
MicroRNA expression profile in the spinal cord injured rat neurogenic bladder by next-generation sequencing
Introduction
It has been estimated that 250,000 to 500,000 people worldwide are suffering from spinal cord injury and approximately 81% of these patients experience different levels of neurogenic bladder (NB) which is one of the leading causes of their morbidity (1,2). Specifically, detrusor overactivity and detrusor-sphincter dyssynergia are typical NB urodynamic findings, and are diagnosed in 95% and 68% of spinal cord-injured (SCI) patients, respectively (3). These involuntary bladder contractions could result in urinary incontinence episodes during the storage phase, inefficient urine voiding, and high residual volume during urine voiding, potentially leading to upper urinary tract damage and substantially impacting the health-related quality of life (4). At the same time, the currently available treatment options are not satisfactory.
The exact mechanisms of NB secondary in spinal cord injury have not yet been identified. It is postulated that C fibers mediated new spinal reflex circuits and neurotrophic hormones like nerve growth factor participate in NB after spinal cord injury (5). In recent years, an increased mount of evidence has indicated that microRNAs (miRs) are essential regulators of most physiological and pathological events, which might also include the potential pathophysiology and treatment outcomes of NB. MiRs are non-coding RNAs, approximately 19–23 nucleotides (nt) in length. They participate in epigenetic post-transcriptional control of protein-coding gene expression primarily by reversible translational repression or mRNA destabilization/degradation and thereby inhibit protein synthesis (6).
Recently, Chermansky et al. reported that the elevated expression of miR-221 and miR-125b in detrusor overactivity in the bladder tissue of patients may predict their high risk for undergoing urinary retention following intradetrusor injection of onabotulinumtoxin-A (6). In addition, the combination of upregulated miR-98-5p and downregulated miR-139-5p in the plasm of patients’ overactive bladder (OAB) was found to be a useful biomarker for OAB. However, no correlation was determined between the levels of miRs and OAB symptom score (7). Meanwhile, the above research only concentrated on finding the expression patterns of miRNA particularly in OAB patients. To our knowledge, no studies have comprehensively investigated miR expressions in the SCI rat NB.
Therefore, this study aimed to explore the miR expression profile in the SCI rat bladder by next-generation sequencing (NGS), which may yield molecular targets for NB. Subsequently, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) bioinformatical analyses were also performed to investigate the functions of these miRs. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tau-20-415).
Methods
Ethical approval
All experimental procedures were implemented in compliance with the National Institute of Health Guidelines for the Care and Use of Laboratory Animals (2) and approved by the Institutional Animal Care and Use Committee of Xuanwu Hospital Capital Medical University (No. 20190128).
SCI rat model
Adult female Wistar rats weighing 200–250 g (Beijing Charles River Laboratories Animal Technology Co., Ltd., Beijing, China) were used in this study, as their urethras are shorter and more conducive to bladder evacuation via abdominal compression. The animals were maintained at 20–26 °C and 30–70% relative humidity under a 12-h light/dark cycle with ad libitum access to food and water. The spinal cord was transected at the T9–10 in rats. Under 3% enflurane inhalation in the rats, Th10 laminectomy was performed, and the dura was sharply transected. The spinal cord was completely severed at T10, and Gelfoam (Ethicon) was placed between the cut ends to aid in hemostasis and to prevent the cut ends from healing. The muscle layer and skin were separately sutured. The rats received ampicillin sodium (100 mg/kg intramuscularly) for 5 days after operation. Bladders were emptied 3 times daily by abdominal compression until reflex voiding returned, and once a day afterward. The normal rats were used as the normal control (NC) group.
Bladder collection
Before bladder collection, 15 female Wistar rats that underwent spinal cord transection at T9–10 were randomly divided into SCI-1, SCI-2, and SCI-3 groups (n=5 for each group), and their bladder tissues were collected 1, 2, and 4 weeks after spinal cord transection, respectively. The rats were anesthetized with 3% enflurane inhalation, and a midline laparotomy was performed in the lower abdomen to expose the bladder. Each bladder was surgically removed at the level of the bladder neck and longitudinally cut into halves on ice. The bladder tissues were then stored in liquid nitrogen.
