
Titin mutation in circulatory tumor DNA is associated with efficacy to immune checkpoint blockade in advanced non-small cell lung cancer
Introduction
Immune checkpoint blockade (ICB) has substantially improved the clinical outcomes of advanced non-small-cell lung cancer (NSCLC) (1,2). However, only a minority of patients satisfactorily respond to ICB treatment. Currently, all clinically approved companion diagnostic tests for ICB immunotherapy, including immunohistochemistry of programmed cell death ligand-1 (PD-L1) expression (3), tumor mutational burden (TMB) (4), and microsatellite instability-high (MSI-H) (5), are based on formalin-fixed paraffin-embedded (FFPE) tumor tissue specimens. Despite the availability of these assays, the identification of responders to ICB immunotherapy is not always possible, partially due to the unavailability of archived tissue, refusal of re-biopsy, or lack of sufficient tumor cells in the specimen. Therefore, it is imperative to develop novel predictive biomarkers to expand the number of patients who may benefit from receiving ICB immunotherapy.
Circulatory tumor DNA (ctDNA) has been considered as a promising biomarker in guiding decision-making in numerous tumor types (6-8), and the utility of ctDNA sequencing is under active investigation in the field of ICB immunotherapy (9-11). Accumulating evidence supports that the estimation of TMB from blood ctDNA (bTMB) has application potential in cases where archival tissues are lacking or insufficient for routine tissue-based TMB assessment (12,13). However, the evaluation of bTMB requires sufficient input genomic DNA, which is not always available. Moreover, a cutoff for delineating high or low bTMB and an optimal sequencing pipeline to determine bTMB remain incompletely defined and need further investigation.
Recently, non-synonymous mutations in Titin (encoded by TTN), which is responsible for the passive elasticity of muscle tissue, have been related to higher TMB and improved ICB treatment efficacy (14). However, the usability of TTN mutation in ctDNA in selecting responders has not been investigated. Here, the predictive significance of TTN mutation was investigated in two independent cohorts of patients with NSCLC to determine whether TTN mutation in ctDNA can serve as a biomarker for sensitivity to ICB immunotherapies. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1118).
Methods
Study design and patients
This retrospective study enrolled 29 and 63 patients who were diagnosed with advanced non-small cell lung cancer (NSCLC) and were treated with ICB immunotherapy at Xinqiao Hospital (Army Medical University, Chongqing) and Shanghai Pulmonary Hospital & Thoracic Cancer Institute (Tongji University School of Medicine, Shanghai), respectively. Patients were enrolled from Jul 2017 to Feb 2019. Both males and females were enrolled. Patients older than 80 years were excluded from the study. Clinical response to ICB treatment was evaluated with RESCIST 1.1 criteria using radiological assessments. The median follow-up was 12 months (ranging from 3–21-month for all patients). Plasma from 5 mL of peripheral blood was obtained immediately before the first infusion of ICB treatment for all patients. Meanwhile, we performed TTN validation sequencing using peripheral blood and matched FFPE biopsies in additional 24 NSCLC cases. In these 24 NSCLC cases, matched samples from 6 patients were used for investigating the consistency between panel-based and whole-exome sequencing (WES); the remaining 18 cases were used for comparing blood-based and tissue-based sequencing of TTN gene.
Sample preparation and DNA isolation
Plasma was isolated from the blood via centrifugation at 1,600 ×g for 10 min and then at 16,000 ×g for 10 min. Cell-free (cf) DNA was extracted from 3 to 5 mL of plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen) according to the manufacturer’s instructions. Tumor FFPE DNA was extracted from 5 to 10 sections (5–10 µm thick) using the QIAamp DNA FFPE Tissue Kit (Qiagen), and genomic DNA (gDNA) was extracted from white blood cells (WBCs) using the Blood Genomic DNA Mini Kit (CWBiotech).
