More Information
Submitted: 29 June 2019 | Approved: 15 July 2019 | Published: 16 July 2019
How to cite this article: Amoo Shen Y, Nian W. Cloning and Characterization of a Pseudo-Response Regulator 7 (PRR7) Gene from Medicago Sativa Involved In Regulating the Circadian Clock. J Plant Sci Phytopathol. 2019; 3: 056-061.
DOI: 10.29328/journal.jpsp.1001032
Copyright License: © 2019 Shen Y, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Cloning and Characterization of a Pseudo-Response Regulator 7 (PRR7) Gene from Medicago Sativa Involved In Regulating the Circadian Clock
Yilin Shen and Weifeng Nian*
The High School Affiliated to Renmin University of China, Beijing 100193, People’s Republic of China
*Address for Correspondence: Weifeng Nian, The High School Affiliated to Renmin University of China, Beijing 100193, People’s Republic of China, Tel: +86 18701220351; Email: sdhmk2008@163.com; sylbdfz@126.com
The circadian clock is an endogenous molecular oscillator with a period of about 24 hours, which regulates the physiology and developmental processes of almost all higher plants. Pseudo-response regulators (PRRs) are an important part of the central clock oscillator, together with other clock genes, constituting interlinked transcriptional feedback loops, which partly influence plant growth and development. In this study, a circadian clock-related gene MsPRR7 was cloned from Medicago sativa (alfalfa) by homologous cloning. The full length MsPRR7 gene was 2648 bp in length, with an open reading frame of 2385 bp encoding a protein of 795amino acids. Phylogenetic analysis showed that the MsPRR7 was closely related to PRR7 from the PRR family of Arabidopsis thaliana. Subcellular localization analysis found that MsPRR7 was located in the nucleus. Quantitative reverse-transcription polymerase chain reactions (qRT-PCR) demonstrated that expression of MsPRR7 gene transcripts in leaves was affected by circadian rhythms, and that its expression level increased with an extension of illumination time, reaching a peak around 8–10 hours. These results will provide the experimental basis for further study of the regulation of PRR family genes in alfalfa.
The plant flowering process represents the transition from vegetative to productive growth, and is controlled by complicated innergenetic regulation and the external environment. Flowering is closely related to the heading date and maturity crops, and has been studied for hundreds of years [1]. Currently, studies on Arabidopsis thaliana have revealed five pathways, the vernalization, photoperiod, gibberellin, autonomous, and endogenous pathways [1]. Initially, it was discovered that day length, namely photoperiod, could be perceived by higher plants and impacted their flowering. Over these years, different models have been put forward to explain the relationship between flowering and photoperiod [2,3]. Recently, a growing number of genes have been characterized as effectors of the natural condition or internal regulatory network, which has enhanced our knowledge of the regulators mediating the flowering pathway and plant circadian clock.
In the study of A. thaliana, a long-day model plant, CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1), LATEELONGATED HYPOCOTYL (LHY) and TIMING OF CABEXPRESSION 1 (TOC1) were firstly identified to be central components of the clock or oscillator [4-6]. CCA1 and LHY are two transcription factors with Myb structures, and they are functionally redundant to some extend. The expression profiles of CCA1, LHY and TOC1 exhibit strict periodicity, with TOC1 expression peaking late in the day and early in the night, which stimulates the expression of CCA1 and LHY. However, accumulation of CCA1 and LHY can also inhibit TOC1 expression through direct binding to cis-elements of the TOC1 promoter [7].
With the nucleotide sequencing of A. thaliana, homologous genes of TOC1 were compiled. This small family, the pseudo-response regulators (PRRs), include PRR1 (TOC1), PRR3, PRR5, PRR7 and PRR9 in A. thaliana [8]. All of these PRRs have similar expression patterns in terms of circadian rhythms, referred to as circadian waves of the PRR quintet [8]. In addition, homologs of the five PRRs were also identified in the short-day monocotyledonous Oryza sativa, indicating that there might be evolutionarily conserved molecular mechanisms underlying circadian rhythms [9].
