First Report of Complete Sequence of a blaNDM-13-Harboring Plasmid from an Escherichia coli ST5138 Clinical Isolate
- 1Department of Laboratory Medicine, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China
- 2Public Health Research Institute Tuberculosis Center, New Jersey Medical School, Rutgers University, Newark, NJ, USA
- 3Department of Laboratory Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA
- 4Department of Laboratory Medicine, The Second Affiliated Hospital of Nanchang University, Nanchang, China
- 5Department of Respiratory Medicine, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China
Since the first report of blaNDM-1, 16 blaNDM variants have been identified among Gram-negative bacteria worldwide. Recently, a novel blaNDM variant, blaNDM-13, was identified in the chromosome of an ST101 Escherichia coli isolate from Nepal. Here we first reported plasmid-mediated blaNDM-13 in a carbapenem-resistant E. coli ST5138 clinical isolate associated with hospital-acquired urinary tract infection from China. blaNDM-13 and blaSHV-12 coexisted on the a ~54 Kb self-transferable plasmid. Compared with NDM-1, NDM-13, NDM-3, and NDM-4 had two amino acid substitutions (D95N and M154L), one amino acid substitution (D95N) and one amino acid substitutions (M154L), respectively. Complete plasmid sequencing showed that blaNDM-13-harboring plasmid (pNDM13-DC33) was highly similar to the blaNDM-1-harboring IncX3 plasmid pNDM-HN380, a common blaNDM-harboring vector circulating in China. In accordance with the structure of pNDM-HN380, pNDM13-DC33 consists of a 33-kb backbone encoding plasmid replication (repB), stability partitioning, and transfer (tra, trb, and pil) functions, and a 21-kb antimicrobial resistance region with high GC content between umuD and mpr genes. In conclusion, the present study is the first report of a plasmid-encoded blaNDM-13 and the complete sequence of a blaNDM-13-harboring plasmid (pNDM13-DC33). blaNDM-13 maybe originate from blaNDM-1 located on a pNDM-HN380-like plasmid by sequential mutations.
Introduction
Enterobacteriaceae, particularly Escherichia coli and Klebsiella pneumioniae, are common pathogens causing nosocomial infections. Carbapenems are the choice for the treatment of infections caused by multi-drug resistant Enterobacteriaceae, especially extended-spectrum β lactamase (ESBL)- and/or plasmid-mediated AmpC (pAmpC)-producing organisms (Tzouvelekis et al., 2012). The emergence of carbapenem-resistant K. pneumonia and E. coli producing carbapenemases (KPCs) and metallo-β-lactamases (MBLs) have become a major global health problem due to the limited number of effective antibiotic options to treat the infections caused by these multi-drug resistant Enterobacteriaceae (Tzouvelekis et al., 2012). In 2009, a novel MBL, named New Delhi metallo-β-lactamase-1 (NDM-1), was identified in a K. pneumoniae isolate from a Swedish patient who had returned from India with a urinary tract infection (Yong et al., 2009). Since then, NDM-1-producing Gram-negative isolates have emerged worldwide. NDM-1 was primarily identified in Enterobacteriaceae, especially in E. coli and K. pneumoniae, from the Indian subcontinent, Balkan states, the Arabian peninsula, and North Africa (Nordmann and Poirel, 2014). In China, NDM-1 was initially identified in 4 clonally unrelated Acinetobacter baumannii isolates in 2011(Chen et al., 2011). Subsequently, this clinically important enzyme has spread among many species of Enterobacteriaceae in China (Hu et al., 2013; Liu et al., 2013; Zhang et al., 2013).
Since the first report of NDM-1, 16 NDM variants have been identified among Gram-negative bacteria worldwide (http://www.ncbi.nlm.nih.gov/pathogens/submit_beta_lactamase/). Recently, a novel NDM variant, NDM-13, was reported in a multidrug-resistant E. coli clinical isolate in Nepal (Shrestha et al., 2015). The blaNDM-13 gene, interestingly, was found to locate within the chromosome of an E. coli ST101 isolate. The aim of the present study was to investigate whether blaNDM-13 was located on the plasmids of clinically isolated Enterobacteriaceae. We first found plasmid-mediated blaNDM-13 and completely sequenced a blaNDM-13-harboring plasmid for the first time from a carbapenem-resistant E. coli ST5138 clinical isolate associated with hospital-acquired urinary tract infection in China.
