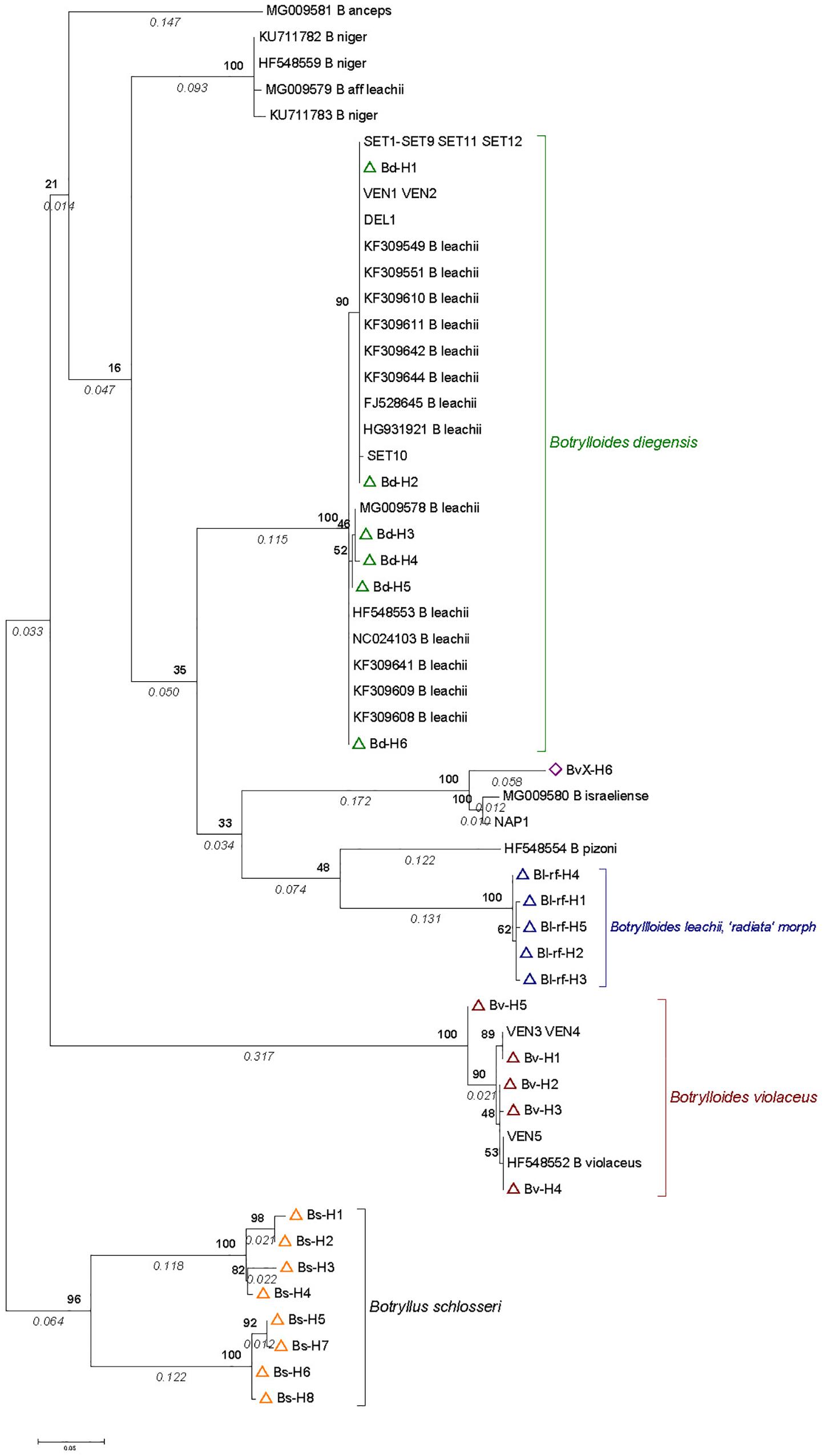
Figure 3. COI phylogenetic tree based on Botrylloides sequences obtained from samples in the English Channel, reference sequences (see Figure 2), and sequences obtained from samples from the Mediterranean Sea (published data or from this study; see details in Supplementary Table S2). The tree was built with MEGA 7 by the Maximum Likelihood Method based on the Hasegawa-Kishino-Yano model with 54% of sites evolutionarily invariable. It is made of 52 sequences over 395 base pairs. They include sequences obtained from Mediterranean Botrylloides either newly collected or obtained from GenBank (see Supplementary Table S2 for codes and details). In addition, we included 6, 5, and 5 haplotypes from our reference sequences for B. diegensis, B. violaceus and B. leachii ‘radiata’ morph (open triangle symbols in green, red and blue, respectively), together with the haplotype named as BvX-H6 (violet diamond) (see details in Supplementary Tables S3, S4). The tree is drawn to scale, with branch lengths measured as number of substitutions per site indicated in italics below the branch. Numbers in bold on nodes indicate percent bootstrap support values (1000 bootstraps).