Steady circadian rhythm is indispensible for plants, and over 89% of transcripts have peak expression at a particular time of day, largely dependent on the environment [10,11]. Transcript profiling also reveals that regulation of flowering, hypocotyl length, cold stress responses, and mitochondrial metabolism showed remarkable changes in the arrhythmic prr9 prr7 prr5 triple mutant [12]. The mutants prr7 and prr9 lengthened the period in constant white light, but there was no change in constant darkness [13]. During daytime, the other PRRs can activate CONSTANS (CO/TOC1), with a double mutant of prr7 and either prr9 or prr5 delaying flowering in A. thaliana [14]. PPR7 was shown to bridge the gap between the circadian system and the temperature responsing system through interaction with HEAT SHOCK FACTOR B2b (HsfB2b), and finally affecting flowering [15]. OsPRR37, the homolog of PRR7, can also affect flowering time of rice, and plays an important role in its photoperiod adaptive response to different latitudes [16]. In addition, homologous genes like SbPRR37 in sorghum [17], Ppd-H1 in barley [18], and TaPRR37 in wheat [19], have been identified in regulating flowering date. However there are differences, with leaf morphology in barley (Hordeum vulgare) and plant height in wheat also being changed [20].
Alfalfa is an important leguminous forage, which enjoys the reputation as‘king of forage’, based on its high yields, rich nutrition, high protein content and good palatability. As a long-day plant, alfalfa tends to be distributed at high latitudes and in sunny areas. However, the mechanism of how alfalfa perceives light signals, and the underlying regulatory pathways, are largely unknown. Notably, the timing of flower is closely related to yields in biomass for energy production [21]. In this current study, we isolated a PRR gene from Medicago sativa. And named it MsPRR7, based on bioinformatics analysis. Subcellular localization and expression were also analyzed to further investigate the function of MsPRR7 in alfalfa. As was the first molecular level insight into the photoperiod of alfalfa, this study provided options for further breeding strategies in alfalfa.
Plant material and growth conditions
Medicago sativa cv. Zhongmu-1 was used as the wild type plant in this study. Seeds were surface sterilized in 70% (v/v) ethanol for 10 min, and washed 4–5 times in sterilized water. Seeds were planted on solidified ½ Murashige and Skoog (MS) medium, and were then placed in a growth chamber (16h light and 8h dark, 22 °C). After 15d, seedlings with good growth were transplanted and cultured at room temperature (20–25 °C) under natural light conditions for 7d.
Plant treatments
In order to examine the expression of the PRR family gene MsPRR7 in alfalfa under different photoperiods, samples of leaf and root tissue were obtained: (1) every 4h from dawn (about 4:30 am); (2) every 4h under dark conditions, following a 24h dark treatment; and (3) every 4h under continuous light conditions, following a 24h light treatment. All plant samples for the preparation of total RNA were immediately frozen with liquid nitrogen and were stored at −80 °C.
Cloning of the MsPRR7 gene and sequence analysis
Total RNA was extracted from cultivar Zhongmu-1 using RNAiso plus (TaKaRa, Japan) and reverse transcribed to cDNA with M-MLV reverse transcriptase according to the instructions of the manufacturer. cDNA was diluted and used for polymerase chain reaction (PCR) amplification. The primers (MsPRR-f: 5’- CTT GAT GGG TGA AGT GCT CA - 3’ and MsPRR-r: 5’- CAG TTG CTG GAA CGA AAT CA - 3) were designed based on a homologous of PRR gene (Mtr_4g061360) from Medicago truncatula, a leguminous model plant. Thirty cycles of a stepped PCR program (98 °C, 10s; 60 °C, 15s; 68 °C, 3 min) were performed, and then the samples were stored at 4 °C. Amplified products were identified on 0.8% agarose gels, and the target fragment was excised and recovered with an Agarose Gel DNA Fragment Recovery Kit Ver.2.0 (TransGen Biotech, China). The recovered product was then ligated into pEASY-T5 (TransGen Biotech, China), generating pEASY-MsPRR, which was transformed into E. coli DH5α (TransGen Biotech, China). Positive single colonies were screened prior to nucleotide sequencing.
