Isoproterenol-Induced Cardiac Diastolic Dysfunction in Mice: A Systems Genetics Analysis
- 1Department of Medicine, University of California, Los Angeles, Los Angeles, CA, United States
- 2Department of Anesthesiology, University of California, Los Angeles, Los Angeles, CA, United States
We examined an isoproterenol heart failure model across a panel of diverse inbred strains of mice, the Hybrid Mouse Diversity Panel (HMDP), using left atrial (LA) and lung weights as well as echocardiogram parameters as surrogates for cardiac diastolic function. We identified gene transcripts that significantly correlated with diastolic function. In addition, we mapped echocardiographic parameters associated with diastolic function. We identified a locus near Tns3-Hus1 to be associated with baseline E/A ratio in mice (p = 1.65E-06), the syntenic region of which was recently associated with E/A ratio in a genome-wide association study (GWAS) meta-analysis of the EchoGen consortium in humans. We also identified a locus near Cdkn2a-Cdkn2b, which is a region syntenic to the human 9p21 locus, to be associated with week 3 A/E ratio (p = 2.15E-06). Our study is the first study to map diastolic dysfunction in mice, in which a locus was found to be shared with a recent human GWAS on diastolic function. Moreover, our cardiac transcriptome correlation and eQTL analysis generated hypotheses for future basic investigations. These results showed that, although technical and physiological challenges limit diastolic function assessment in mice and humans, future investigations examining the genetic architecture of diastolic function among a diverse mouse population, such as the HMDP, in controlled experimental settings, offer distinct advantages in understanding the genetic determinants of diastolic function.
Introduction
Heart failure with preserved ejection fraction (HFpEF) is a clinical syndrome that is characterized by diastolic dysfunction, involving impaired relaxation and compliance of the left ventricle (LV), leading to increased LV filling pressures, heart failure symptoms and an elevated risk of mortality (1–4). Among 5.7 million Americans with heart failure, HFpEF accounts for 50% of all heart failure hospital admissions (5). Although the understanding of HFpEF pathophysiology and risk factors, such as older age, female sex, hypertension, metabolic syndrome, renal dysfunction and obesity, has improved, treatment aimed at altering its natural history has yet to be developed due to the lack of understanding of its molecular underpinnings (6).
The Hybrid Mouse Diversity Panel (HMDP) consists of ~100 well-characterized inbred mouse strains that can be used to study genetic and environmental factors underlying complex traits (7). The HMDP, with a fully annotated genome and an easily controlled environment, can be readily manipulated to create a disease model for in-depth multi-omics molecular characterization at the tissue level. We had previously reported isoproterenol-induced cardiac phenotypes in the HMDP, in which we identified and validated a number of genomic loci associated with systolic heart failure traits via a genome-wide association approach (8–10). Chronic isoproterenol administration in mice has previously been demonstrated to induce diastolic dysfunction, as shown by an upward shift of the pressure-volume relationship (11). Although diastolic function can be assessed by directly measuring LV filling pressures via invasive hemodynamic monitoring (12), it can also be assessed by a number of echocardiographic findings non-invasively. These parameters, E and A velocities, tissue E' velocity, and their derivatives E/A ratio and E/E' ratio, have been used to study diastolic function in mice (13). The present study focuses on diastolic function data and analyses that have not been previously published. In this study we analyzed diastolic function parameters from the heart failure HMDP to identify candidate genomic loci and gene transcripts that may more accurately reflect the pathophysiology of diastolic dysfunction. Finally, we hypothesize the use of a diverse mouse population, such as the HMDP, to help us better understand diastolic dysfunction.
Methods
Assessment of Diastolic Function in Mice
Female mice at a median age of 9 weeks from 104 of the HMDP inbred strains were treated with isoproterenol (30 mg/kg/d) via an intra-abdominally implanted osmotic pump for 21 days as previously described (8–10). Briefly, baseline (prior to isoproterenol pump implantation), week 1, week 2, and week 3 echocardiograms were obtained. Due to tissue E' and A' velocities not being available on the VisualSonics Vevo 770 ultrasonography system used for this study, diastolic function was assessed by E and A velocities and via left atrial and lung weights. At the end of the protocol, heart chambers and organs were dissected and weighed. Left ventricular tissue was frozen in liquid nitrogen and later prepared for gene expression profiling as previously described (8–10).
GWAS and Correlation Analysis
The HMDP mouse strains were previously genotyped using the JAX Mouse Diversity Genotyping Array (14). Mapping of diastolic function traits was performed using the Factored Spectrally Transformed Linear Mixed Models (FaST-LMM) as previously described (15). Gene-trait correlation analyses were performed using WGCNA (16). Additional details of the methods were previously described (8–10). The threshold for genome-wide significance in mice was previously determined through simulation and permutation of strain genotypes in the hybrid mouse diversity panel to be 4.1 × 10−6 by Farber et al. The significance threshold for local expression quantitative trait loci (eQTL) was similarly determined to be 3.63 × 10−3. In our exam of multiple related diastolic function traits, the genome-wide significance threshold for each clinical trait was kept at the nominal p-value of 4.1 × 10−6 without further Bonferroni adjustment to account for each trait tested. We applied the Benjamini-Hochberg (BH) procedure with a false discovery rate of 5% to account for multiple testing in trait-expression correlation analysis.