As for the newly generated sequences, two colonies from Venice (with a clear two-color pattern), all colonies from Sète (with a clear two-color pattern), and one from the Ebro Delta (with a uniform reddish color) grouped with B. diegensis. Another three colonies from Venice (of uniform orange or red coloration) were in the B. violaceus clade. The Botrylloides colony from Miseno Lagoon (NAP1 in Figure 3) clustered with B. israeliense and somewhat less closely with the EC haplotype BvX-H6.
Using GMYC, nine entities were identified among the Botrylloides sequences (Supplementary Table S5), with BvX-H6 assigned to a different entity (‘species’) than B. israeliense plus the sample collected in NAP – both assigned to the same species with a maximum likelihood support of 1. Interestingly the haplotype Bv-H5 was also distinguished from the other B. violaceus haplotypes suggesting a putative additional cryptic species. However, the mPTP analysis distinguished only seven entities (Supplementary Table S5), with BvX-H6, NAP and B. israeliense clustered into a single entity, and Bv-H5 clustered with the other haplotypes of B. violaceus. The other delimited entities were the same in the two analyses (Supplementary Table S5).
Discussion
We used COI sequencing to examine >750 colonies of the genus Botrylloides and Botryllus with a special interest in three species, two introduced species, namely B. violaceus and B. diegensis, and one species putatively native to Europe, namely B. leachii. Combining phylogenetic analyses and barcoding, we showed that COI is a robust barcode for distinguishing the three species. We also confirmed the presence of the two NIS in the EC. In addition, this study went well beyond our expectations: it pointed out (1) the risk of mis-identification between the two NIS when rapidly identified in the field based on simple external criteria (colony color patterns), (2) the presence of errors in databases, with reference data available for B. leachii (native) at the time of the analysis (last checked in May 2019) actually being sequences corresponding to B. diegensis (introduced in the study regions), (3) the presence of B. diegensis, probably previously mistaken as B. leachii, in the Mediterranean Sea, based on our custom reference database, thus pointing to the need to revise all previous reports of B. leachii, as it has likely been confused with B. diegensis and (4) the existence of a cryptic divergent lineage, displaying a single-color pattern similar to B. violaceus.
Thus the molecular findings indicate that single-color colonies lacking larvae cannot be identified to species level with confidence from external appearance alone (see brief discussion in Bishop et al., 2015a), at least until additional distinguishing characters are recognized. Preliminary morphological observations based on external features of the species investigated are presented in Supplementary Figure S3. Identification by internal morphology, i.e., microscopical examination of the zooids and brooded larvae, requires laboratory facilities and training, and would be very time-consuming if large numbers of specimens were to be processed, for instance to estimate the relative abundance of the different species in multiple samples. In addition, some key morphological features, such as gonads and larvae, may be present only seasonally. Recourse to molecular means of identification appears a fruitful option, particularly for colorless preserved specimens that may not be reproductive. Furthermore, clear distinctions based on internal anatomy are not yet very consistently reported in the literature, given the current level of taxonomic confusion. Morphological characterization of specimens that have been pre-sorted into putative taxonomic categories based on DNA sequence information may help to achieve the full clarification of any differences in internal anatomy. However, discovery of reliable differences in zooidal morphology can prove challenging in this genus. Saito et al. (1981) distinguished B. simodensis Saito and Watanabe, 1981 from B. violaceus on the basis of reproductive characteristics and the color of live colonies, while preserved, non-reproductive colonies were very hard to separate on the basis of zooidal morphology. Atsumi and Saito (2011) were subsequently able to distinguish two additional species from B. simodensis based on differences in reproductive seasonality and the color of living colonies, backed up by mtDNA sequence information, despite them sharing very similar zooidal features.
