
The mutation profiles of cell-free DNA in patients with oesophageal squamous cell carcinoma who were responsive and non-responsive to neoadjuvant chemotherapy
Introduction
Oesophageal cancer was identified as the leading cause of cancer-related death in China. Oesophageal cancer has two main subtypes, oesophageal squamous cell carcinoma (OSCC) and oesophageal adenocarcinoma (1). OSCC accounts for 90% of the cases of oesophageal cancer (2). Neoadjuvant chemotherapy (NAC) is the most common treatment approach for patients with resectable oesophageal cancer. The results of meta-analyses indicated that NAC significantly reduced the 3-year mortality and locoregional recurrence rates when compared with surgery alone (3,4). Clinical evidence indicated that 19% of patients randomly assigned to receive chemotherapy had major objective regressions (7% complete radiographic regression and 12% partial radiographic regression) (5). For non-responsive patients, there is a risk of disease progression and a loss of the chance for surgery. Therefore, determining how to select patients who should receive NAC is a vital clinical problem.
Recently, to avoid invasive and repeated tissue biopsies, the analysis of circulating cell-free DNA (cfDNA) was applied for screening, diagnosis, predicting response to therapy, monitoring the tumour burden and early diagnosis of relapse (6-8). Hsieh et al. reported that circulating cfDNA could predict the outcome of OSCC patients undergoing oesophagectomy. Higher cfDNA levels were associated with tumour relapse and shorter disease-free survival (DFS) after oesophagectomy in OSCC patients (9). This study aimed to explore the plasma DNA mutation profile in OSCC patients who responded and those who did not respond to NAC and to screen for molecular biomarkers to predict the efficacy of neoadjuvant chemotherapy.
We present the following article in accordance with the REMARK Guideline.
Methods
Patients
To investigate the impact of NAC on the survival of patients with operable advanced OSCC, we registered a clinical study in ClinicalTrails.Gov (NCT02395705). The clinical study has been enrolled and follow-up has not been completed, so there is no median follow-up data for the above cases. All the patients in this retrospective study were from the patients enrolled in the above clinical study. In this retrospective analysis, 22 stage IIA to IIIB OSCC patients were recruited from the Affiliated Cancer Hospital of Zhengzhou University (Henan Cancer Hospital) between April 2015 and July 2016 according to the inclusion criteria. All subjects received NAC and were stratified in two clinical response groups. Seven cases reached progressive disease (PD) or stable disease (SD), and 15 cases reached partial response (PR) or complete response (CR). This study aimed to screen for molecular biomarkers to predict the efficacy of NAC by exploring the plasma DNA mutation profile in OSCC patients between responders and non-responders.
Inclusion criteria
❖ Histologic diagnosis of squamous cell thoracic oesophageal cancer stage IIA to IIIB [7th Union for International Cancer Control (UICC)-TNM];
❖ Patients must not have received any prior anticancer therapy for oesophageal cancer;
❖ Aged 18 to 75 years old;
❖ Without operative contraindication;
❖ Absolute white blood cells count ≥4.0×109/L, neutrophil ≥1.5×109/L, platelets ≥100.0×109/L, hemoglobin ≥90 g/L, and normal liver and kidney functions, total bilirubin (TBIL) ≤1.5 N (upper limit of normal), aspartate aminotransferase (AST) ≤2.5 N, alanine aminotransferase (ALT) ≤2.5 N, prothrombin time (PT) ≤1.5 N, normal range of activated partial thromboplastin time (APTT), and endogenous creatinine clearance rate (CRE) ≤1.5 N;
❖ Patients must not have been diagnosed with other cancer and must not have received any prior anticancer therapy except for prostate cancer with greater than 5 years of DFS;
❖ Expected R0 resection;
❖ ECOG 0–2;
❖ Signed informed consent document on file;
❖ No metastatic lymph node in cervical by color Doppler sonography.