Results
One-hundred and four classical and recombinant inbred mouse strains of the HMDP underwent intraperitoneal implantation of isoproterenol mini-pumps. Global gene expression of the left ventricular tissue was performed as previously reported (8–10). Diastolic function was assessed by echocardiographic measures, such as E and A velocities, and left atrial and lung weights, which reflect cumulative impact of isoproterenol on LV filling pressures over time, leading to atrial volume increase, pulmonary congestion, and edema (17). Tissue velocity measurement was not available on the ultrasonography system used for the study.
Diastolic Function Assessment Across the HMDP Population
On average across the HMDP strains, left atrial weight at the time of sacrifice had increased by 78.8% among isoproterenol treated vs. control mice (4.29 ± 1.69 mg vs. 2.4 ± 0.583 mg; p = 9.33E-20), indicating significant left atrial remodeling due to isoproterenol-induced elevated filling pressures over 3 weeks (Supplementary Table 1A). Of note, ejection fraction (EF) did not change on average between baseline and week 3 of isoproterenol. These findings are consistent with an isoproterenol-induced HFpEF model on average among HMDP mice. In terms of echocardiographic measures of diastolic function, average E and A velocities increased with isoproterenol during the first week and, thereby, remained relatively unchanged for the rest of isoproterenol treatment (Supplementary Table 2). E/A ratio did not change significantly on average (Supplementary Table 1B). There was a wide distribution of E/A ratio among HMDP strains at week 3 of isoproterenol with only a minority of strains with E/A ratio < 1, indicating varying degrees of diastolic dysfunction (Figure 1). There is no significant correlation between E/A and fibrosis (Supplementary Figure 1), indicative of intrinsic relaxation properties rather than extracellular fibrotic processes as the primary driver of the E/A ratio phenotype.
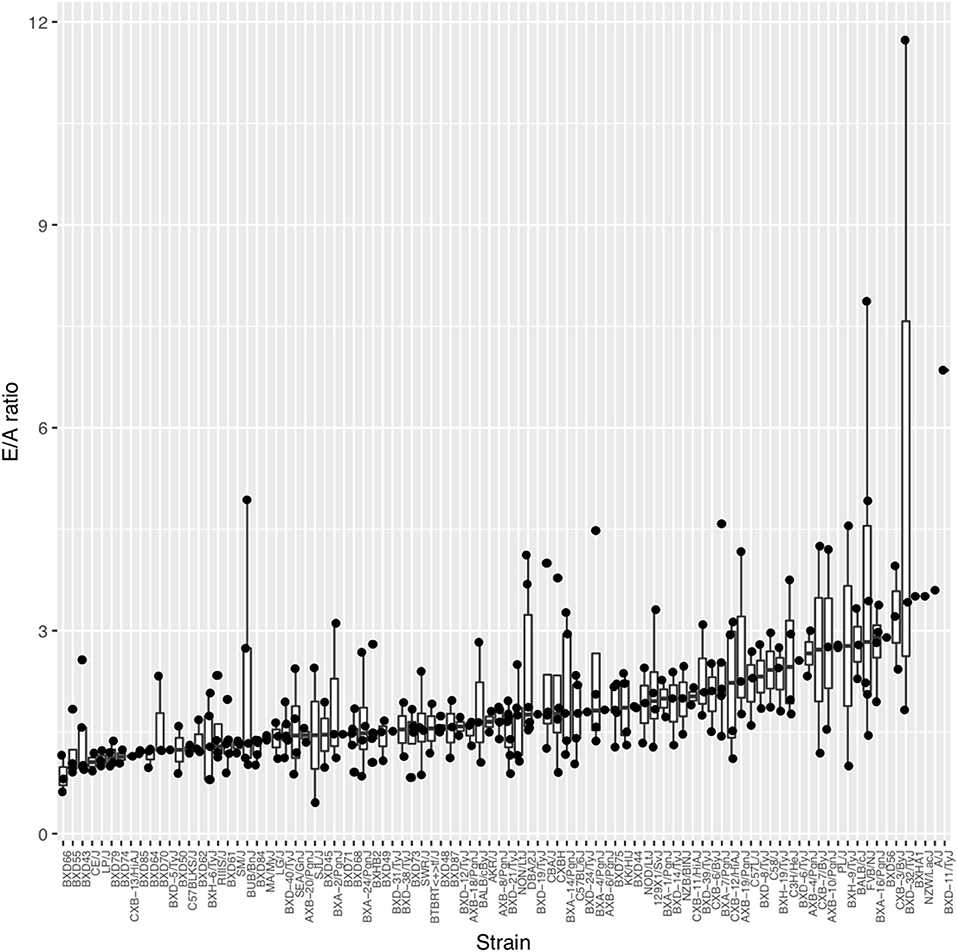
Figure 1. The spectrum of diastolic function as measured by E/A ratio across the Hybrid Mouse Diversity Panel (HMDP) strains. Boxplots represent the intra-strain variation in E/A ratio. Strains are ordered by median E/A ratio.