Bioinformatics Analysis of MsPRR7 sequence
The structure of the cloned sequence was analyzed with software tools associated with the NCBI database (http://www.ncbi.nlm.nih.gov/), obtaining a putative open reading frame (ORF) for further comparative analysis. Multiple sequence alignment of the PRR family in alfalfa and other species was conducted by DNAMAN 6.0. Protein functional domains were analyzed using the InterPro database (http://www.ebi.ac.uk/interpro/). We found homologous proteins in NCBI/GenBank and performed sequence alignment analysis using ClustalX. Phylogenetic tree analysis of MsPRR7 in alfalfa was performed using MEGA7 software (http://megasofware.net/).
Quantitative real-time PCR analysis
Total RNA was isolated from tissue samples using a Spectrum Plant Total RNA Kit (TaKaRa, Japan), and was converted to cDNA with M-MLV reverse transcriptase. Using cDNA as template, PCR was performed by a ABI7300 Real-Time System (Applied Biosystems, USA) with SYBR Green I (TaKaRa, Japan), following the manufacturers’ instructions. Alfalfa ACTIN-7 (XM_003602497.3) was utilized as a reference gene. The following pairs of primers were used separately to amplify each of the cDNA samples: PRR-qPCR-f,5’-AGT GGT GCA TAC AAA GCC TGAA -3’ and PRR-qPCR-r,5’-CCC CTT TAC GCA TTG GAT TGTT-3’ were used to amplify MsPRR7; and Actin-f,5’-CAA AAG ATG GCA GAT GCT GAG GAT-3 and Actin-r,5’-CAT GAC ACC AGT ATG A CG AGG TCG-3’ were used to amplify alfalfa ACTIN-7. Relative gene expression levels were determined using the 2−≥≥CT method.
Subcellular localization studies
For experiments with a green fluorescent protein (GFP) fusion expression vector, primers GFP-f and GFP-r were designed based on the ORF sequences of pSAT6 and the MsPRR7 gene. The primer sequences were GFP-f,5’-CAC CAT TTA CGA ACG ATA GCA TGA ACA ATA ATG TTG-3’, and GFP-r,5’-CTA GTC AGA TCT ACC ATC CCG CAT TTT GAT TGG TA-3’. A target fragment was amplified and recovered covering the sequences for the open reading frame of MsPRR7. After Nco1 and Sma1 digestion, the open reading frames of MsPRR7 without its