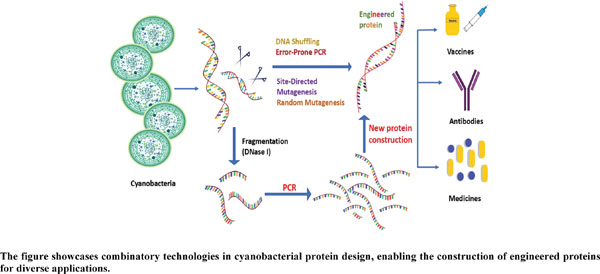
Abstract
Cyanobacteria have emerged as a microbial cell factory to produce a variety of bioproducts, including peptides and proteins. Cyanobacteria stand out among other organisms due to their photoautotrophic metabolism and ability to produce a wide range of metabolites. As photoautotrophic hosts can produce industrial compounds and proteins by using minimal resources such as sunlight, atmospheric carbon dioxide, and fewer nutrients, cyanobacteria are cost-effective industrial hosts. Therefore, the use of protein engineering tools for rational protein design, and the desired modification of enzyme activity has become a desirable undertaking in cyanobacterial biology. Protein engineering can improve their biological functions as well as the stability of their intracellular proteins. This review aims to highlight the success of protein engineering in the direction of cyanobacterial biotechnology and outlines the emerging technologies, current challenges, and prospects of protein engineering in cyanobacterial biotechnology.
Keywords: Cyanobacteria, protein engineering, directed evolution, site-directed mutagenesis, computer-aided protein design, cyanobacterial biotechnology.
[http://dx.doi.org/10.1093/bfgp/ell021] [PMID: 16772275]
[http://dx.doi.org/10.1007/s10811-010-9626-3]
[http://dx.doi.org/10.1016/j.jacme.2015.02.004]
[http://dx.doi.org/10.1021/acssynbio.0c00610] [PMID: 33684287]
[http://dx.doi.org/10.1186/s13068-019-1623-4] [PMID: 31890019]
[http://dx.doi.org/10.1007/s00253-021-11707-y] [PMID: 34882254]
[http://dx.doi.org/10.1038/s41598-017-08641-3] [PMID: 28827777]
[http://dx.doi.org/10.3390/molecules26010247] [PMID: 33466486]
[http://dx.doi.org/10.2174/0929866529666220314111105] [PMID: 35289251]
[http://dx.doi.org/10.1002/cbic.201500280] [PMID: 26661585]
[http://dx.doi.org/10.3109/07388551.2012.716810] [PMID: 22985113]
[http://dx.doi.org/10.1104/pp.18.01534] [PMID: 30626612]
[http://dx.doi.org/10.1038/nrg3927] [PMID: 26055155]
[http://dx.doi.org/10.1016/j.sbi.2021.11.001] [PMID: 34922207]
[http://dx.doi.org/10.1016/j.mib.2006.03.003] [PMID: 16621678]
[http://dx.doi.org/10.1007/s10529-015-1905-2] [PMID: 26153348]
[http://dx.doi.org/10.1021/ja300209u] [PMID: 22404648]
[http://dx.doi.org/10.1038/nbt1097] [PMID: 15908942]
[http://dx.doi.org/10.1002/anie.200901488] [PMID: 19591176]
[http://dx.doi.org/10.1021/ja9030215] [PMID: 19645470]
[http://dx.doi.org/10.1093/protein/gzs097] [PMID: 23223807]
[http://dx.doi.org/10.1007/s00216-004-3020-4] [PMID: 15714301]
[http://dx.doi.org/10.1038/nbt.2812] [PMID: 24509759]
[http://dx.doi.org/10.1016/B978-044452114-9/50004-9]
[http://dx.doi.org/10.1021/acs.chemrev.1c00260] [PMID: 34297541]
[http://dx.doi.org/10.1016/j.biortech.2018.06.057] [PMID: 29957289]
[http://dx.doi.org/10.1016/j.molp.2018.12.007] [PMID: 30580039]
