Repeatability of proton magnetic resonance spectroscopy of the brain at 7 T: effect of scan time on semi-localized by adiabatic selective refocusing and short-echo time stimulated echo acquisition mode scans and their comparison
Introduction
Proton magnetic resonance spectroscopy (MRS) using an ultra-high magnetic field (UHF) like 7 Tesla (7 T) is considered more advantageous over use of lower magnetic fields (1-4). An increased signal-to-noise ratio (SNR) and spectral separation in a UHF are highly advantageous in MRS because they enable a large reduction in signal averaging and provide better spectral analysis despite an increase in the time of repetition (TR) due to specific absorption rate (SAR) limitations (2).
By and large, MRS sequences may be classified into two categories depending on how the signal is acquired: point resolved spectroscopy (PRESS) (5) and stimulated echo acquisition mode (STEAM) (6) sequences. The disadvantages of a UHF include broader linewidths and stronger chemical shift displacement error, sometimes leading to the complete cancellation of the signal (e.g., lactate). From this viewpoint, PRESS that uses non-adiabatic refocusing pulses is inferior to semi-localized by adiabatic selective refocusing (sLASER) and STEAM (7,8) spectroscopy. sLASER uses two pairs of adiabatic refocusing radiofrequency (RF) pulses to have a wider bandwidth and to be less sensitive to B1-inhomogeneity, and the technique can yield a large signal with relatively short echo time (TE) and small voxel displacement (9). Its use at 7 T improves the precision of quantification, as indicated by reduced Cramér-Rao lower bounds (CRLBs) compared to those at 3 T (3).
However, a sequence using several RF pulse events and that involves refocusing increases SAR, particularly at 7 T, and leads to a longer TE. These factors will lead to a longer TR and a longer scan time. A STEAM sequence that uses three 90-degree pulses is often better suited to 7 T by virtue of the reduced SAR compared to sLASER. In addition, STEAM provides a shorter TE than sLASER due to its inherent method of acquisition. Moreover, asymmetric RF pulses can be used to implement short-TE STEAM (sSTEAM) that facilitates the investigation of metabolite peaks with short T2 values (2) and/or J-coupling (8) and to reduce chemical shift displacement error using a wider excitation bandwidth than that used in PRESS or its variant, sLASER. In STEAM, however, these advantages come at the expense of SNR, acquiring only half of the available signal that can be obtained using sLASER (10).
The reliability of MRS has been investigated to evaluate its precision of quantification (4,11-14). A recent study on measurement repeatability compared sLASER to sSTEAM at 4 T (7) and 7 T (15). At 4 T, however, both the chemical shift size and SAR limitations were smaller than those at 7 T, and observations at 7 T only involved glutamate. In addition, less signal averaging is required at 7 T than at 4 T for both sSTEAM (2) and sLASER, but the effect of less signal averaging has seldom been investigated in terms of repeatability at 7 T.
The aims of the current study were (I) to examine effect of the scan time on the repeatability of brain metabolite concentrations quantified with both sLASER and sSTEAM, and (II) to compare the repeatability of the two sequences.
Methods
Subjects
Sixteen healthy volunteers (10 males and 6 females, mean age: 25 years, age range: 20–38 years) who had no known history of neuropsychiatric disorders or substance abuse were scanned using a 7-T whole-body scanner (MAGNETOM 7T, Siemens Healthineers, Erlangen, Germany) equipped with a whole-body gradient system (70 mT/m maximum amplitude, 200 mT/m/ms maximum slew rate). A single-channel transmit and 32-channel receiver head coil (Nova Medical, MA, USA) was used. This study was approved by the institutional review board and was conducted in accordance with the Helsinki Declaration. Written informed consent was obtained from all subjects.
MR acquisition
Subjects were scanned twice with an off-magnet interval between scans to investigate test-retest repeatability. All scanning was performed by the same operator. For scans, the ears and back of the head were covered with dielectric pads containing a CaTiO3 suspension in D2O (40% v/v) to mitigate B1+ inhomogeneity (16,17). A three-dimensional (3D) gradient echo T1-weighted image (so-called “VIBE”) was acquired to position an MRS voxel using the following parameters: TR = 4.5 ms, TE = 2.05 ms, flip angle = 16o, 0.8-mm isotropic spatial resolution, and bandwidth = 490 Hz/pixel.
