The Arsenic Methylation Cycle: How Microbial Communities Adapted Methylarsenicals for Use as Weapons in the Continuing War for Dominance
- 1Department of Cellular Biology and Pharmacology, Herbert Wertheim College of Medicine, Florida International University, Miami, FL, United States
- 2Institute of Environmental Remediation and Human Health, Southwest Forestry University, Kunming, China
Arsenic is toxic and widely distributed in the environment. Most microorganisms have evolved mechanisms to use methylarsenicals as weapons in microbial warfare. This has created cycles of arsenic methylation and demethylation, which constitutes an important part of arsenic biogeochemical cycles and environmental health. Arsenic methylation forms methylarsenite [MAs(III)], arsenobetaine, arsenosugars and the new identified methylarsenical antibiotic, arsinothricin (AST). Bacteria use MAs(III) and AST as antibiotic to obtain a competitive advantage over other bacteria. Microbes produce toxic MAs(III) by (1) methylation of arsenite [As(III)] or (2) reduction of methylarsenate [MAs(V)]. In air, MAs(III) is oxidized to MAs(V) by oxygen, which conferring methylation to be considered as a detoxification pathway in aerobes. MAs(V) also can be re-reduced to MAs(III) by other bacteria, which can enable its survival and dominance with more competitive advantage than other sensitive bacteria. The different composition and diversity of microbial community could generate the sustained pool of MAs(III). These complex interactions have an emergent property., that is, overall scheme of arsenic-containing antibiotics emerges from interaction of multiple species. In response to production of the antibiotic MAs(III), other members of microbial communities have evolved at least four mechanisms for MAs(III) resistance: (1) ArsH detoxifies MAs(III) by oxidation to less toxic MAs(V); (2) ArsI degrades MAs(III) by cleavage of the C-As bond to form less toxic As(III); (3) the ArsP efflux permease confers selective resistance to MAs(III); and (4) the unrelated ArsK efflux permease confers resistance to both trivalent inorganic and organoarsenicals. Environmental application of anthropogenic aromatic arsenicals further fuels bacterial warfare and selection for novel resistance mechanisms. In another microbial adaptation to use environmental arsenic as a weapon, some soil bacteria synthesize a methylarsenate analog of the amino acid glutamic acid termed AST; AST shows a broad-spectrum antibiotic action against both Gram-negative and Gram-positive bacteria. In response to the environmental challenge presented by AST, arsN genes evolved to detoxify it by acetylation of the α-amino group. Thus, to battle for surviving and thriving in the microbial community, life has to adapted and use environmental arsenic as a weapon. Both MAs(III) and AST are natural products with antibiotic-like properties, both are toxic methylarsenicals produced by some microbe and further kill off its competitors, and resistances have arisen for both.
Introduction
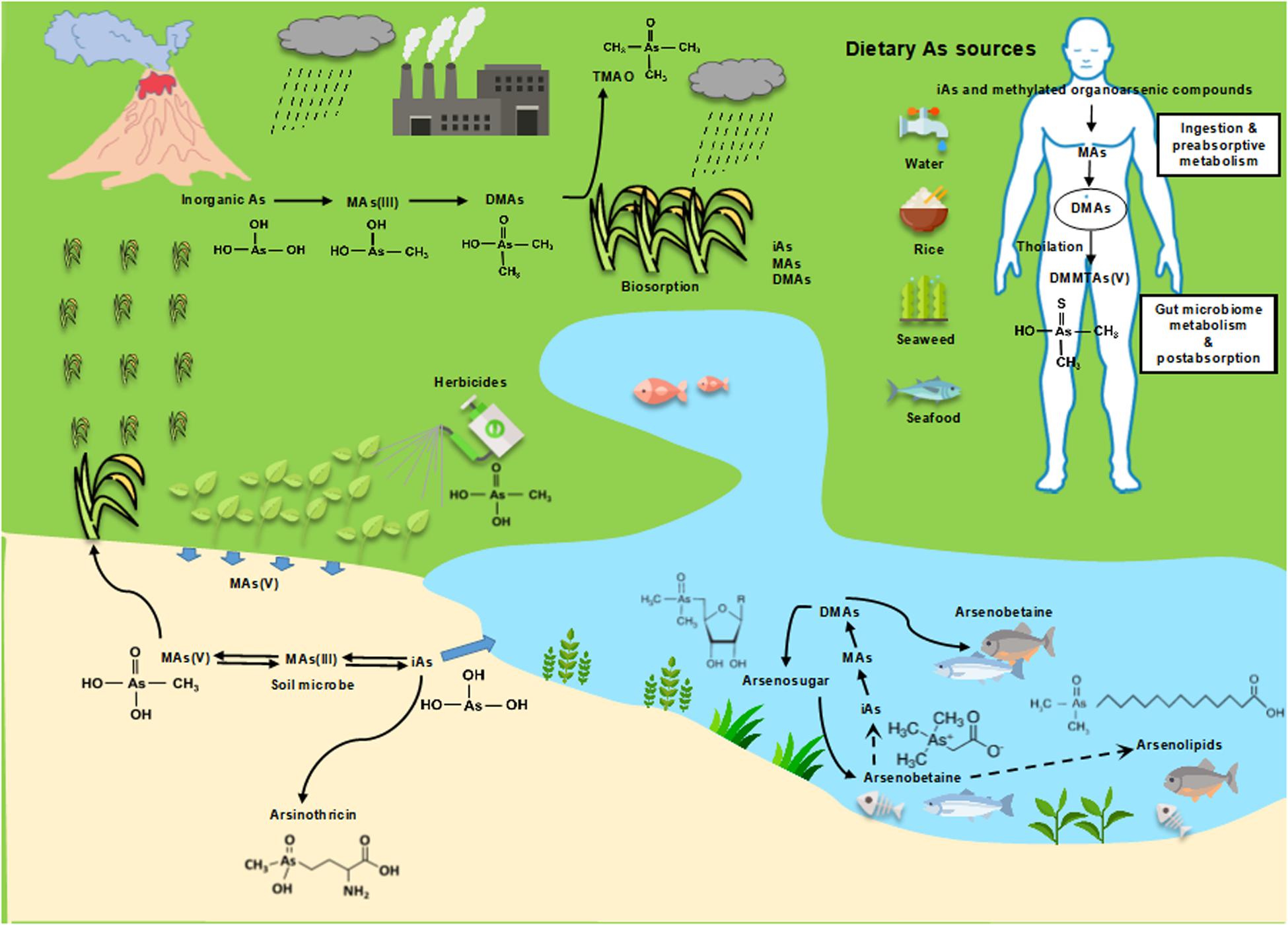
In the environment arsenic often occurs in combination with either inorganic or organic substances to form a wide variety of compounds. Most inorganic arsenic compounds are found in soil, sediments and groundwater. The two biologically-relevant oxidation states of inorganic arsenic are trivalent arsenite [As(III)] and pentavalent arsenate [As(V)]. Arsenate has three pKa values of 2.19, 6.94, 11.5, so in neutral solution the chemical species is primarily the anion As(OH)2O–1. Although the trivalent form is often termed “arsenite” in the literature, that is a misnomer because it implies that it is the anion As(OH)2O–1. However, with three pKa values of 9.23, 10, and 12.13, the chemical species in neutral solution is actually the undissociated trihydroxy acid form As(OH)3. Inorganic arsenic species occurs either naturally or as a result of mining, melting or some applications of arsenic-containing in industry (Mandal and Suzuki, 2002). Some organic arsenicals are found in fish and shellfish and tend to be much less toxic than inorganic arsenic. The various inorganic and organic arsenic compounds are naturally occurred and interlinked through complex biotic and abiotic transformations. A wide variety of organoarsenic compounds are found naturally as a consequence of biomethylation and other biosynthetic pathways. SAM, which is a conjugate of nucleotide adenosine and amino acid methionine, is an essential substrate/cofactor in numerous enzyme-catalyzed reactions, especially the organisms having SAM-dependent enzymes (Roje, 2006). SAM is a universal biological cofactor that transfer of its methyl group to substrates such as lipids, proteins, DNA and other small molecules (inorganic arsenic, chloride, bromide) (Thomas et al., 2004). These methylation processes catalyzed by SAM-dependent methyltransferases have asignificantly role in many essential biological pathways, including biosynthesis, protein repair, signal transduction, metabolism and detoxification (Bauerle et al., 2015). Microbial arsenic metabolism is of special interest as inorganic arsenic biotransformations produce a number of organoarsenical species. These, in turn, enter into metabolic pathways, resulting in hundreds of organoarsenical compounds produced and released into the environment (Taylor et al., 2017).
A common feature of SAM enzymes is the methylation of carbon or phosphorous atoms found in lots of primary and secondary metabolites. As an analog of phosphate, many arsenic compounds with methyl groups are also present in naturally, including MAs(V), DMAs(V), and TMAs(V)O (Ye et al., 2012). In addition, three additional classes of methylated arsenicals have been detected and identified in food: arsenosugars, arsenolipids, and AB. These organoarsenical species are primarily found in marine animals and algae (Chen et al., 2019a). With the continual development of advanced analytical methods, more and more methylated arsenicals are being identified. For example, after exposure to inorganic arsenic, human urine was analyzed and further identified several novel sulfur-containing methylated arsenic metabolites, i.e., MMMTAsV and DMMTAsV (Raml et al., 2007). These organic thioarsenicals have also been detected in the urine and feces of animals in laboratory conditions. The methylarsenical antibiotic AST is generated by bacterium Burkholderia gladioli strain GSRB05, isolated from the rice rhizosphere (Kuramata et al., 2016). In all of these there is a C-As bond with at least one methyl group. Arsenic methylation contributes to the diversity of organoarsenic compounds, which have environmental effects. For example, methylated organoarsenicals play a significant role in controlling host bacterial populations and dominance in complex environments. Both AB and the similar nitrogen−containing compound glycine betaine protect against osmotic stress and a wide variety of temperature extreme environments (Hoffmann et al., 2018). Natural methylated organoarsenicals have already been produced biologically for billion years, however, the major species of arsenic in the environment are inorganic, which implies that most of the methylated species are degraded biologically or abiotically into inorganic arsenic. Thus, microbial arsenic methylation and demethylation play a crucial role in the arsenic biogeochemical cycle.
Two microbially-generated methylarsenicals, MAs(III) and the arsenic-containing amino acid AST, have recently been shown to have antimicrobial properties and have been proposed to be true antibiotics (Chen et al., 2019b; Nadar et al., 2019). First, even though As(III) is itself toxic, MAs(III), the first product during the arsenic methylation pathway catalyzed by the wide-spread enzyme As(III) methyltransferase (orthologs are termed ArsM in microbes and AS3MT in animals), is generally more toxic than inorganic As(III). Biogenically produced MAs(III) fits the definition of “antibiotic” first proposed by Waksman (1947): a toxic organic compound produced by one microbe to kill off its competitors. DMAs(III) may also have antibiotic-like properties, but it is very unstable in an aerobic environment, rapidly oxidizing to DMAs(V), and probably does not play a major role in microbial communities.
Second, the recently identified pentavalent methylated organoarsenical antibiotic AST is as toxic as MAs(III) (Nadar et al., 2019). This is surprising because pentavalent arsenicals are largely less toxic. AST is an broad-spectrum antibiotic and active against both Gram-positive and Gram-negative bacteria. In this review, we will focus on methylated organoarsenicals, in particular how bacteria have adapted to use environmental arsenic as a weapon in the continuing battle for survival and dominance (Figure 1).