As shown by the phylogenetic tree (Figure 2), COI is reliable to distinguish the three accepted species with a clear barcoding gap among them (Supplementary Figure S1). This is in line with previous studies showing that COI can be a relevant barcoding marker for metazoans (Bucklin et al., 2011), notably considering its properties as a rapidly evolving marker (as compared to the nuclear genome). This study also highlights a clear benefit of the barcoding approach, i.e., to enable species identification in groups that lack easy morphological traits to use in taxonomic determination. We carried out extensive sampling of Botrylloides spp. colonies that externally looked like B. violaceus, based on one simple field criterion, namely the single-color patterns of the colonies. The results of COI sequencing and comparison with a newly generated reference dataset clearly indicated that we made a large number of errors in assigning species name in the genus Botrylloides. Error rates varied 0 to 100% (Figure 1) according to the population. We could compare the results of the molecular identifications of the supposedly ‘B. violaceus’ population samples with results of RAS carried by one of us (JB) while sampling for the intended molecular study in the nine populations from the United Kingdom. During the RAS, a semi-quantitative abundance of the distinctive two-color morph B. diegensis was recorded on a four-point scale of abundance. Plotting the percentage of mis-identification as a function of the abundance of two-color B. diegensis shows that the highest mis-identification rates were observed where two-color B. diegensis colonies were relatively common (Figure 4). From this plot, it is clear that the typical two-color morph is easily identified as compared to B. violaceus. It also confirms that the distinctive ‘B. diegensis’ two-color morph is associated with a conspecific uniformly colored morph. Although the number of sites where the two morphs were both identified is low, the correlation further suggests that these two morphs within B. diegensis occur in relatively constant proportions from place to place. Interestingly, the single-color morph of B. diegensis shares haplotypes with the two-color morph (Supplementary Table S1 compared to Supplementary Table S3). In addition, colonies of the same color can be found in both single-color B. diegensis and in B. violaceus.
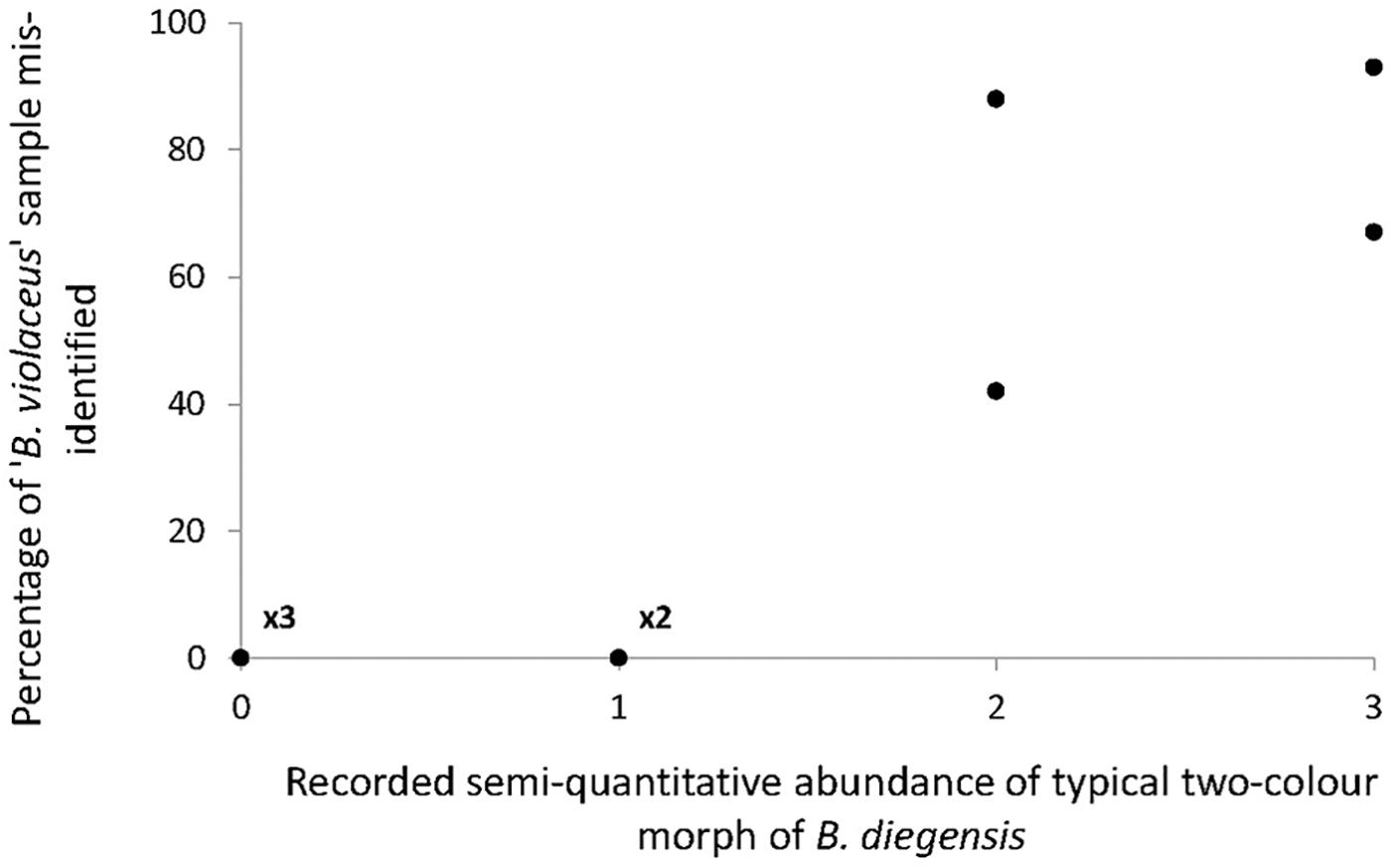
Figure 4. Relationship between the percentage of B. violaceus mis-identification (i.e., individuals that were assigned to B. diegensis with barcoding) and the abundance of typical two-color morph of B. diegensis in the nine study populations on the English coast. The four-point abundance scale was obtained for B. diegensis during rapid assessment surveys carried out at the same time as sampling for molecular analyses.