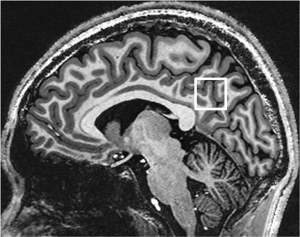
The MRS voxel of 20×20×20 mm3 was positioned at the posterior cingulate cortex (PCC) in the mid-sagittal plane on T1-weighted images. The anterior-inferior corner of the voxel was placed at the splenium of the corpus callosum as shown in Figure 1. Consistent voxel placement among brains was confirmed by the same neuroradiologist for all scans. B0 shimming of first- and second-order terms was achieved using a fast, automatic shimming technique by mapping along projections with an echo-planar imaging readout (FASTESTMAP, Siemens prototype sequence) (18). RF transmit amplitude was adjusted for the localized MRS voxel using the sSTEAM pulse sequence described below. The RF pulse transmit amplitude for water suppression was adjusted using the corresponding pulse sequence and TR.
Proton MR spectra were acquired using sLASER (TE = 32 ms) and sSTEAM (TE = 5 ms and mixing time = 45 ms) pulse sequences (Siemens prototype sequences) with water suppression using the variable pulse power and optimized relaxation delays (VAPOR) technique and outer volume suppressions (OVS) to improve localization (19). In sLASER, TR was 6.5 s due to SAR limitations, and 32 and 48 averages (sLASER×32 and sLASER×48; scan time = 3 min 40 s and 5 min 19 s, respectively) were used to calculate the SNR. In sSTEAM, TR was 4 s, and 32, 48, and 64 averages (sSTEAM4×32, sSTEAM4×48, and sSTEAM4×64; scan time = 2 min 24 s, 3 min 28 s, and 4 min 32 s, respectively) in addition to TR of 8 s for full T1 recovery with 32 averages (sSTEAM8×32; scan time = 4 min 48 s). The scan times listed above include preparation time. See Table 1 for an overview. Other parameters such as a spectral width of 6 kHz and 2,048 data points remained the same for both sequences. Spectral raw data were averaged using scanner software without any corrections or filters. Water spectra were also acquired from the same MRS voxel without OVS to eliminate the magnetization transfer effect (20) with an average of 4 scans after a single preparatory scan. The interval between replicate measurements was approximately 1 h, although the interval was around 5 h for one volunteer.

Full table
Spectrum analysis
Analysis was performed with LCModel version 6.3-1L, which uses a priori knowledge of the spectral components to fit metabolite resonances (21). A basis-set for STEAM of TE = 0 (a standard option of LCModel) was used in sSTEAM as described in a previous study (8), and that for sLASER with TE = 32 ms was developed in-house with numerical simulations using density matrix theory. The same 16 metabolites were quantified with the two basis-sets: N-acetylaspartate (NAA), N-acetylaspartylglutamate (NAAG), alanine (Ala), γ-aminobutyric acid (GABA), aspartate (Asp), creatine (Cr), phosphocreatine (PCr), glutamine (Gln), glutamate (Glu), glutathione (GSH), myo-inositol (Ins), scyllo-inositol (sIns), lactate (Lac), phosphocholine (PCh), glycerophosphocholine (GPC), and taurine (Tau). For Cho, Cr, and NAA, the total amounts of tCho (= GPC + PCh), tCr (= Cr + PCr), and tNAA (= NAA + NAAG) were used because peaks for Cho, Cr, and NAA were poorly separated. The spectral width for analysis was 0.2–4.0 ppm. Eddy current correction and water-scaling for quantification were performed using the spectra for water (20,22).
The precision of quantification was evaluated using CRLBs. Metabolites quantified with CRLBs >50% were classified as not detected, and metabolites quantified with mean or median CRLBs ≤20% were included for further analysis (4).
The SNR for NAA (SNRNAA) and Glu (SNRGlu) peaks was measured as the height of the singlet methyl peak of NAA at 2.01 ppm and that of the multiplet Glu peak at 2.35 ppm from the baseline (23) divided by the noise, which was calculated with a maximum peak-to-peak distance in the 0.2–0.6 ppm range in residuals from LCModel fitting divided by 2.5. SNR was analyzed in terms of the square root of the scan time.