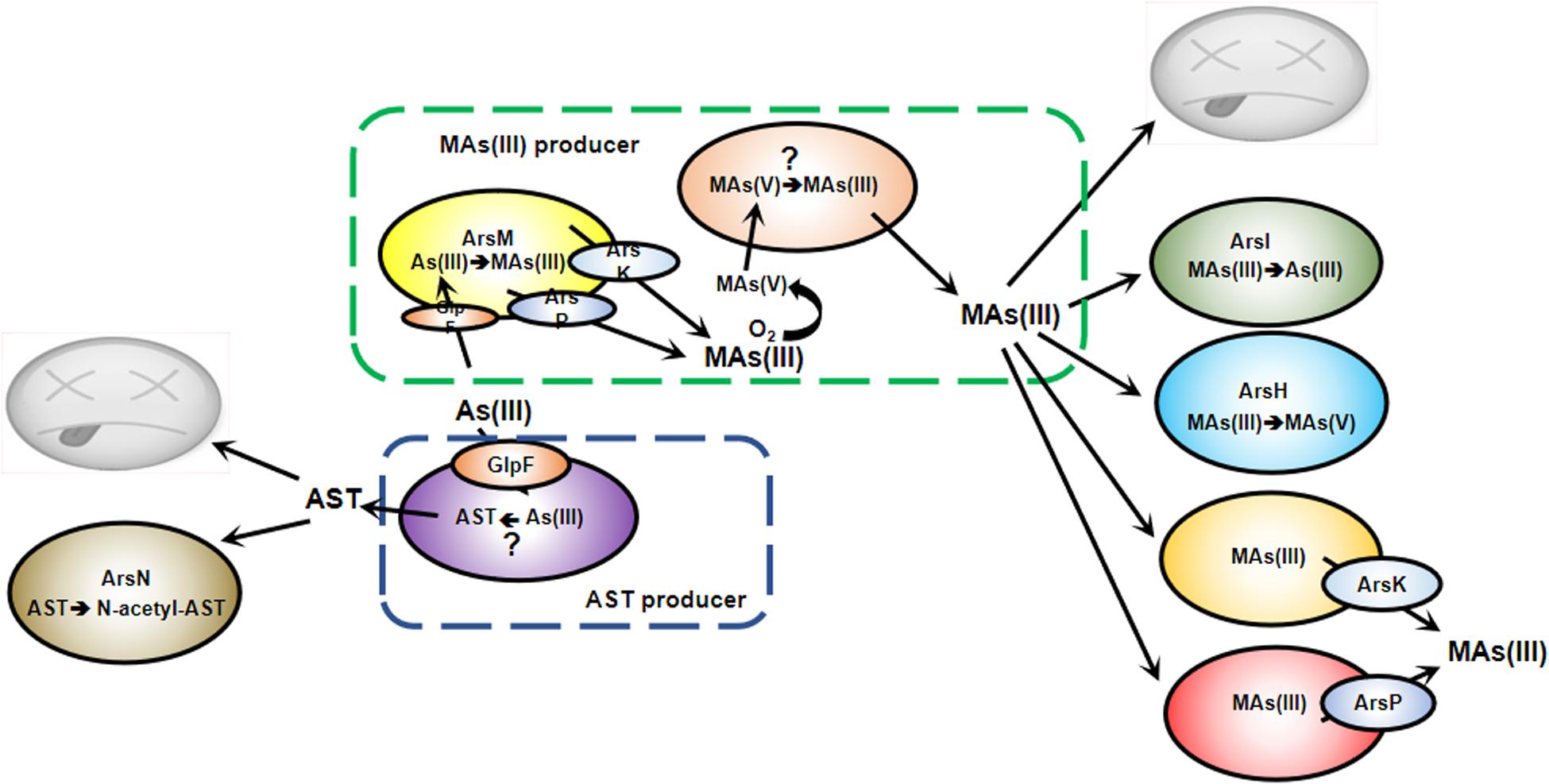
Figure 1. Antibiotic action of MAs(III) and AST in microbial communities. Bacteria are in a constant struggle to survive and thrive in microbial jungles. Bacteria with ArsM methylate As(III) to MAs(III), which is extruded by ArsP or ArsK. In anaerobic environments, this gives those bacteria a competitive advantage over MAs(III) sensitive bacteria. In an aerobic environment, MAs(III) is either oxidized to MAs(V) or demethylated to less toxic As(III), conferring MAs(III) resistance. Both biogenic and anthropogenic MAs(V) can be reduced to MAs(III) by other soil bacteria to kill MAs(III) sensitive community members. However, some bacteria shield themselves from MAs(III) toxicity by expression of arsI, arsH, arsK or arsP genes. Bacteria shield themselves from AST toxicity by expression of arsN.
Environmental Prevalence of Organoarsenicals
Simple Methylarsenicals
Biogeochemical cycling of arsenic results in substantial environmental flux of both inorganic and methylated arsenicals (Figure 2). A number of inorganic and thiolated species are found in nature, as are organoarsenicals in lower amounts. The arsM gene is wide-spread in microbes, including bacteria, archaea, algae, protists, and fungi (Chen et al., 2017), and there is continual production of methylated species in the biosphere, which makes it curious that steady-state amounts are not higher. Microbial degradation might explain at least in part this discrepancy; a recent study suggests that marine DMAs(V) turns over every 8.1 days (Giovannoni et al., 2019).
In addition to microbial methylation, humans and most common laboratory animals extensively methylate inorganic arsenic, and the metabolites are excreted primarily in the urine, which demonstrates the universality of these reactions (Thomas et al., 2004). The cytotoxicity of some arsenic species was analyzed in T24 cells, which was derived from a human urinary bladder carcinoma that lacks the ability of As(III) methylation. In T24 cells, the relative toxicities were determined to be MAs(III) > DMAs(III) ≈ DMMTAs(V) ≈ As(III) ≫ AsV > DMAs(V) > MAs(V) (Moe et al., 2016). Most of the inorganic arsenic, As(III) and As(V), is found in urine primarily as MAs(V) and DMAs(V). In individuals with rapid methylation, DMAs(V) is the major species found in urine. AS3MT is the primary enzyme of arsenic methylation in humans. Analyses of the urinary arsenic profile hints at the methylation capacity of exposed individuals. Most studies investigating the association between urinary arsenic profiles and human diseases measure primarily arsenate and arsenite and methylated MAs(V) and DMAs(V) (Tseng, 2007). Historically arsenic methylation has been considered a detoxification process because those pentavalent methylated arsenicals are relatively less toxic. However, MAs(III) has clearly been shown to be the first product of AS3MT catalysis (Dheeman et al., 2014) and can be further easily oxidized to MAs(V) when exposed to air (Le et al., 2000a). MAs(III) is more toxic than As(III) in Chang human hepatocytes (Petrick et al., 2000). DMAs(III) is the likely second product, but its instability in air results in rapid oxidation to DMAs(V). The kinetics of arsenic methylation was determined with the purified human AS3MT. The catalytic rates of the first two steps of methylation, As(III) to MAs(III) and MAs(III) to DMAs (the presumed product, DMAs(III) were determined (Dong et al., 2015). In the presence of GSH, the Km for wild-type human AS3MT enzymes was measured as approximately 1.6 μM for As(III) and 0.7-0.8 μM for MAs(III). Thus the affinity for the product, MAs(III), is higher than the substrate As(III), so it is difficult for MAs(III) to be released from the active site until it is methylated a second time to DMAs(III). During the continuous methylation, the conformational was changed slowly between the first and second methylation steps (Dheeman et al., 2014). This is the rate-limiting step in arsenic methylation and was shown to be the result of slow reorientation of the methyl group from the SAM-binding site to solvent (Packianathan et al., 2018). Thus it seems more likely that inorganic arsenic methylation is not a detoxication process in animals but rather activates arsenic into more toxic and potentially carcinogenic species (Thomas et al., 2004). Consistent with this concept, with careful handling MAs(III) and DMAs(III) have also been found in urine of individuals chronically exposed to inorganic arsenic via drinking water (Le et al., 2000a). MAs(III) is also found in liver cytosol and hepatocytes of arsenic-exposed rats (Le et al., 2000b).
Methylarsenate, DMAs(V) and TMAs(V)O have also been detected in many field plants (Meharg and Hartley-Whitaker, 2002). Among these plants, rice has been intensively studied for arsenic speciation as it is the staple food in the world’s population. MAs(V) and DMA(V) have often been detected in vegetative tissues and grains of rice plants (Heitkemper et al., 2001). However, no ArsM orthologs have been found in higher plants, only algae, and the source of the methylated arsenic species appears to be microbial in origin (Lomax et al., 2012). As mentioned previously, most of the environmental methylated arsenicals is sourced from ArsM-catalyzed methylation As(III) by prokaryotic and eukaryotic microbes (Zhu et al., 2014). To date, more and more bacterial strains have been identified that are capable of reducing MAs(V), including the environmental isolate Burkholderia sp. MR1, P. putida KT2440, S. putrefaciens 200, and Sinorhizobium meliloti RM1021 (Chen et al., 2019b). However, the genes/enzymes involved in MAs(V) reduction have still not been identified. In addition to the biological pathways are anthropogenic sources of MAs(V). MAs(V) has been used in the United States as the herbicide MSMA, although its use is now restricted to cotton fields, new golf courses and highway medians. DMAs(V) was used during the war in Vietnam to kill rice as the herbicide Agent Blue. Even today both MAs(V) and DMAs(V) are used as herbicides and fungicides in rice and cotton, to increase the farmer’s profitin Eastern Asia. Even though the levels of MAs(V) are low in soil, their historical use as pesticides, as well as their presence in poultry litter that is used as fertilizer with animal manure, result in arsenic contamination in specific soils (Zhao et al., 2013).
Thiolorganoarsenicals
In addition to methylation, the occurrence, structure identification and biological function of thiolated organoarsenicals has recently attracted attention. Analyses of human urine after inorganic arsenic exposure revealed several sulfur-containing arsenic metabolites, including MMMTAs(V), the sulfure counterpart of MAs(V), and DMMTAs(V), the sulfur counterpart of DMAs(V). Pentavalent thiolated arsenicals are much more toxic than their oxoarsenical counterparts and its kinetic behavior and cytotoxicity are similar to that of the trivalent oxoarsenicals (Fan et al., 2018). Thioarsenicals are formed from methylated species such as MAs(V) and DMAs(V), but it is not clear whether biological production is enzymatic. SRB in the human gut are shown to be involved in thiolating methylarsenicals (Rubin et al., 2014). SRB of human gastrointestinal origin produce H2S, which can thiolate methylarsenicals chemically. This suggests that formation of thioarsenicals is non-enzymatic but does not eliminate the possibility of enzymatic pathways of synthesis. Environmental microbes such as Shewanella putrefaciens also thiolate methylated arsenicals, producing MMMTAs(V) and DMMTAs(V) (Rubin et al., 2014; Chen and Rosen, 2016). However, its mechanism is similarly, poorly understood.
Complex Organoarsenicals
More complex methylarsenicals, including arsenosugars, arsenolipids and arsenobetaine are naturally present in marine environments. These methylated organoarsenic compounds are generally considered non-toxic (Taylor et al., 2017).
Arsenosugars Are the Most Widespread and Are the Major Arsenic-Containing Compound in Marine Algae (Typically 2–50 mg Arsenic/kg Dry Mass) (Niegel and Matysik, 2010)
Arsenosugars are carbohydrates in which a di- or tri-methylated arsenic moiety is incorporated into a ribofuranoside, which contains glycerol, phosphate, sulfate or sulfonate (Thomas and Bradham, 2016). Although more than 20 different arsenosugars have been identified as natural products, most of the arsenic found in arsenosugars is associated with just a few compounds (Chen et al., 2019a). The pathway for arsenosugar biosynthesis has still not been fully elucidated, but DMAs synthesis catalyzed by ArsM has been shown to be the precursor of arsenosugars (Xue et al., 2017). The initial step in the pathway of arsenosugar biosynthesis is the methylation of inorganic As(III) by ArsM to MAs(III) and DMAs(III) (Marapakala et al., 2012). Since DMAs(III) is oxidized in air, it is not clear whether trivalent or pentavalent DMAs is the substrate for the first committed step in arsenosugar biosynthesis, but the addition of the deoxyribose moiety is catalyzed by the radical SAM enzyme ArsS, leading to form arsenosugars and possibly other complex organoarsenicals through additional as-yet unidentified steps (Xue et al., 2019). This biotransformation of toxic inorganic arsenic into non-toxic arsenosugars is considered as a detoxification pathway, enable seaweed to sequester high amounts of arsenic in a non-toxic form (Caumette et al., 2012). Arsenosugars can be metabolized by humans. It appears to be metabolized to several arsenic compounds, with DMAs(V) as the major metabolite, constituting 67% of the total excreted arsenic (Francesconi et al., 2002).
Another Group of Methylated Organoarsenic Compounds in Seafood are Arsenolipids, Which Contain Arsenic and a Long Aliphatic Chain.
These natural products are present in fish and algae. found in lipid extracts of algae and fish. The pathway for arsenolipid biosynthesis remains unclear, but most likely they are made by the algae, which are eaten by the fish. This group of compounds includes arsenic-containing fatty acids (AsFA), hydrocarbons (AsHC), glycophospholipids (AsPL) and phosphatidylcholines (AsPCs) (Chen et al., 2019a). To date, about 50 arsenolipids have been identified, including arsenic-containing hydrocarbons, fatty acids, phospholipids, phosphatidylcholines, and fatty alcohols, and more and more unidentified arsenic compounds will likely be discovered due to development the chemical measurement technology (Taylor et al., 2017). Like arsenosugars, arsenolipids also are metabolized in humans and finally degraded into DMAs.