Even before becoming the barcoding marker of choice for many groups, COI had been extensively used in phylogeographic studies, thus to investigate intra-specific diversity and evolutionary history (Avise, 2000; Beheregaray, 2008; Hickerson et al., 2010). Such population-based investigations often revealed divergent lineages, which, through an integrative taxonomy approach, were sometimes assigned to new species (Pante et al., 2015). Botryllus and Botrylloides are complex taxa in which species status has already been debated (Brunetti, 2009; Reem et al., 2018). For instance, Bock et al. (2012) showed that at least three divergent clades that might be cryptic species characterize B. schlosseri. Griggio et al. (2014) further indicated that the clade A (as defined in Bock et al., 2012) may in turn be undergoing speciation. Our eight reference sequences for this accepted species were obtained from specimens collected in Brittany and south-western United Kingdom. They are also indicative of the presence of two divergent clades in the W English Channel (Figure 2), which are corresponding to clade A and E described by Bock et al. (2012). These authors also showed they co-occur in three populations (namely Brest, Falmouth and Brixham) from EC. Similarly, we found one haplotype, which we called BvX-H6, from colonies externally somewhat similar to B. violaceus, that showed a very large sequence divergence from the other studied Botrylloides species, with a divergence similar (ca. 20%) to the one measured between reference sequences of B. violaceus and B. diegensis (Table 2 and Supplementary Figure S1).
Two populations from United Kingdom, namely Southsea and Gosport, where the first dataset revealed the presence of BvX-H6 were sampled a second time in June 2012. Out of the 13 and 35 specimens examined, 77 and 24% specimens showed this particular haplotype in Southsea and Gosport, respectively. In Gosport, the other specimens were all B. diegensis and in Southsea they were all B. violaceus (Supplementary Figure S2). As part of another population-genetics study, we used the microsatellite markers specifically developed to target B. violaceus (Molecular Ecology Resources Primer Development Constorium et al., 2010) on the colonies assigned here to B. violaceus (with haplotypes Bv-H1 to Bv-H5) and to this new clade. Interestingly, whereas all the colonies with COI haplotypes Bv-H1 to Bv-H5 were easily genotyped with these microsatellites, no PCR product could be obtained for any of the BvX-H6 colonies (data not shown, FV, CR, JB unpublished data). This amplification failure strongly suggests that the colonies with haplotype BvX-H6 represent a distinct cryptic species in the EC. The clustering of this form with the Botrylloides colony from Miseno Lagoon (Italy) and with Botrylloides israeliense (Figure 3) indicates that the same divergent clade is represented in the Mediterranean Sea, but the reported divergence between the two geographical regions (ca. 6%), based on comparison of just 395 base pairs, leaves it unclear whether the EC colonies with haplotype BvX-H6 should be regarded as a somewhat divergent lineage of B. israeliense or as a sister species which is possibly undescribed. Discrepancies between the outcome of the GYMC and mPTP species delimitation methods left us with uncertainties regarding the status of this new lineage. The external appearance of B. israeliense shown by Reem et al. (2018: Figure 1b) does not suggest close kinship with the entity represented by BvX-H6 (Supplementary Figure S3, panels 7 and 8), but further molecular studies, including new species delimitation analyses with other markers, ideally coupled with detailed morphological comparison, will be necessary to resolve the status of this lineage and the exact relationship between the EC and Mediterranean populations.
Another facet of this work has been the documentation of some errors in public databases. Such errors have been pointed out previously (e.g., Harris, 2003; Vilgalys, 2003, and references herein). In our case, this issue was assessed by using simple identification searches in the BOLD system and in the NCBI portal. The outcome is illustrated in the Mediterranean Sea with a phylogenetic tree in Figure 3, where it is clear that all our B. diegensis references are mixed with sequences named B. leachii in Genbank. Further, new sequences obtained confirmed that B. diegensis in the Mediterranean can also have a two-color pattern (Venice or Sète specimens) or a single reddish coloration (Ebro Delta). We failed to find any sequence, either published or new, of Mediterranean B. leachii. Considering that this is the Botrylloides species most commonly mentioned in this sea, our results suggest that the presence of B. diegensis might have been strongly underestimated in the Mediterranean region. We also confirm the presence of B. violaceus in the Venice Lagoon, with external aspect identical to single-color colonies of B. diegensis.
