DNAJB4 identified as a potential breast cancer marker: evidence from bioinformatics analysis and basic experiments
Introduction
Breast cancer (BC) is the most common malignant tumor in women worldwide (1,2). Although some studies have pointed out that BC can be prevented (3-6), its morbidity and mortality have rapidly climbed to the top of the list of female tumors in recent years (2). In 1894, Halsted put forward the classical radical mastectomy, which emphasized the complete resection of the tumor (7). But the success rate of BC treatment was still worrying (8). With the deepening of research, Fisher, an American scholar, first proposed that BC was a systemic disease in the 1970s (9-11). This theory provided a theoretical basis for the application of anthracyclines and taxanes in postoperative adjuvant chemotherapy for BC, and laid the foundation for modern comprehensive treatment of BC. At present, the standard process of clinical diagnosis and treatment of BC is the combination of surgery, chemotherapy, radiotherapy, endocrine therapy and targeted therapy after medical imaging detection, tumor marker detection and pathological biopsy (12). Compared with the treatment of BC in the 1980s, the current treatment regimen has improved the prognosis of patients with BC to some extent (2). This method is effective in the treatment of early BC, but it is not effective in patients with advanced BC or distant metastasis (13). Since most of the patients diagnosed with BC for the first time are advanced BC (14-16), the above-mentioned schemes have been difficult to meet the current needs of BC diagnosis and treatment. Targeted therapy as an important part of BC treatment, a large number of studies have shown that HER-2 overexpression of BC patients with HER-2 targeted drugs trastuzumab and pertuzumab can significantly improve the prognosis of patients (17,18). However, HER-2 overexpression BC accounts for only 14% of clinical BC patients (2). Finding new and effective targets for BC therapy has become an urgent problem to be solved in the process of clinical diagnosis and treatment of BC.
In recent years, with the rapid development of gene sequencing technology and the establishment of a large number of tumor database, using bioinformatics technology to screen potential tumor diagnosis and treatment targets has become an important branch of contemporary medicine (19-21). Through bioinformatics techniques, Ge et al. (22) found that ZGRF1 may be associated with the poor prognosis of triple negative BC. Li et al. (23) reported that mitochondrial proteins interacting with ZW10 can be used as prognostic biomarkers of human BC. In this study, differentially expressed genes (DEGs) including DNAJB4 were screened by analyzing the gene chip downloaded from GEO data. Because DNAJB4 was at the core of the PPI network we built, and it was valued by the research team. However, due to the limitations of contemporary sequencing technology and the great heterogeneity of patients in different data sets, the current bioinformatics analysis results still need to be verified by laboratory data. Although DNAJB4 has been reported in previous studies on BC, the studies were mostly limited to the relationship between DNAJB4 expression level and clinical prognosis or tumor drug resistance (24,25). There were few studies on the biological behavior and related signal pathways of DNAJB4 in BC cells. Therefore, this study intends to use the combination of bioinformatics technology and basic experimental technology to identify the biological processes (BP) in BC, in which DNAJ4 was involved, and predicted its related signaling pathways, so as to provide new ideas for the clinical diagnosis and treatment of BC. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/gs-20-431).
Methods
Data sources
Chip data
Gene Expression Omnibus database (GEO), is a gene expression database created and maintained by The National Center for Biotechnology Information (NCBI). Founded in 2000, it contains high-throughput gene expression data submitted by research institutions around the world, and the chip data of all database are open, free and shared to researchers all over the world. We downloaded two microarrays including GSE15852 (26) from HG-U133A Affymetrix Human Genome U133A Array (Platform GPL96) and GSE42568 (27) from HG-U133_Plus_2 Affymetrix Human Genome U133 Plus 2.0 Array (Platform GPL 570) of GEO. In the GSE15852 data set, we selected 86 gene expression profiles, including 43 normal breast tissue samples and 43 primary tumor samples from BC patients. In the GSE42568 data set, we selected 121 gene expression profiles, including 17 normal breast tissue samples and 104 primary tumor samples from BC patients.
Characteristics of patients and specimens
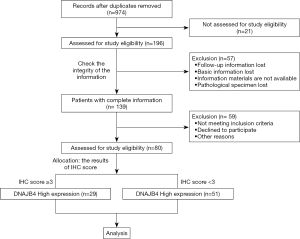
This study adopted the purposive sampling method to choose the patient, who were treated in the First Affiliated Hospital of Wannan Medical College. All patients were admitted and treated according to Guidelines of Chinese society of Clinical Oncology (BC). The inclusion criteria were established as follows: (I) no neoadjuvant chemotherapy or neoadjuvant endocrine therapy before operation; (II) the surgical method accepted by patients was modified radical mastectomy or breast-conserving surgery; (III) methylene blue lymph node tracing method was used during the operation; and the scope of axillary lymph nodes were dissected on the affected side included at least area I and II. (IV) The routine pathology report confirmed that it was invasive BC. (V) The clinical staging of BC is in stage I-II, according to the AJCC staging guidelines. (VI) The postoperative chemotherapy and radiotherapy were completed on time according to the postoperative BC pathological classification. Exclusion criteria: (I) incomplete basic information; (II) bilateral BC; (III) recurrent secondary operation of BC; (VI) complicated with other malignant tumors; (V) history of exposure to radiation and carcinogenic chemicals.