Metabolite concentrations were scaled using the unsuppressed water peak as an internal reference. To correct the water content, tissue fractions in the MRS voxel were calculated (20). T1-weighted images were segmented into gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) using the FMRIB Software Library (FSL: https://fsl.fmrib.ox.ac.uk/fsl/fslwiki). The MRS voxel was registered on segmented T1-weighted images using the software Gannet (http://www.gabamrs.com) to extract the tissue fractions in the voxel. The water concentration in the MRS voxel was calculated assuming a water concentration of 43,300 millimole (mM) for GM, 35,880 mM for WM, and 55,556 mM for CSF (24).
Statistical analysis
The mean or median of CRLBs and metabolite concentrations were calculated using both repeated scans (i.e., n=32). The repeatability of measurement was evaluated using intrasubject coefficients of variation (CVs) derived from the standard deviation of the two measurements divided by their mean. The mean or median of CVs were calculated for all subjects (n=16). To identify significant differences in CRLBs, metabolite concentration, and repeatability, i.e., CVs, among scan sequences and parameters, one-way ANOVA or the Friedman test was used depending on the normality of data distribution (Shapiro-Wilk test). A P value <0.05 was considered significant after a post-hoc test. The CVs for metabolite concentrations were evaluated in conjunction with values related to the precision of quantification: the SNR and CRLBs using mean values from a set of two scans. Metabolite concentrations were measured in mM without correcting for relaxation, and their statistical comparisons of different sequences or TR values were not conducted.
Results
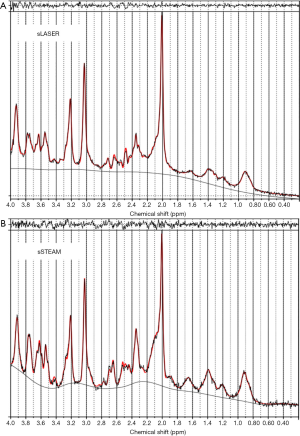
LCModel fitting and residuals of representative MR spectra from sLASER and sSTEAM of the PCC in the same brain are shown in Figure 2. Compared to singlet peaks at 2.0 (methyl groups in NAA and NAAG), 3.0 (methyl groups in Cr and PCr), 3.2 (methyl groups in GPC and PCh), and 3.9 ppm (methylene groups in Cr and PCr), J-coupled peaks at 2.3–2.8 and 3.3–3.8 ppm according to sSTEAM were higher than those according to sLASER. J-coupled peaks at 2.35 (a methylene group in Glu) and 3.75 ppm (methine groups in Glu, Gln, and GSH) were more separated and lower in sLASER than in sSTEAM. Small fitting errors around J-coupled peak regions were noted in residuals with both sequences.
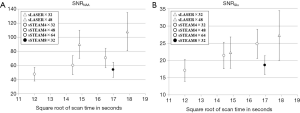
An analysis of SNR indicated that both SNRNAA and SNRGlu increased by the square root of the scan time for most protocols. However, sSTEAM8×32 had a distinctively lower SNR than sSTEAM4×64, which was acquired in a similar scan time (Figure 3). Even when only 32 averages were used and scan time was nearly halved, sSTEAM4×32 resulted in an SNR comparable to that of sSTEAM8×32, suggesting that a longer TR contributed little to improving SNR for both NAA and Glu. SNRNAA of sLASER×32 and sSTEAM4×48, both acquired in a similar scan time, were 90.0 and 60.4, respectively, representing a SNR that nearly 50% higher in sLASER×32 than in sSTEAM4×48, while SNRGlu was 22.3 and 21.4, respectively. Advantages of sLASER over sSTEAM were not noted.