The case of B. diegensis and B. violaceus is a particularly acute issue as these are two NIS, widespread in the northern hemisphere. The lack of reliability of public databases for notorious NIS is an important shortcoming for their early detection and thus effective management (Darling et al., 2017). In this context and for the specific case examined here, it is noteworthy that this mis-identification of B. diegensis occurred during a survey of Mediterranean harbors with the specific aim of examining the distribution of NIS in these well-known introduction hotspots (López-Legentil et al., 2015), and during collections by JB and FV in the EC, specifically targeting B. violaceus in marinas for population-genetic studies. We now have evidence for the presence of B. diegensis in Venice (Italy), Delta of Ebro (Spain), along the Catalan coast (Spain) and in Sète (France). Nevertheless, B. diegensis has not been formally reported in the Mediterranean, although Brunetti and Mastrototaro (2017) mention personal communication by A. and W. Bay-Nouailhat indicating its presence in the W Mediterranean. B. diegensis might be not only present but common and widespread along the Mediterranean coast, although commonly mis-identified as B. leachii. Confusion between B. diegensis and B. leachii might have contributed to statements that these species share very similar morphology (e.g., Van Name, 1945), and Brunetti (2009) synonymized them, although Brunetti and Mastrototaro (2017) treat them as separate. There is little doubt that the distinctive two-color morph now attributed to B. diegensis is a relatively recent arrival in NW Europe, rather than a long-established, potentially native, species such as B. leachii. The very striking two-color morph is not illustrated or mentioned in accounts of Botrylloides in less recent identification guides to the region (e.g., Eales, 1939; Berrill, 1950; Barrett and Yonge, 1958; Millar, 1970; Picton, 1985; Hayward and Ryland, 1990; Gibson et al., 2001). Furthermore, the continuing spread of the distinctive two-color morph to new regions of the English coast from an apparently recent origin on the south coast has been documented in repeated field surveys since 2004 (e.g., Wood et al., 2016). RAS rely substantially on clearly defined field characteristics, which may be elusive in some groups; detailed molecular and morphological studies are called for before field identification can be deemed reliable for the important ascidian genus Botrylloides.
Data Availability Statement
The haplotypes generated by Sanger sequencing have been deposited in GenBank (accession numbers MK978800–MK978824 for the EC sampling and reference sequences and MN076465–MN076483 for the Mediterranean sampling).
Author Contributions
FV and JB contributed to the conception and design of the study. FV, JB, SB, and XT provided the samples and/or carried out the sampling. CR, SB, and XT performed the molecular work or provided the sequence data. FV and CR made the analyses. FV wrote the first draft of the manuscript with substantial contributions of JB for some sections and XT for the Mediterranean part of the manuscript. All authors contributed to the manuscript revision, and read and approved the submitted version.
Funding
This work was part of the Interreg IVA project ‘Marinexus’ financed under the European Regional Development Fund. And also received further support from the AquaNIS2.0 project funded by the Fondation TOTAL, and from project CTM2017-88080 (MCIU/AEI/FEDER/UE) of the Spanish Government.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Anna Yunnie, Christine Wood, Laurent Lévêque, and the divers of the Marine Operations department at the Roscoff Biological Station for their help in sampling and preliminary sorting of the samples in EC. We also thank the marina operators for their permission to conduct the experiment. We are grateful to the following people for helping with the Mediterranean samples or sending specimens to be used as reference: Maria Casso, Franck Gentil, Gretchen Lambert, Adam Langenbacher, Julia Nunn, Mari Carmen Pineda Mark Rius, Kirsty Smith Paolo Sordino, and Davide Tagliapietra. Susanna López-Legentil sequenced the specimen from Miseno Lagoon.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00615/full#supplementary-material
References
Alder, J., and Hancock, A. (1912). The British tunicata: an unfinished monograph. Vol. 3. Aggregata (Ascidiae Compositae). Ray Soc. Publications 93, 1–113.
Appeltans, W., Ahyong, S. T., Anderson, G., Angel, M. V., Artois, T., Bailly, N., et al. (2012). The magnitude of global marine species diversity. Curr. Biol. 22, 2189–2202. doi: 10.1016/j.cub.2012.09.036
Arenas, F., Bishop, J. D. D., Carlton, J. T., Dyrynda, P. J., Farnham, W. F., Gonzalez, D. J., et al. (2006). Alien species and other notable records from a rapid assessment survey of marinas on the south coast of England. J. Mar. Biol. Assoc. U. K. 86, 1329–1337. doi: 10.1017/s0025315406014354
Atsumi, M. O., and Saito, Y. (2011). Studies on Japanese botryllid ascidians. V. A new species of the genus Botrylloides very similar to Botrylloides simodensis in morphology. Zool. Sci. 28, 532–542. doi: 10.2108/zsj.28.532
Avise, J. C. (2000). Phylogeography: the History and Formation of Species. Cambridge, MA: Harvard University Press.
