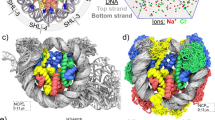
Abstract
The present study contributes to the understanding of DNA compaction in the cell nucleus at the nucleosomal level. The interaction between DNA and histones affects key processes of cell life, including replication and transcription. This interaction can be described in terms of a free energy profile. In this paper, we calculated the free energy profile during DNA unwrapping from the histone octamer. The calculations were carried out using the MM/PBSA method. The calculated profiles are in good agreement with experimental data published earlier. The obtained results indicate the applicability of the technique for studying the effect of posttranslational modifications of histones and histone variants on nucleosome energy, which is important for understanding the mechanisms of transcriptional regulation in chromatin.
Similar content being viewed by others
References
Shaytan, A.K., Landsman, D., and Panchenko, A.R., Nucleosome adaptability conferred by sequence and structural variations in histone H2A-H2B dimers, Curr. Opin. Struct. Biol., 2015, vol. 32, pp. 48–57.
Brower-Toland, B.D., Smith, C.L., Yeh, R.C., Lis, J.T., Peterson, C.L., and Wang, M.D., Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA, Proc. Natl. Acad. Sci. U.S.A., 2002, vol. 99, no. 4, pp. 1960–1965.
Thåström, A., Gottesfeld, J.M., Luger, K., and Widom, J., Histone-DNA binding free energy cannot be measured in dilution-driven dissociation experiments, Biochemistry, 2004, vol. 43, no. 3, pp. 736–741.
Mihardja, S., Spakowitz, A.J., Zhang, Y., and Bustamante, C., Effect of force on mononucleosomal dynamics, Proc. Natl. Acad. Sci. U.S.A., 2006, vol. 103, no. 43, pp. 15871–15876.
Ettig, R., Kepper, N., Stehr, R., Wedemann, G., and Rippe, K., Dissecting DNA-histone interactions in the nucleosome by molecular dynamics simulations of DNA unwrapping, Biophys. J., 2011, vol. 101, no. 8, pp. 1999–2008.
Zhang, B., Zheng, W., Papoian, G.A., and Wolynes, P.G., Exploring the free energy landscape of nucleosomes, J. Am. Chem. Soc., 2016, vol. 138, no. 26, pp. 8126–8133.
Kollman, P.A., et al., Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models, Acc. Chem. Res., 2000, vol. 33, no. 12, pp. 889–897.
Davey, C.A., Sargent, D.F., Luger, K., Maeder, A.W., and Richmond, T.J., Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution, J. Mol. Biol., 2002, vol. 319, no. 5, pp. 1097–1113.
Banks, D.D. and Gloss, L.M., Equilibrium folding of the core histones: The H3-H4 tetramer is less stable than the H2A-H2B dimer, Biochemistry, 2003, vol. 42, no. 22, pp. 6827–6839.
Armeev, G.A., Shaitan, K.V., and Shaytan, A.K., Nucleosome structure relaxation during DNA unwrapping: Molecular dynamics simulation study, Moscow Univ. Biol. Sci. Bull., 2016, vol. 71, no. 3, pp. 141–144.
Guy, A.T., Piggot, T.J., and Khalid, S., Singlestranded DNA within nanopores: Conformational dynamics and implications for sequencing; A molecular dynamics simulation study, Biophys. J., 2012, vol. 103, no. 5, pp. 1028–1036.
Pronk, S., Páll, S., Schulz, R., Larsson, P., Bjelkmar, P., Apostolov, R., Shirts, M.R., Smith, J.C., Kasson, P.M., van der Spoel, D., Hess, B., and Lindahl, E., GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit, Bioinformatics, 2013, vol. 29, no. 7, pp. 845–854.
Humphrey, W., Dalke, A., and Schulten, K., VMD: Visual molecular dynamics, J. Mol. Graphics, 1996, vol. 14, no. 1, pp. 33–38.
Forties, R.A., North, J.A., Javaid, S., Tabbaa, O.P., Fishel, R., Poirier, M.G., and Bundschuh, R., A quantitative model of nucleosome dynamics, Nucleic Acid Res., 2011, vol. 39, no. 19, pp. 8306–8313.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © A.K. Gribkova, G.A. Armeev, A.K. Shaytan, 2017, published in Vestnik Moskovskogo Universiteta, Seriya 16: Biologiya, 2017, Vol. 72, No. 3, pp. 164–168.
About this article
Cite this article
Gribkova, A.K., Armeev, G.A. & Shaytan, A.K. Investigation of histone-DNA binding energy as a function of DNA unwrapping from nucleosome using molecular modeling. Moscow Univ. Biol.Sci. Bull. 72, 142–145 (2017). https://doi.org/10.3103/S009639251703004X
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S009639251703004X