Arsenobetaine (2-Trimethylarsoniumylacetate or AB) Is an Arsenic-Containing Analog of the Amino Acid Derivative Trimethylglycine (Glycine Betaine) and Is Highly Stable and Resistance to Breakdown or Further Metabolism
Arsenobetaine is one of the major organoarsenic compounds found in aquatic organisms. While arsenosugars are found in seaweed, AB is the major arsenic species in most marine animals, include seafood for human consumption, accounting for >80% of total ingested arsenic (Button et al., 2011). AB is similar in physiological properties to glycine betaine with respect to osmotic and thermal stress protection. AB has also been found in marine deep-sea hydrothermal vent ecosystems. Hence, AB may serve as a protectant against the detrimental effects of high salinity and high temperature for microbes that populated by some highly specialized organisms (Hoffmann et al., 2018). No genes for AB biosynthesis have been identified. Currently, there are two proposed pathways for arsenobetaine biosynthesis and both of them rely mostly on detection of potential biosynthetic precursors and intermediates. One possible pathway is via the oxidation of arsenocholine (AC) following breakdown of trimethylated arsenosugars (Caumette et al., 2012; Hoffmann et al., 2018). A second proposed pathway is via reaction of DMAs(III) with glycoxylate. Unlike the biotransformation of arsenosugars and arsenolipids, arsenobetaine is rapidly excreted unaltered in human urine (Thomas and Bradham, 2016; Taylor et al., 2017), and no adverse health effects have been associated with arsenobetaine exposure.
AST, a Newly Discovered Antibiotic
Recently a new compound, arsinothricin [2-amino-4-(hydroxymethylarsinoyl) butanoate, AST], was shown to be synthesized by the rice rhizosphere bacterium Burkholderia gladioli strain GSRB05 (Kuramata et al., 2016). AST shows broad-spectrum antibiotic activity and is effective against both Gram-positive and Gram-negative bacteria, including World Health Organization priority pathogens such as carbapenem-resistant Enterobacter cloacae and Mycobacterium bovis BCG, a causative agent of tuberculosis related to M. tuberculosis. AST is a non-proteogenic amino acid analog of glutamate. Unlike most pentavalent organoarsenicals, AST is as toxic as trivalent MAs(III). The explanation is that its antimicrobial activity results not from the reactivity of the arsenic atom but from its chemical nature as a glutamate analog. It mimics that γ-acylphosphoglutamate intermediate in the glutamine synthetase (GS) reaction. Glutamine synthetase is a crucial enzyme in formation of nitrogen compounds by incorporation of the ammonia from urea. When microbial GS is inhibited, urea builds up and kills sensitive bacteria. AST is similar to 2-amino-4-(hydroxymethylphosphinyl) butanoic acid, or phosphinothricin (PT), an antibiotic produced by Streptomyces species. Like AST, PT is a non-proteogenic amino acid analog of glutamate that inhibits GS. PT is not useful as an antibiotic but has broad-spectrum herbicide activity and is sold under the name Glufosinate. At least in E. coli, AST is taken up better than PT and shows 15-fold higher toxicity compared with PT.
Enzymes and Transporters of Methylarsenical Biotransformations
Arsenic was present in soil and water before the origin of life, so it was imperative that the first organisms had mechanisms to tolerate this pervasive metalloid. The first ars operons evolved in an anoxic atmosphere and must have had either arsB or acr3 genes encoding unrelated As(III) efflux permeases that confer tolerance to high levels of arsenic. Another early adaptation that gave bacteria the ability to use environmental arsenic to obtain a competitive advantage over other bacteria evolved at least twice, once about 3billon years ago (BYa), before the Great Oxygenation Event (GOE), and a second time about 2.5 BYa, when the atmosphere became oxidizing. The first event was the evolution of the ArsM As(III) SAM methyltransferase. In the absence of oxygen, MAs(III) would have been a stable and potent antibiotic. Like most antibiotics, it would have required a way to get out the cells of the producer, and the ArsP MAs(III) efflux permease also appears to have evolved prior to the GOE. Other bacteria acquired the arsP gene and became resistant to MAs(III) - an early example of antibiotic resistance (Chen et al., 2019b). Although cells with arsM would also have produced DMAs(III), when arsP is co-expressed, it extrudes MAs(III) before it can be methylated a second time (Chen et al., 2019b), so the more stable MAs(III) would have been the major species with antibiotic activity. After the GOE, MAs(III) was oxidized to less toxic MAs(V). To take advantage of the new availability of MAs(V), other members of microbial communities evolved and sustained the ability to reduce it back to MAs(III), enable them a competitive advantage over sensitive microbes. Since this cycle of methylation, oxidation and reduction requires multiple species of bacteria, these complex interactions are an emergent property of the microbial community. In addition to MAs(III), AST is generated by other members of microbial communities. To survive under environmental stress of MAs(III) and AST, organisms have developed metabolic strategies for organoarsenical detoxification. First, once the atmosphere became oxidizing, arsenic methylation by ArsM became an As(III) detoxification mechanism because MAs(III) was oxidized in air to MAs(V). Second, the arsP gene was acquired by other bacteria to confer MAs(III) tolerance by extrusion. Third and fourth, the availability of atmospheric oxygen allowed the evolution of new genes/enzymes for the oxidative metabolism of MAs(III). One is the ArsI MAs(III) demethylase, a dioxygenase that catalyzes the cleavage the C-As bond and produces less toxic As(III). Another is the ArsH MAs(III) oxidase that detoxifies the trivalent species by transforming it into the pentavalent species. Finally, resistance to the other organoarsenical antibiotic, AST, arose as the ArsN N-acetyltransferase. These enzymes and transporters will be described in more detail in this section.
ArsM/AS3MT: As(III) S-Adenosylmethionine Methyltransferases
The stepwise biomethylation of inorganic arsenite is catalyzed by As(III) SSAM methyltransferase (EC 2.1.1.137), which is termed alternatively ArsM in microbes or AS3MT in animals. The evolutionary history of this enzyme has created a paradoxical view of its physiological role: is its function arsenic detoxification, activation of arsenic into a toxin or creation of a primordial antibiotic? In extant aerobic microbes, it is undoubted that arsenic methylation isa detoxification process because the trivalent products are rapidly oxidized to less toxic pentavalent species in air, but the physiological role of biomethylation in animals is still unclear.
As mentioned above, the process of arsenic methylation was originally proposed to be a detoxification mechanism in animals, especially humans, where trivalent inorganic arsenic is biotransformed to relatively less toxic pentavalent methylated species. In the 1940s Challenger proposed a chemically plausible scheme for the inorganic arsenic methylation (Challenger, 1951). In his original proposal, in animals, As(V) would enter cells (since shown to be via phosphate transport systems), be initially reduced to As(III) and further biotransformed by oxidative methylation with the addition of a methyl group from the methyl donor SAM to form MAs(V). MAs(V) would then be reduced to MAs(III), which would be the substrate for addition of a second methyl group via an oxidative reaction to yield DMAs(V). In this original proposal the substrates are trivalent, and the products would be pentavalent. This concept was supported by the fact that the pentavalent methylated species were thought to be rapidly excreted, adding to detoxification (Gebel, 2002). At that time the enzymes that catalyze the reaction or reactions were unknown. It was later shown by Styblo and Thomas that the mammalian liver enzyme AS3MT is responsible for methylation of inorganic arsenic (Lin et al., 2002). ArsM was later identified as the microbial ortholog (Qin et al., 2006). Whether AS3MT detoxifies inorganic arsenic or activates it to a more potent toxin and potential carcinogen depends on whether the products are indeed the less toxic pentavalent species or the trivalent metabolites MAs(III) and DMAs(III), which are reactive and highly toxic, and would increase arsenic toxicity (Hughes et al., 2011). The pentavalent state of arsenic in both inorganic and methylated species is an important determinant of toxicity in a wide range of biological systems. Generally, trivalent arsenicals are more toxic than the pentavalent (Le et al., 2000a). Therefore, if the major methylated intracellular products are trivalent, the methylation would increase the toxicity and potentially the carcinogenicity of arsenic. The trivalent species can initiate toxic effects such as DNA-damage (Mass et al., 2001), or be indirectly genotoxic (Hirano et al., 2004). The detection of trivalent arsenic, such as MAs(III) in human populations exposed to inorganic arsenic in their drinking water, has provoked considerable interest in the mechanism of arsenic biomethylation process (Valenzuela et al., 2005). Indeed, the linkage between the biomethylation process and intermediates of methylated metabolites containing either trivalent or pentavalent arsenic remains a critical question in the study of arsenic methylation as a process of detoxification or not. The presence of trivalent species in urine suggests that the products of AS3MT are trivalent, rather than pentavalent. Both purified human AS3MT and algal ArsM were shown to produce MAs(III) as the first methylation product (Marapakala et al., 2012; Dheeman et al., 2014). Presumable the second methylation product is DMAs(III), but this has not been confirmed because DMAs(III) is unstable.
To disclose the enzymatic mechanism of ArsM/AS3MT is essential for understanding its parallel roles in arsenic detoxification and carcinogenesis. To better understand the methylation reaction, the crystal structure of the thermophilic red alga Cyanidioschyzon merolae CmArsM was solved at 1.6 Å (Ajees et al., 2012), and subsequently with and without substrates and products such as MAs(III) (Figure 3). The arsenic binding site is composed of conserved cysteine residues that coordinate trivalent arsenicals, while pentavalent arsenicals cannot bind to AS3MT or ArsM. ArsM/AS3MT has four conserved cysteine residues based on the alignment of mammalian and algal sequences (Li et al., 2016). All four single mutants almost lost As(III) methylation activity, but, when either of the first two cysteines was mutated to serine, the enzyme lost the ability of first methylation step [As(III) to MAs(III)], but could have the ability of the second methylation step [MAs(III) to DMAs(III)] (Dheeman et al., 2014). The role of the cysteine residues in the arsenic methylation process has been well investigated through a combination of structural, biochemical and molecular biological analyses. From the crystal structure, it clearly shows ArsM/AS3MThas three domains, an N-terminal SAM binding domain, a C-terminal domain of unknown function and a middle As(III) binding domain. The role of two conserved cysteine residues is shown in a structure with bound PhAs(III). These two conserved cysteine residues closest to the N-terminus are disulfide bonded with each other, and the rest two conserved cysteines bind the arsenic atom, similar to the bound MAs(III) shown in Figure 3Marapakala et al. (2015). The first cysteine pair appears to have a different role in arsenic methylation catalytic cycle that is only required during the first methylation step [As(III) to MAs(III)] but not the second [MAs(III) to DMAs(III)]. A disulfide bond cascade was proposed to be part of the catalytic cycle of both CmArsM and human AS3MT (Dheeman et al., 2014; Marapakala et al., 2015). In the first methylation step, a disulfide bond is formed between the first two cysteines and S-methyl group from SAM is transferred to As(III) to form an enzyme-bound MAs(V) moiety. To proceed to the second methylation step, the arsenic has to be re-reduced to be trivalent again, and the role of the first two conserved cysteine residues is to reduce the MAs(V) intermediate. Reduction of the arsenic results oxidatizes the cysteines, which form a disulfide bond. To continue to the next methylation step, the disulfide bond has to be reduced. The small redox protein thioredoxin had been shown to be involved in the reaction, but its role was unknown (Thomas et al., 2004). In the proposed disulfide bond cascade, the role of thioredoxin would be reduce the disulfide bond between the first two cysteine residues (Dheeman et al., 2014). Since the first cysteine pair is not required for methylating MAs(III) to DMAs(III), that reaction likely uses an alternative combination of cysteine residues for reduction of an enzyme-bound DMAs(V) intermediate.
