
Identification and integrated analysis of hepatocellular carcinoma-related circular RNA signature
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common malignancy and the main cause of cancer-related death in the world (1). Owing to the lack of early diagnostic biomarkers with high specificity and sensitivity and the high frequency of tumor metastasis, the overall survival of patients with HCC is very poor (2). Therefore, identifying novel biomarkers and therapeutic targets is of great significance in improving the diagnosis and treatment of HCC and could lead to a better understanding of the pathology of HCC.
Accumulating evidence has shown that the dysregulation of circRNAs may contribute to tumorigenesis and cancer progression in humans (3). CircRNA is a newly discovered non-coding RNA with a special structure, which is characterized by a covalent closed-loop without 5’ caps or 3’ tails (4). In recent years, numerous pieces of evidence have revealed that circRNAs could be involved in gene regulation by acting as miRNA sponges, regulating splicing and transcription, or interacting with RNA-binding proteins (RBPs) (5). These findings suggest the potential regulatory role of circRNAs in biologic development and the pathogenesis and progression of diseases, especially types of cancer (6). Aberrant expression of circRNA also played an important role in the initiation and development of HCC (7). Huang et al., for example, found that the activation of the mTOR signaling pathway by circRNA-100338/miR-141-3p/RHEB axis is closely associated with poor prognosis of hepatitis B-related HCC (8). Another study reported that cicrRNA_101368 could function as a sponge of miR-200a to inhibit miR-200a expression, thus promoting the migration of HCC cells through HMGB1/RAGE signaling of miR-200a downstream (9).
In this study, we analyzed the expression profiles of circRNAs, miRNAs, and mRNAs of HCC downloaded from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO). Differentially expressed that circRNAs (DEcircRNAs) were identified and validated. After predicting the sponging of miRNAs by circRNA and miRNA target genes, we established a circRNA-miRNA-mRNA network and a circRNA-miRNA-hub gene network for HCC. To predict the possible mechanisms and function of circRNAs in HCC, we also performed functional enrichment analyses. These results could improve our understanding of the regulatory mechanisms of circRNAs in the pathogenesis and progression of HCC and provide novel therapeutic targets or biomarkers for HCC.
Methods
Microarray data and RNA sequencing data
The microarray data of two circRNA expression profiles were collected from the GEO database (GSE94508 and GSE97332). RNA-sequencing (RNA-seq) data were downloaded from the TCGA database. miRNA sequencing data included 375 liver cancer tissues and 50 normal tissues, and the mRNA sequencing data included 374 liver cancer tissues and 160 normal tissues. We used the Limma R package to explore the significantly differentially expressed genes between tumorous and normal tissues. The cut-off value was |log2FC| >1 and FDR <0.05 (FC, fold change; FDR, false discovery rate). Due to the public availability of data on the GEO and TCGA databases, ethical approval or informed consent was not required for this study.
Cell culture and validation of overlapping circRNAs with real-time Quantitative PCR
SMMC-7721 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) at 37 °C in a 5% CO2 atmosphere in a humidified incubator.
Total RNAs from SMMC-7721 cells were extracted using TRIzol (Invitrogen) according to the manufacturer’s protocol. cDNA was synthesized from total RNA using M-MLV RT (Promega). Then real-time qPCR was applied with SYBR Master Mixture (Takara) on the LightCycler 480 II (Roche) according to standard protocols. The primer sequences for GAPDH acted as the control in these experiments. Primers used for detecting circRNAs were synthesized from GeneChem (Shanghai). Primer sequences were listed in Table 1. The quantity of each mRNA was calculated using the 2−ΔΔCt method, and data were normalized using GAPDH as the loading control.
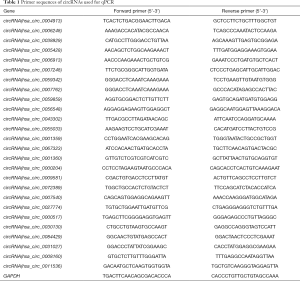
Full table
Predictions of target miRNA and miRNA target genes
The Circular RNA Interactome (CircInteractome) (10) and Cancer-Specific CircRNA (CSCD) (11) were used to predict target miRNAs and their miRNA binding sites (MREs). These target miRNAs were further selected by DEmiRNA based on TCGA, and the screened miRNAs were identified as potential target miRNAs of the DEcircRNAs.
Interactions between miRNA and mRNA were predicted based on the TargetScan (12), miRTarBase (13), and miRDB (14) databases. Only the target mRNAs recognized by the three databases were considered as candidate mRNAs. Moreover, intersections with the DEmRNAs were identified to screen out the hub genes targeted by the DEmiRNAs.
Construction of the CeRNA network
The circRNA-miRNA-mRNA regulatory network was constructed by using a combination of circRNA-miRNA pairs and miRNA-mRNA pairs and visualized by Cytoscape 3.6.1.
GO and KEGG pathway enrichment analysis
mRNAs in the ceRNA network were evaluated by GO (15) annotation and KEGG (16) pathway analyses with the clusterProfiler package in R. P value <0.05 served as the cut-off point to assess the functional pathways.
Gene functional analysis
GSEA (17) is a computational method for assessing whether a set of genes defined by a priori show statistically significant, consistent differences between two biological states. GSEA was performed to analyze the enrichment of datasets between high- and low-expression groups of hub genes. A false discovery rate (FDR) <25% and nominal P value <5% were set as the cut-off criteria. In addition, GSVA (18) was used to find the most relevant pathways of hub genes, according to the gene sets files from the KEGG databases.
