
Detection and recognition of ultrasound breast nodules based on semi-supervised deep learning: a powerful alternative strategy
Introduction
Breast cancer is one of the most common cancers in women worldwide and alone accounts for 30% of all new cancer cases in women (1,2). The incidence rates of breast cancer increased from 2006 to 2015 by approximately 0.3–0.4% per year among non-Hispanic white and Hispanic women and by 1.8% per year among Asian/Pacific Islander women. From 1990 to 2016, the mortality rate of breast cancer in the USA decreased from approximately 30% to 20%, and the decline in breast cancer mortality over the past three decades is primarily due to early detection and treatment (1).
Although the pathological examination is considered the gold standard for breast cancer, it is inefficient and inconvenient (3). X-ray mammography is recommended for women, beginning at the age of 40 years (4). However, mammographic density is associated with a higher risk of breast cancer (5), and more than 50% of women have dense breast tissue (6,7). X-ray mammography’s sensitivity is reduced to 57–71% for women with dense breast tissue (8). Personalized breast cancer screening has been proposed, with tiered use of different imaging modalities and techniques (9). Further studies may prove that ultrasonographic breast screening is efficient and beneficial, especially for women with dense breast tissue (10).
The challenges in breast ultrasound stem from the complexity of images, including noise, artifacts, and low contrast. Manual analysis by sonographers is time-consuming, subjective, and can lead to unintended misdiagnoses due to fatigue (10). Thus, computer-aided detection or artificial intelligence (AI) is essential for improving both the false positives and false negatives of screening breast ultrasound and reducing the biopsy rate (11).
Computer-aided diagnosis of breast cancer based on traditional image analysis has been studied for decades (12); for example, fractal dimension estimation (13), computation of the area of breast lesions based on region growth (14), and classification of breast tumors (15). However, these traditional methods lack robustness because they rely on hand-crafted features. Recently, deep learning powered by advances in large labeled datasets and computing capability has achieved revolutionary breakthroughs in ultrasound image analysis, including classification (16), image quality assessment (17), standard plane detection (18), localization (19), image segmentation (20) and so on.
In breast ultrasound studies, Byra et al. (21) and Yap et al. (22) proposed a convolutional neural network for breast lesion detection, and Cheng et al. applied the denoising autoencoders for classifying breast lesions in ultrasound images (23). Han et al. also proposed a deep learning framework to differentiate malignant and benign nodules (24). Recently, Qi et al. proposed a state-of-the-art method with multiscale kernels and skip connections to diagnose breast nodules on ultrasonography images (25).
Deep learning has achieved high accuracy, but the methods above are all based on supervised learning, which requires a large number of labeled images (26). However, the reality in clinical practice is that only a small number of labeled images and a larger number of unlabeled images are available. Data annotation always requires much time and careful preparation, thus greatly increasing the time and economic costs. More importantly, it is often necessary to label the data differently according to the specific medical application, further resulting in less labeled data. Therefore, reducing the dependence on labeled data of thousands or millions of images and developing medical AI more efficiently are still difficult (27).
For example, more efficient learning methods, semi-supervised learning not requiring a lot of labeled data are assumed to provide a low-cost and powerful alternative approach (27). However, whether semi-supervised methods can be applied to medical images on a large scale is unclear. If semi-supervised learning is as accurate as supervised learning in the medical domain, then medical AI trained by semi-supervised learning may be more economical and faster in development by reducing the amount of data annotations needed. Further exploration is needed to determine whether semi-supervised learning using a small number of labeled images and a large number of unlabeled images can achieve satisfactory performance in medical image analysis, such as based on semi-supervised detection (28), magnetic resonance imaging image segmentation (29,30), data augmentation (31), and histology image classification (32).