MiRs microarray procedures
Three samples of each group and 3 replicates of each tissue were used for RNA sequencing. Libraries were constructed using the NEBNext® Multiplex Small RNA Library Prep Set for Illumina (Set 2; New England BioLabs, Inc., Ipswich, MA, USA). Briefly, total RNA was isolated by using Trizol (Invitrogen; Thermo Fisher Scientific, Inc., USA). The quantity and integrity of RNA yield were assessed by using the Qubit®2.0 (Life Technologies, USA) and Agilent 2200 TapeStation (Agilent Technologies, USA) separately. Enriched fragments were sequenced by HiSeq 2500 Sequencing System (Illumina Inc., San Diego, CA, USA) with single-end 50 bp at Ribobio Co. Ltd (Ribobio, China).
Data processing and bioinformatic analysis
The raw reads were filtered to obtain clean reads by removing those with an adaptor sequence or those with a percentage of unknown bases more than 10%, low-quality reads, and smaller than 17 nt reads by FASTQC. The clean reads obtained were mapped to reference genome by Burrows-Wheeler Aligner (BWA) software. miRDeep2 software was used to identify the known mature miRNA based on miRBase21 (www.miRBase.org) and predict novel miRNA. Databases of Rfam12.1 (www.rfam.xfam.org) and pirnabank (www.pirnabank.ibab.ac.in) were used to identify ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA) and PIWI-interacting RNA (piRNA) by Basic Local Alignment Search Tool (BLAST). The miRNA expression was counted and normalized by reads per million (RPM) values [PRM = (number of reads mapping to miRNA/number of reads in clean data) ×106]. Differential expression between samples was calculated by DESeq2 algorithm according to the criteria of log2 |fold change| >1 and P<0.05. TargetScan, miRDB, miRTarBase, and miRWalk were used to predict the genes targeted by selected miRNA. KOBAS was used for further GO and KEGG pathway analyses.
Quantitative real-time polymerase chain reaction (qRT-PCR)
qRT-PCR analysis was performed to verify the accuracy of the microarray assays. MiR expressions were determined by the CFX ConnectTM Real-Time PCR detection system (Bio-Rad Laboratories, CA, USA) in triplicate and calculated using the 2−ΔΔCt method. U6 was used as an internal reference for miRs. Primers for qRT-PCR were provided by Bulge-Loop miRNA qRT-PCR Primer Sets (Guangzhou RiboBio Co., Ltd., Guangzhou, China). Total RNA was extracted from samples of the bladder by using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., USA). cDNAs were synthesized from 2 µg of total RNA using a mixture of Oligo-dT and random primers or specific primers with M-MLV reverse transcriptase (Promega Corporation, USA). The following thermocycling conditions were used: 95 °C for 1 minute, followed by 40 cycles at 95 °C for 10 seconds, 60 °C for 20 seconds, and 70 °C for 10 seconds.
Statistical analysis
Differential expression analysis of miRs obtained from NGS was performed by the DESeq2 (v. 1.16.1), which was an algorithm to examine differences between groups by using a generalized linear model and assuming a negative binomial distribution of RNA-Seq reads. Statistically, differences in the levels of miRs verified by qRT-PCR between groups were determined by analysis of variance (ANOVA) using SPSS 21.0 (SPSS, Inc., Chicago, IL, USA). Data are presented as mean ± standard deviation (SD). The differentially expressed miRNAs were identified using the following thresholds: P<0.05; log2 |fold change| >1. A value of P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of differentially expressed miRs between SCI and NC groups
Compared with the NC group, 96, 28 and 51 miRs were significantly downregulated in the bladders of the SCI-1, SCI-2, and SCI-3 groups, respectively, and 133, 49 and 76 miRs were significantly upregulated in the rat bladders of SCI-1, SCI-2 and SCI-3 groups, respectively (Tables 1-3). Specifically, miR-21-5p was the most significantly upregulated miR in all the SCI groups. Moreover, 206 new miRs were identified in the bladder and are shown in Table S1.