Target capture and sequencing
A custom-designed panel with DNA probes covering the entire coding sequence of TTN was developed on the BOKE TargetCap™ platform. For each sample, 20–100 ng cfDNA or 200–500 ng FFPE DNA and 500 ng gDNA was used for library preparation and quantification according to KAPA HyperPrep protocols (R). Pools of four libraries were hybridized to the capture panel at 65 °C for 16 h. Washing, recovery, and amplification were conducted sequentially according to the manufacturer’s standard procedures (BOKE). Next, the libraries were purified using AMPure XP (Beckman) and quantified using the Qubit™ dsDNA HS Assay Kit (Thermo Fisher). xGen Exome Research Panel v1.0 (Integrated DNA Technologies) was used to generate WES data for tumor tissue DNA, according to the manufacturer’s standard procedures. The libraries were sequenced on an Illumina Novoseq6000 instrument in PE150 mode (Illumina).
Quality control and variant calling
Adapter sequences were trimmed from the raw reads using Trimmomatic (15). The reads were then aligned with the human reference genome (hg19) using BWA (16). Duplicated reads were removed using Picard (http://broadinstitute.github.io/picard/). Mapped reads were realigned to the genome using the Genome Analysis Tool Kit (17). Somatic and germline mutations were observed using Mutect2 and GATK’s HaplotypeCaller (3.7) with a paired workflow and GATK, respectively. Variants were annotated using ANNOVAR (18) and an in-house developed code. An in-house script was used to verify the identity concordance of paired samples. Somatic copy number alterations were detected using GATK (17).
Germline variant filtering
Germline variants determined by GATK in WBC samples were first filtered using a threshold of minimum coverage of 50× and an allele frequency of >30%. Variants outside coding regions and synonymous mutations annotated with ANNOVAR (18) were filtered out. Further, variants with a minor allele frequency of >0.1% in the ExAC database were considered less functional and ignored in downstream analysis.
Somatic variant filtering
Somatic mutations detected in cfDNA samples were filtered according to the following parameters: (I) ≥4 allele reads support the mutation; (II) >0.5% allele frequency; (III) <4 supporting reads in the WBC control; (IV) mutation frequency is 5 times higher than in the WBC control; (V) mutations do not occur more than two times in the PoN; (VI) no significant strand bias (GATK parameter FS >60 for single nucleotide polymorphisms and FS >200 for indels). Similar filtering rules were applied for somatic mutations from FFPE samples, except that allele frequency had to be >5% and mutation frequency had to be 8 times higher than in the WBC control. Mutations within non-coding regions and synonymous mutations were removed for downstream analysis.
Statistics
The number of mutations in TTN was defined as the sum of missense, nonsense, and indels in the coding region of TTN. The correlation between plasma-based panel-assessed TTN mutation and FFPE-based WES-assessed TMB was determined via the Pearson correlation coefficient. Discrete variables (partial response, stable disease, or disease progression) were summarized as percentages. Kaplan-Meier curves and the log-rank test were used for survival analysis. The hazard ratio (HR) and 95% confidence interval (CI) were estimated using a Cox proportional hazards regression model. P<0.05 was considered statistically significant. Basic statistical analysis was performed using Excel in Office 365 or GraphPad Prism software, version 5.0 (GraphPad Software Inc.).
Availability of data and materials
All raw sequencing data are accessible upon request to the corresponding authors. The clinical characteristic and survival outcomes can be found in the supplementary tables.
Study approval
The trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board of XinQiao Hospital, Army Medical University, Chongqing (Identifier: 2017YD104-01) and Shanghai Pulmonary Hospital & Thoracic Cancer Institute, Tongji University School of Medicine, Shanghai (Identifier: 82072568). Individual consent for this retrospective analysis was waived.