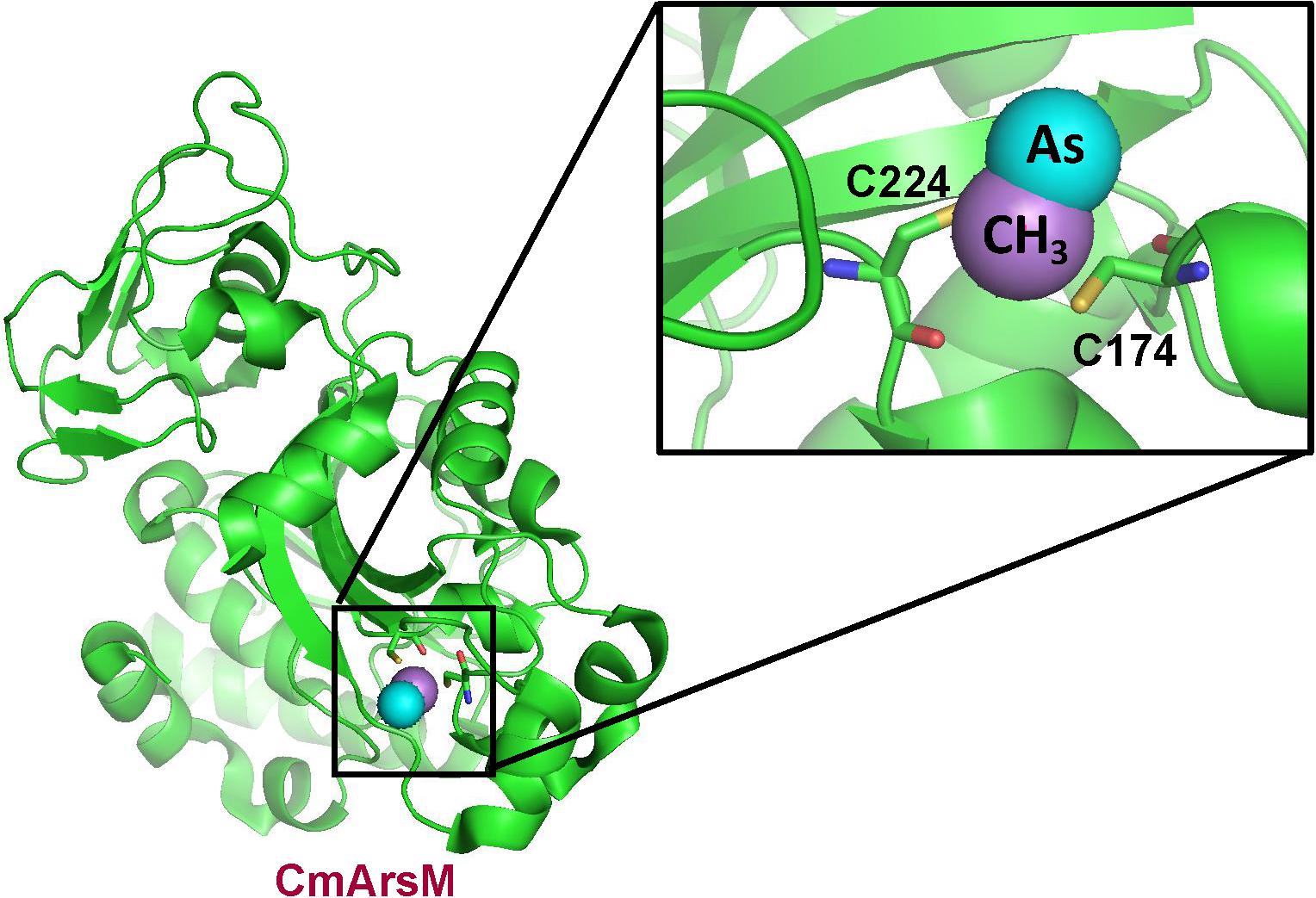
Figure 3. Structure of C. merolae CmArsM with MAs(III) bound to conserved cysteine residues 174 and 224. The CmArsM-MAs(III) structural model was built on the pdb id 4KW7 structure. The backbone of the CmArsM structure is shown in green ribbon. Cysteine residues are shown in stick form, with the sulfur atoms in yellow. The details of the MAs(III) binding site are shown in the expanded view. The methyl group is colored purple, and the arsenic atom is colored cyan.
To date, more than 6000 sequences for genes encoding ArsMs are collected in the NCBI database, with most of them from prokaryotic and eukaryotic microbes. In recent years a combination of structural and functional data have become available that support the idea that the products are toxic, supporting our hypothesis that the ArsM enzyme evolved not for detoxification but to produce a potent antimicrobial that bacteria used to gain a competitive advantage for survival and dominance over MAs(III)-sensitive members of complex microbial communities.
MAs(III) Efflux Permeases
ArsP, an MAs(III)-Specific Efflux Permease
To use MAs(III) as an effective antibiotic, producing cells would need to extrude the toxic compound into the medium for two reasons. First, this would protect the producers from their own toxic products, and second, antibiotic secretion is a prerequisite to killing competitors. Neither of the previously identified arsenic efflux permeases, ArsB or Acr3, transports MAs(III) or confers resistance. A putative membrane permease (ArsP), found in a four-gene ars operon in Campylobacter jejuni RM1221, was associated with elevated resistance to pentavalent roxarsone (Shen et al., 2014). Synthetic aromatic arsenicals such as Rox(V), 4-aminobenzenearsenate, p-ASA(V), and 4-nitrobenzenearsenate, Nit(V) have been widely used in animal husbandry as antimicrobial growth promoters for decades (Nachman et al., 2013). ArsP functions as an efflux transporter specific for extrusion of roxarsone and contributes to the resistance to roxarsone in C. jejuni. It was further identified the substrates of ArsP are actually trivalent organoarsenicals (Chen et al., 2015b). Arsenite-hypersensitive E. coli strain AW3110 expressing C. jejuni arsP, conferred high resistance to MAs(III) and Rox(III) but not to inorganic As(III) or pentavalent organoarsenicals. Control cells of E. coli AW3110 without CjarsP accumulated high level of trivalent organoarsenicals, while cells with CjarsP did not. Everted membrane vesicles prepared from the strain with CjarsP-expression accumulated MAs(III) with the energy derived from oxidation of NADH, consistent with ArsP catalyzing efflux from cells. The transport of As(III), MAs(V) and pentavalent aromatic arsenicals were not observed. The transport property of ArsP showed its selective for the natural product MAs(III) over synthetic trivalent aromatic arsenicals such as Rox(III). With 315 amino acid residues, CjArsP is smaller than ArsB or Acr3 and is predicted to have only eight transmembrane helices (TMs), which is atypical for bacterial permeases (Garbinski et al., 2019). Consistent with a role in MAs(III) efflux, cells that express both arsM and arsP are more resistant to MAs(III) than cells expressing either alone, as would have been expected for the original MAs(III) producers, Lateral gene transfer of arsP to MAs(III)-sensitive bacteria could have spread resistance to MAs(III).
ArsK, a Broad Substrate Efflux Permease
Recently a second MAs(III) efflux permease, ArsK, was found in an arsenic gene island in the chromosome of Agrobacterium tumefaciens GW4 (Shi et al., 2018) as well as in a Bacillus sp. strain (Jia et al., 2019). ArsK orthologs can be found in most of arsenic-resistant bacteria, but not in archaea, fungi, plants or animals. It is in a distinct cluster that is phylogenetically divergent from other arsenic transporters. It is regulated by an ArsR repressor and could be induced As(III), antimonite Sb(III), Rox(III), and MAs(III). As(V) also induced, but that might require reduction to As(III) since no ArsR repressor has been found to respond to pentavalent arsenicals. ArsK shows a broad substrates selectivity, which confers resistance to As(III), Sb(III), Rox(III), and MAs(III) by reducing cellular accumulation of these compounds. It does not confer resistance to As(V), DMAs(III) or Cd(II). ArsK is novel in that it exhibits broad substrate specificity, including both trivalent inorganic and organic species. ArsK has three conserved cysteine residues, but whether they are involved in ArsK function has not been investigated. It is not clear whether ArsK evolved in conjunction with ArsM or independently as a resistance mechanism for MAs(III).
Once the atmosphere because oxidizing, ArsM-catalyzed methylation reversed its function to detoxification because the methylated products would be oxidized in air to the less toxic pentavalent species. Members of aerobic microbial communities, taking advantage of the presence of MAs(V), evolved pathways to reduce MAs(V), regenerating environmental MAs(III) to give them a competitive advantage. As described below, other members of the aerobic microbial communities evolved several different mechanisms for resistance to MAs(III) in addition to the spread of the ArsP and ArsK efflux permeases. This level of complexity, where the properties of the community cannot be predicted from the activity of each individual specie, is an emergent property of these microbial communities: one community member generates a compound that a second community member transforms into an antibiotic that a third community member detoxifies (Chen et al., 2019b).
Enyzmyes That Confer Resistance to MAs(III) or AST
ArsI, a C–As Bond Lyase
ArsM orthologs are wide-spread and are found in microbes, which implies that methylated arsenicals are constantly being synthesized and entering the biosphere. In addition, abundant amounts of organoarsenicals such as MSMA were released into the environment as herbicides and pesticides during the last 50 years. However, the vast majority of arsenic in soil and water is inorganic, with only minor amounts of methylated species present except in specific areas of recent herbicide application (Huang et al., 2011). If microbes expressing As(III) SAM methyltransferases and catalyze arsenic methylation are ubiquitous, why isn’t the majority of the arsenic in the environment methylated species? A reasonable explanation is that methylated species are rapidly degraded.
A two step pathway of MSMA degradation by sequential reduction of MAs(V) to MAs(III) and demethylation of MAs(III) to As(III) was demonstrated in Florida golf course soil. The two steps are performed by different members of aerobic microbial communities: [MAs(V)] reduction and demethylation (Yoshinaga et al., 2011). The arsI gene was cloned from the MAs(III)-demethylating bacterium Bacillus sp. MD1 by selection for resistance to MAs(III) (Yoshinaga and Rosen, 2014). Expression of the arsI gene confers resistance to MAs(III). Purified ArsI is an Fe2+-dependent dioxygenase with C–As lyase activity. It catalyzes the C–As bond cleavage, transforming highly toxic MAs(III) into less toxic As(III). Its ortholog cloned from Thermomonospora curvata, TcArsI, was crystallized with Ni(II) bound to three amino acids that form the metal binding site (Nadar et al., 2014). The enzyme is not active with Ni(II), but the structure shows what the binding site would look like with bound Fe(II). The enzyme has a vicinal cysteine pair in a flexible loop that was shown to be the MAs(III) binding site (Figure 4). The structure of a mutant TcArsI protein (Y100H/V102F) was crystallized in a sequential series of conformations. Arranging the structures in a logical sequence shows that the loop binds MAs(III) in solvent and moves sequentially toward the metal binding site. This suggests that ArsI catalyzed reaction is controlled by a loop-gating mechanism (Nadar et al., 2016). In the open state without ligand, the loop is exposed to solvent, where it can capture MAs(III) substrate and moves toward the active site, where it forms a closed state that orients the C–As bond for dioxygen addition and cleaves it to As(III). In the reaction, one atom of dioxygen is bond to the arsenic atom of MAs(III) and the other oxygen atom is bond to the carbon, forming the products As(OH)3. The other product is a one-carbon product, either formaldehyde or methanol.
