
MicroRNA-34a deficiency leads to impaired wound closure by augmented inflammation in mice
Introduction
Skin wound healing is one fundamental physiological process with a series of complex and well-orchestrated cellular and molecular events (1,2). The whole wound healing process comprises three sequential and overlapping phases: inflammatory phase, proliferative phase and remodeling phase. And appropriate transition from one phase to the next phase is key for proper healing (3-5). In particular, the transition from inflammatory phase to the proliferative phase plays critical role and determinates the outcome of healing (3). Resolution of inflammation in time facilitates the transition of the wound into the proliferative phase, and then promotes wound healing. In contrast, the constant inflammatory state results in failure to enter the proliferative phase, leading to chronic nonhealing wounds characterized by excessive and persistent inflammation (6,7). Thus, after skin injury, resolution of inflammation is one key step for successful healing. Although many factors were identified as key participators regulating wound inflammation resolution, both the cellular and molecular mechanisms of inflammation resolution during skin wound healing are still not fully elucidated.
MicroRNAs are endogenous small non-coding RNAs, which play key roles in diverse physiologic and pathologic processes (8). Recently, the great potential of miRNAs as therapeutic targets for correcting obstacles to avoid healing discords like chronic wounds or keloids has attracted broad interest in investigating their roles in cutaneous wound healing (9,10). Several microRNAs were identified as key regulators of inflammation resolution during healing (11-13). For example, miR-132 was highly upregulated in keratinocytes during the inflammatory phase and peaked in subsequent proliferative phase, which is a critical regulator of skin wound healing facilitating inflammation resolution by targeting HB-EGF (11). To further understanding mechanisms of inflammation resolution during wound healing, more candidate microRNAs are worth exploring.
In this study, miR-34a, the high abundant member of microRNA-34 family in mouse skin, was found significantly downregulated in the inflammatory phase and back to normal levels in the proliferative phase during healing. Both topical knockdown miR-34a levels in wounds by antagomir gel and systematic knockout miR-34a using KO mice resulted in impaired wound healing. Further histopathological investigations showed excessive inflammation in the wounds with miR-34a deficiency. Moreover, over-activated IL-6/STAT3 signal pathway was identified in the wounds of miR-34a KO mice. Our findings indicate that dynamic expression of miR-34a is critical for limiting the inflammatory response and facilitating normal wound healing in mice.
Methods
Animals
All mice related experiments were approved by Institutional Animal Care and Use Committee of Third Military Medical University (TMMU, Chongqing, China). miR-34a knockout mice were obtained from Professor Jun Peng (Department of Hematology, Qilu Hospital, Shandong University, Jinan, China). To minimize potential interference, both sex and age matched wild-type littermates were used as control for miR-34 KO mice. C57BL/6J mice used for detecting miR-34a expression pattern and topical knockdown experiments were obtained from the Institute of Zoology (Chinese Academy of Sciences, Beijing, China). All the mice were bred and maintained at the Center of Experimental Animal, TMMU.
Wound healing studies
To examine the expression pattern of miR-34a during wound healing, the 8 to 12-week-old C57BL/6J mice were anesthetized with 1% pentobarbital sodium (30 mg/kg) and two 6.0 mm-diameter circular full-thickness excisional wounds were made in the shaved dorsal skin. At 0, 3, 7 and 14 days after wounding mice were euthanized, the wound tissues were harvested with a 8-mm biopsy punch for either RNA isolation or in situ hybridization assays.
To knockdown miR-34a in the topical wound sites, antagomir-34a in Pluronic F-127 gel with a concentration of 5 um were used immediately following wound creation as described previously (14). To evaluate the effects of miR-34a knockout on wound healing, the wounds were made on the back skin of both the miR-34a KO and wild-type mice. And then, the wound healing process was digitally photographed at different time points. Wound area measurement was performed as described previously (15,16).
Quantitative real-time PCR
For detecting microRNAs, Bulge-Loop miRNA qRT-PCR Primer Set (Ribobio Co.) was used according to manufacturer’s instructions. For regular quantitative real-time PCR, the method was performed as described previously. Primer pairs used are listed as following: IL-1β F: TCTCGCAGCAGCACATCA; IL-1β R: CACACACCAGCAGGTTAT; IL-6 F: TGGGAAATCGTGGAAATGAG; IL-6 R: CTCTGAAGGACTCTGGCTTTG; TNF-α F: CCCGGGCTCAGCCTCTTCTCATTC; TNF-α R: GGATCCGGTGGTTTGCTACGACGT; IL-10 F: CAACATACTGCTAACCGACTCCT; IL-10 R: TGAGGGTCTTCAGCTTCTCAC; TBP F: AAGGGAGAATCATGGACCAG; TBP R: CCGTAAGGCATCATTGGACT.
