Abstract
Introduction: Adverse drug reactions (ADRs) are responsible for the failure of a significant portion of investigative drugs trials and the major reason for the withdrawal of drugs from clinical research. A number of ADRs are caused by the (undesired) interaction of drugs with key proteins involved in normal biological processes. Identification of these ADR-related proteins facilitates the design of drugs with fewer adverse effects by rationally avoiding unwanted interaction with these proteins.
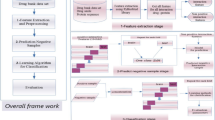
Method: This work explores the use of a statistical learning method, support vector machines (SVMs), for the identification of potential ADR-related proteins. A SVM classification system was trained and tested by using 759 ADR-related proteins of different species and 2280 non-ADR-related proteins.
Results:93.9% of the ADR-related proteins and 98.2% of non-ADR-related proteins were correctly classified.
Discussion: The SVM is potentially useful for facilitating the identification of ADR-related proteins. The development of methods to identify ADR indications of ADR-related proteins are progressing well, an example of which is the web-based ADR-related protein prediction tool SVMDART, which can be accessed at http://jing.cz3.nus.edu.sg/cgi-bin/dart.cgi.


Similar content being viewed by others
References
van de Waterbeemd H, Gifford E. ADMET in silico modelling: towards prediction paradise? Nat Rev Drug Discov 2003 Mar; 2 (3): 192–204
Johnson DE, Wolfgang GH. Predicting human safety: screening and computational approaches. Drug Discov Today 2000 Oct 1; 5 (10): 445–54
Kennedy T. Managing the drug discovery/development interface. Drug Discov Today 1997; 2 (10): 436–44
Rockey WM, Elcock AH. Progress toward virtual screening for drug side effects. Proteins 2002 Sep 1; 48 (4): 664–71
Greene N. Computer systems for the prediction of toxicity: an update. Adv Drug Deliv Rev 2002 Mar 31; 54 (3): 417–31
Barratt MD, Rodford RA. The computational prediction of toxicity. Curr Opin Chem Biol 2001 Aug; 5 (4): 383–8
Durham SK, Pearl GM. Computational methods to predict drug safety liabilities. Curr Opin Drug Discov Devel 2001 Jan; 4 (1): 110–5
Pumford NR, Halmes NC. Protein targets of xenobiotic reactive intermediates. Annu Rev Pharmacol Toxicol 1997; 37: 91–117
Wallace KB, Starkov AA. Mitochondrial targets of drug toxicity. Annu Rev Pharmacol Toxicol 2000; 40: 353–88
Rang HP, Dale MM, Ritter JM. Pharmacology. 4th ed. New York: Churchill Livingstone, 1999
Klaassen CD. Casarett & Doull’s toxicology: the basic science of poisons. 6th ed. New York: McGraw-Hill, 2001
Karchin R, Karplus K, Haussler D. Classifying G-protein coupled receptors with support vector machines. Bioinformatics 2002 Jan; 18 (1): 147–59
Cai CZ, Han LY, Ji ZL, et al. SVM-Prot: web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res 2003 Jul 1; 31 (13): 3692–7
Ji ZL, Han LY, Yap CW, et al. Drug adverse reaction target database (DART): proteins related to adverse drug reactions. Drug Saf 2003; 26 (10): 685–90
Drug adverse reaction target database [online]. Available from URL: http://bidd.nus.edu.sg/group/drt/dart.asp [Accessed 2005 Dec 6]
Bock JR, Gough DA. Predicting protein-protein interactions from primary structure. Bioinformatics 2001 May; 17 (5): 455–60
Ding CH, Dubchak I. Multi-class protein fold recognition using support vector machines and neural networks. Bioinformatics 2001 Apr; 17 (4): 349–58
Yuan Z, Burrage K, Mattick JS. Prediction of protein solvent accessibility using support vector machines. Proteins 2002 Aug 15; 48 (3): 566–70
Cai YD, Liu XJ, Xu XB, et al. Support vector machines for prediction of protein domain structural class. J Theor Biol 2003 Mar 7; 221 (1): 115–20
Bateman A, Birney E, Cerruti L, et al. The Pfam protein families database. Nucleic Acids Res 2002 Jan 1; 30 (1): 276–80
Vapnik V, Chervonenkis A. Theory of pattern recognition. Moscow: Nauka, 1974
Burges CJC. A tutorial on support vector machines for pattern recognition. Data Mining Knowl Disc 1998; 2: 121–67
Trotter MWB, Buxton BF, Holden SB. Support vector machine in combinatorial chemistry. Measure Control 2001; 34 (8): 235–9
Burbidge R, Trotter M, Buxton B, et al. Drug design by machine learning: support vector machines for pharmaceutical data analysis. Comput Chem 2001 Dec; 26 (1): 5–14
Cortes C, Vapnik V. Support vector networks. Machine Learning 1995; 20: 273–97
Baldi P, Brunak S, Chauvin Y, et al. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics 2000 May; 16 (5): 412–24
Acknowledgements
This work is supported by National Natural Science Foundation of China (NSFC grant no. 30400573). The authors do not have any potential conflicts of interest in relation to this article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ji, Z.L., Han, L.Y., Zheng, C.J. et al. Prediction of Putative Adverse Drug Reaction-Related Proteins from Primary Sequence by Support Vector Machines. Int J Pharm Med 19, 317–322 (2005). https://doi.org/10.2165/00124363-200519050-00009
Published:
Issue Date:
DOI: https://doi.org/10.2165/00124363-200519050-00009