stop codon and GFP from the pSAT6 vector were recombined to create a new fusion expression vector pSAT6-MsPRR7-GFP. This was introduced into E coli DH5α, and single colonies were screened by PCR prior to being sequenced. The recombinant plasmid together with pBI121, acting as an empty vector, were transformed into Agrobacterium tumefaciens strain GV3101, and then infiltrated into Nicotiana benthamiana leaves which is about 5–6 weeks old. Concentrated A. tumefaciens was resuspend with antibiotic free infiltration medium and injected into the leaves [22]. Fluorescent signal from GFP was observed byLSM880 confocal microscopy (Zeiss, Germany) after incubation at 22 °C for 48h.
Cloning and sequence analysis of alfalfa MsPRR7
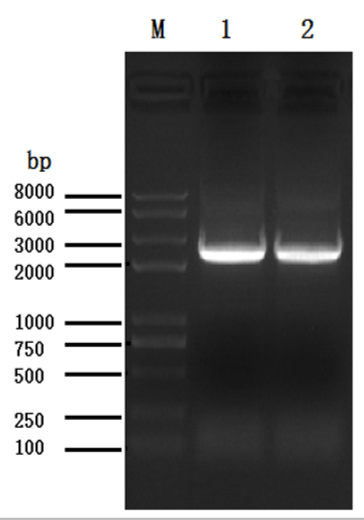
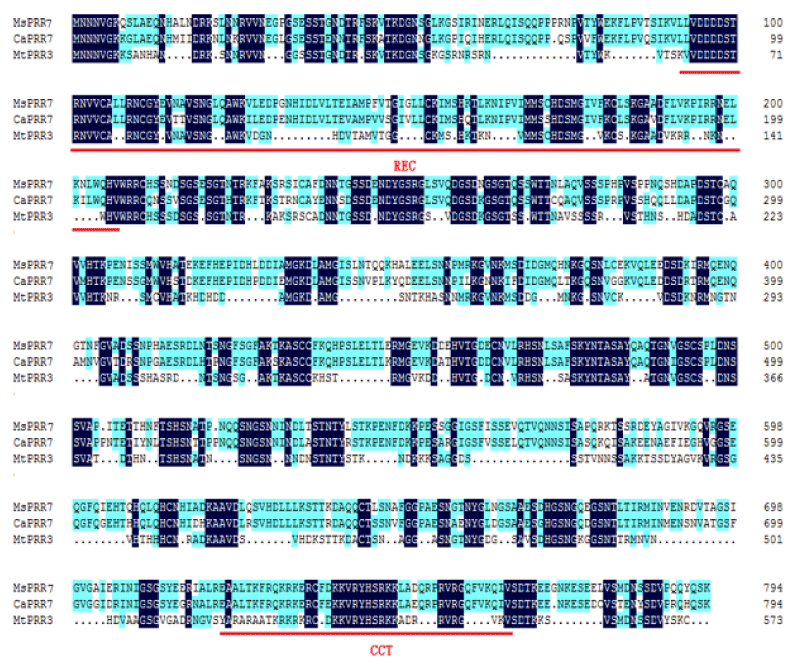
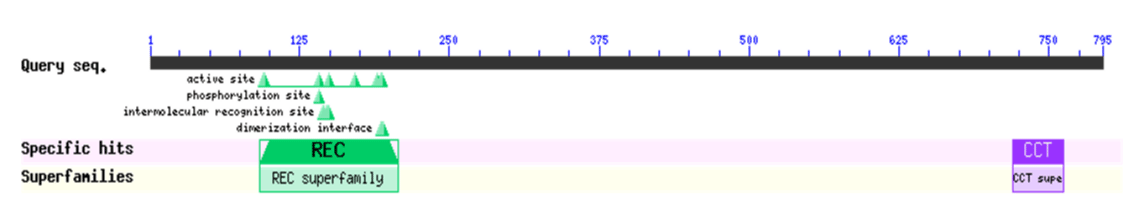
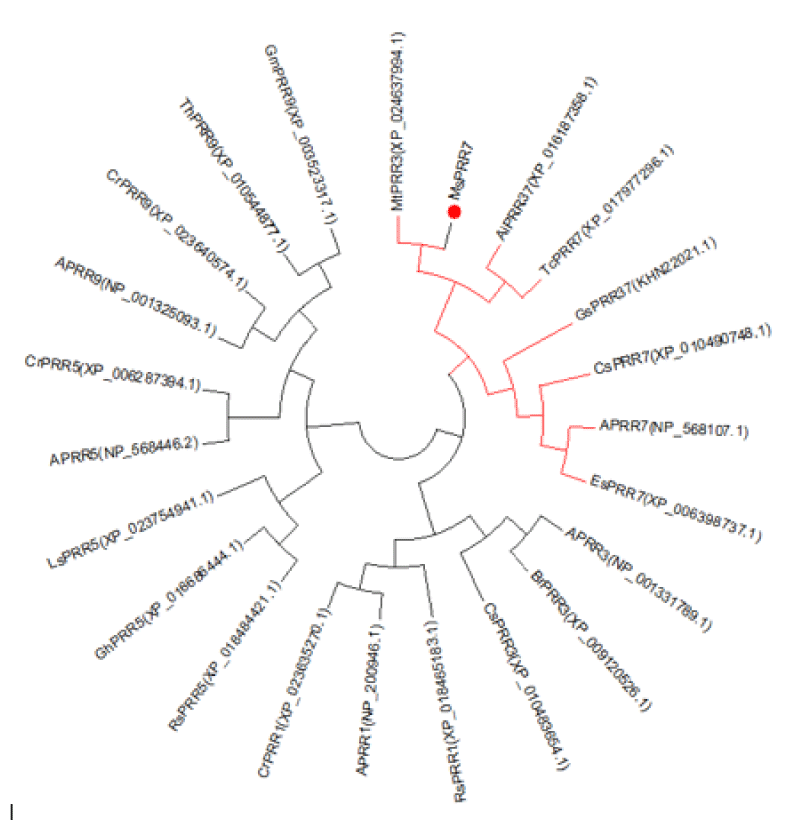
Based on the sequence of the M.truncatula PRR3 gene (XP_024637994.1), we designed a primer to isolate the PRRs of Medicago sativa (Figure 1). We amplified a novel sequence with a similar electrophoretic mobility as a fragment covering MsPRR7. Nucleotide sequencing the of the nucleic acid fragment showed that it was 2385 bp in length, and coded for a protein of 795 amino acids (Figure 2). Its amino acid sequence showed 95% and 80% identity with those of M.truncatula (XP_024637994.1) and Cicer arietinum (XP_012572769.1), respectively (Figure 3). A REC signal receiver domain at the N-terminal and a CCT motif located at the C-terminal were also conserved. These two domains were highly similar to those of other PRR protein structures (Figure 4). In addition, a phylogenetic tree was constructed using the mega7 neighbor-joining method to evaluate the relationship to PRR genes in different species. The putative protein from M. sativa had a high level of sequence similarity to those derived from MsPRR7, AtPRR7 (NP_001331789.1), the obroma cacao PRR7 (XP_017977296.1) and Arachis ipaensis PRR37, indicating homology (Figure 5). As a result, the isolated M.sativa gene was named MsPRR7.