Exclusion criteria
❖ Multiple primary cancer;
❖ The subject cannot understand and sign the informed consent form (ICF);
❖ Patients with concomitant hemorrhagic disease;
❖ Patients who cannot undergo the operation for any unexpected reason;
❖ Inability to use gastric conduit after esophagectomy due to a prior surgery;
❖ Pregnant or breast-feeding;
❖ Patients are diagnosed as or suspected to be allergic to cisplatin or paclitaxel.
Randomization
The investigators took the responsibility to enroll the patients. First, the eligibility criteria are confirmed. Second, patients are randomized to the NAC or surgery alone group.
Randomization and masking
The randomization numbers were generated by a centrally located computer. Patients were randomly assigned (1:1). All the randomized numbers were sealed into envelopes and sent to local sites. After the patient signs the written consent form, the envelope is opened, and the randomized group is unsealed. The trial is unmasked.
Treatment
Neo-adjuvant chemotherapy group (cisplatin and paclitaxel)
- Paclitaxel, 175 mg/m2, d1, cisplatin, 25 mg/m2, d2–4; 3 weeks, 2 cycles;
- Paclitaxel, 87.5 mg/m2, d1, d8, cisplatin, 25 mg/m2, d2–4; 3 weeks, 2 cycles;
- Paclitaxel, 175 mg/m2, d1, cisplatin, 75 mg/m2, d1; 3 weeks, 2 cycles.
Plasma cfDNA extraction
Blood samples were collected in cfDNA BCT tubes containing a formaldehyde-free preservative reagent that could stabilize nucleated blood cells (10,11). Plasma was separated by centrifugation (1,600 g for 10 min) and was then transferred into 1.5 mL Eppendorf tubes (Axygen, Corning, NY, USA) for further centrifugation at 16,000 g for 10 min at 4 °C. The plasma cfDNA was isolated from 1–2 mL of plasma with a MagMAXTM Cell-Free DNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA). Blood cell DNA was extracted with a TIANamp Blood DNA Kit (TIANGEN, Beijing, China). Tumour DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) specimens with a black PREP FFPE DNA Kit (Analytikjena, Jena, Germany). The concentration of DNA was quantified with a Qubit DNA dsDNA Assay Kit (Thermo Fisher Scientific).
Library preparation
For cfDNA samples, 10–50 ng DNA was used for library construction. Extracted DNA (200–500 ng) was sheared into 200-bp fragments with a Covaris M220 Focused-ultrasonicator. Libraries were prepared with KAPA hyper preparation kit (Kapa Biosystems, Wilmington, MA, USA). The ligated fragments were then amplified for 6–11 PCR cycles, and the libraries were purified with agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA) for a double size selection. Quality control of the libraries was performed with a Qubit DNA dsDNA assay kit (Thermo Fisher Scientific) a 2100 Bioanalyzer with the DNA 1000 Kit (Agilent, Santa Clara, CA, USA).
Hybridization capture and sequencing
NimbleGen SeqCap Hybridization and Wash kit (Roche NimbleGen, Switzerland) were used according to the technical note for targeted region selection. Hybridization probe can capture bulk library DNA from 8–12 indexed Illumina libraries. The probe library was designed through the NimbleDesign portal (Version 2) using genome build hg19 NCBI Build 37.1/GRCh37. All hybridization and wash operations were conducted with the NimbleGen SeqCap Hybridization and Wash Kit (Roche NimbleGen, Basel, Switzerland) following the manufacturer’s manual. After hybridization, two 50 µL library amplification PCR were performed for 14 amplification cycles respectively, and then pooled and purified with Agencourt AMPure XP beads (Beckman-Coulter). Finally, quantification and length determination of the prepared library were performed before sequenced on an Illumina HiSeq X Ten using 150-bp paired-end runs
Bioinformatic pipeline
The paired-end sequencing was performed by Illumina HiSeq X-Ten. The hg19 reference genome was used for read mapping with BWA 0.7.12 (default parameters). Resulting alignments were sorted, filtered and indexed using SAMtools. In order to distinguish somatic single nucleotide polymorphism (SNP) from indel mutations, the obtained BAM files from both blood cell and plasma samples for each patient were processed for pairwise variant calling using VarScan (v2.4.2). For each patient, a so-called pair-wise variant calling procedure was employed, using a blood cell sample as normal control to help eliminate germline mutations in the corresponding plasma samples. In blood cell samples, somatic variants were called based on a minimum coverage of 8, and in plasma samples it was 6. The P value threshold to call a somatic site was 0.05, and variants with ≤90% strand bias were kept for further study. The refGene and Exome Aggregation Consortium database were included in Annovar packages, and the Annovar packages were used for variant annotation to obtain population and gene region information. Finally, non-synonymous mutations in exon regions were retained. The mutation burden of each patient was defined as the average mutation allele frequency of each gene calculated by the obtained somatic mutation.