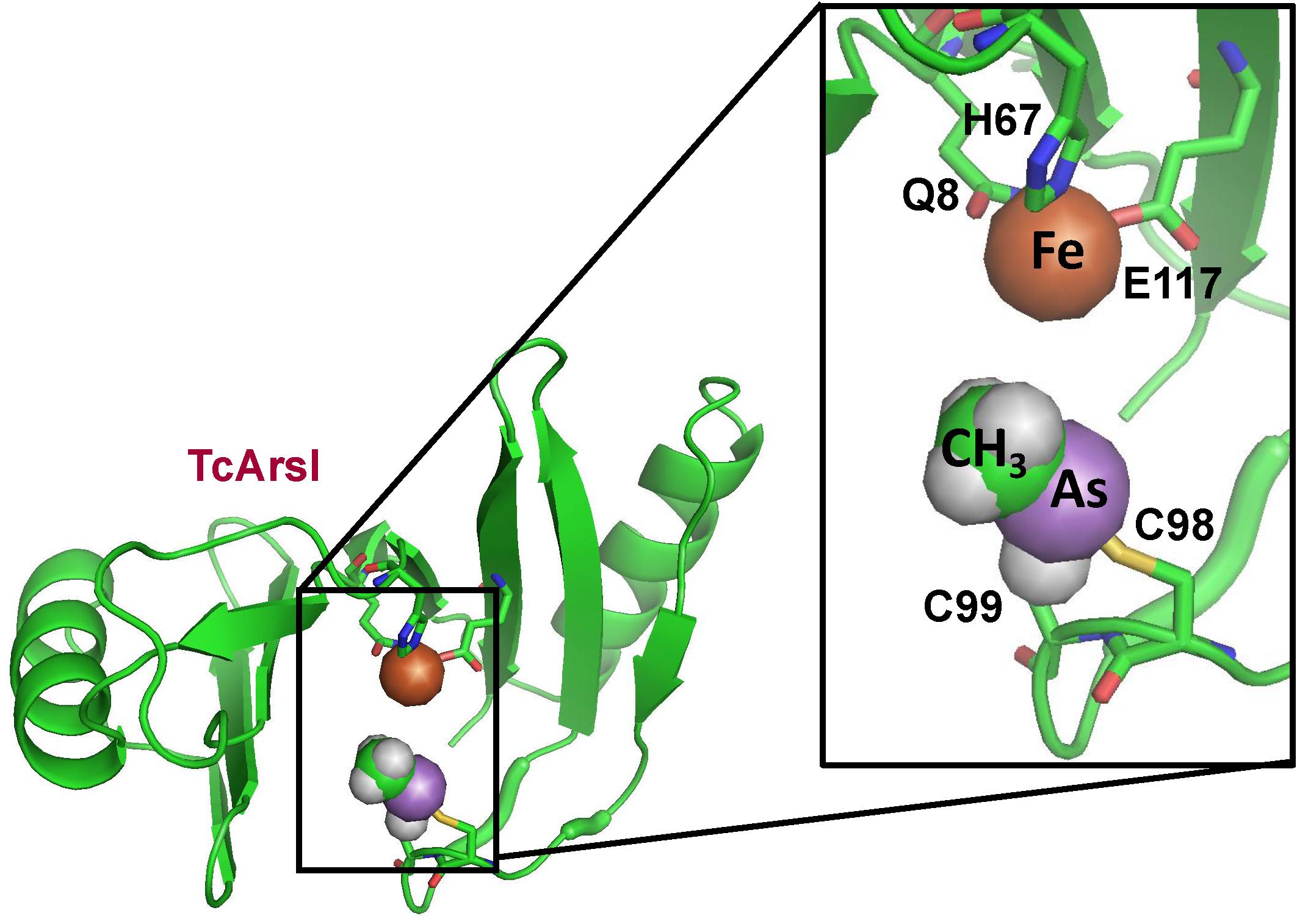
Figure 4. Structure of T. curvata TcArsI with MAs(III) bound to conserved residues Cys174 and Cys224. The TcArsI structural model was built on pdb id 5C4P. The catalytic iron atom is bound to the conserved triad of Gln8, His67 and Glu117. The backbone of the TcArsI structure is shown in green ribbon. Residues in the binding sites are shown in stick form, with the sulfur atoms in yellow, nitrogen in blue and oxygen in red. The details of the MAs(III) binding site are shown in the expanded view. The methyl group is colored green, the arsenic atom is colored purple, and the iron atom is colored orange.
ArsI also can cleave the C–As bond in a broad substrates of trivalent synthetic aromatic arsenicals, such as Rox(III), Nit(III), and p-ASA(III) (Yoshinaga and Rosen, 2014). By electron spray ionization mass spectrometry (ESI-MS) the products of Rox(III) biodegradation by ArsI were shown to be As(III) and 2-nitrohydroquinone (Pawitwar et al., 2017). Thus ArsI incorporates one atom from dioxygen onto the aromatic ring of these aromatic arsenic compounds and the other onto the arsenic atom. arsI genes are widely distributed in aerobic bacteria. To date, about 1305 putative ArsI orthologs have been identified in 963 aerobic bacterial species. Recently, a new arsI from the cyanobacterium Nostoc sp. PCC 7120 was cloned and characterized (Yan et al., 2015). Marine cyanobacteria are responsible the GOE and for most of the oxygen in the present-day atmosphere. The wide-spread existence of MAs(III) degrading enzymes in marine microbes provides new insights into their significant role in the global arsenic biogeochemical cycle.
ArsH, a Methylarsenite Oxidase
ArsH encodes an NADPH-dependent MAs(III) oxidase (Chen et al., 2015a). The arsH gene confers resistance to MAs(III) and is widely distributed in bacterial ars operons. Currently more than 9000 ArsH-related protein sequences are present in the NCBI database. Most (97.7%) of ArsH sequences are found in bacteria, with a few in fungi (2.2%) (Chen et al., 2015a). Sinorhizobium meliloti ArsH purifies as a tetramer, and the crystal structure of ArsH shows that ArsH is a dimer of dimers (Ye et al., 2007; Figure 5). The crystal structure shows it has eight molecules in the asymmetric unit in two identical tetramers, and each tetramer is a functional unit. The structure shows no binding site for substrates, so the physiological function of ArsH could not be deduced from its structure.
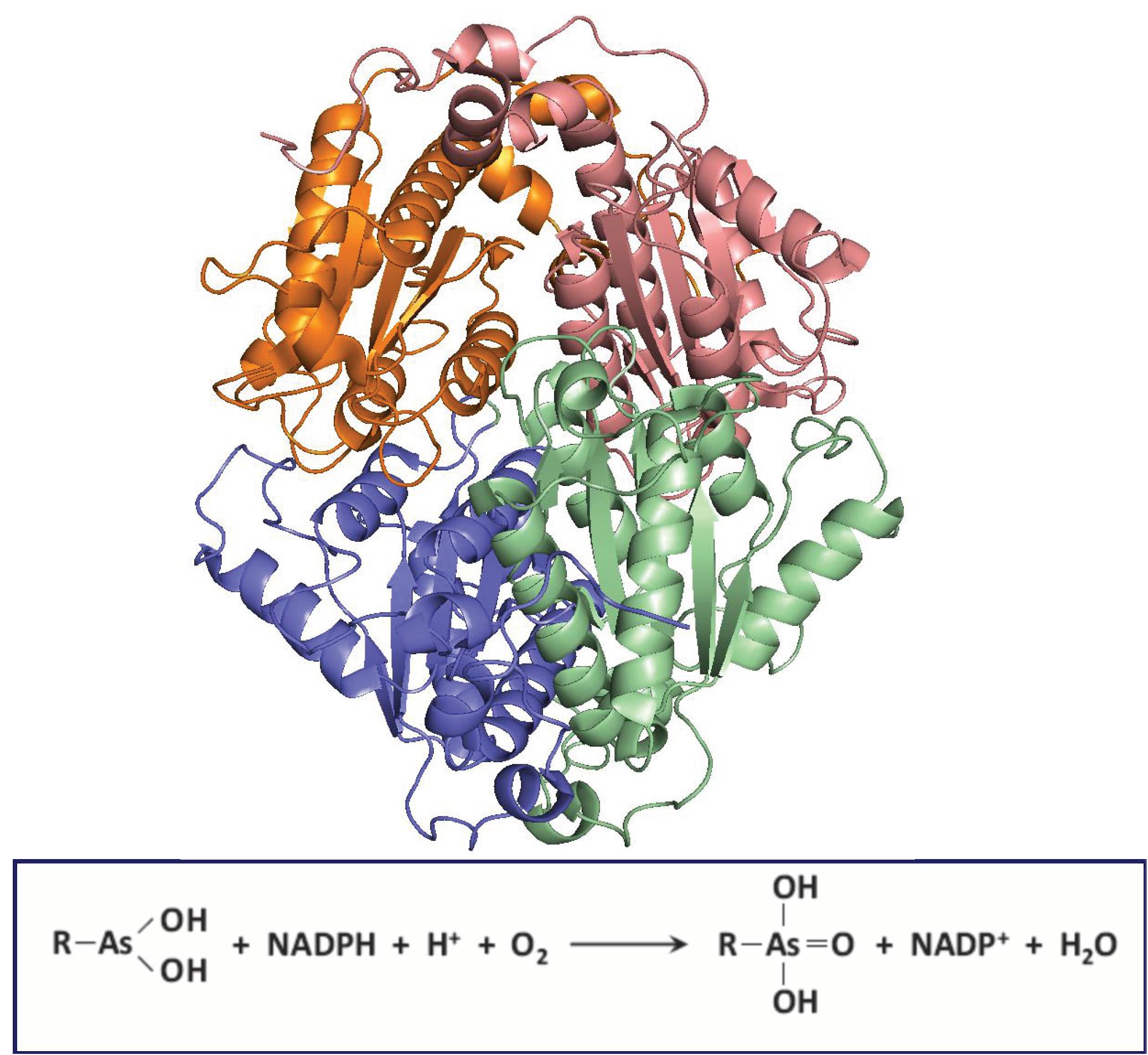
Figure 5. Structure of S. meliloti ArsH. ArsH (pdb id 2Q62) is a homotetramer that uses NADPH and O2 to oxidize MAs(III) to MAs(V). Each subunit in the homotramer is colored differently.
ArsH was originally found in an ars operon in Yersinia enterocolitica. It was suggested to confer resistance to inorganic arsenic in Y. enterocolitica (Neyt et al., 1997) and in S. meliloti (Yang et al., 2005), although the data were not strong. Neither the arsH genes from Acidothiobacillus ferrooxidans or the cyanobacterium Synechocystis conferred resistance to inorganic arsenic (Butcher et al., 2000; Lopez-Maury et al., 2003). Thus it appears that inorganic arsenicals are not substrates. However, the arsH from P. putida strain VTw33 was recently reported to be involved in arsenite oxidation in the periplasm rather than arsenate reduction, although it is not clear that this is a major physiological function (Chang et al., 2018). Purified ArsH was shown to have NADPH-dependent FMN reduction of azo dyes and also can reduce O2 to H2O2 but did not neither oxidize As(III) nor reduce As(V). Synechocystis ArsH has been identified as a quinone reductase (Hervas et al., 2012) and is able to reduce chromate and ferric iron, but the related arsenic resistance was not observed (Xue et al., 2014).
Even though ArsH has reductase activity toward azo dyes and some metals, its presence in ars operons indicated its an arsenic-related function. While the resistance to inorganic arsenic was not confirmed, the physiological role of arsH was shown to have the ability to oxidize toxic MAs(III) and other trivalent organoarsenicals to relatively less toxic pentavalent species (Chen et al., 2015a). Pseudomonas putida KT2440 has two copies of ars operons, each with an arsH gene (Paez-Espino et al., 2015). Deletion of both ars operons in P. putida renders the organism sensitive to MAs(III) and PhAs(III) (but not to Rox(III). The P. putida double ars deletion was 10-fold more resistant to these organoarsenicals than E. coli. An arsH deletion of S. meliloti similarly, became sensitive to trivalent organoarsenicals, indicating that expression of the arsH gene is responsible for resistance to high toxic trivalent organoarsenicals. Heterologous expression of arsH cloned from P. putida ars operon (ars1) in the arsenite hypersensitive E. coli strain AW3110, in which the ars operon was deleted (Carlin et al., 1995), conferred the resistance to MAs(III), PhAs(III), and Rox(III). The fact that ArsH detoxifies trivalent organoarsenicals by oxidation indicates that the physiological function of ArsH is not a reductase but rather a trivalent organoarsenical oxidase. It catalyzes a reaction not unlike a mixed function oxidation, using both NADPH and molecular oxygen (Figure 5). The expression of ArsH alone is sufficient to detoxify the activated trivalent forms of the herbicide MSMA and the poultry antimicrobial growth promoter roxarsone without any extra ars genes. Purified ArsH from either S. meliloti or P. putida are colored yellow and show a typical flavoprotein absorption spectrum (Chen et al., 2015a). S. meliloti and P. putida ArsH contains two predicted FMN binding sites with the amino acid sequence G15SLRTVSYS, and G42STRERSFS, respectively, but no MAs(III) binding site can be recognized from the sequence or structure.
ArsN, an AST-Selective N-Acetyltransferase
As described above, AST is an arsenic-containing non-proteinogenic amino acid analog of glutamate, produced by a rice rhizosphere bacterium (Kuramata et al., 2016). It is an antibiotic that is equally active against both Gram-negative and Gram-positive bacteria, including several pathogenic species (Nadar et al., 2019). Resistance to AST is conferred by the arsN1 gene found in many ars operons. The arsN genes can be classified into two clades based on the sequence similarity. The arsN1 genes in Clade 1 encode proteins more closely related to phosphinothricin N-acetyltransferases, which N-acetylates phosphinothricin using acetyl-CoA. Whereas products of the arsN2 genes from Clade 2 are more closely related to glutamate N-acetyltransferases (N-acetylglutamate synthases) (Nadar et al., 2019). ArsN1 is an N-acetyltransferase that inactivates AST by acetylation of the α-amino group with an acetyl group from acetyl CoA, which prevents binding to enzyme glutamine synthetase. In contrast, the arsN2 gene does not confer AST resistance The structure of P. putida ArsN1 was solved with bound AST and the acetyl donor acetyl CoA (Figure 6). ArsN1 is a homodimer with the AST binding site formed by amino acid residues from both subunits.