Materials and Methods
Bacterial Strain
From Mar, 2014 to Oct, 2014, a total of 87 carbapenem-resistant Enterobacteriaceae (CRE) isolates causing clinical infections isolated from various specimens of patients at the First Affiliated Hospital of Wenzhou Medical University in Wenzhou, east China, were investigated for carbapenemase genes. The isolates were identified as E. coli by an automated microbiology analyzer (bioMe'rieux, Marcy l'Etoile, France) in accordance with the manufacturer's instructions.
Antimicrobial Susceptibility Testing
Gram-negative susceptibility (GNS) card on the Vitek system (bioMe'rieux, Marcy l'Etoile, France) was performed initially for antimicrobial susceptibility testing. Disk diffusion method was used for further confirmation and antimicrobial susceptibility results were interpreted according to the criteria recommended by Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2014). The E-test method was used for the determination of minimum inhibitory concentrations (MICs) of imipenem and meropenem for the E. coli isolate and its transconjugant. E. coli ATCC 25922 was used as control strain for antimicrobial susceptibility testing.
Detection of Carbapenemases and Extended-Spectrum β-Lactamases (ESBLs)
The modified Hodge test (MHT) was further performed on a Mueller-Hinton agar plate with ertapenem as substrate for the detection of carbapenemases as described previously (CLSI, 2014). MBLs were determined using a double-disc synergy test (Peleg et al., 2005). ESBLs were tested using the CLSI-recommended confirmatory double disk combination (CLSI, 2014).
Detection of Resistance Genes
The carbapenemase genes responsible for carbapenem resistance, including blaKPC, blaGES, blaSPM, blaIMP, blaVIM, blaSPM, and blaNDM, were detected using PCR and DNA sequencing as described previously (Queenan and Bush, 2007; Nordmann et al., 2011). ESBLs genes were detected in accordance with the method described previously (Andrade et al., 2010). PCR products were analyzed by electrophoresis in 1% agarose gels and were sequenced on both strands.
Transferability of Plasmids with Carbapenem Resistance
In order to determine whether carbapenem resistance was transferable in E.coli DC33 strain, filter mating conjugation was performed using E. coli 600 as the recipient as previously described (Wang et al., 2004). Plasmid DNA of E. coli DC33 strain was extracted with the QIAGENPlasmid Midi kit (Hilden, Germany) according to the manufacturer's instructions. The plasmid extracts were transferred into E. coli DH5α by using chemical transformation and transformants were selected on Luria-Bertani agar plates containing imipenem (0.5 μg/ml).
Multi-Locus Sequence Typing (MLST)
Multi-Locus Sequence Typing (MLST) was performed on E. coli DC33 using amplification of internal fragments of the seven housekeeping genes including adk, fumC, gyrB, icd, mdh, purA, and recA of E. coli according to MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli).
Determination of blaNDM-13 Location
The total bacterial DNA of E. coli DC33 was first prepared in agarose plugs, digested with S1 nuclease and further separated by pulsed-field gel electrophoresis (PFGE), as described previously (Chen et al., 2011). Then, the DNA bands were transferred horizontally to a nylon membrane (Millipore). A digoxigenin-labeled blaNDM-13 probe was used to hybridize with DNA bands and a nitro-blue tetrazolium/5-bromo-4-chloro-3′-indolylphosphate color detection kit (Roche Applied Sciences) was applied to detected hybridization signals.
Sequencing a blaNDM-13-Harboring Plasmid from the Transconjuguant of E. coli DC33 Strain
In order to completely characterize the plasmid from the transconjugant of E. coli DC33 (designated as pNDM13-DC33), pNDM13-DC33 was isolated, purified, and sequenced using the Illumina MiSeq platform. The sequencing reads were de novo assembled, gaps between contigs were closed, open reading frames (ORFs) were predicted, and annotations were performed as described previously (Chen et al., 2013).