Diastolic Function Correlation With Left Ventricular Gene Transcripts
We examined isoproterenol-treated transcripts that were most significantly correlated with diastolic function traits such as left atrial weight, lung weight, E and A velocities (Table 1). Many of the significantly correlated gene transcripts have been reported to be expressed in the heart but their specific roles in heart failure have not been previously explored. A few of the notable gene transcripts with known roles in the heart are described as follows. Adamts2, which was significantly correlated with lung weight (r = 0.459, p = 5.49e−06), has been previously reported by our group to be a driver of cardiac hypertrophy (8) and confirmed by another group using gain- and loss-of-function genetic mouse model of Adamts2 to negatively regulate cardiomyocyte hypertrophy via the PI3K/AKT-dependent signaling pathway (18). Vav2, which was correlated with E velocity at week 3 (r = 0.469, p = 4.12e−06), is a signal transduction molecule that activates Rho/Rac GTPases, the knockout of which in mice causes tachycardia, hypertension, defects in the heart, arterial walls and kidneys via chronic stimulation of the renin/angiotensin II and sympathetic nervous systems (19). Tmod4, a muscle-specific member of the tropomodulin family of actin-regulatory proteins (20), was found to be negatively correlated with left atrial weight (r = −0.466, p = 3.69e−06). Moreover, Jmjd1a, a histone 3 lysine 9 (H3K9)-specific demethylase that binds all 3 myocardin family members, was positively correlated with A velocity at week 3 with borderline significance (r = 0.457, p = 7.69e−06, BH adjusted p = 0.0549) and is known to be upregulated in myocardium from patients with hypertrophic heart disease (21). Finally, Col6a1, which was correlated with lung weight with borderline significance (r = 0.451, p = 7.95e−06, BH adjusted p = 0.0549) has previously been shown, when absent, to be protective following myocardial infarction by limiting infarct size, apoptosis, aberrant remodeling and fibrosis (22).
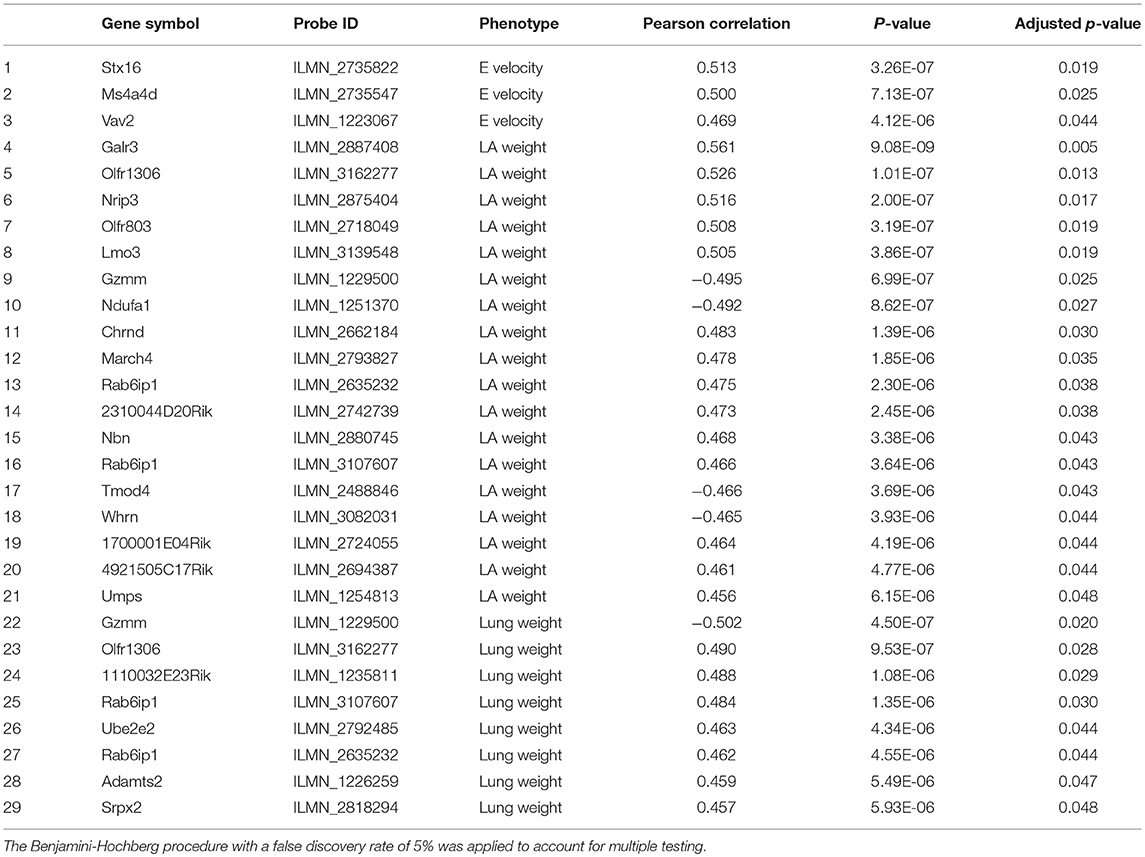
Table 1. Significantly correlated isoproterenol-induced transcripts with diastolic function parameters including left atrial weight, lung, E and A velocity and E/A ratio.