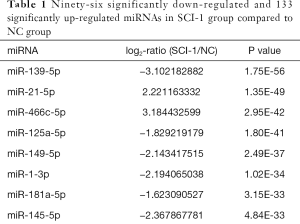
Full table
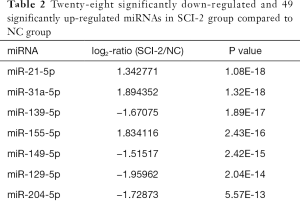
Full table
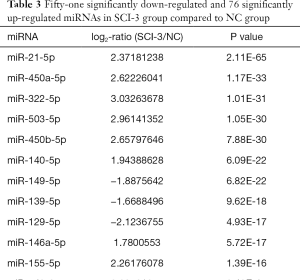
Full table

Full table
Identification of differentially expressed miRs between SCI groups
Compared with the SCI-1 group, 81 and 65 miRs were significantly downregulated in the rat bladders of the SCI-2 and SCI-3 groups, respectively, and 40 and 33 miRs were significantly upregulated in the rat bladders of the SCI-2 and SCI-3 groups respectively (Tables 4,5). Compared with the SCI-2 group, 5 miRs were significantly downregulated, and 21 miRs were significantly upregulated in the rat bladders of the SCI-3 groups (Table 6).
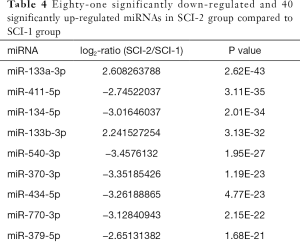
Full table
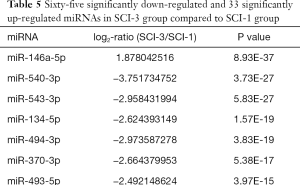
Full table
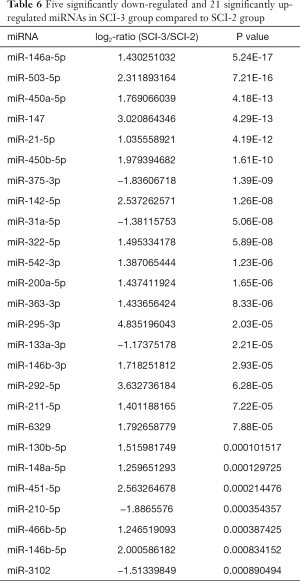
Full table
GO and KEGG analyses of signaling pathways and genes targeted by these differentially expressed miRs
A large set of essential signaling pathways were targeted by these miRs, including PI3K-Akt, MAPK, Rap1, and cGMP-PKG signaling pathways, along with the tight junction, metabolic pathways, regulation of actin cytoskeleton and pathways in cancer, as shown in Figure 1. Meanwhile, Figure 2 shows the genes targeted by these miRs. For example, Smad7 and Smad5 were targeted by miR-21-5p, while nfat5 was targeted by miR-146a-5p, miR-139-5p, and miR-132-3p.
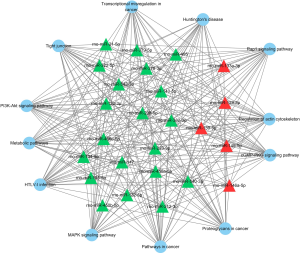
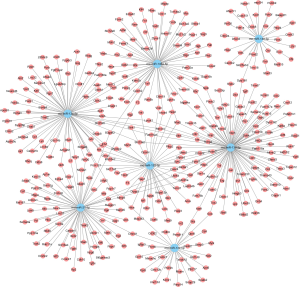
Validation of miR expression by qRT-PCR
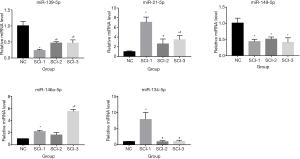
The levels of 5 differentially expressed miRs, including miR-139-5p, miR-21-5p, miR-149-5p, miR-146a-5p, and miR-134-5p, were assessed by qRT-PCR to verify the results of microarray assays. As shown in Figure 3, miR-139-5p was significantly downregulated in all SCI groups compared to the NC group, and miR-21-5p was significantly upregulated in all SCI groups. Furthermore, miR-149-5p was also significantly downregulated in all SCI groups. Nevertheless, miR-146a-5p was significantly upregulated by 5.48-, 2.51-, and 3.46-fold in the SCI-3 group compared to the NC, SCI-1, and SCI-2 groups, respectively, and miR-134-5p was significantly upregulated in the SCI-1 group compared to the NC, SCI-2, and SCI-3 groups.