Results and Discussion
Carbapenemases and ESBLs Production and Detection of Resistance Genes
Among 87 CRE isolates, 7 were positive for blaNDM. After sequencing, E. coli strain DC33 was found to harbor blaNDM-13. E. coli strain DC33 was isolated from a urine culture of a 64-year-old male hospitalized for prostatic hyperplasia in July, 2014. After hospitalized, the patient had the symptom of urinary tract infection. Subsequently, many white cells were found in urine sample under microscope. E. coli strain DC33 was isolated when the patient was hospitalized on day 8. E. coli DC33 was weakly positive for the MHT assay, but β-lactamase activity was inhibited by EDTA, indicating that E. coli DC33 produced a MBL. E. coli DC33 was also positive for CLSI-recommended confirmatory double disk combination test for detecting ESBLs. The results of detection of ESBL genes using PCR showed that E. coli DC33 was also positive for blaSHV while was negative for other resistance genes tested. After DNA sequencing, blaSHV was found to blaSHV-12.
Antimicrobial Susceptibility Testing
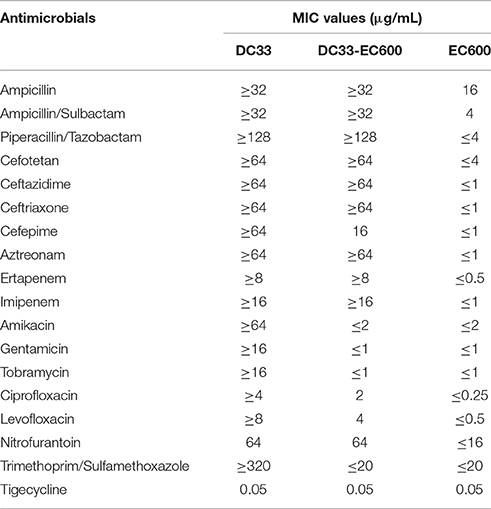
Escherichia coli DC33 exhibited resistance to all antimicrobials tested except tigecycline determined initially by Gram-negative susceptibility (GNS) card on the Vitek system (Table 1), including ampicillin, ampicillin/sulbactam, amikacin, aztreonam, cefotetan, ceftazidme, ceftriaxone, cefepime, ciprofloxacin, ertapenem, gentamicin, imipenem, levofloxacin, nitrofurantoin, piperacillin/tazobactam, tobramycin, and trimethoprim/sulfamethoxazole, The E. coli IOMTU558 carrying blaNDM-13 located on the chromosome from Nepal was highly resistant to all β lactams tested including ampicillin, ampicillin/sulbactam, cefepime, cefoselis, cefotaxime, cefoxitin, cefpirome, ceftazidime, ceftriaxone, cephradine, doripenem, imipenem, meropenem, and moxalactam (Shrestha et al., 2015). The E. coli IOMTU558 was also resistant to other antibiotics including ciprofloxacin, gentamicin, kanamycin, levofloxacin, and tobramycin, but susceptible to amikacin, colistin, fosfomycin, and minocycline (Shrestha et al., 2015). Tigecycline MICs for E. coli DC33 and E. coli IOMTU558 were 0.05 and 2 μg/ml (Shrestha et al., 2015). The antimicrobial susceptibility pattern of E. coli DC33 was further confirmed by disk diffusion method. The resistance of E. coli DC33 to imipenem and meropenem was further corroborated by E-test method.
MLST
MLST result showed E. coli DC33 belonged to ST5138, a single locus variant of ST617. Although ST5138 has been deposited in E. coli MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli), no study about E. coli ST5138 isolate is published. In our previous study, coexistence of blaNDM-1 and blaCMY-42 was found among E. coli ST167 clinical isolates in our hospital (Zhang et al., 2013). As ST5138 was a single-locus variant of S167, we speculate that E. coli DC33 harboring blaNDM-13 is genetically related to E. coli ST 167 isolates carrying blaNDM-1 found in our previous study (Zhang et al., 2013). Recently, a Chinese study found an increasing prevalence of E. coli ST167 clinical isolates carrying both blaNDM-1 and blaNDM-5 on the conjugative IncX3 plasmid in various parts of China (Huang et al., 2016). Therefore, increasing emergence of blaNDM variants among E. coli ST167 and ST167 variants clinical isolates should be of concern. Up to now, blaNDM-13 was only reported in Nepal (Shrestha et al., 2015). The present study is the second report of this novel blaNDM variant.