Figure 1: Amplification of the MsPRR7 gene. Lane 1, the PCR product of MsPRR7 CDS region amplified with cDNA of Medicago truncatula; lane 2, the PCR product of amplification with the designed primer of Medicago sativa.
Figure 2: The nucleotide and amino acid sequence of MsPRR7.
Figure 3: Alignments of PRRs domain between MsPRR7 and other PRRs proteins.
Figure 4: Conserved domain analysis of MsPRR7 protein.
Figure 5: Phylogenetic tree of MsPRR7 and PRRs proteins in other species.
MsPRR7 was located in the cell nucleus
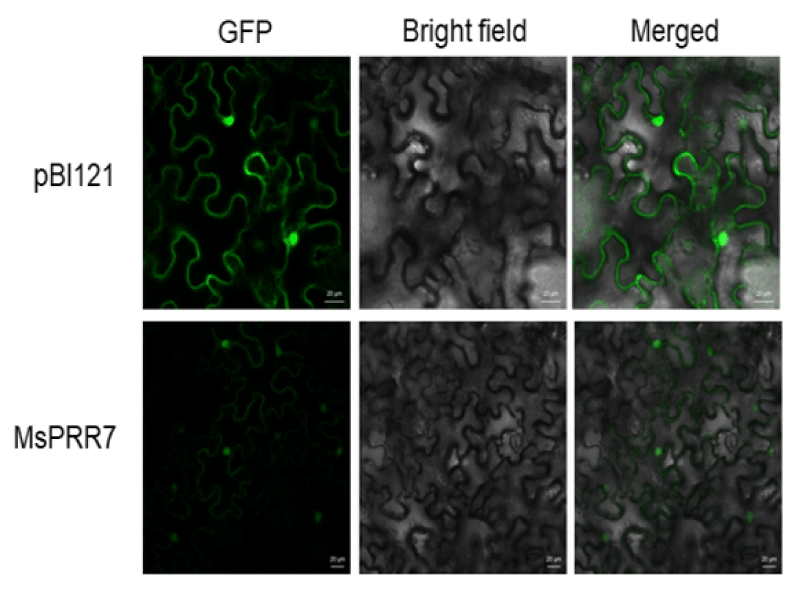
PSORT prediction software suggested that MsPRR7 protein would be mostly located in the nucleus. To confirm the subcellular distribution, we constructed pSAT6-MsPRR7-GFP to express a fusion protein of MsPRR7 with GFP in leaves. N. benthamiana leaves transfected with empty vector pBI121 showed green fluorescent signals in cell walls, nuclei and cell membranes. By contrast, the fluorescence signals of leaf cells transfected with pSAT6-MsPRR7-GFP appeared more frequently in the nucleus, especially in merged images (Figure 6). These results indicated that MsPRR7 was localized to the nucleus, consistent with the software prediction and previous results [23].
Figure 6: Subcellular localization analysis.