Discussion
To the best of our knowledge, this study is the first to comprehensively investigate the miRs expression profile in SCI rat NB. The present results showed that compared with the NC group, 96, 28, and 51 miRs were downregulated in the rat bladders of SCI-1, SCI-2, and SCI-3 groups, respectively, and 133, 49, and 76 miRs were upregulated in the rat bladders of SCI-1, SCI-2, and SCI-3 groups, respectively. Specifically, miR-21-5p was the most significantly upregulated miR in all SCI groups. In addition, 121 miRs (SCI-1 vs. SCI-2), 98 miRs (SCI-1 vs. SCI-3), and 26 miRs (SCI-2 vs. SCI-3) were of significantly different expression. Moreover, 206 new miRs were identified in the bladder. Furthermore, a large set of genes implicated in essential signaling pathways were targeted by these miRs, including PI3K-Akt, MAPK, Rap1, and cGMP-PKG signaling pathways, along with tight junction and metabolic pathways.
These essential signaling pathways have been previously implicated in bladder dysfunction. miR‐139‐5p may inhibit epithelial-mesenchymal transition (EMT) and fibrosis by targeting the lysophosphatidic acid receptor 4 via the PI3K-Akt signaling pathway (8). Also, the activation of the PI3K-Akt signaling pathway may play a pivotal part in bladder ischemia, which might be a mediating variable in the development of detrusor overactivity or fibrosis (9). Furthermore, collagen expression and bladder hypertrophy were regulated by nerve growth factor through the Akt and MAPK pathways (10). In addition, activation of the cGMP-PKG signaling pathway may result in bladder relaxation or reduce phasic contractions in rat bladder strips (11,12). Whether the signaling pathways targeted by these miRs can exhibit these functions mentioned above in vivo will be explored in further studies.
Among these miRNAs, miR-139-5p was the most significantly downregulated miR in the SCI-1 group. Though its level was inclined to follow an increasing trend within 2 weeks after spinal cord injury and thereafter remained stable, its level was also significantly lower than that of the NC group. Specifically, compared with NC group, the expression of miR-139-5p was downregulated by 4.22-fold in the SCI-1 group, while its level was upregulated by 1.98- and 1.94-fold in the SCI-2 and SCI-3 groups compared to the SCI-1 group, with no statistical difference between the SCI-2 and SCI-3 groups. These results were confirmed by the qRT-PCR findings.
Recently, Fırat et al. also reported that miR‐139‐5p was significantly downregulated in OAB patient plasma (7). Rho-associated coiled-coil-containing protein kinase 2 (ROCK2) was identified as a direct target of miR-139-5p and was effective in the ROCK2/myosin-light chain (MLC) and cholinergic pathway. It was proven that contractions of the bladder detrusor in humans are primarily mediated by M3 receptors and depend on inhibiting MLC phosphatase by the activation of ROCK, leading to increased sensitivity to Ca2+ (13). Previous data have suggested that the inhibition of ROCK could ameliorate or reverse detrusor overactivity (14,15). By considering the previous data showing that upregulated RhoA/ROCK signaling is one of the factors that contribute to the development of detrusor overactivity, we speculate that a reduced level of miR‐139‐5p may upregulate the expression of ROCK2, resulting in or aggravating detrusor overactivity. Moreover, miR‐139‐5p was found to inhibit EMT and fibrosis by targeting lysophosphatidic acid receptor 4 via the PI3K-Akt signaling pathway (8).
In addition, miR-21-5p was the most significantly upregulated miR in all SCI groups. Indeed, miR-21-5p has been reported to act as an oncogene through inhibiting cellular apoptosis by targeting tumor suppressor genes (16). However, it should be noted that the overexpression of miR-21-5p abnormally activates transforming growth factor-β1 (TGF-β1) and Hedgehog signaling pathways, promoting tumor invasion by the induction of EMT. It is widely accepted that, TGF-β1 signaling pathway plays a pivotal role in EMT and fibrogenesis. Recently, the upregulated expression of miR-21-5p was reported to be involved in renal, myocardial, pulmonary, and peritendinous fibrosis and may serve as an alternative target to directly inhibit this fibrosis (17-20). Further research indicates that miR-21-5p overexpression may enhance TGF-β1-induced EMT by inhibiting Smad7 (21). Moreover, proliferation, migration, and pro-fibrotic activities of fibroblasts were found to be promoted by miR-21-5p through reducing Smad7 expression (18). More specifically, the increase of intracellular miR-21-5p induced fibroblasts differentiation into myofibroblasts and the overexpression of extracellular matrix (ECM) and fibrogenic markers. Moreover, tissue inhibitor of metalloproteinases (TIMPs) which was implicated in collagen synthesis and accumulation during fibrosis were also targeted by miR-21-5p (22).