Results
Development of a sequencing panel for detecting TTN mutation in ctDNA
We developed a customized panel of oligonucleotide probes targeting the TTN gene (Table S1) to determine the mutation status of TTN in ctDNA via high-throughput sequencing. This customized panel was designed to cover the whole coding sequencing (CDS) of TTN. By sequencing tumor tissues from six NSCLC patients, technical validation of the sequencing panel confirmed a sufficient depth of coverage of the sequencing pipeline (Figure 1A, Figure S1), supporting the reliability of the assay to identify mutated sites in the TTN exons. Subsequently, we investigated the effectiveness of the TTN panel by comparing the results with parallel WES results based on tumor tissues from six treatment-naïve patients with NSCLC. The concordance between the panel-based and WES-based TTN mutations in tumor tissue are summarized in Figure 1B. The mutation status was consistent between panel-based and WES-based sequencing results, measured either by the total number of mutated sites (Figure 1B, left panel) or by the allele frequency of each mutated site (Figure 1B, right panel) in the TTN coding region. Furthermore, a positive correlation was found between the panel-based total number of mutated sites and WES-based TMB (Figure 1C, R2=0.9027, **P=0.0037), which has been previously established in an analysis based on The Cancer Genome Atlas (TCGA) (14). Finally, the performance of TTN panel-based sequencing of ctDNA was examined using tumor tissue samples and peripheral blood samples from an additional 18 patients with NSCLC (Figure 1D). In brief, for these 18 cases, all ten mutation sites detected via panel-based ctDNA sequencing were successfully recapitulated using tumor tissue-based sequencing. In contrast, 14 of 16 ctDNA TTN mutation-negative patients (87.5%) were verified via tumor tissue-based sequencing, suggesting a reasonable false negativity rate of our pipeline in detecting TTN mutation in ctDNA.

TTN mutation in ctDNA predicts responsiveness to ICB treatment in patients with NSCLC
To investigate whether TTN mutation in ctDNA allows identifying patients with a great likelihood to benefit from ICB immunotherapy, two independent cohorts of patients who were diagnosed with advanced NSCLC and treated with ICB immunotherapy were enrolled in this retrospective analysis (29 patients in cohort 1 and 63 patients in cohort 2 (Tables S2,S3). The overall positivity rate of TTN mutation in ctDNA was 28.26% (26/92) for all the patients enrolled (10/29 or 34.5% for cohort 1 and 16/63 or 25.4% for cohort 2), which was comparable to the reported frequency of TTN mutation occurrence (14) (2,928/9,654, 30.33%) in TCGA database.
Compared to TTN mutation-negative patients (absence of detectable TTN mutation by panel-based ctDNA sequencing), TTN mutation-positive patients (with detectable TTN mutation) demonstrated a superior objective response to ICB treatment (Figure 2A,B,C). The objective response rates for the TTN mutation-positive and negative patients were 40.0% (4/10) and 15.8% (3/19) for cohort 1, 50.0% (8/16) and 23.4% (11/47) for cohort 2, and 46.2% (12/26) and 21.2% (14/66) for the combined cohorts, respectively. In addition, TTN mutation-positive patients were associated with a durable progression-free survival (PFS) compared to TTN mutation-negative patients (Figure 2D,E,F). The median PFS was 212 days for positive patients and 113 days for the negative patients (Figure 2D, *P=0.0335; HR =0.40; 95% CI, 0.18–0.90) in cohort 1. In cohort 2, the median PFS was 334.5 and 147 days for patients with and without detectable TTN mutation, respectively (Figure 2E, P=0.0916; HR =0.52; 95%, 0.27–1.01). Overall, the median PFS for the positive and negative patients was 212 and 135 days, respectively, (Figure 2F, *P=0.0154; HR =0.49; 95% CI, 0.29–0.82) in the combined cohort. When designating patients achieving CR/PR or SD for more than 6 months as responders, we found that the proportion of responders were significantly higher in TTN mutant patients in cohorts 1, 2 and the combined cohort (Figure S2, *P=0.021 for cohort 1, *P=0.041 for cohort 2 and *P=0.002 for combined cohort). Taken together, these observations suggest that the ctDNA TTN mutation may be a promising biomarker to identify potential responders to ICB immunotherapy.