In this study, two datasets comprising 8,966 breast nodules from two hospitals were used to confirm that the semi-supervised method could be applied to ultrasound images with large noise issues. The proposed semi-supervised method’s performance was compared with that of an existing supervised learning method and five human sonographers. The proposed method had two stages. The first was locating the bounding box of nodules from the ultrasound images using a faster region-based convolutional neural network (Faster R-CNN) (33), and the located image was then inputted to a semi-supervised classifier called the mean teacher model (34) for recognition. The main contributions were as follows:
- An accurate semi-supervised recognition method for breast nodules was developed, and its performance was compared with that of supervised methods and sonographers;
- The study proved that the semi-supervised method and 4,396 unlabeled nodules could always achieve higher performance than the supervised method when the number of labeled images (800 labeled nodules) was insufficient. Moreover, the semi-supervised method was comparable to the supervised learning method trained on a massive labeled dataset (5,196 labeled nodules);
- The semi-supervised method might also develop an expert-level recognition system for benign and malignant nodules, thus significantly reducing the cost of data annotation and demonstrated the potential of semi-supervised deep learning in medical applications.
Methods
Dataset
The dataset was collected from two hospitals: The Third Affiliated Hospital of Xi’an Jiaotong University (Dataset-A) and The First Affiliated Hospital of Xi’an Jiaotong University (Dataset-B). The institutional review boards approved the two hospital’s study, and a general research authorization was obtained, allowing for retrospective reviews. From the hospital information systems, technicians randomly selected ultrasound images of patients who underwent ultrasound breast examination in the two hospitals and had a pathological diagnosis during 2010–2019. The number of images from each week did not exceed 50 to ensure the randomness of the data, and the images of 9,012 nodules were obtained. The technicians did not participate in the follow-up study. The histopathological findings are shown in Table 1.
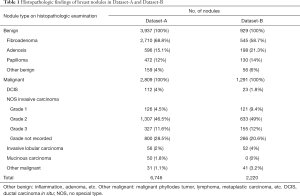
Full table
Dataset review and annotation
Two sonographers reviewed Dataset-A and Dataset-B with 10 years of clinical experience. They manually labeled the location of the breast nodule with bounding boxes and marked them as benign (non-cancer) or malignant (cancer) using a label software, LabelImg (35). The intersection of their labeled bounding boxes was used for the nodule’s labeled location, and the benign or malignant status of the nodule was obtained from pathological reports for type annotations. In total, 4,100 labeled malignant nodules and 4,866 labeled benign nodules (non-cancer) were included in the two datasets, and 46 nodules were removed because of poor image quality (Table 2).
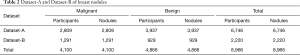
Full table
Proposed network framework
The proposed network framework consisted of two parts: a detection network for breast nodules and a classifier network for benign and malignant nodules. Considering that nodules exist only in a small section of the ultrasound images, the Faster R-CNN was first used to detect the nodules’ bounding boxes. The Faster R-CNN included a feature extractor network and a regional proposal network (RPN). The network of 16 layers from the Visual Geometry Group (VGG16) was selected as the feature extractor because of its excellent feature learning performance (36). The original ultrasound image (>1,000×1,000 pixels) was input to VGG16, and then the RPN was used to generate possible bounding boxes of the nodules (Figure 1).
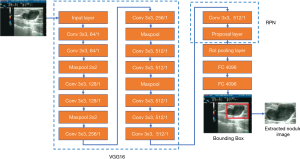
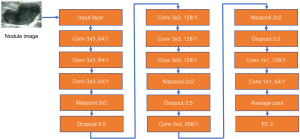
The bounding box’s nodule image was scaled to 128×128 pixels and input to the subsequent classifier network. The classifier network was composed of a multilayer convolutional neural network including three consecutive modules, where each module included some convolution layers, a maximum pooling layer, and a dropout layer, as shown in Figure 2. The network output included benign and malignant categories.