Importantly, previous study suggests that miR-21-5p is upregulated by TGF-β1 via activation of Smad3 rather than Smad2 (23). In a normal state, Smad-3 activation can induce the expression of Smad7, which forms a negative feedback mechanism (24). Nevertheless, in pathological situations, the expression of Smad7 was found to be suppressed, and the negative feedback damaged, which may due to the upregulated expression of miR-21-5p. In contrast, the conditional knockout of Smad2 could enhance miR-21-5p expression (23). Further research is needed to explore the mechanisms underlying the interactions between miR-21-5p and the TGF-β1 signaling pathway. Also, bladder fibrosis after spinal cord injury may bear responsibility for the high intravesical pressures, low bladder compliance, bladder wall stiffness and vesicoureteral reflux (25). Currently, however, there is no effective method for preventing bladder fibrosis. Therefore, it is also of great significance to investigate the functional role of miR-21-5p in bladder fibrosis after spinal cord injury.
The differentially expressed miRs between SCI groups were also investigated in this study and showed distinct patterns of expression over time. Specifically, we found that miR-146a-5p was upregulated by 5.48-, 2.51-, and 3.46-fold in the SCI-3 group compared to the NC, SCI-1, and SCI-2 groups respectively, while miR-146a-5p was upregulated by 2.19-fold in the SCI-1 group compared to the NC group. In addition, miR-134-5p was significantly upregulated in the SCI-1 group compared to the NC, SCI-2, and SCI-3 groups as revealed by NGS and qRT-PCR. Other studies have found that miR-146a-5p could attenuate hepatic fibrosis by negatively regulating the PTPRA-SRC signaling pathway or inhibiting the profibrogenic effects of TGF-β1 and lipopolysaccharide (26,27). Furthermore, transcription factor twist1 directly targeted by miR-134-5p was also implicated in EMT and fibroblast activation and tissue fibrosis in a TGF-β/Smad3-dependent manner (28).
Suprasacral spinal cord injury can abruptly disrupt intraspinal pathways and result in the “spinal shock” phase, during which the bladder is often atonic and areflexic and typically present with overflow incontinence (29). However, the relative concentration of collagen in rat bladders was reported to be significantly decreased in the first 10 days after spinal cord injury (30), which may be in agreement with the expression of miR-134-5p to a certain extent. Therefore, it is reasonable to presume that miR-134-5p might play a role in it. After spinal shock, hypermechanosensitive C-fiber bladder wall afferents were activated gradually and urodynamic findings were mainly characterized by detrusor overactivity or detrusor-sphincter dyssynergia. miR-146a-5p may be involved in this stage of NB due to that it was significantly upregulated in the SCI-3 group compared to the SCI-1 group. In summary, it will be very interesting and meaningful to investigate the relationship between the dynamic change of these differentially expressed miRs and the different stages of NB.
In this study, we investigated the differentially expressed miRs between groups by NGS and qRT-PCR. Furthermore, 206 new miRs were identified in the bladder, and a large set of genes implicated in essential signaling pathways targeted by these miRs were identified. Nevertheless, this study also has some limitations. Firstly, the interactions between miRs and mRNA were not explored. Thus, further experimental studies are needed to verify the proposed interactions and their roles in NB in the future. Secondly, bladder tissues collected 8 weeks or more after spinal cord transection may be required to investigate those miRs that might function at that time. For example, Wang et al. reported that there was no significant difference in the expression of bladder miR-1949 between rats without spinal cord injury and those collected at 3 months following spinal cord injury, while it was significantly increased after the third month (31). Thirdly, more samples or human bladder tissue should be analyzed to confirm our results.