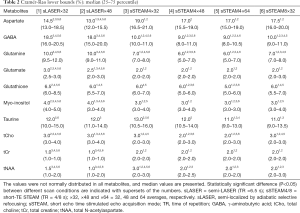
The following metabolites had CRLBs larger than 50% (percentage in all measurements of sLASER and sSTEAM, respectively): Ala (76.6% and 29.7%), Cr (0% and 3.1%), GPC (3.1% and 0%), Lac (48.4% and 46.9%), NAAG (32.8% and 7.8%), PCh (93.8% and 11.7%), PCr (1.6% and 0%), and sIns (29.7% and 51.6%). These metabolites were excluded from further analysis. Median values for the remaining metabolites did not exceed 20%, but CRLBs failed to follow a normal distribution. Median CRLBs were ≤10%, except for Asp in both sequences and GABA in sLASER. Median CRLBs for tCr and tNAA were significantly lower in sLASER than in sSTEAM, whereas the opposite was true for GABA, Gln, Glu, Ins, and tCho; however, the difference was ≤2% except for GABA and Gln (Table 2). Significant differences in CRLBs for Asp, GSH, and Tau were noted in the two sequences only under some scanning conditions. Signal averaging significantly lowered CRLBs for Asp, Glu, GSH, Ins, and tCho in sLASER and for GABA, Gln, GSH, Ins, Tau, tCho, and tNAA in sSTEAM4 (at least between ×32 and ×64). Some metabolites had significantly lower CRLBs in sSTEAM4×64 than in sSTEAM8×32, but the difference was ≤1%, whereas many metabolites had CRLBs in sSTEAM4×32 that were comparable to those in sSTEAM8×32.
Full table
Tissue fractions of GM, WM, and CSF in the MRS voxels were 74.5%±2.7%, 18.2%±3.0% and 7.3%±2.1%, respectively. The water concentration in the MRS voxel was 46,260±1,438 mM (mean ± SD), and the individual water concentration was used to correct the metabolite concentration. Significant differences between sLASER×32 and sLASER×48 in Gln, Glu, and tCr concentrations and between sSTEAM ×32 and sSTEAM ×64 in the tCr concentration were noted, but the differences were 3.8%, 1.5%, 1.4%, and 2.2%, respectively (Table 3).
Full table
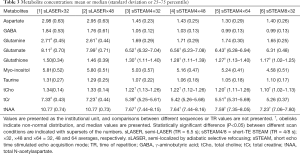
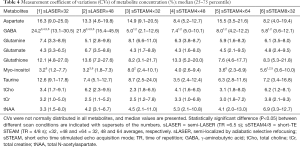
Measurement repeatability was indicated by the CV in the metabolite concentration. CVs failed to follow a normal distribution. Median CVs for Gln, Glu, Ins, tCho, tCr, and tNAA were ≤10% for both scan sequences and under all conditions, and median CVs for Asp, GABA, GSH, and Tau were ≥10% only in some scans (Table 4). The CV for GABA was significantly higher in sLASER (21.8–24.2%) than in sSTEAM (5.0–8.0%), whereas the CV for Ins was significantly higher in sSTEAM (4.0–8.0%) than in sLASER (3.2%) under some conditions. There was no significant difference in the CV for any other metabolite, and repeatability was considered comparable. In addition, an increase in signal averaging did not contribute to significantly lower CVs except for Ins, and its median CV in sSTEAM4×64 (3.6%) was significantly lower than that in sSTEAM8×32 (5.6%). CVs for tNAA and Glu concentrations were not correlated with mean values of SNRNAA and SNRGlu, respectively, except for Glu in STEAM4×32 (r=0.71, P=0.002).
Full table
Discussion
This study compared sLASER and sSTEAM sequences in order to evaluate the repeatability of MRS at 7 T to quantify levels of numerous brain metabolites. sLASER resulted in a higher SNR for NAA compared to sSTEAM. However, sLASER did not necessarily result in lower CRLBs or metabolite CVs. Basically, the sequences resulted in similar levels of CRLBs and CVs. However, when results of using a similar scan time were compared, CRLBs were significantly lower in sSTEAM than in sLASER by ≥4% for GABA, Gln, and GSH, whereas the opposite was true for Asp; the CV for GABA was significantly lower (≥14%) in sSTEAM than in sLASER. A longer scan time resulted in significantly lower CRLBs for some metabolites, but the differences were mostly ≤1%, and there were no significant differences in its effect on CVs.
A previous study compared the Glu concentration according to sLASER and sSTEAM at 7 T and found that sLASER resulted in better measurement reproducibility (15), but the current study found no significant differences in the CV for Glu. The previous study scanned a relatively small number of subjects (n=8) twice with different scan protocols: sLASER (TE = 28 ms, 16 averages, TR = 5 s) and sSTEAM (TE = 7.8 ms, 128 averages, TR = 2 s). The short TR of sSTEAM (2 s) might have impaired longitudinal relaxation and diminished measurement reproducibility.