In situ hybridization
In situ hybridizations of paraffin-embedded skin sections to detect miR-34a were performed according to previous report (14).
Histology and morphometric analysis
H&E sections of the central portion of the wounds were made for histology and morphometric analyses as previously described (15,17). In brief, epidermal thickness was measured at five different positions per mouse and averaged to one data set. Wound width was determined as the distance between the wound margins, which were defined by the last hair follicles. The percentage of re-epithelialization was calculated as distance covered by epithelium dividing the wound width.
Immunohistochemistry
IHC staining in skin tissues were performed as previously described (18). Briefly, tissue sections were stained with rabbit anti-MPO (1:200, Thermo), mouse anti-F4/80 (1:200, BioLegend), rabbit anti-IL-6 (1:600, Proteintech), and mouse anti-p-STAT3 (1:200, CST). Then, DAB chromogenic system was used for final chromogen. Images of areas of interest were collected by Olympus IX73-A21PH microscope (Olympus, Japan). Epidermal p-STAT3 positive cells were quantitified as the number of positive cells per mm migrating epidermis. Quantifications for MPO, F4/80, IL-6, and stromal p-STAT3 positive cells were performed by using Image J by calculating 6-8 40× high power field photos per mouse.
Western blots
Western blots were performed according to our previous publication (19). In brief, protein lysates from wound tissues were extracted using Complete Lysis-M buffer containing both protease inhibitors and phosphatase inhibitors. The protein samples were resolved by SDS-PAGE, transferred onto PVDF membrane, blocked with 5% non-fat milk and incubated with primary antibodies at 4 °C overnight. Primary antibodies used include rabbit anti-p65 (1:1,000, CST), rabbit anti-p-p65 (1:1,000, CST), rabbit anti-STAT3 (1:1,000, CST), mouse anti-p-STAT3 (1:1,000, CST), rabbit anti-IL-6R (1:1,000, Proteintech), and mouse anti-actin (1:1,000, Beyotime, Hangzhou, China). Membranes were incubated with relevant IRDye conjugated secondary antibodies for one hour at room temperature (1:5000, Rockland Immunochemicals Inc., Gilbertsville, PA). The infrared fluorescence image was obtained using Odyssey infrared imaging system (Odyssey CLx, Li-Cor Bioscience, Lincoln, NE).
Statistical analysis
Experimental data were analyzed using GraphPad Prism 5.0 software. Data were presented as mean ± SD. Statistical analysis was performed by Student’s t tests (the U-test was performed to test differences between independent groups at different time points during wound healing). A P value <0.05 was considered to be statistically significant.
Results
Characterization of miR-34a expression during skin wound healing
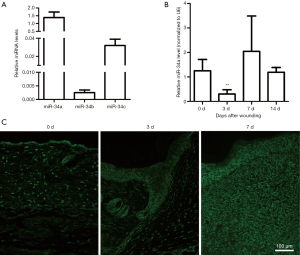
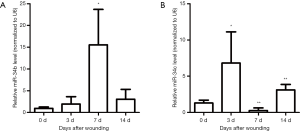
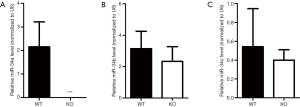
Recent studies have shown that the miR-34 family plays important roles as one tumor suppressor (20,21). It has been reported that tumor suppressors may have special meaning for tissue wound repair and regeneration (22,23). To investigate the possible contributions of miR-34 family members during skin wound healing, their basic expression levels in intact mouse skin tissue samples were examined using real-time RT-PCR. As shown in Figure 1A, miR-34a was strongly expressed in the intact mouse skin samples, whereas miR-34c was expressed at lower levels, and miR-34b was expressed at lowest levels. Then, we examined the dynamic expression of the three miR-34 family members during skin wound healing process. Our results indicated that the three members had diverse dynamic expression during healing (Figure 1B and Figure S1). Although both miR-34c and miR-34b had significantly upregulated in certain time points after injury, their expression levels were still far below miR-34a. So, our subsequent studies focused on miR-34a. Results of real time RT-PCR showed miR-34a expression level was significantly downregulated in day 3 after wounding, then recovered in day 7 and day 14 (Figure 1B). Further examination by in situ hybridization confirmed the results of PCR, which indicated the signal intensity of ISH fluorescence of wounds from day 3 was relatively weaker (Figure 1C).