Conclusions
Several miRs were differentially expressed in the SCI rat NB, and may potentially serve as new molecular targets for NB.
Acknowledgments
ZHS would like to thank Miss Xiaolin Liu for her valuable understanding and assistance.
Funding: This work was supported by the Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (No. ZYLX201801).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-415
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-415
Peer Review File: Available at http://dx.doi.org/10.21037/tau-20-415
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-415). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All experimental procedures were implemented in compliance with the National Institute of Health Guidelines for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of Xuanwu Hospital Capital Medical University (No. 20190128).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Przydacz M, Chlosta P, Corcos J. Recommendations for urological follow-up of patients with neurogenic bladder secondary to spinal cord injury. Int Urol Nephrol 2018;50:1005-16. [Crossref] [PubMed]
- Shang Z, Jia C, Yan H, et al. Injecting RNA interference lentiviruses targeting the muscarinic 3 receptor gene into the bladder wall inhibits neurogenic detrusor overactivity in rats with spinal cord injury. Neurourol Urodyn 2019;38:615-24. [Crossref] [PubMed]
- Hu HZ, Granger N, Jeffery ND. Pathophysiology, Clinical Importance, and Management of Neurogenic Lower Urinary Tract Dysfunction Caused by Suprasacral Spinal Cord Injury. J Vet Intern Med 2016;30:1575-88. [Crossref] [PubMed]
- Oliveira R, Coelho A, Franquinho F, et al. Effects of early intravesical administration of resiniferatoxin to spinal cord-injured rats in neurogenic detrusor overactivity. Neurourol Urodyn 2019;38:1540-50. [Crossref] [PubMed]
- Hamid R, Averbeck MA, Chiang H, et al. Epidemiology and pathophysiology of neurogenic bladder after spinal cord injury. World J Urol 2018;36:1517-27. [Crossref] [PubMed]
- Chermansky CJ, Kadow BT, Kashyap M, et al. MicroRNAs as potential biomarkers to predict the risk of urinary retention following intradetrusor onabotulinumtoxin-A injection. Neurourol Urodyn 2018;37:99-105. [Crossref] [PubMed]
- Fırat E, Aybek Z, Akgun S, et al. Exploring biomarkers in the overactive bladder: Alterations in miRNA levels of a panel of genes in patients with OAB. Neurourol Urodyn 2019;38:1571-8. [Crossref] [PubMed]
- Jiang C, Tong Z, Fang WL, et al. Microrna-139-5p inhibits epithelial-mesenchymal transition and fibrosis in post-menopausal women with interstitial cystitis by targeting LPAR4 via the PI3K/Akt signaling pathway. J Cell Biochem 2018;119:6429-41. [Crossref] [PubMed]
- Yang JH, Siroky M, Yalla S, et al. Mitochondrial stress and activation of PI3K and Akt survival pathway in bladder ischemia. Res Rep Urol 2017;9:93-100. [Crossref] [PubMed]
- Chung CW, Zhang QL, Qiao LY. Endogenous nerve growth factor regulates collagen expression and bladder hypertrophy through Akt and MAPK pathways during cystitis. J Biol Chem 2010;285:4206-12. [Crossref] [PubMed]
- d'Emmanuele di Villa Bianca R, Mitidieri E, Fusco F, et al. Urothelium muscarinic activation phosphorylates CBS(Ser227) via cGMP/PKG pathway causing human bladder relaxation through H2S production. Sci Rep 2016;6:31491. [Crossref] [PubMed]
- Artim DE, Kullmann FA, Daugherty SL, et al. Activation of the nitric oxide-cGMP pathway reduces phasic contractions in neonatal rat bladder strips via protein kinase G. Am J Physiol Renal Physiol 2009;297:F333-40. [Crossref] [PubMed]