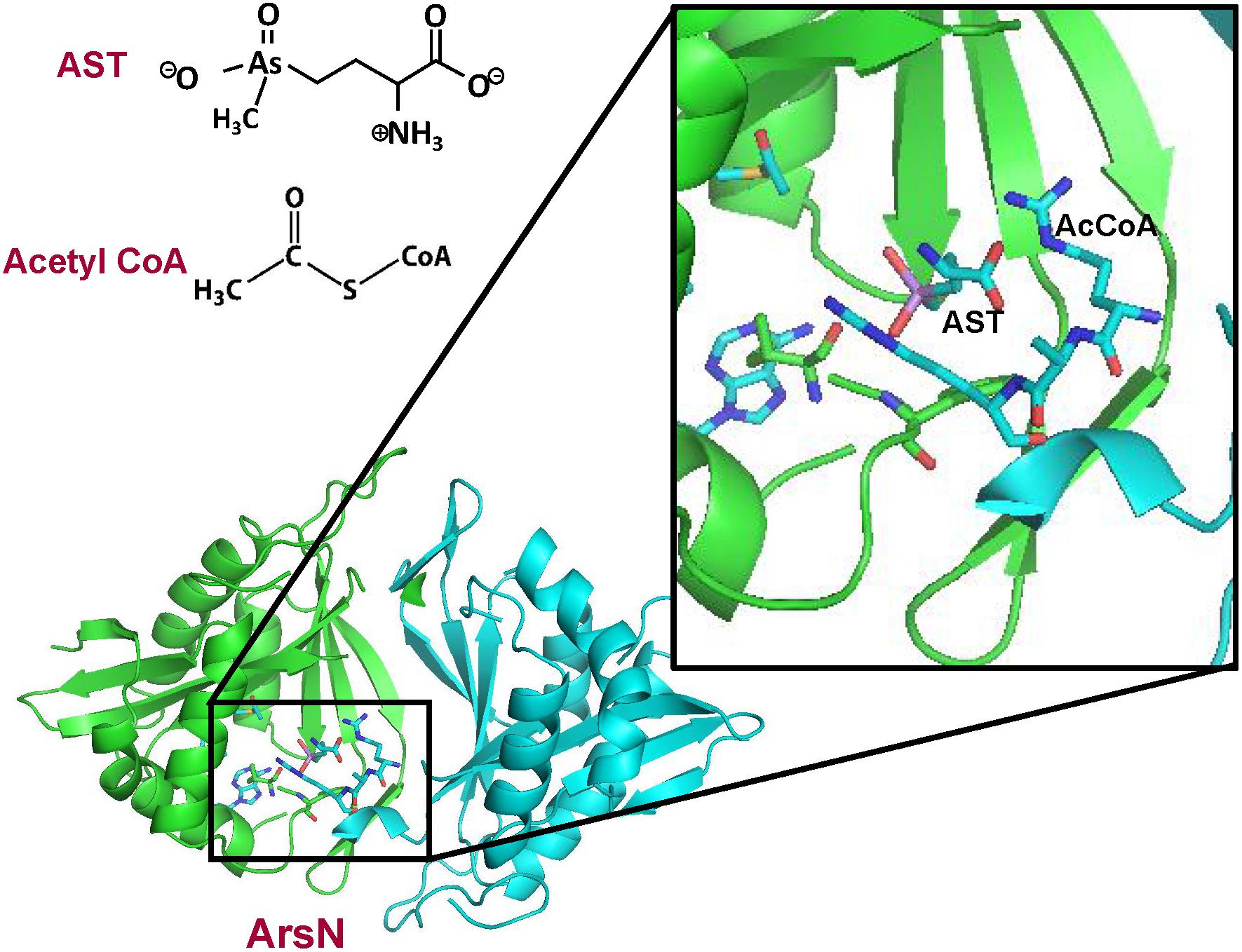
Figure 6. Structure of P. putida ArsN with bound AST and acetylCoA. The ArsN structural model with bound AST and acetylCoA was built on pdb id 5WPH ArsN is a homodimer in which the active site is composed of residues from both subunits. The backbones of the two subunits of the ArsN1 homodimer structure are shown in green and cyan ribbon, respectively. AcetylCoA, AST and residues in the binding sites are shown in stick form. with the sulfur atoms in yellow, nitrogen in blue and oxygen in red. The details of the binding sites are shown in the expanded view.
Organoarsenical Antibiotics and Antimicrobials
The word “arsenic” evokes fear of poison, so the use of arsenic-containing antibiotics and drugs might be looked on with trepidation by the public. However, for centuries arsenicals have been used as traditional medicines in China and, more recently, as antimicrobials and anti-cancer agents in Western medicine. Atoxyl (arsanilic acid or aminophenyl arsonic acid) was the first synthetic organoarsenical drug introduced as the late 19th century as an antimicrobial for treatment of tropical diseases, but its toxicity prevented its general use. In 1908, Paul Erlich was awarded the Nobel Prize in Physiology or Medicine, in part for the development of the atoxyl derivative Salvarsan (arsphenamine) for treatment of syphilis. In organic arsenite in the form of arsenic trioxide (Trisenox or ATO) is currently used as a chemotherapeutic drug for treatment for a type of acute myeloid leukaemia, acute promyelocytic leukaemia (APL) (Ramaekers et al., 2019). Synthetic organoarsenicals such as melarsoprol have been used for treatment of tropical parasitic diseases such as African trypanosomiasis (Chagas Disease). Related antimonials including Pentostam were used for treatment of leshmanial diseases such as kala-azar. Atoxyl and other derivatives of phenylarsonic acid such as roxarsone have been added to animal feed since the 1920s for prevention of infectious diseases in poultry, such as roxarsone, although it is currently no longer used in many countries such as the United States because of potential risk to the environment (Chen et al., 2019a).
The evolution of ArsM is predicted to have happened more than 3 Bya in an anoxic world with appearance of free oxygen in the Earth’s atmosphere before the accumulation of oxygen in the atmosphere (2.45–2.32 Bya) during the Great Oxygenation Event (GOE) (Chen et al., 2017). This calls into question that whether the primordial role of ArsM played the role as a detoxification pathway: methylation of arsenite definitely increases toxicity by producing highly toxic intermediates: MAs(III) and DMAs(III), which would have been thermodynamically stable in the oxygen-free environment that existed before the GOE. Thus, ArsM would not be performed as a detoxification enzyme unless these products can be converted to less toxic MAs(V) and DMAs(V) by sufficient oxygen. Thus we propose that ArsM was originally evolved to produce extremely toxic trivalent methylarsenicals to function as antibiotics to kill competitors in the primordial environment enrich with inorganic arsenic; after the GOE the trivalent species were non-enzymatically oxidized to pentavalent forms by oxygen, changing ArsM into a detoxifying enzyme (Li et al., 2016). Life has not only adapted effectively to their own environments enrich with arsenic, but also can utilize arsenic as an offensive weapon to gain a competitive advantage over sensitive members of the complex microbial communities (Chen et al., 2019b). The original antibiotic producer (ArsM hosts) may have not only defended itself by extrusion of its own toxic products MAs(III) with methylarsenite efflux permeases (ArsP or ArsK) (Chen et al., 2015b; Shi et al., 2018) but also put the toxin outside of the cells, where it could kill its sensitive neighbors (Figure 1). There is some evidence that suggests that arsP usually found to co-exists with arsM, primarily in anaerobes, which might be an ancestral feature in lineages of anaerobes. The evolution of these two correlated MAs(III) resistance gene supports our hypothesis that ArsP evolved for detoxification of MAs(III) in ArsM hosts (Chen et al., 2017).
After the GOE, microbes adapted to the presence of MAs(V) evolving new pathways of continual re-reduction to MAs(III), conferring them a competitive advantage over MAs(III) sensitive bacteria. The environmental pressure of MAs(V) reduction drove the convergent evolution of multiple MAs(III) detoxification mechanisms (Figure 7A). In particular, the availability of oxygen allowed the rise of oxygen-dependent MAs(III) resistance enzymes, including ArsH, which uses O2 to enzymatically oxidize MAs(III) to MAs(V) and ArsI, which degrades MAs(III) by cleavage of the methyl group to form less toxic As(III), adding dioxygen across the C-As bond. In a second adaptation, AST is synthesized by soil bacteria, although the biosynthetic pathway and intermediates remain largely unknown, and, in response other bacteria evolved the arsN gene to detoxify AST by acetylation of the α-amino group (Figure 7B).
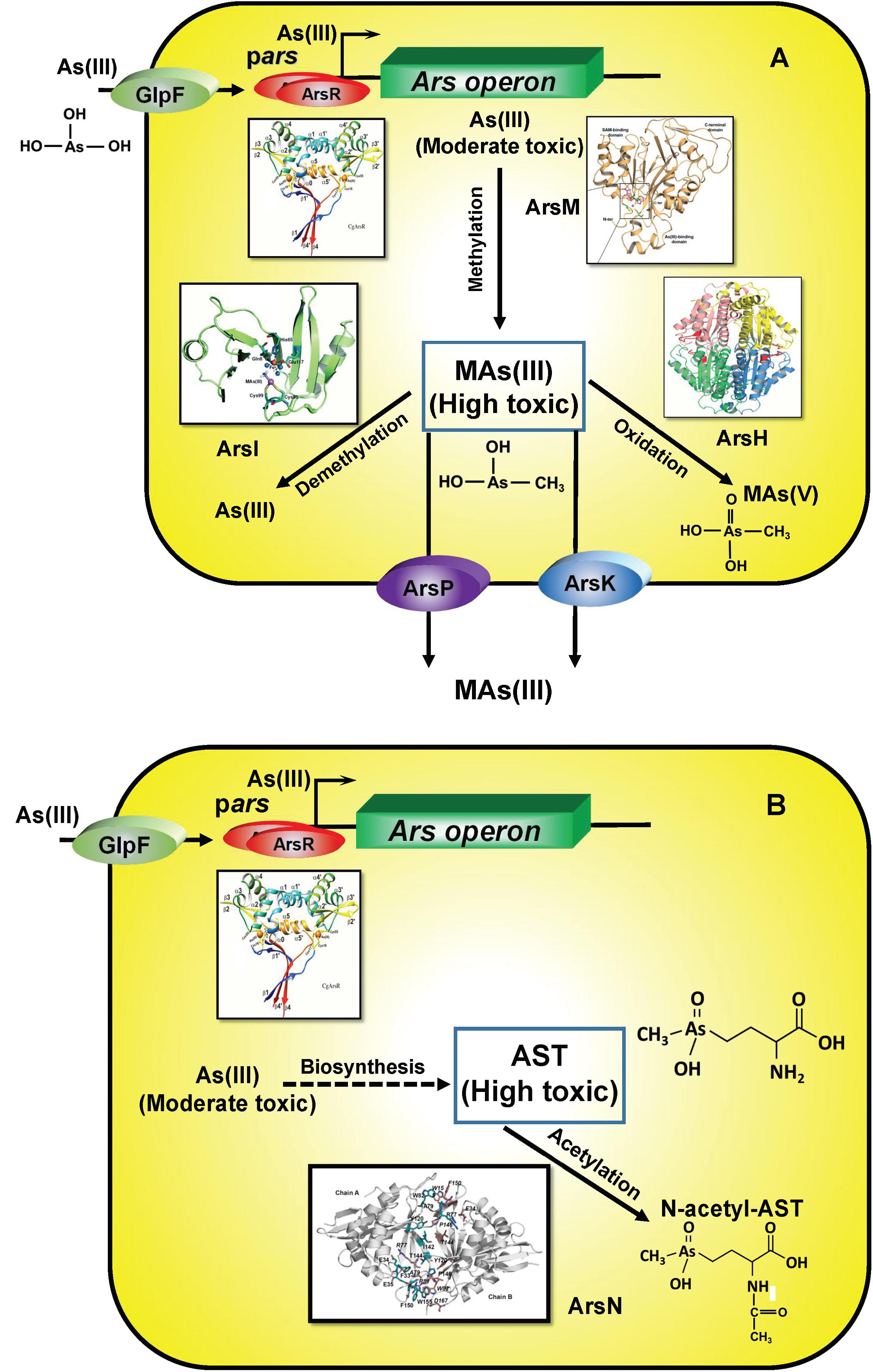
Figure 7. Enzymes involved the detoxification of methylated MAs(III) (A) and AST (B). As(III) is taken up by aquaglyceroporins such as GlpF. In ars operons, transcription is controlled by the ArsR repressor, which senses either As(III) or MAs(III). Crystal structures of relevant enzymes are shown.