[http://dx.doi.org/10.3390/md9091607] [PMID: 22131961]
[http://dx.doi.org/10.1186/1754-6834-7-69] [PMID: 24920957]
[http://dx.doi.org/10.1016/j.algal.2012.03.001]
[http://dx.doi.org/10.1038/s41598-017-17831-y] [PMID: 29235513]
[http://dx.doi.org/10.1093/nar/gnh028] [PMID: 14872057]
[http://dx.doi.org/10.1080/09168451.2019.1687281] [PMID: 31690227]
[http://dx.doi.org/10.1021/acs.jafc.9b06254] [PMID: 31755253]
[http://dx.doi.org/10.1016/B978-0-444-64114-4.00012-1]
[http://dx.doi.org/10.1073/pnas.0901522106] [PMID: 19528653]
[http://dx.doi.org/10.1002/pro.2202] [PMID: 23225662]
[http://dx.doi.org/10.3390/polym10080910] [PMID: 30960835]
[http://dx.doi.org/10.1016/S0141-8130(02)00010-7] [PMID: 11911900]
[http://dx.doi.org/10.1016/j.biortech.2016.05.014] [PMID: 27213577]
[http://dx.doi.org/10.1186/1472-6750-7-25]
[http://dx.doi.org/10.1093/nar/gnh110] [PMID: 15304544]
[http://dx.doi.org/10.1021/acssynbio.5b00018] [PMID: 25901796]
[http://dx.doi.org/10.1007/s00253-010-2603-6] [PMID: 20422179]
[http://dx.doi.org/10.1128/AEM.01056-10] [PMID: 20709836]
[http://dx.doi.org/10.1016/j.ymben.2015.09.010] [PMID: 26410450]
[http://dx.doi.org/10.3389/fpls.2020.00237] [PMID: 32194609]
[http://dx.doi.org/10.1021/acssynbio.7b00214] [PMID: 28858481]
[http://dx.doi.org/10.1021/acs.jafc.7b00002] [PMID: 28128561]
[http://dx.doi.org/10.1021/acssynbio.7b00431] [PMID: 29397685]
[http://dx.doi.org/10.1002/bit.26988] [PMID: 30963538]
[http://dx.doi.org/10.1007/s10811-020-02186-1]
[http://dx.doi.org/10.1016/j.ymben.2018.07.005] [PMID: 30017797]
[http://dx.doi.org/10.1073/pnas.0707148104] [PMID: 18025466]
[http://dx.doi.org/10.1007/s00253-018-9093-3] [PMID: 29802477]
[http://dx.doi.org/10.1016/j.algal.2020.102158]
[http://dx.doi.org/10.1080/07391102.2020.1821782] [PMID: 32990187]
[http://dx.doi.org/10.1002/pmic.201500043] [PMID: 26010509]
[http://dx.doi.org/10.1002/cbic.201402128]
[http://dx.doi.org/10.1093/bioinformatics/btaa1071] [PMID: 33367682]
[http://dx.doi.org/10.1016/j.abb.2022.109196] [PMID: 35339426]
[http://dx.doi.org/10.1093/protein/gzu035] [PMID: 25169579]
[http://dx.doi.org/10.1016/j.jbi.2022.104016] [PMID: 35143999]
[http://dx.doi.org/10.1002/anie.200500767] [PMID: 15929154]
[http://dx.doi.org/10.1093/pcp/pcu075] [PMID: 24904028]
[http://dx.doi.org/10.1016/S0006-291X(02)00434-5] [PMID: 12054744]
[http://dx.doi.org/10.1016/j.tibtech.2010.12.003] [PMID: 21211860]
[http://dx.doi.org/10.1046/j.1365-2958.2003.03825.x] [PMID: 14651639]
[http://dx.doi.org/10.1073/pnas.1701083114] [PMID: 28739907]
[http://dx.doi.org/10.3390/toxins13100711] [PMID: 34679003]
[http://dx.doi.org/10.3389/fimmu.2021.626616] [PMID: 34025638]
[http://dx.doi.org/10.1007/978-1-4939-6887-9_14]
[http://dx.doi.org/10.1006/abio.1994.1060] [PMID: 8179197]
[http://dx.doi.org/10.1016/j.talanta.2015.12.028] [PMID: 26838417]
[http://dx.doi.org/10.1016/j.jbiotec.2005.08.006] [PMID: 16150509]