Beheregaray, L. B. (2008). Twenty years of phylogeography: the state of the field and the challenges for the Southern Hemisphere. Mol. Ecol. 17, 3754–3774. doi: 10.1111/j.1365-294X.2008.03857.x
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., et al. (2013). GenBank. Nucleic Acids Res. 41, D36–D42. doi: 10.1093/nar/gks1195
Berrill, N. J. (1950). The tunicata, with an account of the British species. Ray Soc. Publications 133, 1–354.
Bishop, J. D. D., Roby, C., Yunnie, A. L. E., Wood, C. W., Lévêque, L., Turon, X., et al. (2013). The Southern Hemisphere ascidian Asterocarpa humilis is unrecognised but widely established in NW France and Great Britain. Biol. Invasions 15, 253–260. doi: 10.1007/s10530-012-0286-x
Bishop, J. D. D., Wood, C. A., Lévêque, L., Yunnie, A. L. E., and Viard, F. (2015a). Repeated rapid assessment surveys reveal contrasting trends in occupancy of marinas by non-indigenous species on opposite sides of the western English Channel. Mar. Pollut. Bull. 95, 699–706. doi: 10.1016/j.marpolbul.2014.11.043
Bishop, J. D. D., Wood, C. A., Yunnie, A. L., and Griffiths, C. A. (2015b). Unheralded arrivals: non-native sessile invertebrates in marinas on the English coast. Aquat. Invasions 10, 249–264. doi: 10.3391/ai.2015.10.3.01
Bock, D. G., MacIsaac, H. J., and Cristescu, M. E. (2012). Multilocus genetic analyses differentiate between widespread and spatially restricted cryptic species in a model ascidian. Proc. R. Soc. B: Biol. Sci. 279, 2377–2385. doi: 10.1098/rspb.2011.2610
Bock, D. G., Zhan, A., Lejeusne, C., MacIsaac, H. J., and Cristescu, M. E. (2011). Looking at both sides of the invasion: patterns of colonization in the violet tunicate Botrylloides violaceus. Mol. Ecol. 20, 503–516. doi: 10.1111/j.1365-294X.2010.04971.x
Bouckaert, R., Vaughan, T. G., Barido-Sottani, J., Duchêne, S., Fourment, M., Gavryushkina, A., et al. (2019). BEAST 2.5: An advanced software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 15:e1006650. doi: 10.1371/journal.pcbi.1006650
Brunetti, R. (2009). Botryllid species (Tunicata, Ascidiacea) from the Mediterranean coast of Israel, with some considerations on the systematics of Botryllinae. Zootaxa 2289, 18–32. doi: 10.11646/zootaxa.2289.1.2
Bucklin, A., Steinke, D., and Blanco-Bercial, L. (2011). DNA barcoding of marine metazoa. Annu. Rev. Mar. Sci. 3, 471–508. doi: 10.1146/annurev-marine-120308-080950
Campbell, M. L., Gould, B., and Hewitt, C. L. (2007). Survey evaluations to assess marine bioinvasions. Mar. Pollut. Bull. 55, 360–378. doi: 10.1016/j.marpolbul.2007.01.015
Carlton, J. T. (2009). “Deep invasion ecology and the assembly of communities in historical time,” in Biological Invasions in Marine Ecosystems, eds G. Rilov, and J. A. Crooks, (Berlin: Springer), 13–56. doi: 10.1007/978-3-540-79236-9_2
Cohen, A. N., Harris, L. H., Bingham, B. L., Carlton, J. T., Chapman, J. W., Lambert, C. C., et al. (2005). Rapid assessment survey for exotic organisms in southern California bays and harbors, and abundance in port and non-port areas. Biol. Invasions 7, 995–1002. doi: 10.1007/s10530-004-3121-1
Comtet, T., Sandionigi, A., Viard, F., and Casiraghi, M. (2015). DNA (meta)barcoding of biological invasions: a powerful tool to elucidate invasion processes and help managing aliens. Biol. Invasions 17, 905–922. doi: 10.1007/s10530-015-0854-y
Darling, J. A., Galil, B. S., Carvalho, G. R., Rius, M., Viard, F., and Piraino, S. (2017). Recommendations for developing and applying genetic tools to assess and manage biological invasions in marine ecosystems. Mar. Policy 85, 54–64. doi: 10.1016/j.marpol.2017.08.014
Dias, J. P., Fotedar, S., Munoz, J., Hewitt, M. J., Lukehurst, S., Hourston, M., et al. (2017). Establishment of a taxonomic and molecular reference collection to support the identification of species regulated by the Western Australian prevention list for introduced marine pests. Manag. Biol. Invasions 8, 215–225. doi: 10.3391/mbi.2017.8.2.09
Eales, N. B. (1939). The Littoral Fauna of Great Britain. A Handbook for Collectors. Cambridge: Cambridge University Press.