We obtained the BC specimens and adjacent tissues (3 cm from the edge of the cancer edge) of the BC patients included in this study from the specimen library of the Pathology Department of the First Affiliated Hospital of Wannan Medical College. All specimens were fixed with 10% formalin during routine resection and routinely dehydrated and preserved by paraffin embedding. The paraffin-embedded tissue block is stored in the normal temperature paraffin-embedded specimen library by the pathology specialist.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of the First Affiliated Hospital of Wannan Medical College (No. 2019070, the registration number of ethics board) and informed consent was taken from all the patients.
BC cell lines
MCF10A, MBA-MD-231 and MDA-MB-436 BC cell lines were purchased from Precell Biotechnology Co., Ltd. (https://www.procell.com.cn/). MCF10A human normal breast cells were cultured in DMEM (11995065, Gibco, USA) +5% Horse Serum (16050-122, Gibco, USA) +20 ng/mL EGF (PHG0311, Gibco, USA) +0.5 µg/mL hydrocortisone (C-130, Sigma, USA) +10 µg/mL insulin (91077C-100MG, Sigma, USA) +1% NEAA (0823, ScienCell, USA) +1% P/S (SV30010, Hyclone, USA) medium. MBA-MD-231 human BC cell lines were cultured in Leibovitz’s L-15 (11415-064, Gibco, USA) +10% FBS (10099-141, Gibco, USA) +1% P/S (SV30010, Hyclone, USA) medium. The culture condition of MDA-MB-436 BC cell line was Leibovitz’s L-15 (11415-064, Gibco, USA) +10 µg/mL insulin +16 µg/mL glutathione (16100, Thermo, USA) +15% FBS (10099-141, Gibco, USA) +1% P/S (SV30010, Hyclone, USA). Except for the identity provided by Procell Biotechnology Co., Ltd., the cell line has not been independently certified. All cells were cultured in 5% CO2 humidifier at 37 °C, and mycoplasma contamination was detected in all cell lines.
Experimental method
Screening of DEGs
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r) is an interactive network analysis tool officially released by GEO database, which relies on two R packages, GEOquery (28) and LIMMA (29). It is mainly used to analyze and obtain biological information contained in gene chips of GEO database. In this study, GEO2R was used to screen DEGs between normal breast tissue and BC. The P value was adjusted by Benjamini and Hochberg [false discovery rate (FDR)] method, and the adj. P value was used as the basis to distinguish statistically significant and false positive genes. In the study, when a gene symbol has a unique probe set, it is considered to be statistically significant. On the contrary, probe sets lacking of corresponding gene symbols or genes with not only one probe set were removed or averaged, respectively. In the present study, when |Log2FC| >1 and adj. P value <0.01 is considered to be statistically significant.
KEGG and GO enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) (30) is a bioinformatics database with annotations and information links as its main functions. The database quotes Gene Ontology (GO) (31) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (32) established by the Kanhisa Laboratory of the Bioinformatics Center of Kyoto University, Japan. DAVID database also integrates analysis tools, so this tool can find out the most significantly enriched biological annotations from thousands of related annotations at the statistical level, which are large-scale lists of genes or proteins, and provide comprehensive biological function annotations for corresponding genes to help users extract biological information.
Construction of protein-protein interaction (PPI) network
The interaction between the most significant module and hub genes network proteins in cells can reveal the function of proteins at the molecular level, which is essential for the understanding of cell development, differentiation and apoptosis, as well as other biological regulatory mechanisms and life activities. It provides an important theoretical basis for exploring the pathogenesis of major diseases, the treatment and prevention of diseases and the development of new drugs. In this study, the PPI network was constructed using the Search Tool for the Retrieval of Interacting Gene (STRING) database (https://string-db.org) (33), based on the DEGs determined by the researchers to identify relationships between these DEGs and the functional groups. Cytoscape (version 3.7.2) (34), an open software, was used to visualize the PPI networks. Molecular Complex Detection (MCODE) (version 1.6) (35), a plug-in of Cytoscape, was used to filter out the most significant module from the PPI network with degree cut-off =2, node score cut-off =0.2 and K-Core =2.