Handling of macromolecules in an analysis is an important issue, especially when TE is short. This study included default macromolecule basis sets in its analysis. In LCModel, constrained regularization (25), which is nearly model-free, can be useful since it shows that the regularized LCModel baseline can automatically account well for a strong MM signal in high-quality data, with no prior knowledge of (or biasing assumptions about) the MM signal (26). Several previous studies suggested that a short TE resulted in a precision of quantification equivalent to or better than that of MRS with a long TE when measuring Glu, Gln, GSH, and GABA (4,12,27,28). Based on such studies, Wijtenburg et al. (29) hypothesized that very short echo time (TE) acquisitions would have comparable reproducibility to long TE acquisitions. Focusing on GSH, they found that short-TE STEAM had the lowest mean test-retest CV, followed closely by PRESS and SPECIAL while MEGA-PRESS was the worst. Spectral analysis was performed using LCModel; macromolecule signals in the short TE sequences were handled by automatically including a basis set of macromolecules and accounting for their presence within the spectrum. These findings are consistent with results of short-TE STEAM analyzed using LCModel.
The SNR for NAA and Glu increased with the number of averages in both sLASER and sSTEAM4. The sLASER resulted in a higher SNR for NAA compared to sSTEAM, but the SNR for Glu was almost the same for the two. Compared to SNRNAA, SNRGlu is presumed to be lower due to a lower metabolite concentration, a shorter T2 relaxation time, and J-coupling. A point also worth noting is that the peak height at 2.35 ppm contains macromolecules. However, the effect of the latter two of three factors is considered negligible in sSTEAM acquisition (7,8), and SNRGlu resulted in almost the same values under those conditions. sSTEAM4×64 resulted in a higher SNRGlu than sSTEAM8×32, although sSTEAM8×32 was acquired over a slightly longer time. This is because the number of averages has a larger effect on SNR than the difference in T1 relaxation. The T1 value for Glu at 7 T was around 1.2 s (30).
CRLBs indicate the precision of quantification (the lower, the better). Median CRLBs were ≤10% except for Asp and GABA. Signal averaging significantly lowered CRLBs for Asp, Glu, GSH, Ins, and tCho in sLASER and CRLBs for GABA, Gln, GSH, Ins, Tau, tCho, and tNAA in sSTEAM4 (at least between ×32 and ×64), but the differences were ≤2%. The effect of averaging was limited, which is probably attributable to many metabolites that had relatively low CRLBs. This finding agrees with the results of a previous study which found that less averaging was required to lower CRLBs at 7 T than at 4 T (2) due to a larger chemical shift separation of metabolite peaks at UHF. There were small differences in median CRLBs between the two sequences (≤2%) except for Asp, GABA, and Gln. The median CRLB for Asp was higher in sSTEAM, whereas the median CRLBs for GABA and Gln were higher in sLASER. Despite such differences, the two sequences yielded comparable results for most metabolites.
Metabolite concentrations were measured without correcting for relaxation, which was considered impractical for clinical scans, and differences between the two sequences in some metabolite concentrations were noted. In addition, a study comparing sLASER and sSTEAM at 4 T (7) noted such differences and attributed them to differences in TE, relaxation time, basis-set, and errors in chemical shift size. There were significant differences in Gln, Glu, and tCr concentrations measured by sLASER×32 and sLASER×48, but the mean difference was 0.1, 0.12, and 0.1 mM or 3.8%, 1.5%, and 1.4%. The only difference between sSTEAM4×32 (5.39 mM) and sSTEAM4×64 (5.51 mM) was the tCr concentration, which differed 0.12 mM or 2.2%. Metabolite concentrations were consistent within each sequence.
Measurement repeatability was evaluated using the CV in the metabolite concentration, and CVs were mostly lower than 5% for Glu, Ins, tCho, tCr, and tNAA, whereas CVs were higher for Asp, GABA, Gln, GPC, GSH, and Tau. Among J-coupled metabolites, Glu and Ins had lower CRLBs and CVs, presumably because they were present in higher concentrations. Another reason might be the detectability of dominant peaks at 2.34/2.36 and 3.53 ppm for Glu and Ins, respectively, at 7 T. The largest difference between the two sequences was noted for GABA, and its median CV was much smaller in sSTEAM. GABA has a much shorter T2 value (e.g., 63 ms) (31) than Glu (124 ms at the occipital lobe) (30) and other metabolites (121–170 ms) (32). This difference caused less signal decay during TE in sSTEAM than in sLASER and may be one reason for the higher CRLB for GABA than for Glu. However, sLASER×32 yielded comparable results to sSTEAM4×32 except for GABA, so the former can be an appropriate choice.