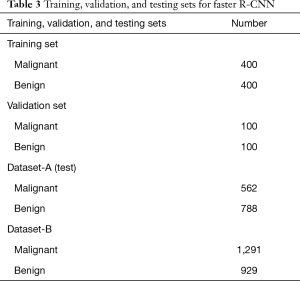
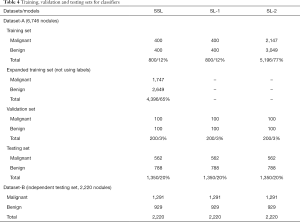
SSL and SL were used to represent semi-supervised learning and supervised learning, respectively. Three versions of the classifier network (Figure 2) were obtained based on different training datasets and learning methods (SSL and SL), including two SL versions (SL-1 and SL-2) and an SSL version. Dataset-A was randomly divided into a training set, validation set, and testing set (Tables 3,4). Table 4 lists the 3 cases of the classifiers’ training, including a small number of labeled nodule images, a small number of nodule images plus a large number of unlabeled images, and a large number of labeled nodule images. The 20% of the data in Dataset-A and all in Dataset-B (as an independent testing set) were used for testing.
Full table
Full table
The SL-1 model was trained on a small number of labeled nodule images (15%, 1,000) for SL. The labeled nodules were divided into a training set (800 nodules) and a validation set (200 nodules). However, the SL-2 model was trained on a large number of labeled nodule images (80%, 5,396 nodules), which were also divided into a training set (n=5,196) and a validation set (n=200). The SSL model was the proposed semi-supervised model trained on the same training set (n=800) and validation set (n=200) with SL-1. However, the remaining nodules (4,396 nodules) were also used for SSL, but their labels were ignored (as unlabeled).
The SSL model was trained using the mean teacher learning strategy (34), including a student network and a teacher network (Figure 3). The teacher network was considered as the SSL model to recognize breast nodules. The two networks had the same network architecture as the classifier network (SL-1 and SL-2). The teacher network provided the pseudo-labels for unlabeled nodules, which were used to calculate mean square error (consistency cost) with predicted labels by the student network. The cross-entropy between the predicted labels by the student network and the real labels was calculated as the classification cost. The sum of consistency cost and classification cost, as the total cost, was used to train the student network in each iteration. However, the weights
of teacher network at training step t were updated by way of the exponential moving average. We defined , where θt was the weights of the student network at training step t, and α was the smoothing coefficient.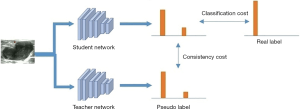
Faster R-CNN fine-tuning for ultrasound images
The training, validation, and testing sets of the Faster R-CNN are shown in Table 3. The bounding boxes of 800 labeled nodules were used to train the Faster R-CNN (these nodules came from the training set in Dataset-A, shown in Table 4). The network was initialized using a Faster R-CNN pre-trained model, which was trained on ImageNet, downloaded from the link on the Faster R-CNN website (37), and then fine-tuned by ultrasound images.
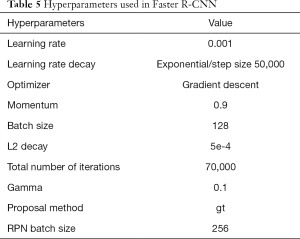
The ultrasound images in the validation set were used for hyperparameter selection. We followed the tuning process to achieve the generalized performance and tried some different parameter values, including learning rate (0.01, 0.001, 0.0005), batch size (16, 64, 128), and L2 decay (0.001, 0.0005, 0.000). Other parameter values were consistent with the recently published Faster R-CNN code. The hyperparameter settings with the highest average similarity on the validation set were selected.
The hyperparameters are listed in Table 5. In order to prevent the model from overfitting early, the training was divided into two stages. Firstly, the feature extraction layers of the VGG16 network were frozen, while the ultrasound images updated the weights of the RPN. After 5,000 iterations, the VGG16 was allowed to be updated. The initial learning rate was set to 0.001, which decreased exponentially. The batch size was 128, the optimizer was a gradient descent optimizer, momentum was 0.9, and the number of iterations was 70,000.