Topical inhibition of miR-34a by antagomir resulted in delayed skin wound healing
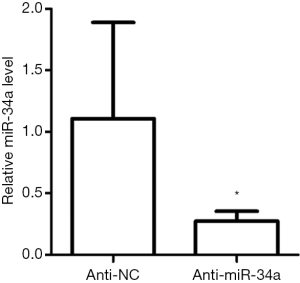
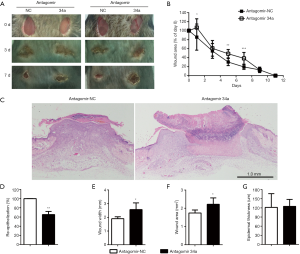
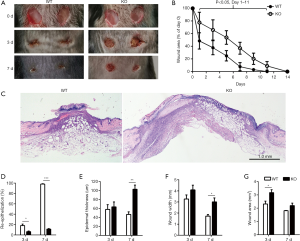
As a well-known pleiotropic molecule, there are extensively reported that miR-34a has tumor suppressive functions like inhibiting cell stemness, proliferation, and migration, promoting apoptosis, and participating in immunological regulation (24-30). So, we speculated that the abundant miR-34a may have certain role in modulating skin wound healing. To investigate the in vivo effects of miR-34a on wound healing, endogenous miR-34 a was downregulated by administrating Pluronic gel with antagomir-34a to the wounds. The availability of antagomir gel for local inhibition of target miRNA in wounds have been reported by others and us previously (14,31). In this experiment, topical antagomir-34a treatment inhibited the miR-34a expression levels effectively examined by real time RT-PCR (Figure S2). The antagomir-34a treated wounds showed slower healing for day 1, day 5 and day 7 time points compared with the autologous antagomir-NC treated control wounds, although all the wounds healed before day 11 (Figure 2A,B). Further morphometric analyses of the histopathological H&E sections confirmed the gross observation. Local knockdown of miR-34a delayed wound re-epithelialization (Figure 2C,D), showed larger wound widths and wound areas (Figure 2E,F), although without impacts on epidermal thickness (Figure 2G).
miR-34a knockout mice showed impaired wound closure
Further experiments on miR-34a KO mice were implemented to confirm the effects of miR-34a on skin wound healing. miR-34a was undetectable in the skin samples of miR-34a KO mice (Figure S3A). And miR-34a knockout had no effects on the expression levels of both miR-34b and miR-34c in skin samples (Figure S3B,C). In line with the results of topical knockdown of miR-34a on wound healing, wounds of miR-34a KO mice showed significantly impaired healing from the day 1 to day 11 after wounding (Figure 3A,B). Histologically, miR-34a KO wounds had shorter epithelial migrating tongues by day 3 and day7, and thicker epidermis by day 7, compared to WT controls (Figure 3C,D,E). Meanwhile, both wound width and wound area were significantly larger in miR-34a KO wounds (Figure 3F,G).
Loss of miR-34a enhanced wound inflammation
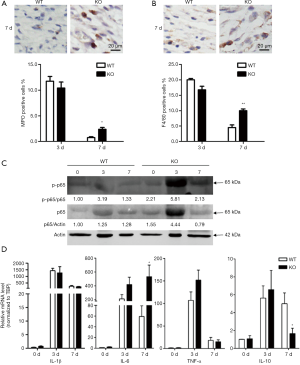
miR-34a downregulated in day 3 after injury and then restored to the normal levels (Figure 1B,C), which suggested miR-34a was specifically reduced in the inflammatory phase during wound healing. And further local knockdown miR-34a using antagomir or systematic knockout miR-34a in mice resulted in impaired healing with larger residual crusta (Figures 2,3), which was a feature of excessive inflammatory response. So, we examined the infiltration of neutrophils and macrophages after wounding, which are critical inflammatory cells for proper healing. As shown in Figure 4A and B, the accumulations of both MPO-positive neutrophils and F4/80-positive macrophages were significantly increased in miR-34a KO mice compared to WT mice in wounds of day 7 after wounding. Further WB for examining both p65 and p-p65 expression levels corroborated the enhanced inflammatory response in wounds of miR-34a KO mice compared to WT mice (Figure 4C). To explore the possible mechanism of enhanced inflammatory response in wounds of miR-34a KO mice, real-time RT-PCR for classical cytokines expression levels were implemented (Figure 4D). During wound healing process, miR-34a knockout mice had little effect on IL-1β expression levels and showed increased TNF-α expression in day 3 wounds but without significant difference. However, IL-6 expression was downregulated in day 7 wounds of WT mice, while sustained in those of KO mice. In addition, IL-10 expression showed a sharp reduction in day 7 wounds of KO mice.