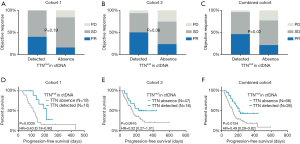
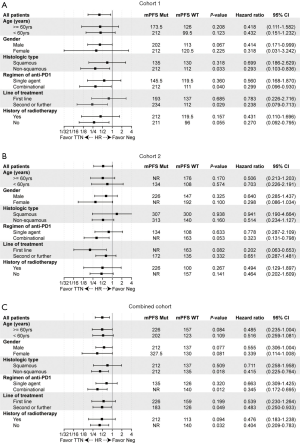
In the univariate Cox proportional hazards regression model, only TTN mutation status was significantly associated with PFS (Table S4, HR =0.40; 95% CI, 0.18–0.90; *P=0.03) in cohort 1. In cohort 2, the pathologic type (squamous vs. non-squamous carcinoma), treatment regimen (single agents or combinational regimen), and TTN mutation status tended to be significantly associated (with P≤0.15) with survival outcomes (Table S4, pathological type, HR =0.51, 95% CI, 0.25-1.05, P=0.12; regimen of treatment, HR =1.71, 95% CI, 0.87-3.38, P=0.09; TTN mutation, HR =0.52, 95% CI, 0.27-1.02, P=0.09). By examining the combined analysis, the regimen and TTN mutation status remained significantly associated with PFS with ICB therapy (Table S4, regimen of treatment, HR =1.64, 95% CI, 0.97–2.58, *P=0.05; TTN mutation, HR =0.49, 95% CI, 0.29–0.82, *P=0.02). Variables with P value less than 0.15 were included in the multivariate Cox proportional hazards regression model. The mutation status showed the strongest association with PFS in each cohort (cohort 1, HR =0.39, 95% CI, 0.15–1.00, *P=0.05; cohort 2, HR =0.56, 95% CI, 0.25–1.23, P=0.11; combined cohort, HR =0.47, 95% CI, 0.26–0.85, *P=0.01).
Predictive significance of TTN mutation in subgroups
We further explored the clinical relevance of the TTN mutation in different subgroups of patients (Figure 3). This stratified analysis showed that patients with positive TTN mutation tended to benefit more from the improved benefits of ICB treatment in most stratified subgroups. The predictive significance seemed more informative in female patients, patients with non-squamous NSCLC, or patients who received a combinational regimen (Figure 3C). Although the effects for most stratifications were not statistically significant, probably due to the small population size, the numerical trends of better survival for TTN mutation-positive patients were observed in nearly all the subgroups of each cohort.
Discussion
In the past decades, tumor-associated neoantigens, which arise from non-synonymous somatic mutations, have been recognized as the main targets of cytotoxic T lymphocytes in the tumor microenvironment (19,20). These tumor cell-recognizing T cells can be potentiated by the administration of ICB agents, either by reinvigorating their exhausted cytolytic activity (21,22) or by depleting immunoregulatory cell types (23). The quantity and quality of the neoantigen repertoire significantly contribute to clinical response to ICB treatment in patients with NSCLC (24). There is currently no consensus on how to extract biologically meaningful neoantigens from massive genomic data (25). Due to this shortcoming, TMB is still the best alternative measure of neoantigens. However, accurate estimation of the TMB remains a technical challenge and is not always possible in clinical practice. In the present study, we developed a non-invasive, cost-effective, and easy-to-accomplish method to detect TTN mutation in peripheral blood samples. Using this method, we demonstrated that the detectable TTN mutation in ctDNA was associated with significant benefit from ICB treatment in patients with advanced NSCLC.
In addition to MSI-H (5), KRAS/TP53 (26), POLE (27), or STK11 (28), high TMB (≥10 mutations/MB) has been approved as a biomarker for predicting responsiveness to ICB immunotherapy in patients with unresectable or metastatic disease. In the KEYNOTE-158 trial, the objective response rate for TMB-high patients was 29%, including 4% of patients with complete response and 25% of patients with partial response. As for response duration, 57% of patients exhibited a response duration of more than one year, and half of the patients experienced response of more than 2 years. These data supported that high TMB is a practical predictive biomarker. However, there are two major issues with the clinical application of high TMB. First, high TMB was determined using an FDA-approved companion diagnostic test, FoundationOneCDx, which counts non-synonymous mutations in tumor tissues, whereas the implementation of TMB assessment is limited by the availability of nucleic acid materials or the heterogeneity of the tumor microenvironment (29). Second, as a continuous variable, there is no absolute cutoff value to define high TMB, making it difficult for clinicians to make decisions in some cases. The cutoff of 10 mutations/MB to define high TMB was selected rather arbitrarily. Based on this definition, only 15.89% of patients (120 of 755 patients with evaluable TMB) were classified as TMB-high, and inevitably, a considerable portion of the patients must have missed the opportunity to receive ICB immunotherapy. Therefore, it is necessary to improve the strategy for selecting optimal candidates who will benefit from ICB therapy.