Expression analysis of MsPRR7 under different photoperiods
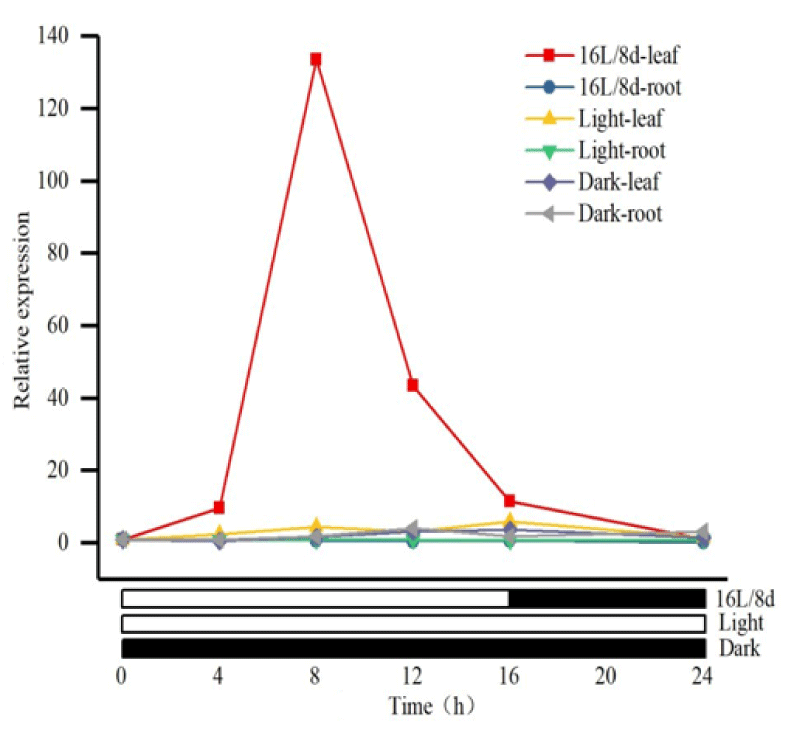
To clarify the expression pattern of MsPRR7 in alfalfa, real-time PCR was performed to assess transcript levels in roots and leaves. Under a 16h light/8h dark photoperiod, at room temperature with natural light conditions, and with relative expression of MsPRR7 under dark treatment being normalized to 1.0, the expression level of MsPRR7 in leaves increased with the extension of illumination time. A peak expression was reached when the illumination was extended for about 8h, with a value 133.5 times higher than that at 0h of extended illumination. As expected, the expression level of MsPRR7 in the roots did not change significantly with fluctuations of photoperiod or continuous light or darkness. In addition, under conditions of complete darkness, the expression of MsPRR7 in leaves and roots did not change significantly with extension of illumination time. However, after continuous illumination treatment, the MsPRR7 appeared to show two peak values (Figure 7). The above results indicated that the MsPRR7 gene exhibited circadian expression, being strongly affected by photoperiod.
Figure 7: Relative expression of MsPRR7 under various photoperiod.
Circadian clocks allow plants to precisely regulate growth and developmental processes with daily and seasonal changes in the environment. As an important part of the central clock oscillator, PRR genes, together with other clock genes, provide interlinked transcriptional feedback loops that maintain periodic oscillation consistent with the external environment. To date, PRRs, especially PRR7, have been shown to have conserved functions in the regulation of flowering and the response of plants to daylight. However, the genetic interplay and regulatory relationships among these genes/loci remain largely unknown. In this current study, we have identified and characterized a novel PRR gene from alfalfa.
The PRR family genes encode response regulators that belong to aubiquitous two-component signal transduction system in plants. They can transmit information on photoperiod to plants and determine flowering time. Gao, et al. [16] supported the opinion that PRR37 expression correlates with rice grain yield under different photoperiod conditions. This effect was not determined for alfalfa and MsPRR7; however, lowering time is an important trait for alfalfa, and often late-maturing cultivars have higher yields.