Full table
Classifier network training
The classifier networks (SL-1 and SL-2 models) performed SL using 800 and 5,196 labeled nodules, respectively, while the SSL model used 800 labeled and 4,396 unlabeled nodules, as shown in Table 4. In SSL, the teacher network was initialized with the student network. By closing the teacher network pipeline for calculating pseudo-labels with unlabeled data, supervised training can be achieved. The 200 labeled nodules in the validation set were used for the hyperparameter selection.
The hyperparameters near the hyperparameter settings provided by Tarvainen (34,38) were used to obtain satisfactory results in the validation set. Data augmentation such as image flipping with uniform distribution (0.0, 2.0) and random translation with scale 2 was used. Some different parameter values were tried, including learning rate (0.005, 0.003, 0.001), batch size (64.0, 128.0), iteration number (40,000.0, 100,000.0), ramp-up length (5,000.0, 25,000.0, 40,000.0), and ramp-down length (5,000.0, 25,000.0, 40,000.0). Other parameters were consistent with the previously published study (34), in which the convolutional kernels were initialized by Gaussian distribution with a mean of 0 and a standard deviation of 0.05.
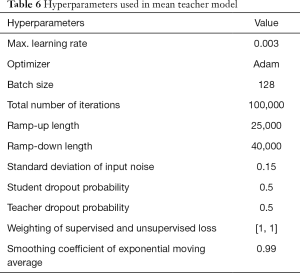
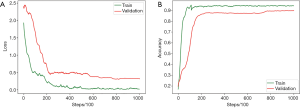
The optimal parameters with the highest recognition accuracy in the validation set were selected. The parameter values are listed in Table 6. The batch size was set to 128. The maximum learning rate was set to 0.003, and the ramp-up and down strategies were used in training. The learning rate slowly grew to the maximum, was maintained, and then started to decline. The change in the learning rate speed was determined by the parameters: ramp-up length, ramp-down length, the total number of iterations, and current training step (38). The total number of iterations was initially set to 100,000, but the training would be stopped early if the validation set’s accuracy could not be improved. In SSL, the same proportion of labeled and unlabeled nodules was maintained in each batch because of the imbalance between the labeled and unlabeled images. The student network weights were updated in each step, but the teacher network used the exponential moving average to update the weights, and the smoothing coefficient was set to 0.99. The training curves of the SSL model are shown in Figure 4.
Full table
These codes were implemented in Python version 3.6.9 (39) and Tensorflow version 1.15.0 (40) and were trained and tested on the servers with an NVIDIA Tesla V00 Graphic Processing Unit, 128 GB memory, and two Intel Xeon Gold 5122 central processing units.
Results
The 20% data of Dataset-A and all the data of Dataset-B were used for performance evaluation. The results included the detection accuracy of nodule location, and sensitivity [true positive/(true positive + false negative)], specificity [true negative/(true negative + false positive)], accuracy [(true positive + true negative)/all samples], and area under the curve (AUC) of the recognition of benign or malignant status.
Detecting the position of breast nodules
The average similarity criterion (ASC) was used to evaluate the detection accuracy, which was defined as
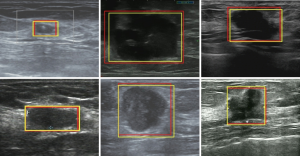
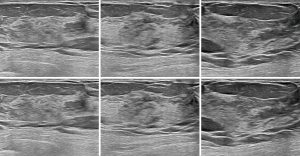
, where R1 was the location of the bounding box given by the sonographers, but R2 was the bounding box predicted by Faster R-CNN. The ASC was the intersection of R1 and R2, divided by their union. The mean and standard deviation of ASC on the testing sets in Dataset-A (testing set, 1,350) and Dataset-B (2,220) were 0.88±0.03 and 0.86±0.02, respectively.The mean value of ASC on the two testing sets was higher than 0.85, indicating that the detection network locations were very consistent with the locations given by the sonographers. Figure 5 shows some bounding boxes of nodules, where the yellow bounding box represents the box given by the sonographers, and the red represents the box predicted by Faster R-CNN, which could achieve accurate detection of breast nodules in ultrasound images. This detection network was also effective for normal breast images, as shown in Figure 6.