Selection and analysis of HUB gene
The free Cytoscape plug-in CytoHubba (36) is used to identify central genes from the most significant module. The BP of HUB genes were analyzed and visualized using Biological Networks Gene Oncology tool (37) (Bingo) (version 3.0.3) plug-in of Cytoscape. The overall survival (OS) and disease-free survival (DFS) of Hub gene were analyzed by Kaplan-Meier method in Kaplan-Meier Plotter (38) (http://kmplot.com/analysis/index.php?p).
Immunohistochemistry
The breast carcinomas and their adjacent normal tissues were fixed with formalin (10%), embedded in paraffin and cut into thick sections of 4 µm. According to the manufacturer’s instructions, tissue sections were stained with DNAJB4 antibody (1:50, Abcam, USA), secondary antibody kit (SP-9000, ZSGB-BIO, China), DAB kit (ZLI-9017, ZSGB-BIO, China). Two experienced pathologists examined the immunostaining results and divided them into five grades according to the percentage of positive cells (PP), that is, ≤25% for 1 score, 26–50% for 2 scores, 51–75% for 3 scores, and 76–100% for 4 scores. According to the staining intensity (SI) of nucleus or cytoplasm, it was divided into four grades: 0 score for no staining, 1 scores for weak staining, 2 scores for moderate staining and 3 scores for strong staining. Finally, immunohistochemical staining is considered positive when SI and PP take more than 3 scores (39).
Production and transfection of lentiviral vector
Three lentiviral vectors of DNAJB4-shRNA were designed to knock down the expression of DNAJB4 in MDA-MB-231 and MDA-MB-436 cells. The sequences of DNAJB4 specific siRNA1 (siDNAJB4-1) are: 5'-GGGAATTGAGAAAGGAGCTTCAGAT-3'; siRNA2 (siDNAJB4-2) are: 5'-GAGGCATTGTGTGGCTGCTCAATTA-3'; siRNA3 (siDNAJB4-3) are: 5'-TGAGGTGTCCTTCCCAGATACTATA-3'; control virus vector siRNA is: 5'TTCTCCGAACGTGTCACGTAA-3'. And the most effective vectors are selected by RT-qPCR (Figure S1). The non-targeted shRNA lentivirus vector is used as the negative control, and the blank control refers to the non-vector. The shRNA lentivirus vector was purchased from Hanbio Biotechnology Company (Shanghai, China) (Figure S2, Table S1).
RT-qPCR
The total RNA of these collected cells was extracted using TRIzol reagent (15596026, Invitrogen, USA). Samples were treated with chloroform (20%), vortexed briefly, and incubated at room temperature for 15 min. Samples were then centrifuged at high speed at 4 °C for 15 min. The aqueous phase was moved to a new tube and an equal volume of isopropanol was added. Samples were incubated at room temperature for 10 min, followed by centrifugation at high speed at 4 °C for 10 min. Pellets were washed in 95% ethanol, dried and resuspended in nuclease-free water. RevertAid First Strand cDNA Synthesis Kit (K1622, Thermo, USA) was used to synthesize cDNA, according to the manufacturer’s instructions. Using DNAJB4 and β-actin specific primers made by Sangon Biotech (Shanghai, China), qPCR was performed in LightCycler96 (Roche, USA) according to TB Green® Premix Ex Taq™ II (RR820A, Takara, Japan) instructions. The fold change in the expression of DNAJB4 was calculated by the 2−ΔΔCT formula, and the GAPDH mRNA level was used as a control. The sense primer of DNAJB4 is 5'-CCAGCAGACATTGATTTTTATCATT-3'. The antisense primer of DNAJB4 is 5'-CCATCCAGTGTTGGTACATTAATT-3'. The sense primer of β-actin is 5'-AGCACTGTGTTGGCGTACAG-3', and antisense primer of β-actin is 5'-TCCCTGGAGAAGAGCTACGA-3' (Figure S2, Table S1).
Western bolting
Using lysate (PMFS: RIPA =1:100) (BOSTER Biological Technology, China) to separate total protein of the collected cells. The protein samples were separated by SDS-PAGE (8%) and transferred to a PVDF membrane (IPVH00010, millipore, USA). Membranes were blocked in 5% non-fat dry milk (1706404, BioRad, USA) dissolved in TBS-T for 1 h and incubated overnight at 4 °C with primary antibodies. The membranes incubated with the corresponding horseradish peroxidase (HRP) (abs20002A, absin, China) coupled with the secondary antibody. ECL reagent (WBKLS0100, Millipore, USA) was used to detect immune response bands.