Microbial communities are particularly important for generation of a sustained pool of MAs(III), these complex interactions are an emergent property of the entire community. To approximate these communities under laboratory conditions, cells of the environment isolate Burkholderia sp. MR1, which reduces MAs(V) to produce toxic MAs(III) (predator), and a MA(III)-sensitive strain of E. coli that constitutively expresses the gene for green fluorescent protein (GFP) (prey) were grown in mixed culture with the addition of MAs(V). Loss of viability by the prey was estimated from GFP fluorescence. When the prey expressed arsI, arsH or arsP from a plasmid, they were resistant to MAs(III) production by Burkholderia sp. MR1. These results clearly show that the behavior of MAs(III) performed as an antibiotic in a community setting: a toxic organic substance generated by one microbe to kill off its competitors. Thus bacterial communities have evolved not only to survival and dominance in the environmental enrich with arsenic through resistance mechanisms but have also evolved at least pathways to use arsenic to obtain a competitive advantage in microbial warfare, synthesis of MAs(III) and AST (Nadar et al., 2019; Figure 1).
Summary
Arsenic is the most ubiquitous toxin throughout the environment in soil and water, and bacteria have evolved mechanisms to use methylated arsenicals as antibiotics to give them a competitive advantage for growth in microbial communities. A number of methylated arsenic species are synthesized by various bacteria, including mono-, di- and trimethylated arsenicals, and other complex organoarsenicals such as arsenosugars, arsenolipids and arsenobetaine. Even fungi, algae and animals, including humans methylate arsenic. Methylation plays an important role in arsenic biogeochemical cycling in terrestrial, aquatic and atmospheric environments. Some methylated organoarsenic compounds showed high toxicity, especially trivalent species such as MAs(III). AST is a special case in being a highly toxic pentavalent methylated arsenical because of its mimicry of the phosphoglutamate intermediate in the glutamine synthetase reaction.
Antibiotic resistance has been resulted in one of the biggest public health threats of our time. It is urgent to explore some new and novel antibiotics to prevent and treat the rise in antibiotic-resistant bacteria worldwide. The antibiotic action of MAs(III) and AST indicates that they are perspective to be progenitors of a new classes of organoarsenical antibiotics. There are still a number of genes in ars operons for which functions related to arsenicals have not been found; it indicates that some other resistance mechanisms might be exist to against additional unknown natural products containing arsenic.
Author Contributions
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by NIH grants R35 GM136211, R01GM55425, and R01 ES023779 to BR and the Natural Science Foundation of China grant 41967023 to JC.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
AB, arsenobetaine; AST, arsinothricin (2-amino-4-methylarsonobutanoic acid); DMAs(III), dimethylarsenitek; DMAs(V), dimethylarsenate; DMMTAs(V), dimethylmonothioarsinic acid MAs(III), methylarsenite; MAs(V), methylarsenate; MMMTAs(V), monomethyl monothioarsonic acid; MSMA, monosodium methylarsenate; Nit(V), nitarsone; pAsA(V), p-arsanilic acid; PhAs(III), phenylarsenite; Rox(III), reduced roxarsone; Rox(V), roxarsone (3-nitro-4 hydroxybenzenearsonic acid); SAM, S-adenosylmethionine; SRB, sulfur reducing bacteria; (TMAs(V)O, trimethylarsine oxide.
References
Ajees, A. A., Marapakala, K., Packianathana, C., Sankaran, B., and Rosen, B. P. (2012). The structure of an As(III) S-adenosylmethionine methyltransferase: insights into the mechanism of arsenic biotransformation. Biochemistry 51, 5476–5485. doi: 10.1021/bi3004632
Bauerle, M. R., Schwalm, E. L., and Booker, S. J. (2015). Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation. J. Biol. Chem. 290, 3995–4002. doi: 10.1074/jbc.R114.607044
Butcher, B. G., Deane, S. M., and Rawlings, D. E. (2000). The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli. Appl. Environ. Microbiol. 66, 1826–1833. doi: 10.1128/Aem.66.5.1826-1833.2000
Button, M., Moriarty, M. M., Watts, M. J., Zhang, J., Koch, I., and Reimer, K. J. (2011). Arsenic speciation in field-collected and laboratory-exposed earthworms Lumbricus terrestris. Chemosphere 85, 1277–1283. doi: 10.1016/j.chemosphere.2011.07.026
Carlin, A., Shi, W., Dey, S., and Rosen, B. P. (1995). The ars operon of Escherichia coli confers arsenical and antimonial resistance. J. Bacteriol. 177, 981–986. doi: 10.1128/jb.177.4.981-986.1995
Caumette, G., Koch, I., and Reimer, K. J. (2012). Arsenobetaine formation in plankton: a review of studies at the base of the aquatic food chain. J. Environ. Monit. 14, 2841–2853. doi: 10.1039/c2em30572k
Chang, J. S., Yoon, I. H., and Kim, K. W. (2018). Arsenic biotransformation potential of microbial arsH responses in the biogeochemical cycling of arsenic-contaminated groundwater. Chemosphere 191, 729–737. doi: 10.1016/j.chemosphere.2017.10.044
Chen, J., Bhattacharjee, H., and Rosen, B. P. (2015a). ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Mol. Microbiol. 96, 1042–1052. doi: 10.1111/mmi.12988
Chen, J., Madegowda, M., Bhattacharjee, H., and Rosen, B. P. (2015b). ArsP: a methylarsenite efflux permease. Mol. Microbiol. 98, 625–635. doi: 10.1111/mmi.13145
Chen, J., Garbinski, L. D., Rosen, B. P., Zhang, J., Xiang, P., and Ma, L. Q. (2019a). Organoarsenical compounds: occurrence, toxicology and biotransformation. Crit. Rev. Environ. Sci. Technol. 50, 217–243. doi: 10.1080/10643389.2019.1619375
Chen, J., Yoshinaga, M., and Rosen, B. P. (2019b). The antibiotic action of methylarsenite is an emergent property of microbial communities. Mo. Microbiol. 111, 487–494. doi: 10.1111/mmi.14169
Chen, J., and Rosen, B. P. (2016). Organoarsenical biotransformations by Shewanella putrefaciens. Environ. Sci. Technol. 50, 7956–7963. doi: 10.1021/acs.est.6b00235
Chen, S. C., Sun, G. X., Rosen, B. P., Zhang, S. Y., Deng, Y., Zhu, B. K., et al. (2017). Recurrent horizontal transfer of arsenite methyltransferase genes facilitated adaptation of life to arsenic. Sci. Rep. 7:7741. doi: 10.1038/s41598-017-08313-210.1038/s41598-017-08313-2
Dheeman, D. S., Packianathan, C., Pillai, J. K., and Rosen, B. P. (2014). Pathway of human AS3MT arsenic methylation. Chem. Res. Toxicol. 27, 1979–1989. doi: 10.1021/tx500313k
Dong, H., Xu, W., Pillai, J. D., Packianathan, C., and Rosen, B. P. (2015). High-throughput screening-compatible assays of As(III) S-adenosylmethionine methyltransferase activity. Anal. Biochem. 480, 67–73. doi: 10.1016/j.ab.2015.04.011
Fan, C. J., Liu, G. L., Long, Y. M., Rosen, B., and Cai, Y. (2018). Thiolation in arsenic metabolism: a chemical perspective. Metallomics 10, 1368–1382. doi: 10.1039/c8mt00231b
Francesconi, K. A., Tanggaard, R., McKenzie, C. J., and Goessler, W. (2002). Arsenic metabolites in human urine after ingestion of an arsenosugar. Clin. Chem. 48, 92–101. doi: 10.1093/clinchem/48.1.92
Garbinski, L. D., Rosen, B. P., and Chen, J. (2019). Pathways of arsenic uptake and efflux. Environ. Int. 126, 585–597. doi: 10.1016/j.envint.2019.02.058
Gebel, T. W. (2002). Arsenic methylation is a process of detoxification through accelerated excretion. Int. J. Hyg. Environ. Health 205, 505–508. doi: 10.1078/1438-4639-00177
Giovannoni, S. J., Halsey, K. H., Saw, J., Muslin, O., Suffridge, C. P., Sun, J., et al. (2019). A parasitic arsenic cycle that shuttles energy from phytoplankton to heterotrophic bacterioplankton. mBio 10:e0246-19. doi: 10.1128/mBio.00246-
Heitkemper, D. T., Vela, N. P., Stewart, K. R., and Westphal, C. S. (2001). Determination of total and speciated arsenic in rice by ion chromatography and inductively coupled plasma mass spectrometry. J. Anal Atomic Spectrom. 16, 299–306. doi: 10.1039/b007241i
Hervas, M., Lopez-Maury, L., Leon, P., Sanchez-Riego, A. M., Florencio, F. J., and Navarro, J. A. (2012). ArsH from the Cyanobacterium Synechocystis sp PCC 6803 Is an efficient NADPH-Dependent quinone reductase. Biochemistry 51, 1178–1187. doi: 10.1021/bi201904p
Hirano, S., Kobayashi, Y., Cui, X., Kanno, S., Hayakawa, T., and Shraim, A. (2004). The accumulation and toxicity of methylated arsenicals in endothelial cells: important roles of thiol compounds. Toxicol. Appl. Pharmacol. 198, 458–467. doi: 10.1016/j.taap.2003.10.023
Hoffmann, T., Warmbold, B., Smits, S. H. J., Tschapek, B., Ronzheimer, S., Bashir, A., et al. (2018). Arsenobetaine: an ecophysiologically important organoarsenical confers cytoprotection against osmotic stress and growth temperature extremes. Environ. Microbiol. 20, 305–323. doi: 10.1111/1462-2920.13999
Huang, J. H., Hu, K. N., and Decker, B. (2011). Organic arsenic in the soil environment: speciation, occurrence, transformation, and adsorption behavior. Water Air Soil Pollut. 219, 401–415. doi: 10.1007/s11270-010-0716-2
Hughes, M. F., Beck, B. D., Chen, Y., Lewis, A. S., and Thomas, D. J. (2011). Arsenic exposure and toxicology: a historical perspective. Toxicol. Sci. 123, 305–332. doi: 10.1093/toxsci/kfr184
Jia, M. R., Tang, N., Cao, Y., Chen, Y. S., Han, Y. H., and Ma, L. Q. (2019). Efficient arsenate reduction by As-resistant bacterium Bacillus sp. strain PVR-YHB1-1: characterization and genome analysis. Chemosphere 218, 1061–1070. doi: 10.1016/j.chemosphere.2018.11.145
Kuramata, M., Sakakibara, F., Kataoka, R., Yamazaki, K., Baba, K., Ishizaka, M., et al. (2016). Arsinothricin, a novel organoarsenic species produced by a rice rhizosphere bacterium. Environ. Chem. 13, 723–731. doi: 10.1071/En14247
Le, X. C., Lu, X., Ma, M., Cullen, W. R., Aposhian, H. V., and Zheng, B. (2000a). Speciation of key arsenic metabolic intermediates in human urine. Anal. Chem. 72, 5172–5177. doi: 10.1021/ac000527u
Le, X. C., Ma, M. S., Lu, X. F., Cullen, W. R., Aposhian, H. V., and Zheng, B. S. (2000b). Determination of monomethylarsonous acid, a key arsenic methylation intermediate, in human urine. Environ. Health Perspect. 108, 1015–1018. doi: 10.1289/ehp.001081015
Li, J., Pawitwar, S. S., and Rosen, B. P. (2016). The organoarsenical biocycle and the primordial antibiotic methylarsenite. Metallomics 8, 1047–1055. doi: 10.1039/c6mt00168h
Lin, S., Shi, Q., Nix, F. B., Styblo, M., Beck, M. A., Herbin-Davis, K. M., et al. (2002). A novel S-adenosyl-L-methionine:arsenic(III) methyltransferase from rat liver cytosol. J. Biol. Chem. 277, 10795–10803. doi: 10.1074/jbc.M110246200
Lomax, C., Liu, W. J., Wu, L., Xue, K., Xiong, J., Zhou, J., et al. (2012). Methylated arsenic species in plants originate from soil microorganisms. New Phytol. 193, 665–672. doi: 10.1111/j.1469-8137.2011.03956.x
Lopez-Maury, L., Florencio, F. J., and Reyes, J. C. (2003). Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp strain PCC 6803. J. Bacteriol. 185, 5363–5371. doi: 10.1128/Jb.185.18.5363-5371.2003
Mandal, B. K., and Suzuki, K. T. (2002). Arsenic round the world: a review. Talanta 58, 201–235. doi: 10.1016/s0039-9140(02)00268-0
Marapakala, K., Packianathan, C., Ajees, A. A., Dheeman, D. S., Sankaran, B., Kandavelu, P., et al. (2015). A disulfide-bond cascade mechanism for arsenic(III) S-adenosylmethionine methyltransferase. Acta Crystallogr Section DStruct. Biol. 71, 505–515. doi: 10.1107/S1399004714027552
Marapakala, K., Qin, J., and Rosen, B. P. (2012). Identification of catalytic residues in the As(III) S-adenosylmethionine methyltransferase. Biochemistry 51, 944–951. doi: 10.1021/bi201500c
Mass, M. J., Tennant, A., Roop, B. C., Cullen, W. R., Styblo, M., Thomas, D. J., et al. (2001). Methylated trivalent arsenic species are genotoxic. Chem. Res. Toxicol. 14, 355–361. doi: 10.1021/tx000251l
Meharg, A. A., and Hartley-Whitaker, J. (2002). Arsenic uptake and metabolism in arsenic resistant and nonresistant plant species. New Phytol. 154, 29–43. doi: 10.1046/j.1469-8137.2002.00363.x
Moe, B., Peng, H., Lu, X., Chen, B., Chen, L. W., Gabos, S., et al. (2016). Comparative cytotoxicity of fourteen trivalent and pentavalent arsenic species determined using real-time cell sensing. J. Enviro. Sci. 49, 113–124. doi: 10.1016/j.jes.2016.10.004
Nachman, K. E., Baron, P. A., Raber, G., Francesconi, K. A., Navas-Acien, A., and Love, D. C. (2013). Roxarsone, inorganic arsenic, and other arsenic species in chicken: a US-Based market basket sample. Environ. Health Perspect. 121, 818–824. doi: 10.1289/ehp.1206245
Nadar, S. V., Yoshinaga, M., Kandavelu, P., Sankaran, B., and Rosen, B. P. (2014). Crystallization and preliminary X-ray crystallographic studies of the ArsI C-As lyase from Thermomonospora curvata. Acta Crystallogr. Section FStruct. Biol. Commun. 70, 761–764. doi: 10.1107/S2053230x14008814
Nadar, V. S., Chen, J., Dheeman, D. S., Galvan, A. E., Yoshinaga-Sakurai, K., Kandavelu, P., et al. (2019). Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic. Commun. Biol. 2:131.