Recognition of benign or malignant type nodules
The training set, validation set, and the testing set from Dataset-A were repeatedly and randomly allocated 10 times to estimate the statistics of classifier networks’ performance according to the proportions shown in Table 4. Dataset-B was used for independent testing to evaluate the generalization ability of classification accuracy across multiple medical centers.
The SSL, SL-1 and SL-2 models in two testing sets shown in Table 4 were tested, as shown in Table 7. The SSL model used the teacher network of mean teacher model as the recognition model, which had the same network structure as the SL-1 and SL-2 models. When 800 labeled nodules were used for supervised training, the AUC of SL-1 was 0.83±0.050, making it difficult to achieve satisfactory clinical practice performance. In contrast, when a large number of unlabeled nodules (4,396.0) were added to SSL for semi-supervised training, the AUC of the SSL model was significantly increased to 0.934±0.026. Moreover, the SSL model’s performance was comparable to that of the SL-2 model trained on 5,196 labeled nodules (0.934±0.026 vs. 0.952±0.027).

Full table
The tests on Dataset-B are shown in Table 7, demonstrating whether the conclusions above were still applicable to the independent testing set. The performance of the SSL model was better than that of the SL-1 model (0.916±0.022 vs. 0.815±0.049) and close to that of the SL-2 model (0.916±0.022 vs. 0.918±0.017). The generalization performance of the SSL model across different datasets was also maintained.
The results showed that when 4,396 unlabeled nodules were added for training, the SSL achieved a significant improvement in performance over the SL-1 model without using unlabeled data (P<10−5). Moreover, no significant difference (P=0.11) was found between SSL using a small number of labeled nodules (800.0) plus a larger number of unlabeled nodules (4,396.0), and the SL-2 model using a massive set of labeled nodules (5,196.0).
Human-AI comparison
Five sonographers (A–E) were recruited to evaluate the performance of SSL in clinical applications, and their years of clinical experience were 3–15 (A: 3 years, B: 4 years, C: 7 years, D: 10 years, and E: 15 years). They independently diagnosed 2,220 breast nodules from Dataset-B.
The AUCs of the five human sonographers, average AUC, and SSL were ranked. The AUC of SSL was 0.922, ranked as second; it was lower only than that of sonographer D. The average AUC of the five sonographers was 0.889, indicating that the diagnostic ability of SSL was better than the average ability of the human sonographers. The SSL could achieve excellent accuracy and was practically useful for breast cancer diagnosis (Figure 7, Table 8).
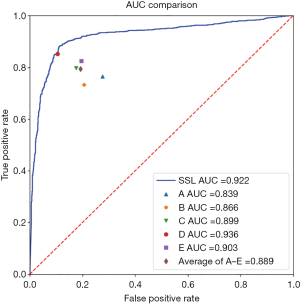
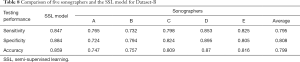
Full table
Comparison with related studies
The SSL was compared with three previous deep learning methods of breast ultrasound analysis; the results are shown in Table 9: SSL performance was comparable to that of previous SL methods.
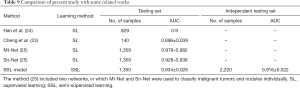
Full table
Discussion
Ultrasound is an important screening method for breast cancer, but accurate diagnosis of breast ultrasonographic images requires years of training, and the challenges have never been overcome due to noise, artifacts, and low contrast in images (10). Computer-assisted systems are expected to provide a powerful approach to reducing the high rates of false positives and false negatives of breast cancer (11).