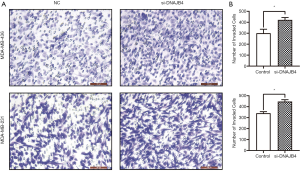
Transwell assay
The transwell invasion assay used 24 transwell chambers (MCEP24H48, Millipore, USA). First, the upper ventricle surface of the basement membrane of the Transwell chamber was coated with Matrigel (354230, Corning, USA). Then, 3×104 cells were suspended in 0.2 mL serum-free medium and added to the insert. 0.5 mL medium containing 20% fetal bovine serum was added into the lower chamber as a chemotactic inducer. After incubating at 37 °C for 48 hours, the cells on the upper surface of the membrane were carefully removed with cotton swabs. The cells on the lower surface were fixed with 100% methanol, and then stained with 0.1% crystal violet (C8470, Solarbio, China). Five random visual fields were selected, each insert was magnified 200×, and the number of cells was counted under an optical microscope (Leica, Germany).
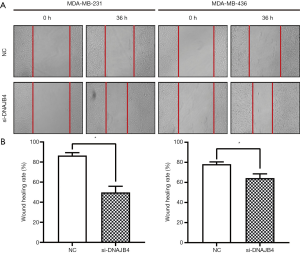
Wound healing experiment
The cells were seeded in a 6-well plate at an appropriate density and cultured until 100% confluence. The yellow straw tip was used to make a straight scratch to simulate the wound. Microscope-based photos were taken at 0 or 36 hours after wounding.
Design of clinical study
This study is a retrospective study, and the patients included in the study were not stratified or matched. The first patient included in this study on January 1, 2012, and the follow-up of the last case ended on November 23, 2019. BC local recurrence or distant metastasis is considered to be the research endpoint. The flow of patients through the study was described in Figure 1.
Statistical analysis
The experimental data statistics of this project were carried out in SPSS 24.0 for Windows. The measurement data were shown as mean ± SD. The measurement data were analyzed by two-tailed one-way ANOVA, and the LSD method was used for pairwise comparison among groups. The survival curve was drawn by Kaplan-Meier method, and the difference of survival data was tested by Logrank test. In the study, the test level α is set to 0.05, a value of P<0.05 was considered as statistically significant.
Results
Identification of DEGs in BC
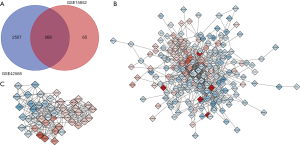
In the GSE15852 and GSE42568 data sets, compared with normal breast samples, GEO2R analysis was used to screen for DEGs in BC samples meeting the criteria that adjP<0.01 and |logFC| ≥1 (2,775 genes were selected in GSE42568 data set and 333 genes were selected in GSE15852 data set). And 268 DEGs were overlapped in the two data sets, as demonstrated in the Wayne diagram (Figure 2A), which includes 73 up-regulated genes and 195 down-regulated genes.
Gene ontology and KEGG pathway analysis of DEGs in breast
The GO and KEGG pathway analysis of DEGs in was carried out by using DAVID online annotation tool. The results of GO analysis showed that the changes in the BP of DEGs were obviously enriched in negative regulation of cell proliferation, lipid metabolic process, cellular response to hormone stimulus, response to drug, positive regulation of protein kinase B signaling, positive regulation of transcription and DNA-templated, response to estrogen. The changes of molecular function (MF) were mainly concentrated in oxidoreductase activity, heparin binding, nucleic acid binding transcription factor activity and ATP binding. The changes of cell composition (CC) were mainly concentrated in membrane raft, cell surface, lateral plasma membrane, bicellular tight junction, apical plasma membrane and membrane. KEGG pathway analysis showed that down-regulated DEGs were mainly enriched in PPAR signaling pathway, AMPK signaling pathway, pathways in cancer; and the up-regulated DEGs were primarily enriched in ECM-receptor interaction; protein digestion and absorption (Table 1).

Full table
PPI network construction and module analysis
The network construction of DEGs was carried out in the online tool of STRING, and the results were visualized in Cytoscape (Figure 2B). The MCODE plug-in in Cytoscape software was used to grab the most significant module (Figure 2C) in the network constructed by DEGs. The genes contained in the most significant module in the PPI network were analyzed by GO and KEGG in the DAVID online tool. The results indicate that the genes in this module were mainly enriched in response to estrogen, mitotic spindle organization, RNA polymerase II transcription factor activity (ligand-activated) (Table 2).