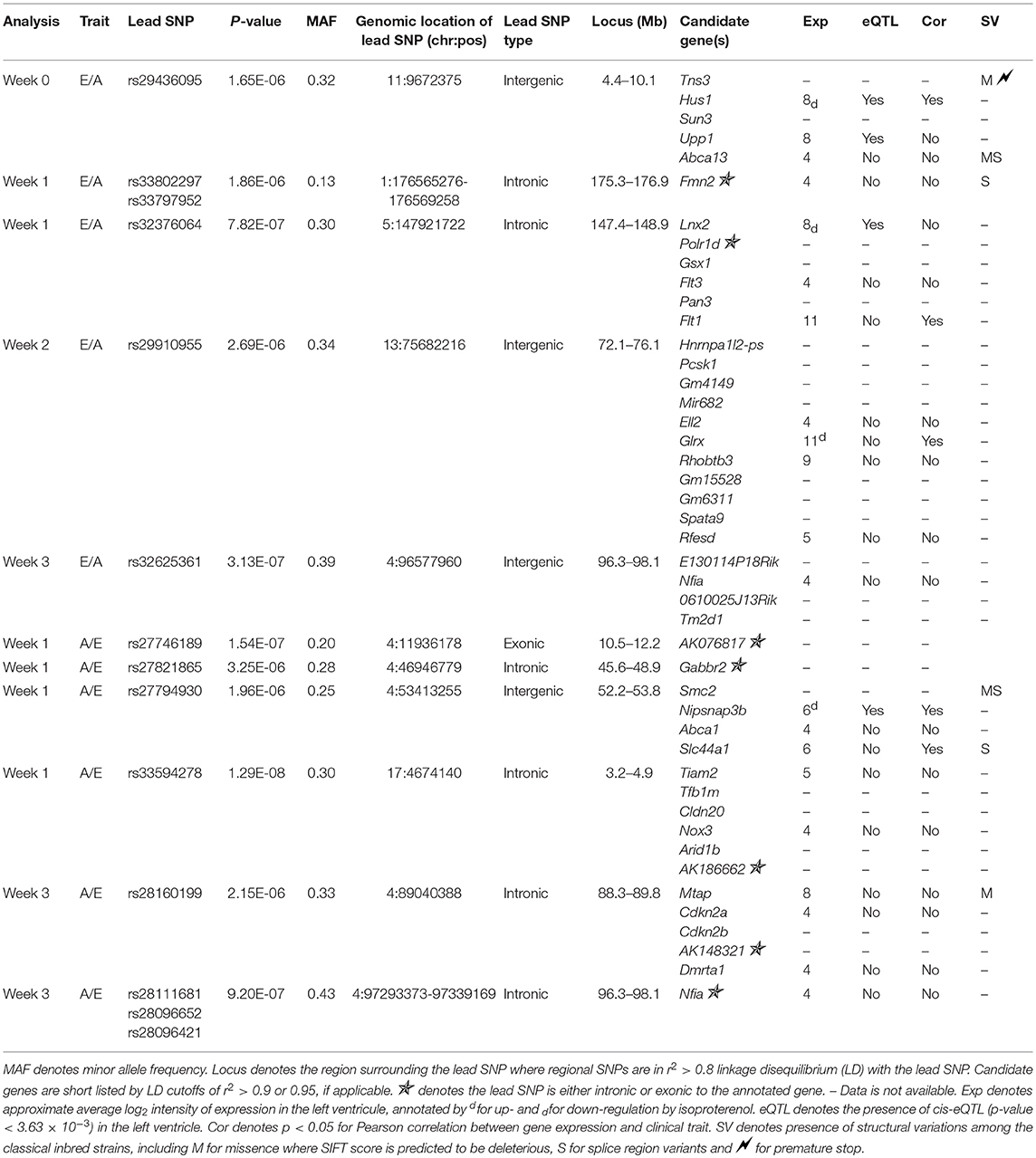
Genome-Wide Association of Diastolic Function Parameters Across the HMDP Population
Assuming that diastolic function cannot be normal under continuous beta-adrenergic stimulation by isoproterenol over 3 weeks, E/A ratio measurements may be treated as a monotonic continuous variable, where the higher the value the more diastolic dysfunction there is. This assumption obviates the need to distinguish normal vs. pseudonormal (grade II) patterns. We mapped E/A ratio and its inverse A/E ratio (in case the inverse value of the E/A ratio performs better on the linear mixed model utilized by FaST-LMM for association testing) across the HMDP on isoproterenol and identified 11 significant loci for diastolic function (Figure 2 and Table 2). Mapping of left atrial weight did not yield significant loci, which could reflect isoproterenol driving the phenotype to the extreme such that there is insufficient variation to map. Here we highlight a few of the more interesting loci.