The CV in the metabolite concentration provides insight into the effects of averaging. In sLASER, 32 and 48 averages resulted in no significant differences. In sSTEAM, the only significant difference between sSTEAM4×64 and sSTEAM8×32 was in the Ins concentration, and the former was more precise. However, the concentration did not differ significantly among the 3 sSTEAM4 scans. This suggests that a larger number of averages with a longer scan time might be affected by field perturbations such as patient motion (33). In theory, a larger number of averages would be better, but this study suggests that 32 averages is acceptable in terms of repeatability for both sLASER and sSTEAM.
A low SAR and short TR in sSTEAM are beneficial at 7 T, something sLASER cannot provide. A recent study at 7 T using a STEAM sequence reduced TR to 3 s and noted a significant difference in patients with first-episode psychosis in comparison to healthy controls (34). In nearly half the scan time, sSTEAM4×32 resulted in metabolite concentrations and CVs comparable to those in sSTEAM8×32 with no significant differences. Median CRLBs were significantly higher in sSTEAM4×32 than in sSTEAM8×32, but the difference was ≤1%. A short scan time of 2.5 minutes was considered sufficient for a reliable and repeatable MRS in the current study. A shorter scan time would be preferable to reduce motion artifacts, and especially when scanning patients. A point also worth noting is that a short TR causes possible T1-weighting of the metabolite concentrations. At 7 T, T1 values for brain metabolites analyzed in this study ranged from 1.14 (GSH) to 2.15 (Tau) s in GM (35). A TR of 4 s is not sufficient for full longitudinal relaxation, but spectra acquired with TRs of 4 and 10 s at 7 T using PRESS indicated that differences between the two TRs in the signal intensity of NAA, Cr, and Cho were very small (32). A TR of 4 s is considered advantageous over a longer TR with fewer averages.
This study has a limitation. Spectra were measured only at the PCC, where RF transmit efficiency is relatively high and water signal line width is narrow (36). The PCC is considered to be an ideal location, and measurement repeatability might decrease at other areas of the brain.
Both sequences were found to quantify brain metabolite concentrations with a high level of repeatability, i.e., low CVs, and to provide highly precise quantification, i.e., low CRLBs, at 7 T. The two sequences resulted in significant differences (≥4%) in Asp, GABA, and Glu concentrations, but CVs were comparable except for GABA, for which sSTEAM was considered to be a better choice.
Acknowledgments
Funding: This work was partly supported by a research grant from Siemens Healthcare K.K., Japan.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form(available at http://dx.doi.org/10.21037/qims-20-517). TO serves as an unpaid editorial board member of Quantitative Imaging in Medicine and Surgery. TO reports grants from Siemens Healthcare K.K., during the conduct of the study; HK reports personal fees from Siemens Healthcare K.K. (Japan), during the conduct of the study; YU reports personal fees from Siemens Healthcare K.K. (Japan), during the conduct of the study; NS was a Siemens Research Collaboration manager in charge of MR Spectroscopy. NS retired in October 2017 and do not currently receive any salary from Siemens. RTS reports personal fees from Siemens Medical Solutions, USA Inc., during the conduct of the study; SA reports personal fees from Siemens Healthineers, during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by institutional review board of Graduate School of Medicine, Kyoto University, and written informed consent was obtained from all subjects.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Michaeli S, Garwood M, Zhu X, DelaBarre L, Andersen P, Adriany G, Merkle H, Ugurbil K, Chen W. Proton T2 relaxation study of water, N-acetylaspartate, and creatine in human brain using Hahn and Carr-Purcell spin echoes at 4T and 7T. Magn Reson Med 2002;47:629-33. [Crossref] [PubMed]
- Tkác I, Öz G, Adriany G, Uğurbil K, Gruetter R. In vivo 1H NMR spectroscopy of the human brain at high magnetic fields: Metabolite quantification at 4T vs. 7T. Magn Reson Med 2009;62:868-79. [Crossref] [PubMed]
- Pradhan S, Bonekamp S, Gillen JS, Rowland LM, Wijtenburg AS, Edden RA, Barker PB. Comparison of single voxel brain MRS AT 3T and 7T using 32-channel head coils. Magn Reson Imaging 2015;33:1013-8. [Crossref] [PubMed]
- Terpstra M, Cheong I, Lyu T, Deelchand DK, Emir UE, Bednařík P, Eberly LE, Öz G. Test-retest reproducibility of neurochemical profiles with short-echo, single-voxel MR spectroscopy at 3T and 7T. Magn Reson Med 2016;76:1083-91. [Crossref] [PubMed]
- Bottomley PA. Spatial Localization in NMR Spectroscopy in Vivo. Ann NY Acad Sci 1987;508:333-48. [Crossref] [PubMed]
- Frahm J, Merboldt KD, Hänicke W, Haase A. Stimulated echo imaging. J Magn Reson (1969) 1985; 64:81-93.