Patients’ blood cell samples were used to construct copy number baseline which were used as a negative control. CNVkit was used to call copy number variation (CNV) from the plasma samples for each patient. Three main sources of bias, the GC content, target footprint size and spacing, and repetitive sequences, which can induce the extraneous variability of sequencing read depth were evaluated and corrected as well.
Statement of ethics approval
This work was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by Ethics Committee of the Affiliated Cancer Hospital of Zhengzhou University (Henan Cancer Hospital) (NO.: 2014ys38) and informed consent was taken from all the patients.
Statistical analysis
Clinical characteristics of patients were summarized using descriptive statistics. Group values were fit to a normal distribution test. Student’s t-test was used to analyze normally distributed data. Nonparametric tests were also used because the data were not normally distributed. P<0.05 was considered statistically significant. Statistical analysis was performed using standard statistical software (SPSS version 16.0, Inc., Chicago, IL, USA). The Python Package Python 3.6-seaborn was used for the copy number clustering and heat map presentation.
Results
Clinical characteristics and their association with cfDNA concentrations of patients with OSCC
A total of 22 patients with OSCC were recruited for the study from the Affiliated Cancer Hospital of Zhengzhou University (Henan Cancer Hospital) between April 2015 and July 2016. Table 1 summarizes the clinical characteristics of the patients. All subjects received NAC and were divided into two groups according to their response to the therapy. Fifteen patients were in the response group, and seven patients were in the non-response group. The cfDNA concentrations were not significantly different between patients who responded and patients who did not respond to NAC (Table 1).
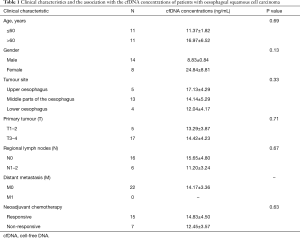
Full table
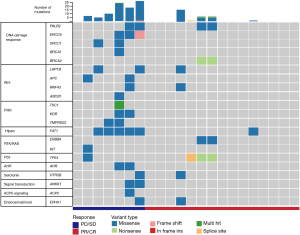
The molecular profile of patients with OSCC in the response and non-response groups
We collected a pretreatment blood sample from patients who responded and patients who did not respond to NAC. The plasma cfDNA and white blood cell DNA were sequenced by a cancer gene-targeted next-generation sequencing (NGS) panel. The significantly mutated genes were identified. As shown in Figure 1, the most frequently mutated genes included PALB2, ERCC6, XRCC1, BRCA1, BRCA2, LRP1B, APC, RNF43, TSC1, KDR, TMPRSS2, FAT1, ERBB4, KIT, TP53, AHR, HTR3B, ANKK1, ACP5, and EPHX1. These mutated genes were involved in the DNA damage response, and the Wnt, PI3K, Hippo, RTK/RAS, P53, AHR, serotonin, ACP5, and endocannabinoid signalling pathways (Figure 1).