Full table
Hub gene selection and analysis
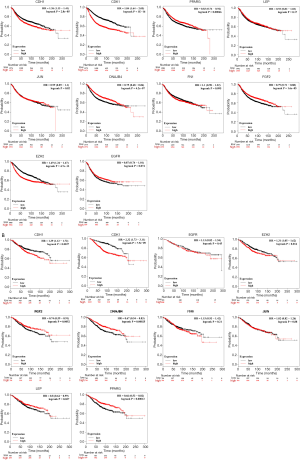
The cytoHubba plug-in in Cytoscape software was used to capture the genes in the most significant module in PPI network which ranked the top 10 and node degree >10 as Hub genes. The gene symbols, full names and functions related to these captured Hub genes were listed in Table 3, in which DNAJB4 has the highest degree of 29. The BP of HUB gene was analyzed and visualized (Figure 3) by the plug-in of Bingo, a biological network gene oncology tool of Cytoscape APPs. The analysis results suggested that the biological functions of the hub genes were mainly focused on regulation of cell cycle process, regulation of nuclear division, cell cycle process, cell cycle, cell division. The analysis of hub genes in the Kaplan-Meier Plotter online tool showed that there was a significant difference in OS between BC patients with CDH1, CDK1, PPARG, LEP, DNAJB4, FGF2 and EZH2 altered and the iterated (Figure 4A); and there is also a significant difference in DFS between patients with and without changes in CDH1, CDK1, PPARG, LEP, DNAJB4, FGF2, and EZH2 (Figure 4B).

Full table

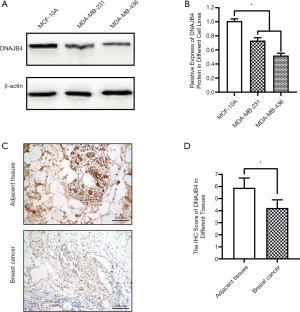
Expression of DNAJB4 in BC cell lines and cancer tissues
Western Bolt was used to detect the expression of DNAJB4 in normal breast cell line MCF10A and BC cell lines. The results showed that the expression of DNAJB4 in MDA-MB-231 and MDA-MB-436 cell lines was significantly lower than that in MCF10A cell line, and the difference was statistically significant (Figure 5A,B). The expression of DNAJB4 was detected by immunohistochemistry on cancer tissues and the adjacent tissues of 80 BC patients (basic demographic characteristics can be found in Table S2). The results revealed that the immunohistochemical staining scores of DNAJB4 in BC was significantly lower than those in adjacent tissues (Figure 5C,D).
Effect of down-regulation of DNAJB4 on migration and invasion of BC cell line
Wound healing assay was used to detect the migration ability of BC cells. The results manifested that 36 hours after the establishment of the wound model, the migration ability of DNAJB4 knockdown MDA-MB-231 and MDA-MB-436 BC cells increased significantly (P<0.05) (Figure 6). The results of Transwell chamber assay found that after interfering with the expression of DNAJB4 in MDA-MB-231 and MDA-MB-436, the number of cells invaded through the chamber was significantly increased compared with the control group, and the difference was statistically significant (P<0.05) (Figure 7).
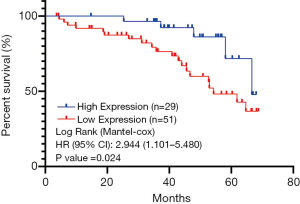
The relationship between the expression of DNAJB4 and the prognosis of BC patients
The survival analysis was performed on the collected follow-up data, and the correlation between the expression level of DNAJB4 protein and the prognosis of BC patients was analyzed by Kaplan-Meier method. Comparison of DFS between the two groups demonstrated that the median follow-up was 64 months (4–68 months), and the total number of events were 25 cases (DFS: 25/80 =31.5%), of which 5 were in the DNAJB4 high expression group (2 lung metastasis, 1 bone metastasis and 2 chest wall recurrence; DFS: 5/29 =17.24%). 20 cases progressed in DNAJB4 low expression group (7 cases of lung metastasis, 3 cases of bone metastasis, 6 cases of liver metastasis, 4 cases of chest wall recurrence; DFS: 20/51 =39.22%). The median survival time of DFS in patients with high expression of DNAJB4 was higher than that in patients with low expression of DNAJB4 (66.45 vs. 54.27 months). Logrank test showed that there was significant difference in DFS between the two groups (HR =2.944, 95% CI: 1.101–5.48, P=0.024) (Figure 8). BC patients with high expression of DNAJB4 had a relatively good prognosis.
Discussion
BC is the leading cause of tumor-related death in women worldwide (2). In recent years, although some progress has been made in the prevention, diagnosis and treatment of BC with increasing researches (6,40,41). However, the pathogenesis of BC is not clear. Besides, the morbidity and mortality continue to rise and the age of patients’ trend to become younger and younger (2,42). BC has the common characteristics of most solid tumors, that is, abnormal genetic and epigenetic changes in cancer occurrence and development (43-45). The concept of molecular subtypes of BC was put forward by Perou’s team (46) in 2000, and it is pointed out that patients with different molecular types of BC have different clinical characteristics and prognosis. With the development of genomics, transcriptome, proteomics and bioinformatics, the conclusion of Perou’s team has been confirmed by most studies. BC-related molecules have also been reported that the overexpression of ERBB2 is high risk of postoperative recurrence and poor prognosis (47,48); and BRCA1/2 gene mutation has a higher risk of BC (49,50). Therefore, the screening and identification of BC-related genes and signal pathways is of great significance for the clinical prevention and treatment of BC. Based on the results of high-throughput sequencing, bioinformatics technology, and its efficiency in the selection of tumor-related candidate genes has made it a popular technology in modern oncology research. However, due to the limitations of contemporary sequencing techniques, the results still need to be verified by biological experiments.