- Öz G, Tkáč I. Short-echo, single-shot, full-intensity proton magnetic resonance spectroscopy for neurochemical profiling at 4 T: Validation in the cerebellum and brainstem. Magn Reson Med 2011;65:901-10. [Crossref] [PubMed]
- Marjańska M, Auerbach EJ, Valabrègue R, Moortele PV, de , Adriany G, Garwood M. Localized 1H NMR spectroscopy in different regions of human brain in vivo at 7 T: T2 relaxation times and concentrations of cerebral metabolites. NMR Biomed 2012;25:332-9. [Crossref] [PubMed]
- Scheenen TWJ, Klomp DWJ, Wijnen JP, Heerschap A. Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med 2008;59:1-6. [Crossref] [PubMed]
- Moonen CTW, Kienlin MV, Zijl PCMV, Cohen J, Gillen J, Daly P, Wolf G. Comparison of single-shot localization methods (steam and press) for In vivo proton NMR spectroscopy. NMR Biomed 1989;2:201-8. [Crossref] [PubMed]
- O’Gorman RL, Michels L, Edden RA, Murdoch JB, Martin E. In vivo detection of GABA and glutamate with MEGA-PRESS: Reproducibility and gender effects. J Magn Reson Imaging 2011;33:1262-7. [Crossref] [PubMed]
- Prinsen H, Graaf RA, de , Mason GF, Pelletier D, Juchem C. Reproducibility measurement of glutathione, GABA, and glutamate: Towards in vivo neurochemical profiling of multiple sclerosis with MR spectroscopy at 7T. J Magn Reson Imaging 2017;45:187-98. [Crossref] [PubMed]
- Deelchand DK, Kantarci K, Öz G. Improved localization, spectral quality, and repeatability with advanced MRS methodology in the clinical setting. Magn Reson Med 2018;79:1241-50. [Crossref] [PubMed]
- Zhang Y, Taub E, Salibi N, Uswatte G, Maudsley AA, Sheriff S, Womble B, Mark VW, Knight DC. Comparison of reproducibility of single voxel spectroscopy and whole-brain magnetic resonance spectroscopy imaging at 3T. NMR Biomed 2018;31:e3898. [Crossref] [PubMed]
- Marsman A, Boer VO, Luijten PR, Pol HE, Klomp DW, Mandl RC. Detection of Glutamate Alterations in the Human Brain Using 1H-MRS: Comparison of STEAM and sLASER at 7 T. Front Psychiatry 2017;8:60. [Crossref] [PubMed]
- Teeuwisse WM, Brink WM, Webb AG. Quantitative assessment of the effects of high-permittivity pads in 7 Tesla MRI of the brain. Magn Reson Med 2012;67:1285-93. [Crossref] [PubMed]
- O’Brien KR, Magill AW, Delacoste J, Marques JP, Kober T, Fautz H, Lazeyras F, Krueger G. Dielectric pads and low- B1+ adiabatic pulses: Complementary techniques to optimize structural T1w whole-brain MP2RAGE scans at 7 tesla. J Magn Reson Imaging 2014;40:804-12. [Crossref] [PubMed]
- Gruetter R, Tkáč I. Field mapping without reference scan using asymmetric echo-planar techniques. Magn Reson Med 2000;43:319-23. [Crossref] [PubMed]
- Tkác I, Starčuk Z, Choi IY, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med 1999;41:649-56. [Crossref] [PubMed]
- Gasparovic C, Song T, Devier D, Bockholt HJ, Caprihan A, Mullins PG, Posse S, Jung RE, Morrison LA. Use of tissue water as a concentration reference for proton spectroscopic imaging. Magn Reson Med 2006;55:1219-26. [Crossref] [PubMed]
- Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med 1993;30:672-9. [Crossref] [PubMed]
- Klose U. In vivo proton spectroscopy in presence of eddy currents. Magn Reson Med. 1990;14:26-30. [Crossref] [PubMed]
- Giapitzakis IA, Shao T, Avdievich N, Mekle R, Kreis R, Henning A. Metabolite-cycled STEAM and semi-LASER localization for MR spectroscopy of the human brain at 9.4T. Magn Reson Med 2018;79:1841-50. [Crossref] [PubMed]
- Provencher SW. LCModel & LCMgui User’s manual. 2016.