Over-activated IL-6/STAT3 signaling in wounds of miR-34a KO mice
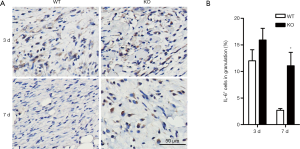
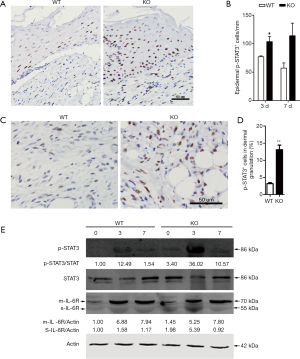
IL-6/STAT3 signaling pathway has showed extensive interactions with miR-34a (32-35), so we focused on this pathway in further investigations. To verify the constant elevated IL-6 expression detected by real-time PCR, immunohistochemical staining was used for semi-quantitative analysis of IL-6-positive cells in granulations, which confirmed the persistent high expression of IL-6 in day 7 wounds of miR-34a KO mice (Figure 5). As STAT3 is one critical component of IL-6 downstream signal pathway, we examined the phosphorylated STAT3 in the wounds by immunohistochemistry. Quantization of p-STAT3-positive epidermal cells indicated more numbers of positive cells in KO mice at both day 3 and day 7 time points, although there was no significant difference in day 7 between the two groups (Figure 6A,B). Moreover, the p-STAT3 positive cells in dermal granulation in wounds of miR-34a KO mice were also significantly higher than those of WT mice in day 7 (Figure 6C,D). In line with the IHC results, WB for the phosphorylated STAT3 showed similar trend that miR-34a KO in wounds promoted STAT3 signaling activation (Figure 6E). As one key miR-34a functional target, IL-6R was also examined. The result indicated elevated sIL-6R (soluble IL-6R) level in day 0 and 3 wounds of KO mice, while less affected mIL-6R (membrane-bound IL-6R) level (Figure 6E).
Discussion
In the current study, we found that the cutaneous abundant miR-34a deficiency resulted in impaired wound healing, which was caused by excessive inflammation response through over-activated IL-6/STAT3 signaling pathway in mice.
Previous studies showed different expression patterns of three miR-34 family members in skin tissues (25,36). Our results indicated that miR-34a was the abundant expression member of the family in skin, which expressed higher than miR-34b and miR-34c in the order of magnitude. This expression feature from intact full-thickness mouse skin samples is in accord with the results from primary mouse keratinocytes and human skin tissues (25,36). Further experiments were performed to examine the dynamic expressions of miR-34 family members during skin wound healing of mice. Although both miR-34b and miR-34c expressed variably, miR-34a was still the abundant member of miR-34 family during healing. Thus, we focused on role of miR-34a in wound healing for next investigations. We found that miR-34a was downregulated specifically in inflammatory phase of wound repair (day 3 after wounding) detected by both real time RT-PCR and ISH. As one tumor suppressor microRNA, the early reduction of miR-34a may facilitate wound healing by promoting keratinocytes proliferation and migration, just like inhibition of p53 and RB at the initial stages of regeneration (22,23). Also, miR-34a plays important roles in immune response, including impairing neutrophils migration (37), inhibiting macrophages efferocytosis (38), modulating dendritic cells differentiation (39), regulating cytokines productions (29), and so on. Thus, the downregulation of miR-34a in inflammatory phase may be beneficial to neutrophils chemotaxis and cytokines producing in this repair stage.