Location of blaNDM-13 Gene and Transferability of Plasmids Carrying blaNDM-13
S1-PFGE result showed that a ~54-Kb plasmid was found in E. coli DC33 (Figure 1). Subsequently, blaNDM-13 gene was found to be located on this plasmid, not on chromosome, which was confirmed by Southern-blot (Figure 1). The blaNDM-13-harboring plasmid of E. coli DC33, designated as pNDM13-DC33, was successfully transferred into recipient E. coli 600 by filter mating conjugation. The antimicrobial resistance patterns of E. coli DC33 and its transconjugant were showed in Table 1. Shrestha et al found that blaNDM-13 was located within the chromosome (Shrestha et al., 2015). However, blaNDM-13 was first confirmed to be located on the plasmid in the present study.
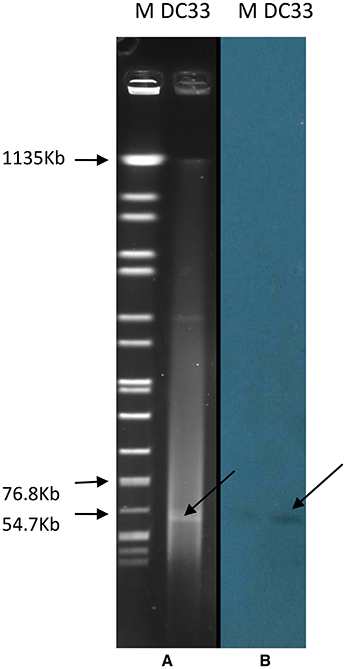
Figure 1. S1-digested plasmid DNA of E. coli DC33 (A). Southern blot hybridization with blaNDM-13 of E. coli DC33 (B). M, Salmonella serotype Braenderup strain H9812.
Complete Sequence of pNDM13-DC33
Plasmid pNDM13-DC33 is 54.035-bp in length, with an average GC content of 49.03% (Figure 2). BLASTn analysis showed that pNDM13-DC33 is similar to pNDM-HN380, an IncX3-type plasmid carrying blaNDM-1 among Enterobacteriaceae isolates in China (Ho et al., 2012), with 100% query coverage and >99.9% nucleotide identity (with 8 single nucleotide polymorphisms, SNPs). In China, IncX3-type plasmids carrying blaNDM variants have been widely found among E. coli clinical isolates with different clones including ST648, ST156, ST131, ST167, and ST3835 clones (Feng Y. et al., 2015; Huang et al., 2016; Wang et al., 2016; Yang et al., 2016). Notably, these similar plasmids have been identified in several hospitals from different geographic regions in China (Wang et al., 2014; Feng J. et al., 2015; Qu et al., 2015), suggesting that pNDM-HN380-like plasmids are common NDM vectors that likely contribute significantly to the dissemination of blaNDM variants in China.

Figure 2. Plasmid structures of pNDM_HN380 (JX104760), pNDM13_DC33(KX094555) and the blaNDM-13 neighboring genetic environment in IOMTU558 (LC012596). Colored arrows represent open reading frames, with dark blue, yellow, green, red, light blue, and orange arrows representing replication genes, mobile elements, plasmid transfer genes, resistancegenes, IS5, and plasmid backbone genes, respectively. Blue shading denotes regions of shared homology among different plasmids.