In this study, we identified 268 DEGs from two mRNA chip datasets (GSE15852 and GSE42568) in the GEO database, including 73 up-regulated genes and 195 down-regulated genes. The annotation results of DAVID database reveal that the DEGs of BC are enriched in the BP of negative regulation of cell proliferation, lipid metabolic process, cellular response to hormone stimulus, response to drug, positive regulation of protein kinase B signaling, positive regulation of transcription and DNA-templated, response to estrogen in the perspective of membrane raft, cell surface, lateral plasma membrane, bicellular tight junction, apical plasma membrane and membrane perspectives. Instead, the annotation results of KEGG pathway suggest that the DEGs were enriched in PPAR signaling pathway, AMPK signaling pathway, pathways in cancer etc. Existing studies have shown that PPAR signaling pathway, AMPK signaling pathway and others all play important roles in tumorigenesis and development (51,52), and the results of this study are consistent with it. The screened DEGs were used to construct a PPI network in the STRING database. The construction results were visualized in Cytoscape software, and the most significant module of DEGs was found out through Cytoscape’s MCODE plugin. The functional annotation of the most significant module showed that its related signaling pathways were mainly concentrated in PPAR signaling pathway, pathways in cancer, AMPK signaling pathway and MAPK signaling pathway. The CytoHubba plug-in was used to grab the genes ranked in the top 10 of the most significant modules as Hub genes, including CDH1, PPARG, EZH2, DNAJB4, LEP, FN1, EGFR, CDK1, FGF2, and JUN. HUB genes were analyzed for GO and KEGG enrichment through the Bingo plug-in in Cytoscape. The results suggest that the BP of HUB genes are mainly enriched in regulation of cell cycle process, regulation of nuclear division, cell cycle process, cell cycle, cell division. In order to clarify the correlation between the expression level of each Hub gene and the prognosis of patients, all HUB genes were placed in the Kaplan-Meier Plotter online tool for survival analysis. The analysis found that there is a significant difference in OS between BC patients with CDH1, CDK1, PPARG, LEP, DNAJB4, FGF2 and EZH2 changed and the unchanged; and there is also a significant difference in DFS between patients with and without changes in CDH1, CDK1, PPARG, LEP, DNAJB4, FGF2, and EZH2.
Among the 10 hub genes, the degree of DNAJB4 was the highest, which was 29. In the survival analysis of Kaplan-Meier Plotter database, it was found that the differential expression of DNAJB4 had a significant effect on both DFS and OS. These results suggest that DNAJB4 may play an important role in the occurrence and development of BC. In order to determine the expression level and role of DNAJB4 in BC, the expression level of DNAJB4 was detected in 80 cases of BC and BC cell lines. The results demonstrate that DNAJB4 was low expressed in both BC and BC cell lines. Further functional experiments of BC cells showed that after down-regulating the expression of DNAJB4 in MDA-MB-231 and MDA-MB-436 BC cell lines by gene interference technique, the invasion and migration ability of MDA-MB-231 and MDA-MB-436 BC cells was significantly enhanced. The DFS data of 80 cases of BC included in this study were tested by Logrank test. It was found that the risk of recurrence of BC patients with low expression of DNAJB4 was 2.944 times higher than that of patients with high expression of BC, and the difference was statistically significant. It should be pointed out that in this study, there is no further study on the specific mechanism of how DNAJB4 inhibits the invasion and metastasis of BC cells, and there is a lack of relevant in vivo experiments, and the relevant conclusions still need to be verified by future studies.
Conclusions
In summary, this study found that DNAJB4 may be a tumor suppressor gene, which is low expressed in BC. Preliminary experimental results indicate that DNAJB4 may be involved in the regulation of invasion and migration of BC cells, and its expression level is related to the prognosis of BC patients. Therefore, further study on the BP and molecular mechanism of DNAJB4 could provide new ideas for the diagnosis and treatment of BC.
Acknowledgments
Special thanks to Yinhua Liu and Lei He (Pathology Department of The First Affiliated Hospital of Wannan Medical College) for tissue sectioning and staining.