To determine the roles of miR-34a during skin wound healing, both topical inhibition of miR-34a by antagomir gel in wounds and systematic knockout of miR-34a in mice models were employed. Unexpectedly, both models showed miR-34a deficiency resulted in impaired wound closure. Recent study reported that elevated miR-34a in wounds by local intradermal injecting mimics also led to delayed healing (36). Those conflictive results displayed the complexity of miR-34a function in skin wound healing. Theoretically, microRNAs serve as one moderate gene regulatory model, which have target genes with differential sensitivity under different cellular contexts (40,41). Studies have shown transgenic expression and deletion of one same miRNA regulate largely distinct sets of target genes (40). So, loss-of-function and gain-of-function could impact different target genes and signaling pathways, then present different phenotypes. That is, the appropriate expression levels of miR-34a, neither high or low, is critical for proper wound healing. Experimentally, well-designed in vivo loss-of-function studies are more convincing than the gain-of-function research. The microRNA mimics should be used with caution for introducing the supraphysiological levels of mature miRNAs and artifactual RNA species led to non-specific changes in gene expression (42). We note that the wounds treated with miR-34a mimics achieved far more than the physiological conditions (36). In addition, indiscriminate upregulating miR-34a levels in all the repairing cells would confuse the actual role of miR-34a during wound healing (36), for miR-34a showed diverse or even opposite effects in different cell types (43,44).
There are reports that miR-34a in keratinocytes can inhibit proliferation, migration, and promote differentiation (24,25,36). Also, reports indicated miR-34a can inhibit angiogenesis (45,46). So, the effect of miR-34a deficiency on delayed healing in mice is a bit surprising to us. In the other hand, miR-34a also have extensive and complexed regulatory roles in inflammatory response (29,37-39,43,44). And persistent inflammatory response with resolution dysregulating could led to chronic non-healing wounds (3,6,47,48). Further investigations indicated the phenotypes of miR-34a-deficiency-related impaired wounds resembled the chronic wounds, which showed excessive inflammatory response and delayed re-epithelialization. Therefore, we speculated that augmented inflammation caused by miR-34a loss-of-function overcame the possible advantages from miR-34a deficiency like promoting keratinocytes proliferation and migration, improving angiogenesis, and so on.
To interpret the augmented inflammation induced by miR-34a deficiency, we identified IL-6/STAT3 signaling pathway was over-activated in KO wounds, which suggested sustaining activation of this pathway may block resolution of inflammation and then resulted in delayed wound closure. It has been reported that IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis, which considers that STAT3 activation can inhibit miR-34a transcription and in turn miR-34a can suppress IL-6R to control the IL-6/STAT3 pathway activation (32). If so, it partly explains the downregulated miR-34a in the early inflammatory phase, which stage showed stronger STAT3 pathway activation. However, the IL-6R protein levels did not show significant increasement in KO wounds in the mIL-6R form except the slight upregulated sIL-6R form in early KO wounds. Although sIL-6R can activate the proinflammatory IL-6 trans-signaling even on cells without IL-6R expression (49), its contribution in the delayed wound closure of miR-34a KO mice needs further investigation with caution.
Conclusions
Taken together, our study revealed the dynamic expression of miR-34a is critical for resolution of inflammation during wound healing in mice. Our findings indicate that miR-34a deficiency augments skin wound inflammation response through over-activated IL-6/STAT3 signaling pathway, and then leads to impaired wound closure. Those results suggest targeted inhibition of miR-34a for tissue repair and/or regeneration should require serious consideration.
Acknowledgments
The authors thank Prof. Jun Peng (Qilu Hospital, Shandong University, Jinan, China) for providing us the miR-34a knockout mice.