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Gibson, R., Hextall, B., and Rogers, A. (2001). Photographic Guide to the Sea and Shore Life of Britain and North-West Europe. Oxford: Oxford University Press.
Griggio, F., Voskoboynik, A., Iannelli, F., Justy, F., Tilak, M.-K., Xavier, T., et al. (2014). Ascidian mitogenomics: comparison of evolutionary rates in closely related taxa provides evidence of ongoing speciation events. Genome Biol. Evol. 6, 591–605. doi: 10.1093/gbe/evu041
Hall, T. A. (1999). BioEdit: a user friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Harris, J. D. (2003). Can you bank on GenBank? Trends Ecol. Evol. 18, 317–319. doi: 10.1016/S0169-5347(03)00150-2
Hayward, P. J., and Ryland, J. S. (1990). The Marine Fauna of the British Isles and North-West Europe. Volume. 2: Molluscs to Chordates. Oxford: Oxford University Press.
Hebert, P. D., Hollingsworth, P. M., and Hajibabaei, M. (2016). From writing to reading the encyclopedia of life. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150321. doi: 10.1098/rstb.2015.0321
Hickerson, M. J., Carstens, B. C., Cavender-Bares, J., Crandall, K. A., Graham, C. H., Johnson, J. B., et al. (2010). Phylogeography’s past, present, and future: 10 years after. Mol. Phylogenet. Evol. 54, 291–301. doi: 10.1016/j.ympev.2009.09.016
Kapli, P., Lutteropp, S., Zhang, J., Kobert, K., Pavlidis, P., Stamatakis, A., et al. (2017). Multi-rate poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics 33, 1630–1638. doi: 10.1093/bioinformatics/btx025
Knowlton, N. (2000). Molecular genetic analyses of species boundaries in the sea. Hydrobiologia 420, 73–90. doi: 10.1007/978-94-017-2184-4_8
Lambert, C. C., and Lambert, G. (2003). Persistence and differential distribution of nonindigenous ascidians in harbors of the Southern California Bight. Mar. Ecol. Prog. Ser. 259, 145–161. doi: 10.3354/meps259145
Lehtiniemi, M., Ojaveer, H., David, M., Galil, B., Gollasch, S., McKenzie, C., et al. (2015). Dose of truth—Monitoring marine non-indigenous species to serve legislative requirements. Mar. Policy 54, 26–35. doi: 10.1016/j.marpol.2014.12.015
Lejeusne, C., Bock, D. G., Therriault, T. W., MacIsaac, H. J., and Cristescu, M. E. (2011). Comparative phylogeography of two colonial ascidians reveals contrasting invasion histories in North America. Biol. Invasions 13, 635–650. doi: 10.1007/s10530-010-9854-0
López-Legentil, S., Legentil, M., Erwin, P., and Turon, X. (2015). Harbor networks as introduction gateways: contrasting distribution patterns of native and introduced ascidians. Biol. Invasions 17, 1623–1638. doi: 10.1007/s10530-014-0821-z
McGlashan, D. J., Ponniah, M., Cassey, P., and Viard, F. (2008). Clarifying marine invasions with molecular markers: an illustration based on mtDNA from mistaken calyptraeid gastropod identifications. Biol. Invasions 10, 51–57. doi: 10.1007/s10530-007-9106-0
Millar, R. H. (1970). British Ascidians (Synopses of the British Fauna, New Series, No. 1). London: Academic Press.