Funding: This work was funded by the Foundation of the Health and Planning Committee of Sichuan Province (No. 17PJ599, 18PJ005); Foundation of the Department of Science and Technology of Sichuan Province (No. 19YYJC0384).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/gs-20-431
Data Sharing Statement: Available at http://dx.doi.org/10.21037/gs-20-431
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-20-431). Each author reports grants from Foundation of the Health and Planning Committee of Sichuan Province (No. 17PJ599, 18PJ005), grants from Foundation of the Department of Science and Technology of Sichuan Province (No. 19YYJC0384), during the conduct of the study.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of the First Affiliated Hospital of Wannan Medical College (No. 2019070, the registration number of ethics board) and informed consents were taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin 2019;69:438-51. [Crossref] [PubMed]
- Solinas C, Aiello M, Migliori E, et al. Breast cancer vaccines: Heeding the lessons of the past to guide a path forward. Cancer Treat Rev 2020;84:101947. [Crossref] [PubMed]
- Ligibel JA. Could the Women’s Health Initiative Breathe New Life Into Breast Cancer Prevention? J Clin Oncol 2020;38:1375-7. [Crossref] [PubMed]
- Anthis NJ, Kavanaugh-Lynch MHE. The Global Challenge to Prevent Breast Cancer: Surfacing New Ideas to Accelerate Prevention Research. Int J Environ Res Public Health 2020;17:1394. [Crossref] [PubMed]
- Tharmapalan P, Mahendralingam M, Berman HK, et al. Mammary stem cells and progenitors: targeting the roots of breast cancer for prevention. EMBO J 2019;38:e100852. [Crossref] [PubMed]
- Halsted WS. I. The Results of Operations for the Cure of Cancer of the Breast Performed at the Johns Hopkins Hospital from June, 1889, to January, 1894. Ann Surg 1894;20:497-555. [Crossref] [PubMed]
- Halsted WS. I. The Results of Radical Operations for the Cure of Carcinoma of the Breast. Ann Surg 1907;46:1-19. [Crossref] [PubMed]
- Fisher B, Ravdin RG, Ausman RK, et al. Surgical adjuvant chemotherapy in cancer of the breast: results of a decade of cooperative investigation. Ann Surg 1968;168:337-56. [Crossref] [PubMed]
- Fisher B. Breast cancer. Postgrad Med 1969;45:73-8. [Crossref] [PubMed]
- Fisher B. Status of adjuvant therapy: results of The National Surgical Adjuvant Breast Project studies on oophorectomy, postoperative radiation therapy, and chemotherapy. Other comments concerning clinical trials. Cancer 1971;28:1654-8. [Crossref] [PubMed]
- Gradishar WJ, Anderson BO, Balassanian R, et al. Breast Cancer, Version 4.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2018;16:310-20. [Crossref] [PubMed]
- Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA 2019;321:288-300. [Crossref] [PubMed]
- Sabik LM, Eom KY, Dahman B, et al. The Impact of Massachusetts Health Reform on Colorectal and Breast Cancer Stage at Diagnosis. Med Care 2020;58:183-91. [Crossref] [PubMed]
- Franzoi MA, Schwartsmann G, de Azevedo SJ, et al. Differences in Breast Cancer Stage at Diagnosis by Ethnicity, Insurance Status, and Family Income in Young Women in the USA. J Racial Ethn Health Disparities 2019;6:909-16. [Crossref] [PubMed]
- Banham D, Roder D, Keefe D, et al. Disparities in breast screening, stage at diagnosis, cancer treatment and the subsequent risk of cancer death: a retrospective, matched cohort of aboriginal and non-aboriginal women with breast cancer. BMC Health Serv Res 2019;19:387. [Crossref] [PubMed]
- Hurvitz SA, Martin M, Jung KH, et al. Neoadjuvant Trastuzumab Emtansine and Pertuzumab in Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer: Three-Year Outcomes From the Phase III KRISTINE Study. J Clin Oncol 2019;37:2206-16. [Crossref] [PubMed]
- Hennessy MA, Morris PG. Optimizing Anti-HER2 Therapy in early breast cancer: updates from the KRISTINE trial. Ann Palliat Med 2020;9:504-9. [Crossref] [PubMed]
- Dai JB, Zhu B, Lin WJ, et al. Identification of prognostic significance of BIRC5 in breast cancer using integrative bioinformatics analysis. Biosci Rep 2020;40:BSR20193678. [Crossref] [PubMed]
- Agarwal S, Kashaw SK. Potential target identification for breast cancer and screening of small molecule inhibitors: A bioinformatics approach. J Biomol Struct Dyn 2020.1-15. [Crossref] [PubMed]
- Jin H, Huang X, Shao K, et al. Integrated bioinformatics analysis to identify 15 hub genes in breast cancer. Oncol Lett 2019;18:1023-34. [Crossref] [PubMed]
- Ge W, Jiang M, Zhang F, et al. ZGRF1 Is Associated with Poor Prognosis in Triple-Negative Breast Cancer and Promotes Cancer Stemness Based on Bioinformatics. Onco Targets Ther 2020;13:2843-54. [Crossref] [PubMed]
- Li HN, Zheng WH, Du YY, et al. ZW10 interacting kinetochore protein may serve as a prognostic biomarker for human breast cancer: An integrated bioinformatics analysis. Oncol Lett 2020;19:2163-74. [Crossref] [PubMed]
- Acun T, Doberstein N, Habermann JK, et al. HLJ1 (DNAJB4) Gene Is a Novel Biomarker Candidate in Breast Cancer. OMICS 2017;21:257-65. [Crossref] [PubMed]
- Uretmen Kagiali ZC, Sanal E, Karayel O, et al. Systems-level Analysis Reveals Multiple Modulators of Epithelial-mesenchymal Transition and Identifies DNAJB4 and CD81 as Novel Metastasis Inducers in Breast Cancer. Mol Cell Proteomics 2019;18:1756-71. [Crossref] [PubMed]
- Pau Ni IB, Zakaria Z, Muhammad R, et al. Gene expression patterns distinguish breast carcinomas from normal breast tissues: the Malaysian context. Pathol Res Pract 2010;206:223-8. [Crossref] [PubMed]
- Clarke C, Madden SF, Doolan P, et al. Correlating transcriptional networks to breast cancer survival: a large-scale coexpression analysis. Carcinogenesis 2013;34:2300-8. [Crossref] [PubMed]
- Davis S. MPS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846-7. [Crossref] [PubMed]
- Smyth GK. limma: Linear Models for Microarray Data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor 2005.