- Provencher SW. A constrained regularization method for inverting data represented by linear algebraic or integral equations. Comput Phys Commun 1982;27:213-27. [Crossref]
- Pfeuffer J, Tkáč I, Provencher SW, Gruetter R. Toward an in Vivo Neurochemical Profile: Quantification of 18 Metabolites in Short-Echo-Time 1H NMR Spectra of the Rat Brain. J Magn Reson 1999;141:104-20. [Crossref] [PubMed]
- Deelchand DK, Marjańska M, Hodges JS, Terpstra M. Sensitivity and specificity of human brain glutathione concentrations measured using short-TE 1H MRS at 7 T. NMR Biomed 2016;29:600-6. [Crossref] [PubMed]
- Wijtenburg SA, Rowland LM, Edden RAE, Barker PB. Reproducibility of brain spectroscopy at 7T using conventional localization and spectral editing techniques: 1 H MRS Reproducibility Study at 7T. J Magn Reson Imaging 2013;38:460-7. [Crossref] [PubMed]
- Wijtenburg SA, Near J, Korenic SA, Gaston FE, Chen H, Mikkelsen M, Chen S, Kochunov P, Hong EL, Rowland LM. Comparing the reproducibility of commonly used magnetic resonance spectroscopy techniques to quantify cerebral glutathione. J Magn Reson Imaging 2019;49:176-83. [Crossref] [PubMed]
- Li N, Li L, Zhang Y, Araneta MF, Johnson C, Shen J. Quantification of in vivo transverse relaxation of glutamate in the frontal cortex of human brain by radio frequency pulse-driven longitudinal steady state. PLoS One 2019;14:e0215210. [Crossref] [PubMed]
- Intrapiromkul J, Zhu H, Cheng Y, Barker PB, Edden RAE. Determining the in vivo transverse relaxation time of GABA in the human brain at 7T. J Magn Reson Imaging 2013;38:1224-9. [Crossref] [PubMed]
- Li Y, Li Y, Xu D, Ozturk-Isik E, Lupo JM, Chen AP, Vigneron DB, Nelson SJ. T1 and T2 Metabolite Relaxation Times in Normal Brain at 3T and 7T. J Mol Imaging Dyn 2012;S1:002.
- Wilm BJ, Duerst Y, Dietrich BE, Wyss M, Vannesjo SJ, Schmid T, Brunner DO, Barmet C, Pruessmann KP. Feedback field control improves linewidths in in vivo magnetic resonance spectroscopy. Magn Reson Med 2014;71:1657-62. [Crossref] [PubMed]
- Wang AM, Pradhan S, Coughlin JM, Trivedi A, DuBois SL, Crawford JL, Sedlak TW, Nucifora FC, Nestadt G, Nucifora LG, Schretlen DJ, Sawa A, Barker PB. Assessing Brain Metabolism With 7-T Proton Magnetic Resonance Spectroscopy in Patients With First-Episode Psychosis. JAMA Psychiatry 2019;76:314-23. [Crossref] [PubMed]
- Xin L, Schaller B, Mlynarik V, Lu H, Gruetter R. Proton T1 relaxation times of metabolites in human occipital white and gray matter at 7 T. Magn Reson Med 2013;69:931-6. [Crossref] [PubMed]
- Emir UE, Auerbach EJ, Moortele P, Marjańska M, Uğurbil K, Terpstra M, Tkáč I, Öz G. Regional neurochemical profiles in the human brain measured by 1H MRS at 7 T using local B1 shimming. NMR Biomed 2012;25:152-60. [Crossref] [PubMed]