ctDNA sequencing is actively and widely used in immuno-oncology (10) and has also been introduced in routine clinical practice. An advantage of the technique is that dynamic monitoring of treatment can be easily achieved using ctDNA sequencing without the limitation of tissue availability (30,31). Using ctDNA-assessed bTMB as a surrogate measurement of tissue TMB has been long investigated in the stratification of patients suitable for ICB therapy (32). However, the protocol and sequencing panel used in bTMB assessment is not fully standardized yet, and its robustness as a predictive biomarker needs to be validated in prospective clinical trials. Most importantly, in contrast to tumor tissue-based TMB estimation, the ctDNA-based TMB estimation cannot effectively avoid interference from germline mutations; thus, it leads to a bias in calculations, especially in patients with a relatively low mutation count. Further, the cutoff for defining the TMB as “TMB-high” needs to be determined. Thus, a ctDNA-based assay with binary readout may provide an alternative solution to overcome the adverse impacts of unreliable tissue supply and technical defects.
Titin is a structural protein found in striated muscles (33). In our previous study, by performing a genome-scale screening, we found that the length of the coding regions for each gene was positively associated with mutation frequencies of the gene in solid tumors (14). Coincidentally, TTN is the gene with the longest coding sequence in the human genome. Moreover, the highest number of mutations was found in the coding regions of TTN, suggesting a high possibility of accumulating somatic mutations in genes with longer coding regions. Furthermore, we demonstrated that tumor-tissue TTN mutation was associated with high TMB, high objective response rate, and durable survival in seven publicly accessible cohorts. These pilot analyses led us to investigate whether TTN mutation in ctDNA could predict the clinical efficacy of ICB therapy in patients with advanced NSCLC.
Our study suggests the value of ctDNA TTN mutation in predicting clinical response in patients with advanced NSCLC. The binary readout (positive/negative) of our assay overcomes the difficulty in identifying a reasonable threshold for selecting patients eligible for ICB immunotherapy. The clinical significance of TTN mutation was confirmed in two independent cohorts of patients receiving ICBs. However, there were three major limitations to our study. (I) Due to limited tissue availability and the high cost of sequencing, the TTN panel for ctDNA sequencing was validated in the paired tumor biopsy and peripheral blood samples of only 18 patients. Hence, the consistency between tumor-based and ctDNA-based TTN mutation needs to be confirmed in a larger cohort. (II) The correlation between ctDNA-based TMB and the number of mutations in TTN CDS should be validated in a larger cohort. (III) The association between improved survival, objective response, and detectable TTN mutation in ctDNA was validated in the two cohorts in a retrospective manner. The performance of PD-L1 expression, MSS or MSI-H status or other established ICB-predictive biomarkers should be compared to the performance of ctDNA TTN mutation in identifying responders to ICB treatment. (IV) TTN is not routinely included in the commercially available sequencing panels. Therefore, publicly accessible data could not be used to test the performance of our biomarker in mixed solid tumor types.
Conclusions
In conclusion, we used ctDNA sequencing to demonstrate that the TTN mutation has the potential to serve as an easy-to-apply predictive biomarker for ICB immunotherapy in patients with advanced NSCLC. However, prospective clinical trials with multiple solid tumor types are required to validate the universal application and robustness of this biomarker.