- Wróbel A, Rechberger T. The Influence of Maxacalcitol, Vitamin D3 Analog, on Detrusor Overactivity in Conscious Rats. Urology 2016;93:224.e7-224.e15. [Crossref] [PubMed]
- Wróbel A, Serefko A, Rechberger E, et al. Inhibition of Rho kinase by GSK 269962 reverses both corticosterone-induced detrusor overactivity and depression-like behaviour in rats. Eur J Pharmacol 2018;837:127-36. [Crossref] [PubMed]
- Wróbel A, Rechberger T. The influence of Rho-kinase inhibition on acetic acid-induced detrusor overactivity. Neurourol Urodyn 2017;36:263-70. [Crossref] [PubMed]
- Ghorbanmehr N, Gharbi S, Korsching E, et al. miR-21-5p, miR-141-3p, and miR-205-5p levels in urine-promising biomarkers for the identification of prostate and bladder cancer. Prostate 2019;79:88-95. [Crossref] [PubMed]
- Nonaka CKV, Macêdo CT, Cavalcante BRR, et al. Circulating miRNAs as Potential Biomarkers Associated with Cardiac Remodeling and Fibrosis in Chagas Disease Cardiomyopathy. Int J Mol Sci 2019;20:4064. [Crossref] [PubMed]
- Cui H, He Y, Chen S, et al. Macrophage-Derived miRNA-Containing Exosomes Induce Peritendinous Fibrosis after Tendon Injury through the miR-21-5p/Smad7 Pathway. Mol Ther Nucleic Acids 2019;14:114-30. [Crossref] [PubMed]
- Wang J, He F, Chen L, et al. Resveratrol inhibits pulmonary fibrosis by regulating miR-21 through MAPK/AP-1 pathways. Biomed Pharmacother 2018;105:37-44. [Crossref] [PubMed]
- Sun L, Xu T, Chen Y, et al. Pioglitazone attenuates kidney fibrosis via miR-21-5p modulation. Life Sci 2019;232:116609. [Crossref] [PubMed]
- Wang JY, Gao YB, Zhang N, et al. miR-21 overexpression enhances TGF-beta1-induced epithelial-to-mesenchymal transition by target smad7 and aggravates renal damage in diabetic nephropathy. Mol Cell Endocrinol 2014;392:163-72. [Crossref] [PubMed]
- Almalki SG, Agrawal DK. Effects of matrix metalloproteinases on the fate of mesenchymal stem cells. Stem Cell Res Ther 2016;7:129. [Crossref] [PubMed]
- Zhong X, Chung AC, Chen HY, et al. Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J Am Soc Nephrol 2011;22:1668-81. [Crossref] [PubMed]
- Lan HY. Diverse roles of TGF-beta/Smads in renal fibrosis and inflammation. Int J Biol Sci 2011;7:1056-67. [Crossref] [PubMed]
- Fry CH, Kitney DG, Paniker J, et al. Fibrosis and the bladder, implications for function ICI-RS 2017. Neurourol Urodyn 2018;37:S7-S12. [Crossref] [PubMed]
- Yuan BY, Chen YH, Wu ZF, et al. MicroRNA-146a-5p Attenuates Fibrosis-related Molecules in Irradiated and TGF-beta1-Treated Human Hepatic Stellate Cells by Regulating PTPRA-SRC Signaling. Radiat Res 2019;192:621-9. [Crossref] [PubMed]
- Zou Y, Cai Y, Lu D, et al. MicroRNA-146a-5p attenuates liver fibrosis by suppressing profibrogenic effects of TGFbeta1 and lipopolysaccharide. Cell Signal 2017;39:1-8. [Crossref] [PubMed]
- Palumbo-Zerr K, Soare A, Zerr P, et al. Composition of TWIST1 dimers regulates fibroblast activation and tissue fibrosis. Ann Rheum Dis 2017;76:244-51. [Crossref] [PubMed]
- Shi P, Zhao X, Wang J, et al. Effects of acute sacral neuromodulation on bladder reflex in complete spinal cord injury rats. Neuromodulation 2013;16:583-9; discussion 589. [Crossref] [PubMed]
- Nagatomi J, Gloeckner DC, Chancellor MB, et al. Changes in the biaxial viscoelastic response of the urinary bladder following spinal cord injury. Ann Biomed Eng 2004;32:1409-19. [Crossref] [PubMed]
- Wang T, Liu Y, Yuan W, et al. Identification of microRNAome in rat bladder reveals miR-1949 as a potential inducer of bladder cancer following spinal cord injury. Mol Med Rep 2015;12:2849-57. [Crossref] [PubMed]