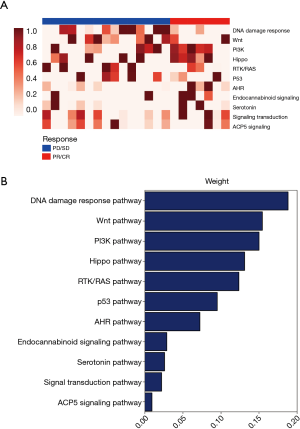
The predictive value of the baseline mutational burden for predicting the response to NAC
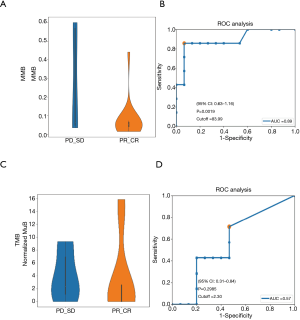
We analysed the pathway score of the driver gene molecular mutation burden (MMB) between the responsive and non-responsive patients. We found that the differential MMB score included the DNA damage response, Wnt pathway, PI3K, Hippo pathway, RTK/RAS pathway, p53 pathway, and AHR pathway (Figure 2A). The differential pathways were ranked in order of decreasing weight of the MMB score (Figure 2B). The MMB of the driver genes was significantly higher in the non-responsive group compared with the responsive group (Figure 3A). Moreover, we used the receiver operation characteristic (ROC) curve to assess the predictive value of the baseline MMB for predicting the response to NAC. The MMB had an area under the ROC curve of 0.89 for predicting the response to NAC (Figure 3B) (AUC =0.89, P=0.0019). In contrast, higher blood tumour mutational burden (TMB) levels were observed in the responsive group than in the non-responsive group (Figure 3C). The TMB had an area under the ROC curve of 0.57 for predicting the response to NAC (Figure 3D) (AUC =0.57, P=0.2985).
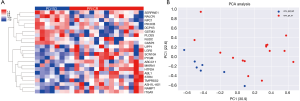
The CNVs of patients with OSCC who were responsive and non-responsive to NAC
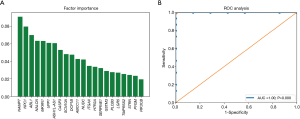
We further investigated the CNV pattern of OSCC patients. The cfDNA and the genomic DNA extracted from white blood cells were analysed in parallel to identify somatic CNVs. We identified 22 genes with CNV patterns that could distinguish patients who responded to NAC from those who did not respond (Figure 4A). Principal component analysis (PCA) showed that the cfDNA CNV profile was different between the baseline cfDNA in patients who responded and patients who did not respond to NAC (Figure 4B). The 22 genes were ranked in order of decreasing importance (Figure 5A). The cfDNA CNVs had an area under the ROC curve of 1.0 for predicting the response to NAC (Figure 5B).
Discussion
NAC is already the standard treatment in Japan and Britain. Our study shed light on potential biomarkers for predicting the response to NAC. We found that the combination of the cfDNA driver gene MMB and CNV profile achieved a good predictive value for the prediction of the response to NAC.
Previous studies have focused on investigating the cfDNA levels as predictive biomarkers for tumour relapse and distant metastasis after oesophagectomy in OSCC patients (9). Few studies have explored the plasma cfDNA molecular profiles of OSCC patients who received NAC. In the current study, we found differences in the molecular alterations between OSCC patients who were responsive and non-responsive to NAC. We found that the differential MMB score affected the DNA damage response, Wnt pathway, PI3K, Hippo pathway, RTK/RAS pathway, p53 pathway, and AHR pathway.
Several studies reported the predictive value of biomarkers for predicting the response to NAC (12,13). According to our data, compared with patients who were non-responsive to NAC, OSCC patients who responded to NAC had a low driver gene MMB at baseline. The MMB had an area under the ROC curve of 0.89 for predicting the response to NAC. In contrast, higher blood TMB levels were observed in the responsive group. The difference between the TMB and MMB may be explained as follows. The TMB was defined as the number of somatic mutations per megabase of the tumour genome examined. It may therefore be speculated that a tumour bearing a higher TMB level may be highly immunogenic and may induce an anti-tumour immune response, leading to a good clinical prognosis. However, the MMB was defined as the sum of the mean variant allele frequency (VAF). The VAF for each somatic mutation was calculated as its variant supporting reads divided by the reference supporting reads. Thus, the TMB may show the overall number of somatic mutations per megabase of the tumour. The MMB showed the individual gene molecular alteration burden of the tumour.