Acknowledgments
Funding: This work was supported by the National Nature Science Foundation of China (The National Science Fund for Distinguished Young Scholars, 81925030; Foundation of Innovative Research Group, 81821003) to B Zhu, and by The Second Affiliated Hospital to Army Medical University (No.2018XLC1008) to B Zhu.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1118
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1118
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1118). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board of Xinqiao Hospital, Army Medical University, Chongqing (Identifier: 2017YD104-01) and Shanghai Pulmonary Hospital & Thoracic Cancer Institute, Tongji University School of Medicine, Shanghai (Identifier: 82072568). Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Suresh K, Naidoo J, Lin CT, et al. Immune Checkpoint Immunotherapy for Non-Small Cell Lung Cancer: Benefits and Pulmonary Toxicities. Chest 2018;154:1416-23. [Crossref] [PubMed]
- Brody R, Zhang Y, Ballas M, et al. PD-L1 expression in advanced NSCLC: Insights into risk stratification and treatment selection from a systematic literature review. Lung Cancer 2017;112:200-15. [Crossref] [PubMed]
- Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol 2019;16:341-55. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Schumacher TN, Scheper W. A liquid biopsy for cancer immunotherapy. Nat Med 2016;22:340-1. [Crossref] [PubMed]
- Gros A, Parkhurst MR, Tran E, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med 2016;22:433-8. [Crossref] [PubMed]
- Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov 2014;4:650-61. [Crossref] [PubMed]
- Snyder A, Morrissey MP, Hellmann MD. Use of Circulating Tumor DNA for Cancer Immunotherapy. Clin Cancer Res 2019;25:6909-15. [Crossref] [PubMed]
- Cabel L, Proudhon C, Romano E, et al. Clinical potential of circulating tumour DNA in patients receiving anticancer immunotherapy. Nat Rev Clin Oncol 2018;15:639-50. [Crossref] [PubMed]
- Goldberg SB, Narayan A, Kole AJ, et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin Cancer Res 2018;24:1872-80. [Crossref] [PubMed]
- Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441-8. [Crossref] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Jia Q, Wang J, He N, et al. Titin mutation associated with responsiveness to checkpoint blockades in solid tumors. JCI Insight 2019;4:e127901 [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164 [Crossref] [PubMed]
- Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015;348:69-74. [Crossref] [PubMed]
- Chen F, Zou Z, Du J, et al. Neoantigen identification strategies enable personalized immunotherapy in refractory solid tumors. J Clin Invest 2019;129:2056-70. [Crossref] [PubMed]
- Park J, Kwon M, Kim KH, et al. Immune Checkpoint Inhibitor-induced Reinvigoration of Tumor-infiltrating CD8(+) T Cells is Determined by Their Differentiation Status in Glioblastoma. Clin Cancer Res 2019;25:2549-59. [Crossref] [PubMed]
- Twyman-Saint Victor C, Rech AJ, Maity A, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 2015;520:373-7. [Crossref] [PubMed]
- Du X, Tang F, Liu M, et al. A reappraisal of CTLA-4 checkpoint blockade in cancer immunotherapy. Cell Res 2018;28:416-32. [Crossref] [PubMed]
- McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463-9. [Crossref] [PubMed]
- Richters MM, Xia H, Campbell KM, et al. Best practices for bioinformatic characterization of neoantigens for clinical utility. Genome Med 2019;11:56. [Crossref] [PubMed]
- Dong ZY, Zhong WZ, Zhang XC, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res 2017;23:3012-24. [Crossref] [PubMed]
- Mehnert JM, Panda A, Zhong H, et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest 2016;126:2334-40. [Crossref] [PubMed]
- Kwack WG, Shin SY, Lee SH. Primary Resistance to Immune Checkpoint Blockade in an STK11/TP53/KRAS-Mutant Lung Adenocarcinoma with High PD-L1 Expression. Onco Targets Ther 2020;13:8901-5. [Crossref] [PubMed]
- Jia Q, Wu W, Wang Y, et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun 2018;9:5361. [Crossref] [PubMed]
- Chen PL, Roh W, Reuben A, et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov 2016;6:827-37. [Crossref] [PubMed]
- Jia Q, Chiu L, Wu S, et al. Tracking Neoantigens by Personalized Circulating Tumor DNA Sequencing during Checkpoint Blockade Immunotherapy in Non-Small Cell Lung Cancer. Adv Sci (Weinh) 2020;7:1903410 [Crossref] [PubMed]
- Wang Z, Duan J, Cai S, et al. Assessment of Blood Tumor Mutational Burden as a Potential Biomarker for Immunotherapy in Patients With Non-Small Cell Lung Cancer With Use of a Next-Generation Sequencing Cancer Gene Panel. JAMA Oncol 2019;5:696-702. [Crossref] [PubMed]
- Linke WA. Titin Gene and Protein Functions in Passive and Active Muscle. Annu Rev Physiol 2018;80:389-411. [Crossref] [PubMed]