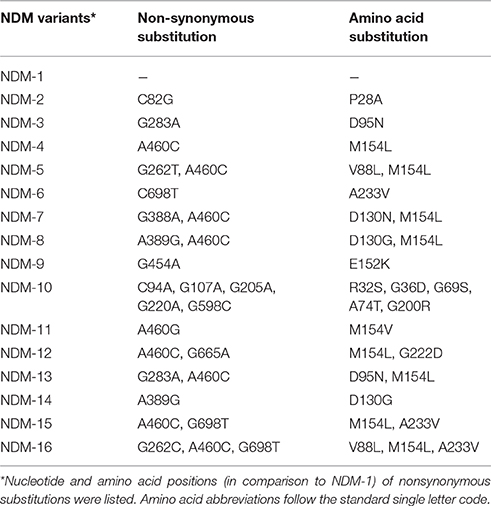
In accordance with the structure of pNDM-HN380, pNDM13-DC33 consists of a 33-kb backbone encoding plasmid replication (repB), stability partitioning, and transfer (tra, trb, and pil) functions, and a 21-kb antimicrobial resistance region with comparatively high GC content between umuD and mpr genes, suggesting that these two regions were likely acquired and genetically distinct. The resistance region of pNDM13-DC33, containing 16 ORFs sequentially organized as IS26, blaSHV-12, ygbI, ygbJ, IS26, insE, groL, cutA1, dsbc, ΔtrpF, bleMBL, blaNDM-13, ΔISAba125, IS5, ΔISAba125, and Tn3 tnpA, was nearly identical to that of pNDM-HN380, but with the exception that they carry different blaNDM variants (pNDM13-DC33 with blaNDM-13 and pNDM-HN380 with blaNDM-1) (Figure 2). Of note, compared with NDM-1, NDM-13, NDM-3, and NDM-4 had two amino acid substitutions (D95N and M154L), one amino acid substitution (D95N) and one amino acid substitutions (M154L), respectively (Table 2). Although NDM-13 (with two substitutions including the D95N and M154L relative to NDM-1) did not show increased hydrolytic activity against carbapenems, cephalosporins, and penicillins, it increased the affinity of NDM-13 for cefotaxime and affected the catalytic activity of the enzyme against cefotaxime (Shrestha et al., 2015). Our finding that blaNDM-13-harboring pNDM13-DC33 closely resembles blaNDM-1-harboring pNDM-HN380 provides evidence that novel blaNDM variants emerge by sequential mutations of a pNDM-HN380-like plasmid carrying blaNDM-1.
The chromosomal organization of the blaNDM-13 gene initially found in the E. coli isolate IOMTU558 from Nepal was similar to that in pNDM13-DC33, except for a 260-bp deletion in ISAba125 of 260 bp (353 to 94 bp upstream blaNDM-13 start codon). In contrast, the corresponding ISAba125 on pNDM13-DC33 was in full-length (1087 bp), but was interrupted by the insertion of an IS5 (at 265 bp upstream blaNDM-13 start codon) (Figure 1). A comparison of the chromosomal organization flanking blaNDM-13 in the E. coli isolate IOMTU558 (Shrestha et al., 2015) with plasmids harboring blaNDM identified the a set of ordered genes, tnpA-IS30-blaNDM-13-bleMBL-trpF-dsbC-cutA-groES-groL, that were nearly identical in plasmid pPMK1 from Nepal, plasmidpKPX-1 from Taiwan, plasmid pNDM-MAR from Morocco, and in an Enterobacter hormaechei CCHB10892 plasmid from Brazil (Villa et al., 2012; Huang et al., 2013; Carvalho-Assef et al., 2014; Stoesser et al., 2014; Shrestha et al., 2015). This finding suggests that the chromosomal copy of blaNDM-13 may be the result of a rare integration event where a region of the plasmid recombined into the E. coli genome.
In conclusion, the present study is the first report of a plasmid-encoded blaNDM-13 and the complete sequence of a blaNDM-13-harboring plasmid (pNDM13-DC33). blaNDM-13 maybe originate from blaNDM-1 located on a pNDM-HN380-like plasmid by sequential mutations. The emergence of novel plasmid-mediated blaNDM variants, originating through the mutations in blaNDM from an epidemic plasmid, poses a concern that NDM variants with different β-lactamases hydrolytic activity will evolve.
Nucleotide Sequence Accession Number
The complete nucleotide sequences of plasmid pNDM13-DC33 has been deposited as GenBank accession no. KX094555.
Ethical Approval
The Ethics Committee of the first Affiliated Hospital of Wenzhou Medical University exempted this study from review because the present study focused on bacteria.