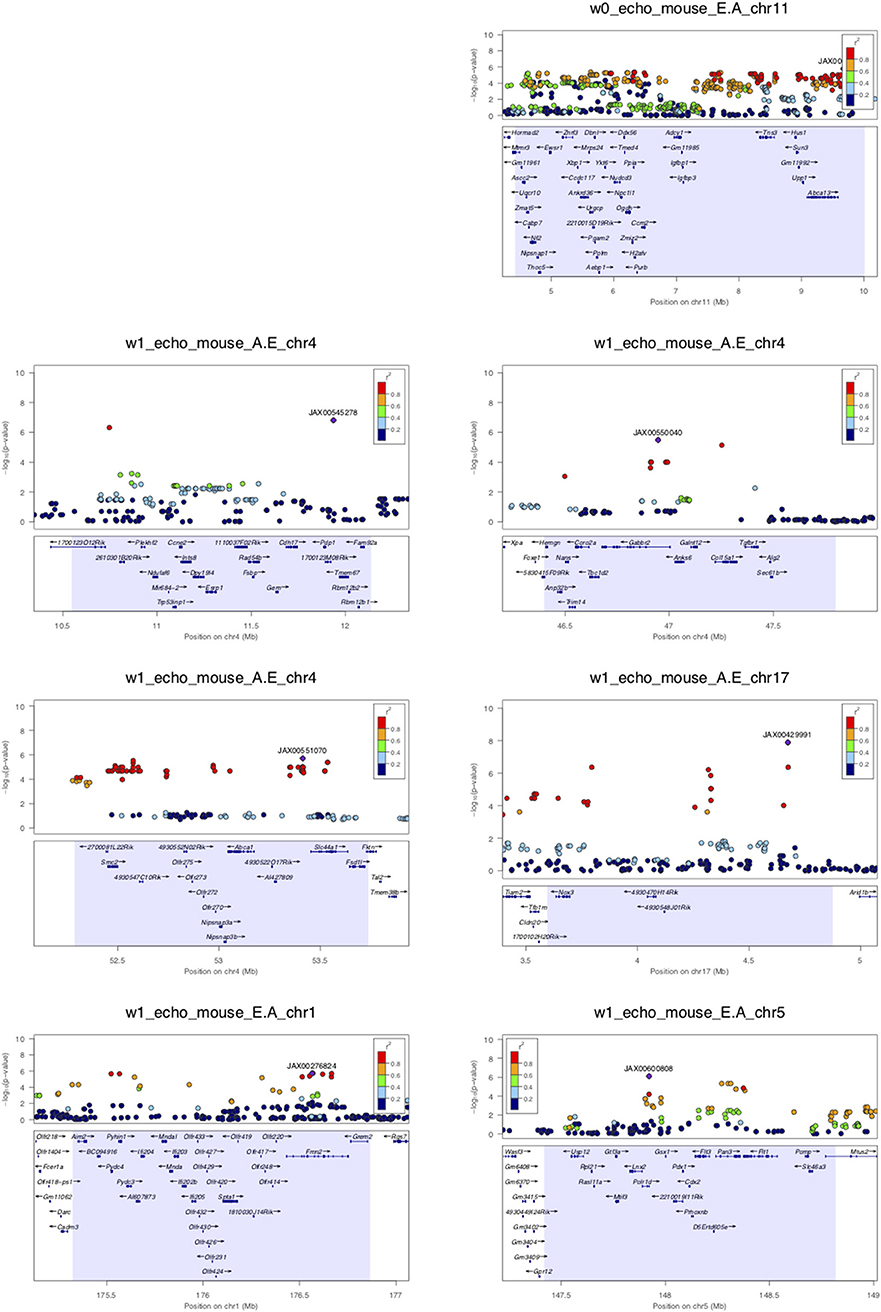
Figure 2. LocusZoom plots for regional visualization of genome-wide association scan results of echocardiographic diastolic function loci. The regional lead single nucleotide polymorphism (SNP) is represented by the purple diamond symbol and annotated by its SNP ID. Surrounding SNPs are color coded according to the strength of pairwise linkage disequilibrium (LD) in terms of the r2 metric to the lead SNP. w0, w1, w2, and w3 denote baseline, week 1, week 2, and week 3 of isoproterenol. E.A denotes E/A ratio. A.E denotes A/E ratio.
Week 0 E/A ratio mapped to a linkage disequilibrium block (r2 > 0.9) around SNP rs29436095 spanning 5 genes. Of note, in a large genome-wide association meta-analysis of studies in the EchoGen consortium, E/A ratio was mapped to a statistically suggestive human locus near Tns3 (p = 2.11e−4), which is syntenic to this region. The AA and GG genotypes at lead SNP rs29436095 conferred a median E/A ratio of 1.66 and 2.11, respectively. Among these genes, Hus1 expression was downregulated with isoproterenol. Hus1 expression under the control condition showed a trend to positive correlation with E/A ratio (cor = 0.193, p = 0.0867) and mapped to the same locus as E/A ratio, indicative of a cis-eQTL that may underly the phenotype of week 0 E/A ratio. These results suggest that Hus1 may play a role in baseline left ventricular diastolic function and is regulated with chronic isoproterenol administration.