CNV, a type of structural variation, was characterized as a duplication or deletion event that was observed in OSCC patients. The implications of CNVs for the treatment and prediction of OSCC patients’ clinical outcomes are currently underappreciated. In this study, PCA analysis showed that the cfDNA CNVs were different between the baseline cfDNA in patients who responded and patients who did not respond to NAC. The cfDNA CNVs had an area of 1.0 under the ROC curve for predicting the response to NAC. These results suggest that the CNV might be used as a biomarker to predict the response to NAC.
In this study, we explored a candidate model to predict the response to NAC based on the baseline cfDNA driver gene MMB and CNV. This model achieved good sensitivity and specificity. However, a prospective cohort study should be designed to validate the clinical predictive value.
Acknowledgments
Funding: This work was supported by grants from the Medical and Health Technology Innovation Project of Chinese Academy of Medical Sciences (2018-12M-3-003) (Spatial-Temporal Mapping Analysis on Chinese Cancer Burden) and National Key Sci-Tech Special Project of China (No. 2018ZX10302207).
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-230
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-230). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This work was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by Ethics Committee of the Affiliated Cancer Hospital of Zhengzhou University (Henan Cancer Hospital) (NO.: 2014ys38) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Tu CC, Hsu PK. The frontline of esophageal cancer treatment: questions to be asked and answered. Ann Transl Med 2018;6:83. [Crossref] [PubMed]
- Iyer R, Wilkinson N, Demmy T, et al. Controversies in the multimodality management of locally advanced esophageal cancer: evidence-based review of surgery alone and combined-modality therapy. Ann Surg Oncol 2004;11:665-73. [Crossref] [PubMed]
- Urschel JD, Vasan H. A meta-analysis of randomized controlled trials that compared neoadjuvant chemoradiation and surgery to surgery alone for resectable esophageal cancer. Am J Surg 2003;185:538-43. [Crossref] [PubMed]
- Kelsen DP, Winter KA, Gunderson LL, et al. Long-term results of RTOG trial 8911 (USA Intergroup 113): a random assignment trial comparison of chemotherapy followed by surgery compared with surgery alone for esophageal cancer. J Clin Oncol 2007;25:3719-25. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015;21:795-801. Erratum in: Nat Med. 2015 Jul;21(7):doi:10.1038/nm0715-827b; Nat Med. 2015 Jul;21(7):827. doi: 10.1038/nm0715-827b. [Crossref] [PubMed]
- Xu RH, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater 2017;16:1155-61. [Crossref] [PubMed]
- Hsieh CC, Hsu HS, Chang SC, et al. Circulating Cell-Free DNA Levels Could Predict Oncological Outcomes of Patients Undergoing Esophagectomy for Esophageal Squamous Cell Carcinoma. Int J Mol Sci 2016;17:2131. [Crossref] [PubMed]
- Norton SE, Lechner JM, Williams T, et al. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin Biochem 2013;46:1561-5. [Crossref] [PubMed]
- Das K, Fernando MR, Basiaga S, et al. Effects of a novel cell stabilizing reagent on DNA amplification by PCR as compared to traditional stabilizing reagents. Acta Histochem 2014;116:55-60. [Crossref] [PubMed]
- Hsu FM, Cheng JC, Chang YL, et al. Circulating mRNA Profiling in Esophageal Squamous Cell Carcinoma Identifies FAM84B As A Biomarker In Predicting Pathological Response to Neoadjuvant Chemoradiation. Sci Rep 2015;5:10291. [Crossref] [PubMed]
- Wen J, Luo K, Liu H, et al. MiRNA Expression Analysis of Pretreatment Biopsies Predicts the Pathological Response of Esophageal Squamous Cell Carcinomas to Neoadjuvant Chemoradiotherapy. Ann Surg 2016;263:942-8. [Crossref] [PubMed]