Deep learning can accurately classify breast nodules, with an AUC even exceeding 0.9, reaching the sonographers’ level (22-25). From a methodological point of view, almost all successful methods currently depend on SL, requiring thousands or more of nodule images and pathological results to ensure the convergence of neural networks. Data annotation aims to associate the samples with the ground truth, thus providing an optimization direction for SL. A huge number of images and reports must be reviewed one by one, followed by sonographers’ manual labeling. Moreover, it is more common clinically that many samples do not have ground truth, such as prospective studies (27). Many patients undergo breast ultrasound but not a pathological examination.
The data preparation and annotations have become one of the most demanding tasks in medical deep learning systems, not just for breast ultrasound. How to efficiently prepare and use medical data is an urgent issue (27). Recently, methods requiring less labeled data have gained attention, such as SSL (29-32).
Some studies have proven that when labeled data is limited, SSL can achieve good results in some medical images (29-32). However, this conclusion has not been evaluated on a large scale, especially for medical images with large amounts of noise, artifacts, and low contrast, for example, breast ultrasound. Whether unlabeled data can improve the recognition accuracy in breast nodules is unclear.
This current study proved SSL performance for a small number of labeled nodules was close to that of SL in massive datasets of labeled nodules. The SSL model trained on 800 labeled nodules and 4,396 unlabeled nodules might be as good as the SL-2 model trained on 5,196 labeled nodules. Moreover, SSL was also comparable to some previous studies of SL on breast ultrasound. Also, we compared the diagnostic accuracy of the SSL model and five sonographers. The SSL model surpassed four of the five sonographers and ranked second, revealing the feasibility of building an excellent expert-level system based on SSL.
SSL might not need as many labeled breast nodules as for previous methods based on SL. The main highlight of the present study was proposing a semi-supervised breast nodule recognition method, which proved that recognition accuracy on large, noisy ultrasound images could be achieved even if the number of labeled nodules was significantly reduced.
The proposed method may also be extended to other medical imaging techniques with better quality images and a higher signal-to-noise ratio than ultrasound images. This study confirmed that SSL has great potential for clinical practice lacking labeled data. The existing SL can be replaced with SSL to alleviate data preparation difficulty in medical computer-assisted systems significantly.
Conclusions
This study proposed a detection and recognition method for breast nodules based on SSL, which was trained on a small amount of labeled data. The proposed method’s performance was as good as that of SL trained on a large number of nodules and was better than the accuracy of four out of five sonographers. The study revealed the application prospects of SSL in the medical imaging domain, greatly reducing the cost and time involved in current medical data annotation. Future studies should aim to reduce the amount of data annotations further and achieve an unsupervised learning method that does not require any annotations.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-12b). The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Institutional Review Board of the Third Affiliated Hospital of Xi’an Jiaotong University, and a general research authorization was obtained allowing for retrospective reviews. Informed consent was waived by the Institutional Review Board.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Leong ASY, Zhuang Z. The changing role of pathology in breast cancer diagnosis and treatment. Pathobiology 2011;78:99-114. [Crossref] [PubMed]