Nadar, V. S., Yoshinaga, M., Pawitwar, S. S., Kandavelu, P., Sankaran, B., and Rosen, B. P. (2016). Structure of the ArsI C-As lyase: insights into the mechanism of degradation of organoarsenical herbicides and growth promoters. J. Mol. Biol. 428, 2462–2473. doi: 10.1016/j.jmb.2016.04.022
Neyt, C., Iriarte, M., Thi, V. H., and Cornelis, G. R. (1997). Virulence and arsenic resistance in yersiniae. J. Bacteriol. 179, 612–619. doi: 10.1128/jb.179.3.612-619.1997
Niegel, C., and Matysik, F. M. (2010). Analytical methods for the determination of arsenosugars-A review of recent trends and developments. Anal. Chim. Acta 657, 83–99. doi: 10.1016/j.aca.2009.10.041
Packianathan, C., Li, J., Kandavelu, P., Sankaran, B., and Rosen, B. P. (2018). Reorientation of the methyl group in MAs(III) is the rate-limiting step in the ArsM As(III) S-adenosylmethionine methyltransferase reaction. ACS Omega. 3, 3104–3112. doi: 10.1021/acsomega.8b00197
Paez-Espino, A. D., Durante-Rodriguez, G., and de Lorenzo, V. (2015). Functional coexistence of twin arsenic resistance systems in Pseudomonas putida KT2440. Environ. Microbiol. 17, 229–238. doi: 10.1111/1462-2920.12464
Pawitwar, S. S., Nadar, V. S., Kandegedara, A., Stemmler, T. L., Rosen, B. P., and Yoshinaga, M. (2017). Biochemical characterization of ArsI: a novel c-as lyase for degradation of environmental organoarsenicals. Environ. Sci. Technol. 51, 11115–11125. doi: 10.1021/acs.est.7b03180
Petrick, J. S., Ayala-Fierro, F., Cullen, W. R., Carter, D. E., and Aposhian, H. V. (2000). Monomethylarsonous acid (MMA(III)) is more toxic than arsenite in chang human hepatocytes. Toxicol. Appl. Pharmacol. 163, 203–207. doi: 10.1006/taap.1999.8872
Qin, J., Rosen, B. P., Zhang, Y., Wang, G., Franke, S., and Rensing, C. (2006). Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 103, 2075–2080. doi: 10.1073/pnas.0506836103
Ramaekers, B. L. T., Riemsma, R., Grimm, S., Fayter, D., Deshpande, S., Armstrong, N., et al. (2019). Arsenic trioxide for treating acute promyelocytic leukaemia: an evidence review group perspective of a NICE single technology appraisal. Pharmacoeconomics 37, 887–894. doi: 10.1007/s40273-018-0738-y
Raml, R., Rumpler, A., Goessler, W., Vahter, M., Li, L., Ochi, T., et al. (2007). Thio-dimethylarsinate is a common metabolite in urine samples from arsenic-exposed women in Bangladesh. Toxicol. Appl. Pharmacol. 222, 374–380. doi: 10.1016/j.taap.2006.12.014
Roje, S. (2006). S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry 67, 1686–1698. doi: 10.1016/j.phytochem.2006.04.019
Rubin, S. S. C. D., Alava, P., Zekker, I., Du Laing, G., and Van de Wiele, T. (2014). Arsenic thiolation and the role of sulfate-reducing bacteria from the human intestinal rract. Environ. Health Perspect. 122, 817–822. doi: 10.1289/ehp.1307759
Shen, Z. Q., Luangtongkum, T., Qiang, Z. Y., Jeon, B., Wang, L. P., and Zhang, Q. J. (2014). Identification of a novel membrane transporter mediating resistance to organic arsenic in Campylobacter jejuni. Antimicrob. Agents Chemother. 58, 2021–2029. doi: 10.1128/Aac.02137-13
Shi, K. X., Li, C., Rensing, C., Dai, X. L., Fan, X., and Wang, G. J. (2018). Efflux transporter ArsK Is responsible for bacterial resistance to arsenite, antimonite, trivalent roxarsone, and methylarsenite. Appl. Environ. Microbiol. 84:e01842-18.
Taylor, V., Goodale, B., Raab, A., Schwerdtle, T., Reimer, K., Conklin, S., et al. (2017). Human exposure to organic arsenic species from seafood. Sci. Total Environ. 580, 266–282. doi: 10.1016/j.scitotenv.2016.12.113
Thomas, D. J., and Bradham, K. (2016). Role of complex organic arsenicals in food in aggregate exposure to arsenic. J. Environ. Sci. China 49, 86–96. doi: 10.1016/j.jes.2016.06.005
Thomas, D. J., Waters, S. B., and Styblo, M. (2004). Elucidating the pathway for arsenic methylation. Toxicol. Appl. Pharmacol. 198, 319–326. doi: 10.1016/j.taap.2003.10.020
Tseng, C. H. (2007). Arsenic methylation, urinary arsenic metabolites and human diseases: current perspective. J. Environ. Sci. Health Part CEnviron, Carcinog. Ecotoxicol. Rev. 25, 1–22. doi: 10.1080/10590500701201695
Valenzuela, O. L., Borja-Aburto, V. H., Garcia-Vargas, G. G., Cruz-Gonzalez, M. B., Garcia-Montalvo, E. A., Calderon-Aranda, E. S., et al. (2005). Urinary trivalent methylated arsenic species in a population chronically exposed to inorganic arsenic. Environ. Health Perspect. 113, 250–254. doi: 10.1289/ehp.7519
Waksman, S. A. (1947). What is an antibiotic or an antibiotic substance? Mycologia 39, 565–569. doi: 10.1080/00275514.1947.12017635
Xue, X. M., Yan, Y., Xu, H. J., Wang, N., Zhang, X., and Ye, J. (2014). ArsH from Synechocystis sp PCC 6803 reduces chromate and ferric iron. Fems Microbiol. Lett. 356, 105–112. doi: 10.1111/1574-6968.12481
Xue, X. M., Ye, J., Raber, G., Francesconi, K. A., Li, G., Gao, H., et al. (2017). Arsenic methyltransferase is involved in arsenosugar biosynthesis by providing DMA. Environ. Sci. Technol. 51, 1224–1230. doi: 10.1021/acs.est.6b04952
Xue, X. M., Ye, J., Raber, G., Rosen, B. P., Francesconi, K., Xiong, C., et al. (2019). Identification of steps in the pathway of arsenosugar biosynthesis. Environ. Sci. Technol. 53, 634–641. doi: 10.1021/acs.est.8b04389
Yan, Y., Ye, J., Xue, X. M., and Zhu, Y. G. (2015). Arsenic demethylation by a C-As lyase in Cyanobacterium Nostoc sp PCC 7120. Environ. Sci. Technol. 49, 14350–14358. doi: 10.1021/acs.est.5b03357
Yang, H. C., Cheng, J., Finan, T. M., Rosen, B. P., and Bhattacharjee, H. (2005). Novel pathway for arsenic detoxification in the legume symbiont Sinorhizobium meliloti. J. Bacteriol. 187, 6991–6997. doi: 10.1128/jb.187.20.6991-6997.2005
Ye, J., Rensing, C., Rosen, B. P., and Zhu, Y. G. (2012). Arsenic biomethylation by photosynthetic organisms. Trends Plant Sci. 17, 155–162. doi: 10.1016/j.tplants.2011.12.003
Ye, J., Yang, H. C., Rosen, B. P., and Bhattacharjee, H. (2007). Crystal structure of the flavoprotein ArsH from Sinorhizobium meliloti. FEBS Lett. 581, 3996–4000. doi: 10.1016/j.febslet.2007.07.039
Yoshinaga, M., Cai, Y., and Rosen, B. P. (2011). Demethylation of methylarsonic acid by a microbial community. Environ. Microbiol. 13, 1205–1215. doi: 10.1111/j.1462-2920.2010.02420.x
Yoshinaga, M., and Rosen, B. P. (2014). A C-As lyase for degradation of environmental organoarsenical herbicides and animal husbandry growth promoters. Proc. Natl. Acad. Sci. U.S.A. 111, 7701–7706. doi: 10.1073/pnas.1403057111
Zhao, F. J., Zhu, Y. G., and Meharg, A. A. (2013). Methylated arsenic species in rice: geographical variation, origin, and uptake mechanisms. Environ. Sci. Technol. 47, 3957–3966. doi: 10.1021/es304295n
Keywords: methylarsenicals, arsenic methylation, methylarsenite, arsinothricin, arsenic-containing antibiotics, antibiotic resistance
Citation: Chen J and Rosen BP (2020) The Arsenic Methylation Cycle: How Microbial Communities Adapted Methylarsenicals for Use as Weapons in the Continuing War for Dominance. Front. Environ. Sci. 8:43. doi: 10.3389/fenvs.2020.00043
Received: 22 October 2019; Accepted: 26 March 2020;
Published: 21 April 2020.
Edited by:
Adrien Mestrot, University of Bern, SwitzerlandReviewed by:
Mahmud Hossain, Bangladesh Agricultural University, BangladeshNadia Valentina Martínez-Villegas, Instituto Potosino de Investigación Científica y Tecnológica (IPICYT), Mexico
Copyright © 2020 Chen and Rosen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barry P. Rosen, brosen@fiu.edu
 Jian Chen
Jian Chen Barry P. Rosen
Barry P. Rosen