Author Contributions
JL, XQ, DZ, ZZ, YC, YG, and SW isolated bacteria and performed the laboratory measurements. FY and LW made substantial contributions to conception and design. LC, YT, and BK revised the manuscript critically for important intellectual content. LC and JL participated in experimental design and data analysis. FY drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported in part by National Institutes of Health (NIH) Grant R01AI090155 (to BK) and R21AI117338 (to LC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Zhejiang Provincial Program for the Cultivation of High-level Innovative Health talents to FY.
References
Andrade, L. N., Minarini, L. A., Pitondo-Silva, A., Climaco, E. C., Palazzo, I. C., Medeiros, M. I., et al. (2010). Determinants of β-lactam resistance in meningitis-causing Enterobacteriaceae in Brazil. Can. J. Microbiol. 56, 399–407. doi: 10.1139/W10-020
Carvalho-Assef, A. P., Pereira, P. S., Albano, R. M., Berião, G. C., Tavares, C. P., Chagas, T. P., et al. (2014). Detection of NDM-1-, CTX-M-15-, and qnrB4-producing Enterobacter hormaechei isolates in Brazil. Antimicrob. Agents Chemother. 58, 2475–2476. doi: 10.1128/AAC.02804-13
Chen, L., Chavda, K. D., Fraimow, H. S., Mediavilla, J. R., Melano, R. G., Jacobs, M. R., et al. (2013). Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob. Agents Chemother. 57, 269–276. doi: 10.1128/AAC.01648-12
Chen, Y., Zhou, Z., Jiang, Y., and Yu, Y. (2011). Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemother. 66, 1255–1259. doi: 10.1093/jac/dkr082
CLSI (2014). Performance Standards for Antimicrobial Susceptibility Testing, 24th Informational Supplement (M100-S24). Wayne, IL: Clinical and Laboratory Standards Institute.
Feng, J., Qiu, Y., Yin, Z., Chen, W., Yang, H., Yang, W., et al. (2015). Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of Citrobacter freundii. J. Antimicrob. Chemother. 70, 2987–2991. doi: 10.1093/jac/dkv232
Feng, Y., Yang, P., Xie, Y., Wang, X., McNally, A., and Zong, Z. (2015). Escherichia coli of sequence type 3835 carrying blaNDM-1, blaCTX-M-15, blaCMY-42 and blaSHV-12. Sci. Rep. 5:12275. doi: 10.1038/srep12275
Ho, P. L., Li, Z., Lo, W. U., Cheung, Y. Y., Lin, C. H., Sham, P. C., et al. (2012). Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg. Microbes Infect. 1, e39. doi: 10.1038/emi.2012.37
Hu, L., Zhong, Q., Tu, J., Xu, Y., Qin, Z., Parsons, C., et al. (2013). Emergence of blaNDM-1 among Klebsiella pneumoniae ST15 and novel ST1031 clinical isolates in China. Diagn. Microbiol. Infect. Dis. 75, 373–376. doi: 10.1016/j.diagmicrobio.2013.01.006
Huang, T. W., Chen, T. L., Chen, Y. T., Lauderdale, T. L., Liao, T. L., Lee, Y. T., et al. (2013). Copy number change of the NDM-1 sequence in a multidrug-resistant Klebsiella pneumoniae clinical isolate. PLoS ONE 8:e62774. doi: 10.1371/journal.pone.0062774
Huang, Y., Yu, X., Xie, M., Wang, X., Liao, K., Xue, W., et al. (2016). Widespread dissemination of carbapenem-resistant Escherichia coli sequence type 167 strains harboring blaNDM-5 in clinical settings in China. Antimicrob. Agents Chemother. 60, 4364–4368. doi: 10.1128/AAC.00859-16
Liu, Z., Li, W., Wang, J., Pan, J., Sun, S., Yu, Y., et al. (2013). Identification and characterization of the first Escherichia coli strain carrying NDM-1 gene in China. PLoS ONE 8:e66666. doi: 10.1371/journal.pone.0066666
Nordmann, P., and Poirel, L. (2014). The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 20, 821–830. doi: 10.1111/1469-0691.12719
Nordmann, P., Poirel, L., Carrër, A., Toleman, M. A., and Walsh, T. R. (2011). How to detect NDM-1 producers. J. Clin. Microbiol. 49, 718–721. doi: 10.1128/JCM.01773-10