Funding: This work was supported by the Chinese National Natural Science Foundation (Grant No. 81372061 to Tao Wang, No. 81725019 to Junping Wang), Military Programs (Grant No. AWS14C002 to Yongping Su), and Natural Science Foundation of Chongqing (Grant No. cstc2016jcyjA0381 to Tao Wang).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.03.161). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The whole study protocol was approved by the Ethics Committee of Third Military Medical University and all the procedures were done according to the Chinese Guidelines for the Care and Use of Laboratory Animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev 2003;83:835-70. [Crossref] [PubMed]
- Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med 2014;6:265sr6. [Crossref] [PubMed]
- Landen NX, Li D, Stahle M. Transition from inflammation to proliferation: a critical step during wound healing. Cell Mol Life Sci 2016;73:3861-85. [Crossref] [PubMed]
- Gurtner GC, Werner S, Barrandon Y, et al. Wound repair and regeneration. Nature 2008;453:314-21. [Crossref] [PubMed]
- DiPietro LA. Angiogenesis and scar formation in healing wounds. Curr Opin Rheumatol 2013;25:87-91. [Crossref] [PubMed]
- Zhao R, Liang H, Clarke E, et al. Inflammation in Chronic Wounds. Int J Mol Sci 2016.17. [PubMed]
- Krzyszczyk P, Schloss R, Palmer A, et al. The Role of Macrophages in Acute and Chronic Wound Healing and Interventions to Promote Pro-wound Healing Phenotypes. Front Physiol 2018;9:419. [Crossref] [PubMed]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol 2007;23:175-205. [Crossref] [PubMed]
- Singhvi G, Manchanda P, Krishna Rapalli V, et al. MicroRNAs as biological regulators in skin disorders. Biomed Pharmacother 2018;108:996-1004. [Crossref] [PubMed]
- Li D, Landen NX. MicroRNAs in skin wound healing. Eur J Dermatol 2017;27:12-4. [Crossref] [PubMed]
- Li D, Wang A, Liu X, et al. MicroRNA-132 enhances transition from inflammation to proliferation during wound healing. J Clin Invest 2015;125:3008-26. [Crossref] [PubMed]
- van Solingen C, Araldi E, Chamorro-Jorganes A, et al. Improved repair of dermal wounds in mice lacking microRNA-155. J Cell Mol Med 2014;18:1104-12. [Crossref] [PubMed]
- Das A, Ganesh K, Khanna S, et al. Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation. J Immunol 2014;192:1120-9. [Crossref] [PubMed]
- Wang T, Zhao N, Long S, et al. Downregulation of miR-205 in migrating epithelial tongue facilitates skin wound re-epithelialization by derepressing ITGA5. Biochim Biophys Acta 2016;1862:1443-52.
- Long S, Zhao N, Ge L, et al. MiR-21 ameliorates age-associated skin wound healing defects in mice. J Gene Med 2018;20:e3022. [Crossref] [PubMed]
- Wang T, Feng Y, Sun H, et al. miR-21 regulates skin wound healing by targeting multiple aspects of the healing process. Am J Pathol 2012;181:1911-20. [Crossref] [PubMed]
- Chen L, Mirza R, Kwon Y, et al. The murine excisional wound model: Contraction revisited. Wound Repair Regen 2015;23:874-7. [Crossref] [PubMed]
- Wang T, Long S, Zhao N, et al. Cell Density-Dependent Upregulation of PDCD4 in Keratinocytes and Its Implications for Epidermal Homeostasis and Repair. Int J Mol Sci 2015;17. [Crossref] [PubMed]
- Fan B, Wang T, Bian L, et al. Topical Application of Tat-Rac1 Promotes Cutaneous Wound Healing in Normal and Diabetic Mice. Int J Biol Sci 2018;14:1163-74. [Crossref] [PubMed]
- Misso G, Di Martino MT, De Rosa G, et al. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids 2014;3:e194. [Crossref] [PubMed]
- Farooqi AA, Tabassum S, Ahmad A. MicroRNA-34a: A Versatile Regulator of Myriads of Targets in Different Cancers. Int J Mol Sci 2017;18. [Crossref] [PubMed]
- Charni M, Aloni-Grinstein R, Molchadsky A, et al. p53 on the crossroad between regeneration and cancer. Cell Death Differ 2017;24:8-14. [Crossref] [PubMed]
- Sage J.. The retinoblastoma tumor suppressor and stem cell biology. Genes Dev 2012;26:1409-20. [Crossref] [PubMed]
- Lefort K, Brooks Y, Ostano P, et al. A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J 2013;32:2248-63. [Crossref] [PubMed]