Week 1 A/E ratio mapped to a locus near SNP rs27794930, which is in an intergenic region between Abca1 and Slc44a1. Nearby SNPs in r2 > 0.95 LD with the lead SNP spanned a ~1.5 Mb window containing many genes. Among these genes, Nipsnap3b expression was upregulated with isoproterenol. Nipsnap3b expression under the control condition was positively correlated with week 1 A/E ratio (ILMN_2657376, cor = 0.32, p = 0.004; ILMN_2592629, cor = 0.25, p = 0.02) and mapped to the same locus as A/E ratio, indicative of a cis-eQTL. Our results suggest that Nipsnap3b may play a role in left ventricular diastolic function at week 1 of isoproterenol and is regulated by chronic isoproterenol administration. Nipsnap3b, which is highly expressed in skeletal muscle and heart, is a member of the NIPSNAP family with putative roles in vesicular trafficking (23, 24).
Week 3 A/E ratio and E/A ratio mapped to a locus near Nfia (Figure 2B). Although there are currently no follow up data regarding the role of nuclear factor 1 a in the heart, this locus has previously been associated with QRS duration on ECG, arrhythmia, and cardiomyopathy (25). Finally, week 3 A/E ratio also mapped to a locus near SNP rs28160199, which is in an intronic region of a non-coding RNA AK148321. Nearby SNPs in r2 > 0.8 LD with the lead SNP spanned across a region syntenic to the human 9p21 locus, containing ANRIL, CDKN2A, and CDKN2B, that has been associated with coronary artery disease, diabetes, and multiple cancers (26, 27).
Discussion
This is the first study to genetically map diastolic function phenotypes in mice, in which a locus was found to be shared with a recent human GWAS on diastolic function. Moreover, we presented cardiac transcriptome correlation and cis-eQTL analyses that generated hypotheses regarding the involvement of the presented candidate genes and loci in the pathophysiology of diastolic dysfunction to be confirmed in follow-up basic investigations.
Recently, a large GWAS meta-analysis of echocardiographic traits, that included 46,533 individuals from 30 studies of the EchoGen consortium, evaluated 16 traits encompassing left ventricular structure, systolic and diastolic function and revealed a single significantly associated locus (28). This locus flagged by a lead intronic SNP rs12440869 on chromosome 15 near gene IQCH was found to be significantly associated with transmitral A-wave peak velocity (meta-analysis p = 1.31e-08). In addition, twenty-three suggestive loci for transmitral E-wave velocity, E/A ratio, deceleration time, isovolumetric relaxation time (IVR), E' velocity, E/E' velocity, diastolic dysfunction with preserved ejection function, and heart failure with preserved ejection function were reported (28). Molecular mechanisms leading to diastolic dysfunction underlying these human loci have yet to be confirmed by basic investigations. Of interest, one of the suggestive loci near Tns3 is syntenic to a locus that week 0 E/A ratio mapped to in our study. The shared locus identified for E/A ratio suggests that a systems genetics approach in mice could be a feasible alternative in dissecting the genetic architecture of diastolic pathophysiology. Moreover, the Tns3 locus contains many genes. We were able to prioritize Hus1 as a putative causal gene based on cardiac tissue transcriptome correlation and cis-eQTL analyses, highlighting another benefit of studying diastolic function using the systems genetics approach in mice.
Although widely accepted in clinical practice and previously reported to accurately quantify the degree of cardiac diastolic performance in mice (13), there are a number of limitations and technical considerations worth noting regarding non-invasive assessment of diastolic function in both humans and mice. An important limitation of diastolic function assessment in mice is the difficulty in obtaining a true apical view to accurately measure mitral annular velocities by tissue Doppler. Also, diastolic function assessment in mice is somewhat complicated by a normal resting mouse heart rates at around 600 beats per minute. In the setting of excess rise, that is, heart rate > 650 bpm, due to stress of activation of the autonomic nervous system, fusion of LV filling early E and late A waves often occurs, making it difficult to assess diastolic function in mice (29). In a study examining diastolic function parameters using multiple mouse models of heart disease, mitral E/A ratio assessment was noted to be technically difficult due to high heart rates; combined measurement of left atrial area and reverse longitudinal strain rate and/or isovolumic relaxation time (IVRT) was felt to be necessary for an overall assessment of diastolic function (30). A major limitation of our study is that, at the time of our echocardiographic assessment, tissue Doppler velocity and longitudinal strain rate were not yet available. We relied on the ratio of mitral inflow E and A velocities, which is invariant to Doppler interrogation angle, as our primary echocardiographic measure of diastolic function for genome mapping. Thus, future diastolic assessment incorporating additional measures may more accurately capture the phenotype of interest.