Peleg, A. Y., Franklin, C., Bell, J. M., and Spelman, D. W. (2005). Dissemination of the metallo-β-lactamase gene blaIMP-4 among gram-negative pathogens in a clinical setting in Australia. Clin. Infect. Dis. 41, 1549–1556. doi: 10.1086/497831
Queenan, A. M., and Bush, K. (2007). Carbapenemases: the versatile β-Lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/CMR.00001-07
Qu, H., Wang, X., Ni, Y., Liu, J., Tan, R., Huang, J., et al. (2015). NDM-1-producing Enterobacteriaceae in a teaching hospital in Shanghai, China: IncX3-type plasmids may contribute to the dissemination of blaNDM-1. Int. J. Infect. Dis. 34, 8–13. doi: 10.1016/j.ijid.2015.02.020
Shrestha, B., Tada, T., Miyoshi-Akiyama, T., Shimada, K., Ohara, H., Kirikae, T., et al. (2015). Identification of a novel NDM variant, NDM-13, from a multidrug-resistant Escherichia coli clinical isolate in Nepal. Antimicrob. Agents Chemother. 59, 5847–5850. doi: 10.1128/AAC.00332-15
Stoesser, N., Giess, A., Batty, E. M., Sheppard, A. E., Walker, A. S., Wilson, D. J., et al. (2014). Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a Nepali hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob. Agents Chemother. 58, 7347–7357. doi: 10.1128/AAC.03900-14
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., and Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
Villa, L., Poirel, L., Nordmann, P., Carta, C., and Carattoli, A. (2012). Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J. Antimicrob. Chemother. 67, 1645–1650. doi: 10.1093/jac/dks114
Wang, L. H., Liu, P. P., Wei, D. D., Liu, Y., Wan, L. G., Xiang, T. X., et al. (2016). Clinical isolates of uropathogenic Escherichia coli ST131 producing NDM-7 metallo-β-lactamase in China. Int. J. Antimicrob. Agents. 48, 41–45. doi: 10.1016/j.ijantimicag.2016.03.009
Wang, M., Sahm, D. F., Jacoby, G. A., and Hooper, D. C. (2004). Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob. Agents Chemother. 48, 1295–1299. doi: 10.1128/AAC.48.4.1295-1299.2004
Wang, X., Xu, X., Li, Z., Chen, H., Wang, Q., Yang, P., et al. (2014). An outbreak of a nosocomial NDM-1-producing Klebsiella pneumoniae ST147 at a teaching hospital in mainland China. Microb. Drug Resist. 20, 144–149. doi: 10.1089/mdr.2013.0100
Yang, R. S., Feng, Y., Lv, X. Y., Duan, J. H., Chen, J., Fang, L. X., et al. (2016). Emergence of NDM-5 and MCR-1-Producing Escherichia coli Clone ST648 and ST156 from A single muscovy duck (Cairina moschata). Antimicrob. Agents Chemother. doi: 10.1128/AAC.01365-16
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new Metallo-β-Lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Keywords: Escherichia coli, blaNDM-13, plasmid
Citation: Lv J, Qi X, Zhang D, Zheng Z, Chen Y, Guo Y, Wang S, Chen L, Kreiswirth BN, Tang Y-W, Chen Z, Hu L, Wang L and Yu F (2016) First Report of Complete Sequence of a blaNDM-13-Harboring Plasmid from an Escherichia coli ST5138 Clinical Isolate. Front. Cell. Infect. Microbiol. 6:130. doi: 10.3389/fcimb.2016.00130
Received: 16 August 2016; Accepted: 28 September 2016;
Published: 13 October 2016.
Edited by:
Philip R. Hardwidge, Kansas State University, USAReviewed by:
Guoqiang Zhu, Yangzhou University, ChinaShannon D. Manning, Michigan State University, USA
Copyright © 2016 Lv, Qi, Zhang, Zheng, Chen, Guo, Wang, Chen, Kreiswirth, Tang, Chen, Hu, Wang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangxing Wang, 38805@163.com
Fangyou Yu, wzjxyfy@163.com
†These authors have contributed equally to this work.
 Jingnan Lv1†
Jingnan Lv1†  Liang Chen
Liang Chen Fangyou Yu
Fangyou Yu