- Oeffinger KC, Fontham ET, Etzioni R, Herzig A, Michaelson JS, Shih YC, Walter LC, Church TR, Flowers CR, LaMonte SJ, Wolf AM, DeSantis C, Lortet-Tieulent J, Andrews K, Manassaram-Baptiste D, Saslow D, Smith RA, Brawley OW, Wender RAmerican Cancer Society. Breast cancer screening for women at average risk: 2015 guideline update from the American cancer society. JAMA 2015;314:1599-614. [Crossref] [PubMed]
- Evans DG, van Veen EM, Howell A, Astley S. Heritability of mammographic breast density. Quant Imaging Med Surg 2020;10:2387-91. [Crossref] [PubMed]
- Kerlikowske K, Zhu W, Tosteson AN, Sprague BL, Tice JA, Lehman CD, Miglioretti DLBreast Cancer Surveillance Consortium. Identifying women with dense breasts at high risk for interval cancer: a cohort study. Ann Intern Med 2015;162:673-81. [Crossref] [PubMed]
- Jo HM, Lee EH, Ko K, Kang BJ, Cha JH, Yi A, Jung HK, Jun JK. Alliance for breast Cancer Screening in Korea (ABCS-K). Prevalence of women with dense breasts in Korea: results from a nationwide cross-sectional study. Cancer Res Treat 2019;51:1295-301. [Crossref] [PubMed]
- Berg WA, Rafferty EA, Friedewald SM, Hruska CB, Rahbar H. Screening algorithms in dense breasts: AJR expert panel narrative review. AJR Am J Roentgenol 2021;216:275-94. [Crossref] [PubMed]
- Cozzi A, Schiaffino S, Giorgi Rossi P, Sardanelli F. Breast cancer screening: in the era of personalized medicine, age is just a number. Quant Imaging Med Surg 2020;10:2401-7. [Crossref] [PubMed]
- Brem RF, Lenihan MJ, Lieberman J, Torrente J. Screening breast ultrasound: past, present, and future. AJR Am J Roentgenol 2015;204:234-40. [Crossref] [PubMed]
- Stower H. AI for breast-cancer screening. Nat Med 2020;26:163. [PubMed]
- Jalalian A, Mashohor S, Mahmud R, Saripan MI, Ramli A, Karasfi B. Computer-aided detection/diagnosis of breast cancer in mammography and ultrasound: a review. Clin Imaging 2013;37:420-6. [Crossref] [PubMed]
- Chen DR, Chang RF, Chen CJ, Ho MF, Kuo SJ, Chen ST, Hung SJ, Moon WK. Classification of breast ultrasound images using fractal feature. Clin Imaging 2005;29:235-45. [Crossref] [PubMed]
- Tianur, Nugroho HA, Sahar M, Ardiyanto I, Indrastuti R, Choridah L. Classification of breast ultrasound images based on posterior feature. Proceedings of 2016 International Conference on Information Technology Systems and Innovation (ICITSI), 2016 Oct 24-27, Bandung, Indonesia. New York: IEEE, 2016.
- Kuo SJ, Hsiao YH, Huang YL, Chen DR. Classification of benign and malignant breast tumors using neural networks and three-dimensional power doppler ultrasound. Ultrasound Obstet Gynecol 2008;32:97-102. [Crossref] [PubMed]
- Pang S, Yu Z, Orgun MA. A novel end-to-end classifier using domain transferred deep convolutional neural networks for biomedical images. Comput Methods Programs Biomed 2017;140:283-93. [Crossref] [PubMed]
- Abdi AH, Luong C, Tsang T, Allan G, Nouranian S, Jue J, Hawley D, Fleming S, Gin K, Swift J, Rohling R, Abolmaesumi P. Automatic quality assessment of echocardiograms using convolutional neural networks: feasibility on the apical four-chamber view. IEEE Trans Med Imaging 2017;36:1221-30. [Crossref] [PubMed]
- Chen H, Wu L, Dou Q, Qin J, Li S, Cheng JZ, Ni D, Heng PA. Ultrasound standard plane detection using a composite neural network framework. IEEE Trans Cybern 2017;47:1576-86. [Crossref] [PubMed]
- Baka N, Leenstra S, Walsum T. Ultrasound aided vertebral level localization for lumbar surgery. IEEE Trans Med Imaging 2017;36:2138-47. [Crossref] [PubMed]
- Yu L, Guo Y, Wang Y, Yu J, Chen P. Segmentation of fetal left ventricle in echocardiographic sequences based on dynamic convolutional neural networks. IEEE Trans Biomed Eng 2017;64:1886-95. [Crossref] [PubMed]
- Byra M, Piotrzkowska-Wrblewska H, Dobruch-Sobcza KK, Nowicki A. Combining Nakagami imaging and convolutional neural network for breast lesion classification. Proceedings of 2017 IEEE International Ultrasonics Symposium (IUS), 2017 Sep 6-9, Washington DC, USA. New York: IEEE, 2017.