- Huang DW, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 2007;8:R183. [Crossref] [PubMed]
- Cheng L, Lin H, Hu Y, et al. Gene function prediction based on the Gene Ontology hierarchical structure. PLoS One 2014;9:e107187. [Crossref] [PubMed]
- Tanabe M, Kanehisa M. Using the KEGG database resource. Curr Protoc Bioinformatics 2012;Chapter 1:Unit1.12.
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Doncheva NT, Morris JH, Gorodkin J, et al. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J Proteome Res 2019;18:623-32. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8 Suppl 4:S11. [Crossref] [PubMed]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005;21:3448-9. [Crossref] [PubMed]
- Nagy Á, Lanczky A, Menyhart O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Friedrichs K, Gluba S, Eidtmann H, et al. Overexpression of p53 and prognosis in breast cancer. Cancer 1993;72:3641-7. [Crossref] [PubMed]
- Kanchan RK, Siddiqui JA, Mahapatra S, et al. microRNAs Orchestrate Pathophysiology of Breast Cancer Brain Metastasis: Advances in Therapy. Mol Cancer 2020;19:29. [Crossref] [PubMed]
- Heil J, Kuerer HM, Pfob A, et al. Eliminating the breast cancer surgery paradigm after neoadjuvant systemic therapy: current evidence and future challenges. Ann Oncol 2020;31:61-71. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Natarajan R, Aljaber D, Au D, et al. Environmental Exposures during Puberty: Window of Breast Cancer Risk and Epigenetic Damage. Int J Environ Res Public Health 2020;17:493. [Crossref] [PubMed]
- Fontes-Sousa M, Amorim M, Salta S, et al. Predicting resistance to endocrine therapy in breast cancer: It’s time for epigenetic biomarkers Oncol Rep 2019;41:1431-8. (Review). [Crossref] [PubMed]
- Oh TG, Wang SM, Muscat GE. Therapeutic Implications of Epigenetic Signaling in Breast Cancer. Endocrinology 2017;158:431-47. [PubMed]
- Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747-52. [Crossref] [PubMed]
- Sun J, Chia S. Adjuvant chemotherapy and HER-2-directed therapy for early-stage breast cancer in the elderly. Br J Cancer 2017;116:4-9. [Crossref] [PubMed]
- Tan YO, Han S, Lu YS, et al. The prevalence and assessment of ErbB2-positive breast cancer in Asia: a literature survey. Cancer 2010;116:5348-57. [Crossref] [PubMed]
- Trainer AH, Lewis CR, Tucker K, et al. The role of BRCA mutation testing in determining breast cancer therapy. Nat Rev Clin Oncol 2010;7:708-17. [Crossref] [PubMed]
- Nelson HD, Huffman LH, Fu R, et al. Genetic risk assessment and BRCA mutation testing for breast and ovarian cancer susceptibility: systematic evidence review for the U.S. Preventive Services Task Force. Ann Intern Med 2005;143:362-79. [Crossref] [PubMed]
- Ren L, Konger RL. Evidence that peroxisome proliferator-activated receptor gamma suppresses squamous carcinogenesis through anti-inflammatory signaling and regulation of the immune response. Mol Carcinog 2019;58:1589-601. [Crossref] [PubMed]
- Ciccarese F, Zulato E, Indraccolo S. LKB1/AMPK Pathway and Drug Response in Cancer: A Therapeutic Perspective. Oxid Med Cell Longev 2019;2019:8730816. [Crossref] [PubMed]