Finally, diastolic function is a product of environmental, genetic, and co-morbid factors in humans and non-invasive parameters of diastolic function assessment are subjected to changes in loading conditions that may be difficult to control in both humans and mice, even under well-defined experimental settings. Quantitative measures of diastolic function that reflect intrinsic elastance, such as end diastolic pressure-volume relationship (EDPVR) and end systolic pressure-volume relationship (ESPVR) derived from invasive hemodynamic monitoring under different loading conditions, are robust and may significantly increase mapping power and accuracy. We hypothesize that genetic mapping of invasive measurements of left ventricular intrinsic elastance in a genetically diverse population, such as the HMDP, will significantly improve the accuracy of mapping for various models of diastolic dysfunction, such as aging and obesity, and accelerate the understanding of diastolic pathophysiology in mice and humans.
Data Availability
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48760.
Ethics Statement
All animal experiments were conducted following guidelines established and approved by the University of California, Los Angeles Institutional Animal Care and Use Committee. This study was carried out in accordance with the recommendations of the University of California, Los Angeles Institutional Animal Care and Use Committee. The protocol was approved by the University of California, Los Angeles Institutional Animal Care and Use Committee.
Author Contributions
JW, YW, and AL contributed to conception and design of the study. JW performed echocardiograms, measured diastolic parameters, prepared samples for RNA isolation, organized the data, performed statistical analysis, and drafted the manuscript. AH-V provided critical revisions for important intellectual content of the manuscript. YW and AL provided critical feedback and mentorship. All authors contributed to manuscript revision and approved the submitted version.
Funding
This study was supported by funding through NIH R01 HL123295 (YW and AL), R01 HL129639 (YW and JW), and K08 HL133491 (JW) as well as UCLA Geffen School of Medicine Cardiovascular Theme Award (JW).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2019.00100/full#supplementary-material
References
1. Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE Jr, Drazner MH, et al. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. (2013) 62:e147–239. doi: 10.1016/j.jacc.2013.05.019
2. Halley CM, Houghtaling PL, Khalil MK, Thomas JD, Jaber WA. Mortality rate in patients with diastolic dysfunction and normal systolic function. Arch Intern Med. (2011) 171:1082–7. doi: 10.1001/archinternmed.2011.244
3. Gharacholou SM, Scott CG, Takahashi PY, Nkomo VT, McCully RB, Fine NM, et al. Left ventricular diastolic function and long-term outcomes in patients with normal exercise echocardiographic findings. Am J Cardiol. (2013) 112:200–7. doi: 10.1016/j.amjcard.2013.03.009
4. Aljaroudi W, Alraies MC, Halley C, Rodriguez L, Grimm RA, Thomas JD, et al. Impact of progression of diastolic dysfunction on mortality in patients with normal ejection fraction. Circulation. (2012) 125:782–8. doi: 10.1161/CIRCULATIONAHA.111.066423
5. Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, et al. Heart disease and stroke statistics−2015 update: a report from the American Heart Association. Circulation. (2015) 131:e29–322.
6. Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE Jr, Colvin MM, et al. 2017 ACC/AHA/HFSA Focused Update of the 2013 ACCF/AHA Guideline for the Management of Heart Failure: a report of the American college of cardiology/american heart association task force on clinical practice guidelines and the heart failure society of America. J Card Fail. (2017) 23:628–51.
7. Lusis AJ, Seldin MM, Allayee H, Bennett BJ, Civelek M, Davis RC, et al. The hybrid mouse diversity panel: a resource for systems genetics analyses of metabolic and cardiovascular traits. J Lipid Res. (2016) 57:925–42. doi: 10.1194/jlr.R066944
8. Rau CD, Romay MC, Tuteryan M, Wang JJ, Santolini M, Ren S, et al. Systems genetics approach identifies gene pathways and Adamts2 as drivers of isoproterenol-induced cardiac hypertrophy and cardiomyopathy in mice. Cell Syst. (2017) 4:121–8 e4. doi: 10.1016/j.cels.2016.10.016
9. Rau CD, Wang J, Avetisyan R, Romay MC, Martin L, Ren S, et al. Mapping genetic contributions to cardiac pathology induced by Beta-adrenergic stimulation in mice. Circ Cardiovasc Genet. (2015) 8:40–9. doi: 10.1161/CIRCGENETICS.113.000732
10. Wang JJ, Rau C, Avetisyan R, Ren S, Romay MC, Stolin G, et al. Genetic dissection of cardiac remodeling in an isoproterenol-induced heart failure mouse model. PLoS Genet. (2016) 12:e1006038. doi: 10.1371/journal.pgen.1006038
11. Brooks WW, Conrad CH. Isoproterenol-induced myocardial injury and diastolic dysfunction in mice: structural and functional correlates. Comp Med. (2009) 59:339–43.