- Yap MH, Pons G, Marti J, Ganau S. Sentis Me, Zwiggelaar R, Davison AK, Marti R. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE J Biomed Health Inform 2018;22:1218-26. [Crossref] [PubMed]
- Cheng JZ, Ni D, Chou YH, Qin J, Tiu CM, Chang YC, Huang CS, Shen D, Chen CM. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci Rep 2016;6:24454. [Crossref] [PubMed]
- Han S, Kang HK, Jeong JY, Park MH, Kim W, Bang WC, Seong YK. A deep learning framework for supporting the classification of breast lesions in ultrasound images. Phys Med Biol 2017;62:7714-28. [Crossref] [PubMed]
- Qi X, Zhang L, Chen Y, Pi Y, Chen Y, Lv Q, Yi Z. Automated diagnosis of breast ultrasonography images using deep neural networks. Med Image Anal 2019;52:185-98. [Crossref] [PubMed]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Willemink MJ, Koszek WA, Hardell C, Wu J, Fleischmann D, Harvey H, Folio LR, Summers RM, Rubin DL, Lungren MP. Preparing medical imaging data for machine learning. Radiology 2020;295:4-15. [Crossref] [PubMed]
- Wang D, Zhang Y, Zhang K, Wang LW. FocalMix: semi-supervised learning for 3D medical image detection. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). 2020 Jun 13-19., Seattle WA, USA. IEEE, 2020:3951-60.
- Ruijsink B, Puyol-Anton E, Li Y, Bai W, Kerfoot E, Razavi R, King AP. Quality-aware semi-supervised learning for CMR segmentation. arXiv: 2009.00584. 2020. Available online: https://arxiv.org/abs/2009.00584
- Li S, Zhang C, He X. Shape-aware semi-supervised 3D semantic segmentation for medical images. arXiv: 2007.10732. 2020. Available online: https://arxiv.org/abs/2007.10732
- Chen C, Qin C, Qiu H, Ouyang C, Wang S, Chen L,Tarroni G, Bai W, Rueckert D. Realistic adversarial data augmentation for MR image segmentation. arXiv: 2006.13322. 2020. Available online: https://arxiv.org/abs/2006.13322
- Shaw S, Pajak M, Lisowska A, Tsaftaris SA, ONeil AQ. Teacher-student chain for efficient semi-supervised histology image classification. arXiv: 2003.08797. 2020. Available online: https://arxiv.org/abs/2003.08797.
- Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell 2017;39:1137-49. [Crossref] [PubMed]
- Tarvainen A, Valpola H. Mean teachers are better role models: weight-averaged consistency targets improve semi-supervised deep learning results. arXiv:1703.01780. 2018. Available online: https://arxiv.org/pdf/1703.01780
- Tzutalin. Labelimg Software. Version 1.0.0 [software]. 2019 Dec 23. Available online: https://github.com/tzutalin/labelImg
- Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv: 1409.1556. 2015. Available online: https://arxiv.org/pdf/1409.1556
- Chan FH. Faster-RCNN_TF. Version 0.4.9 [software]. 2017 Mar 13. Available online: https://github.com/smallcorgi/Faster-RCNN_TF
- Tarvainen A. Mean-teacher. Version 0.4.0 [software]. 2018 May 31. Available online: https://github.com/CuriousAI/mean-teacher
- Python Software Foundation. Python. Version 3.6.9 [software]. 2019 June 18. Available online: https://www.python.org
- Google Inc. Tensorflow. Version 1.15.0 [software]. 2019 Oct 17. Available online: https://pypi.org/project/tensorflow