- Antonini D, Russo MT, De Rosa L, et al. Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Invest Dermatol 2010;130:1249-57. [Crossref] [PubMed]
- Guessous F, Zhang Y, Kofman A, et al. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle 2010;9:1031-6. [Crossref] [PubMed]
- Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 2007;26:731-43. [Crossref] [PubMed]
- Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 2007;26:745-52. [Crossref] [PubMed]
- Jiang P, Liu R, Zheng Y, et al. MiR-34a inhibits lipopolysaccharide-induced inflammatory response through targeting Notch1 in murine macrophages. Exp Cell Res 2012;318:1175-84. [Crossref] [PubMed]
- Zeng H, Hu M, Lu Y, et al. MicroRNA 34a promotes ionizing radiation-induced DNA damage repair in murine hematopoietic stem cells. FASEB J 2019;33:8138-47. [Crossref] [PubMed]
- de Kerckhove M, Tanaka K, Umehara T, et al. Targeting miR-223 in neutrophils enhances the clearance of Staphylococcus aureus in infected wounds. EMBO Mol Med 2018;10. [Crossref] [PubMed]
- Rokavec M, Oner MG, Li H, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest 2014;124:1853-67. [Crossref] [PubMed]
- Li H, Rokavec M, Hermeking H.. Soluble IL6R represents a miR-34a target: potential implications for the recently identified IL-6R/STAT3/miR-34a feed-back loop. Oncotarget 2015;6:14026-32. [PubMed]
- Lin X, Lin BW, Chen XL, et al. PAI-1/PIAS3/Stat3/miR-34a forms a positive feedback loop to promote EMT-mediated metastasis through Stat3 signaling in Non-small cell lung cancer. Biochem Biophys Res Commun 2017;493:1464-70. [Crossref] [PubMed]
- Yokomizo R, Yanaihara N, Yamaguchi N, et al. MicroRNA-34a/IL-6R pathway as a potential therapeutic target for ovarian high-grade serous carcinoma. Oncotarget 2019;10:4880-93. [PubMed]
- Wu J, Li X, Li D, et al. MicroRNA-34 Family Enhances Wound Inflammation by Targeting LGR4. J Invest Dermatol 2020;140:465-476.e11. [Crossref] [PubMed]
- Cao M, Shikama Y, Kimura H, et al. Mechanisms of Impaired Neutrophil Migration by MicroRNAs in Myelodysplastic Syndromes. J Immunol 2017;198:1887-99. [Crossref] [PubMed]
- McCubbrey AL, Nelson JD, Stolberg VR, et al. MicroRNA-34a Negatively Regulates Efferocytosis by Tissue Macrophages in Part via SIRT1. J Immunol 2016;196:1366-75. [Crossref] [PubMed]
- Hashimi ST, Fulcher JA, Chang MH, et al. MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood 2009;114:404-14. [Crossref] [PubMed]
- Jin HY, Oda H, Chen P, et al. Differential Sensitivity of Target Genes to Translational Repression by miR-17~92. PLoS Genet 2017;13:e1006623. [Crossref] [PubMed]
- Hsin JP, Lu Y, Loeb GB, et al. The effect of cellular context on miR-155-mediated gene regulation in four major immune cell types. Nat Immunol 2018;19:1137-45. [Crossref] [PubMed]
- Jin HY, Gonzalez-Martin A, Miletic AV, et al. Transfection of microRNA Mimics Should Be Used with Caution. Front Genet 2015;6:340. [Crossref] [PubMed]
- Liu H, Liu W, Tang X, et al. IL6/STAT3/miR34a protects against neonatal lung injury patients. Mol Med Rep 2017;16:4355-61. [Crossref] [PubMed]
- Zhou J, Shuang O, Li J, et al. miR-34a alleviates spinal cord injury via TLR4 signaling by inhibiting HMGB-1. Exp Ther Med 2019;17:1912-8. [PubMed]
- Kumar B, Yadav A, Lang J, et al. Dysregulation of microRNA-34a expression in head and neck squamous cell carcinoma promotes tumor growth and tumor angiogenesis. PLoS One 2012;7:e37601. [Crossref] [PubMed]
- Zhao T, Li J, Chen AF. MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am J Physiol Endocrinol Metab 2010;299:E110-6. [Crossref] [PubMed]
- Elliott CG, Forbes TL, Leask A, et al. Inflammatory microenvironment and tumor necrosis factor alpha as modulators of periostin and CCN2 expression in human non-healing skin wounds and dermal fibroblasts. Matrix Biol 2015;43:71-84. [Crossref] [PubMed]
- Nunan R, Harding KG, Martin P. Clinical challenges of chronic wounds: searching for an optimal animal model to recapitulate their complexity. Dis Model Mech 2014;7:1205-13. [Crossref] [PubMed]
- Rose-John S.. The Soluble Interleukin 6 Receptor: Advanced Therapeutic Options in Inflammation. Clin Pharmacol Ther 2017;102:591-8. [Crossref] [PubMed]