12. Brutsaert DL, Sys SU, Gillebert TC. Diastolic failure: pathophysiology and therapeutic implications. J Am Coll Cardiol. (1993) 22:318–25. doi: 10.1016/0735-1097(93)90850-Z
13. Respress JL, Wehrens XH. Transthoracic echocardiography in mice. J Vis Exp. (2010) 39:1738. doi: 10.3791/1738
14. Yang H, Ding Y, Hutchins LN, Szatkiewicz J, Bell TA, Paigen BJ, et al. A customized and versatile high-density genotyping array for the mouse. Nat Methods. (2009) 6:663–6. doi: 10.1038/nmeth.1359
15. Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods. (2011) 8:833–5. doi: 10.1038/nmeth.1681
16. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. (2008) 9:559. doi: 10.1186/1471-2105-9-559
17. Nagueh SF, Appleton CP, Gillebert TC, Marino PN, Oh JK, Smiseth OA, et al. Recommendations for the evaluation of left ventricular diastolic function by echocardiography. J Am Soc Echocardiogr. (2009) 22:107–33. doi: 10.1016/j.echo.2008.11.023
18. Wang X, Chen W, Zhang J, Khan A, Li L, Huang F, et al. Critical role of ADAMTS2 (A Disintegrin and Metalloproteinase With Thrombospondin Motifs 2) in Cardiac Hypertrophy Induced by Pressure Overload. Hypertension. (2017) 69:1060–9. doi: 10.1161/HYPERTENSIONAHA.116.08581
19. Sauzeau V, Jerkic M, Lopez-Novoa JM, Bustelo XR. Loss of Vav2 proto-oncogene causes tachycardia and cardiovascular disease in mice. Mol Biol Cell. (2007) 18:943–52. doi: 10.1091/mbc.e06-09-0877
20. Gokhin DS, Lewis RA, McKeown CR, Nowak RB, Kim NE, Littlefield RS, et al. Tropomodulin isoforms regulate thin filament pointed-end capping and skeletal muscle physiology. J Cell Biol. (2010) 189:95–109. doi: 10.1083/jcb.201001125
21. Zhang QJ, Tran TAT, Wang M, Ranek MJ, Kokkonen-Simon KM, Gao J, et al. Histone lysine dimethyl-demethylase KDM3A controls pathological cardiac hypertrophy and fibrosis. Nat Commun. (2018) 9:5230. doi: 10.1038/s41467-018-07173-2
22. Luther DJ, Thodeti CK, Shamhart PE, Adapala RK, Hodnichak C, Weihrauch D, et al. Absence of type VI collagen paradoxically improves cardiac function, structure, and remodeling after myocardial infarction. Circ Res. (2012) 110:851–6. doi: 10.1161/CIRCRESAHA.111.252734
23. Buechler C, Bodzioch M, Bared SM, Sigruener A, Boettcher A, Lapicka-Bodzioch K, et al. Expression pattern and raft association of NIPSNAP3 and NIPSNAP4, highly homologous proteins encoded by genes in close proximity to the ATP-binding cassette transporter A1. Genomics. (2004) 83:1116–24. doi: 10.1016/j.ygeno.2003.12.011
25. Ritchie MD, Denny JC, Zuvich RL, Crawford DC, Schildcrout JS, Bastarache L, et al. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation. (2013) 127:1377–85. doi: 10.1161/CIRCULATIONAHA.112.000604
26. Congrains A, Kamide K, Ohishi M, Rakugi H. ANRIL: Molecular mechanisms and implications in human health. Int J Mol Sci. (2013) 14:1278–92. doi: 10.3390/ijms14011278
27. Holdt LM, Teupser D. Long Noncoding RNA ANRIL: Lnc-ing genetic variation at the chromosome 9p21 locus to molecular mechanisms of Atherosclerosis. Front Cardiovasc Med. (2018) 5:145. doi: 10.3389/fcvm.2018.00145
28. Wild PS, Felix JF, Schillert A, Teumer A, Chen MH, Leening MJG, et al. Large-scale genome-wide analysis identifies genetic variants associated with cardiac structure and function. J Clin Invest. (2017) 127:1798–812. doi: 10.1172/JCI84840
29. Lindsey ML, Kassiri Z, Virag JAI, de Castro Bras LE, Scherrer-Crosbie M. Guidelines for measuring cardiac physiology in mice. Am J Physiol Heart Circ Physiol. (2018) 314:H733–H52. doi: 10.1152/ajpheart.00339.2017
Keywords: diastolic function, genome-wide association, isoproterenol, mouse, gene expression
Citation: Wang J, Huertas-Vazquez A, Wang Y and Lusis AJ (2019) Isoproterenol-Induced Cardiac Diastolic Dysfunction in Mice: A Systems Genetics Analysis. Front. Cardiovasc. Med. 6:100. doi: 10.3389/fcvm.2019.00100
Received: 19 March 2019; Accepted: 08 July 2019;
Published: 31 July 2019.
Edited by:
George W. Booz, University of Mississippi Medical Center School of Dentistry, United StatesReviewed by:
Perry D. Moerland, University Medical Center Amsterdam, NetherlandsPatrick G. Burgon, University of Ottawa, Canada
Copyright © 2019 Wang, Huertas-Vazquez, Wang and Lusis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jessica Wang, jessicawang@mednet.ucla.edu
 Jessica Wang
Jessica Wang Adriana Huertas-Vazquez
Adriana Huertas-Vazquez Yibin Wang
Yibin Wang Aldons J. Lusis
Aldons J. Lusis