Existing studies of PRRs have mainly focused on their regulatory role in the flowering pathway, [23] Showed that red light could inhibit growth of hypocotyls in plants overexpressing PRR5. Similar phenotypes were identified in plants overexpressing PRR1and PRR9 [24,25], however, there have been no comparable studies on plants overexpressing PRR7. Based on a genome-wide association scan (GWAS) in photoperiod-dependent regulation, Ppd-H1, a potentially homologous gene to PRR7, is considered to be one of the determinants of barley leaf morphology [20]. Leaf morphology is an important agronomic trait, which can influence plant photosynthesis and ultimately, biomass. Although we have identified expression patterns of MsPRR7 in alfalfa, whether this gene responses to the red light, could affect alfalfa morphology or how many PRR genes exist in this species are unknown. Future research in these areas might enhance our understanding of circadian clock-related pathways and be helpful in the breeding of alfalfa.
- Srikanth A, Schmid M: Regulation of flowering time: all roads lead to Rome, Cell Mol Life Sci. 2011; 68: 2013-2037. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/21611891
- Pittendrigh CS: Circadian rhythms and the circadian organization of living systems, Cold Spring Harb SympQuant Biol. 1960; 25: 159-184. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/13736116
- Pittendrigh CS: Circadian surfaces and the diversity of possible roles of circadian organization in photoperiodic induction, Proc Natl Acad Sci USA. 1972; 69: 2734-2737. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC427028/
- Wang ZY, Tobin EM: Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression, Cell. 1998; 93: 1207-1217. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/9657153
- Alabadí D, Yanovsky MJ, Más P, Harmer SL, Kay SA. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis, Curr Biol. 2002; 12: 757-761. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/12007421
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, et al. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell. 2002; 2: 629-641. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/12015970
- Mizuno T, Nakamichi N. Pseudo-Response Regulators (PRRs) or True Oscillator Components (TOCs). Plant Cell Physiol. 2005; 46: 677-685. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/15767264
- Matsushika A, Makino S, Kojima M, Mizuno T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell physiol. 2000; 41: 1002-1012. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/11100772
- Murakami M, Ashikari M, Miura K. The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell physiol. 2003; 44: 1229-1236.
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. Plos Genet. 2008; 4: 14. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/18248097
- Hazen SP, Naef F, Quisel T. Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays, Genome Biol. 2009; 10: 17.
- Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, et al. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci USA. 2009; 106: 7251-7256. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/19359492
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol. 2005; 15: 47-54. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/15649364
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005; 46: 1175-1189.PubMed: https://www.ncbi.nlm.nih.gov/pubmed/15951566
- Kolmos E, Chow BY, Pruneda-Paz JL, Kay SA. HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc Natl Acad Sci USA. 2014; 111: 16172-16177. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/25352668
- Gao H, Jin M, Zheng XM, Chen J, Yuan D, et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice, Proc Natl Acad Sci USA. Correction. 2014; 111: 16337–16342. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/25378698
- Yang S, Murphy RL, Morishige DT, Klein PE, Rooney WL, et al. Sorghum phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and SbGHD7, repressors of SbEHD1, SbCN8 and SbCN12. Plos One. 2014; 9: 105352. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4133345/
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005; 310: 1031-1034. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/16284181
- Zhang W, Zhao G, Gao L, Kong X, Guo Z, et al. Functional studies of heading date-related geneTaPRR73, a paralog ofPpd1in common wheat. Front Plant Sci. 2016; 7: 772. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/27313595
- Digel B, Tavakol E, Verderio G, Tondelli A, Xu X, et al.: Photoperiod-H1 (Ppd-H1) controls leaf size. Plant Physiol. 2016; 172: 405-415. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/27457126
- Jung C, Müller AE. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009; 14: 563-573. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/19716745
- Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nature Protocols. 2006; 1: 2019-2025. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/17487191
- Sato E, Nakamichi N, Yamashino T, Mizuno T. Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol. 2002; 43: 1374-1385. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/12461138
- Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, et al. Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 2000; 41: 791-803. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/10945350
- Matsushika A, Imamura A, Yamashino T, Mizuno T. Aberrant expression of the light-inducible and circadian-regulated APRR9 Gene belonging to the circadian-associated APRR1/TOC1 quintet results in the phenotype of early flowering in Arabidopsis thaliana, Plant Cell Physiol. 2002; 43: 833-843. PubMed: https://www.ncbi.nlm.nih.gov/pubmed/12198185