Molecular Ecology Resources Primer Development Constorium, Abdoullaye, D., Acevedo, I., Adebayo, A. A., Behrmann-Godel, J., Benjamin, R. C., et al. (2010). Permanent genetic resources added to molecular ecology resources database 1 August 2009–30 September 2009. Mol. Ecol. Resour. 10, 232–236. doi: 10.1111/j.1755-0998.2009.02796.x
Nunes, A. L., Katsanevakis, S., Zenetos, A., and Cardoso, A. C. (2014). Gateways to alien invasions in the European seas. Aquat. Invasions 9, 133–144. doi: 10.3391/ai.2014.9.2.02
Nydam, M., and Stachowicz, J. J. (2007). Predator effects on fouling community development. Mar. Ecol. Prog. Ser. 337, 93–101. doi: 10.3354/meps337093
Ojaveer, H., Galil, B. S., Campbell, M. L., Carlton, J. T., Canning-Clode, J., Cook, E. J., et al. (2015). Classification of non-indigenous species based on their impacts: considerations for application in marine management. PLoS Biol. 13:e1002130. doi: 10.1371/journal.pbio.1002130
Ojaveer, H., Galil, B. S., Minchin, D., Olenin, S., Amorim, A., Canning-Clode, J., et al. (2014). Ten recommendations for advancing the assessment and management of non-indigenous species in marine ecosystems. Mar. Policy 44, 160–165. doi: 10.1016/j.marpol.2013.08.019
Ordóñez, V., Pascual, M., Fernández-Tejedor, M., and Turon, X. (2016). When invasion biology meets taxonomy: Clavelina oblonga (Ascidiacea) is an old invader in the Mediterranean Sea. Biol. Invasions 18, 1203–1215. doi: 10.1007/s10530-016-1062-0
Pante, E., Puillandre, N., Viricel, A., Arnaud-Haond, S., Aurelle, D., Castelin, M., et al. (2015). Species are hypotheses: avoid connectivity assessments based on pillars of sand. Mol. Ecol. 24, 525–544. doi: 10.1111/mec.13048
Picton, B. E. (1985). Ascidians of the British Isles. A Colour Guide. Ross-on-Wye: Marine Conservation Society.
Pons, J., Barraclough, T. G., Gomez-Zurita, J., Cardoso, A., Duran, D. P., Hazell, S., et al. (2006). Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55:595609.
Ratnasingham, S., and Hebert, P. D. N. (2007). BOLD: the barcode of life data system: barcoding. Mol. Ecol. Notes 7, 355–364. doi: 10.1111/j.1471-8286.2007.01678.x
Reem, E., Douek, J., and Rinkevich, B. (2018). Ambiguities in the taxonomic assignment and species delineation of botryllid ascidians from the israeli mediterranean and other coastlines. Mitochondrial DNA Part A 29, 1073–1080. doi: 10.1080/24701394.2017.1404047
Rocha, R. M., Salonna, M., Griggio, F., Ekins, M., Lambert, G., Mastrototaro, F., et al. (2019). The power of combined molecular and morphological analyses for the genus Botrylloides: identification of a potentially global invasive ascidian and description of a new species. Syst. Biodivers. 1–18. doi: 10.1080/14772000.2019.1649738
Rozas, J., Sanchez-DelBarrio, J. C., Messeguer, X., and Rozas, R. (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497. doi: 10.1093/bioinformatics/btg359
Ryland, J. S. (2015). Gender of the genus Botrylloides Milne Edwards. Zootaxa 3973, 398–400. doi: 10.11646/zootaxa.3973.2.13
Saito, Y., Mukai, H., and Watanabe, H. (1981). Studies on Japanese compound styelid ascidians II. A new species of the genus Botrylloides and redescription of B. violaceus Oka. Publications Seto Mar. Biol. Lab. 26, 357–368. doi: 10.5134/176032
Sambrook, K., Holt, R. H. F., Sharp, R., Griffith, K., Roche, R. C., Newstead, R. G., et al. (2014). Capacity, capability and cross-border challenges associated with marine eradication programmes in Europe: the attempted eradication of an invasive non-native ascidian, Didemnum vexillum in Wales, United Kingdom. Mar. Policy 48, 51–58. doi: 10.1016/j.marpol.2014.03.018
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 101, 11030–11035. doi: 10.1073/pnas.0404206101
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Van Name, W. G. (1945). The north and south American ascidians. Bull. Am. Museum Nat. Hist. 84, 1–476.
Vilgalys, R. (2003). Taxonomic misidentification in public DNA databases. New Phytol. 160, 4–5. doi: 10.1046/j.1469-8137.2003.00894.x
Keywords: non-native species, cryptic species, barcoding, COI, marine, tunicates
Citation: Viard F, Roby C, Turon X, Bouchemousse S and Bishop J (2019) Cryptic Diversity and Database Errors Challenge Non-indigenous Species Surveys: An Illustration With Botrylloides spp. in the English Channel and Mediterranean Sea. Front. Mar. Sci. 6:615. doi: 10.3389/fmars.2019.00615
Received: 22 June 2019; Accepted: 18 September 2019;
Published: 01 October 2019.
Edited by:
Tifeng Shan, Institute of Oceanology, Chinese Academy of Sciences, ChinaReviewed by:
Néstor E. Ardila, EcoMar s.a.s, ColombiaAlfonso Angel Ramos-Esplá, University of Alicante, Spain
Copyright © 2019 Viard, Roby, Turon, Bouchemousse and Bishop. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Frédérique Viard, viard@sb-roscoff.fr
 Frédérique Viard
Frédérique Viard Charlotte Roby
Charlotte Roby Xavier Turon
Xavier Turon Sarah Bouchemousse1
Sarah Bouchemousse1  John Bishop
John Bishop