Results
Identification of differentially expressed circRNAs and miRNAs
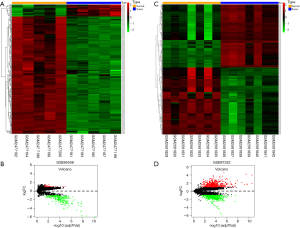
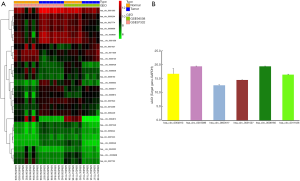
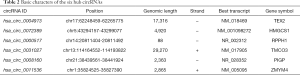
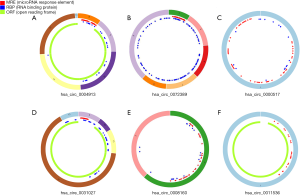
Two circRNA microarray datasets (GSE94508 and GSE97332) were downloaded from the GEO database, and the DEcircRNAs were generated by using the limma R package. A total of 270 DEcircRNAs (25 up-regulated and 245 down-regulated) were identified in the GSE94508 dataset (Figure 1A,B). A total of 869 DEcircRNAs (421 up-regulated and 448 down-regulated) were identified in the GSE97332 dataset (Figure 1C,D). We found that 26 overlapping DEcircRNAs were obtained from two datasets, and these were chosen for further validation. The expressions of these DEcircRNAs in SMMC-7721 cells were detected using qPCR. Among them, the mRNA expression abundance of six DEcircRNAs was validated to be up-regulated in SMMC-7721 cells (Figure 2), indicating that they were appropriate for further research. Moreover, the basic characteristics of the six DEcircRNAs are listed in Table 2. Their basic structural patterns are shown in Figure 3 based on the data from CSCD.
Full table
Based on the TCGA-LIHC database, we obtained 251 differentially expressed miRNAs (DEmiRNAs, 229 up-regulated and 22 down-regulated) using edgeR package analysis with |log2FC| >1 and FDR <0.05 (Figure S1).
The construction of the ceRNA network and functional assessment
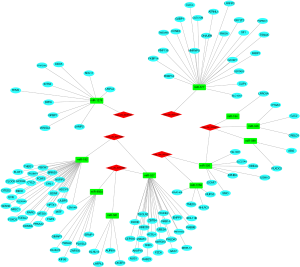
To better comprehend the role of circRNA in miRNA mediated mRNA, we constructed a circRNA-miRNA-mRNA (ceRNA) network. Firstly, using the CircInteractome database, we predicted the miRNAs targeted by the 6 DEcircRNAs and identified 100 circRNAs-miRNAs pairs. After intersecting with the DEmiRNAs, only 13 circRNA-miRNA pairs remained, including 6 circRNAs and 11 DEmiRNAs. We then identified mRNAs targeted by the 11 DEmiRNAs in 3 databases (miRDB, miRTarBase, and TargetScan). Lastly, we established a ceRNA network based on 6 circRNA nodes, 11 miRNA nodes, and 114 mRNA nodes in HCC (Figure 4).
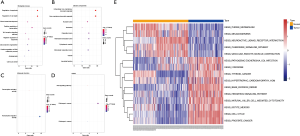
To achieve a better understanding of the underlying biological processes (BP) and pathways associated with the circRNAs-related target genes in the ceRNA network, we applied the GO and KEGG pathway enrichment analyses. The results of BP, cellular component (CC), molecular function (MF), and KEGG pathways are indicated in Figure 5A,B,C,D (P<0.05). In addition, GSVA further confirmed that a high level of circRNAs-related target genes was significantly enriched in 9 KEGG terms (thyroid cancer, pathogenic Escherichia coli infection, hypertrophic cardiomyopathy hcm, lysosome, hedgehog signaling pathway, oocyte meiosis, natural killer cell-mediated cytotoxicity, base excision repair, and cell cycle) (Figure 5E).
The construction of the CircRNA-miRNA-hub gene network and functional enrichment analysis of hub genes
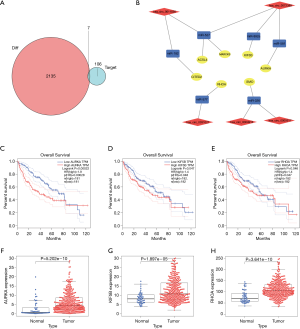
Based on the thresholds of |log2FC| >1 and FDR <0.05, we obtained a total of 2,135 differentially expressed mRNAs (DEGs) from the TCGA-LIHC and GTEx databases (Figure S2). The above results showed that 7 DEGs were involved in the ceRNA network. Then, to further explore the functions of these hub genes in the network in carcinogenesis and the development of HCC, we also established a CircRNA-miRNA-hub gene regulatory network. Firstly, we presented 106 circRNAs-related target genes and 2,135 DEGs in TCGA and GTEx and generated 7 common target genes DEGs (CITED2, ACSL4, MARCKS, KIF5B, AURKA, SMO, and RHOA) using Venn diagram analysis (Figure 6A). Then, Cytoscape was used to construct the circRNA-miRNA-hub gene networks (Figure 6B). Moreover, the Kaplan-Meier curves, which were plotted using the GEPIA database, indicated that higher expression of AURKA, KIF5B, and RHOA were significantly correlated with poorer overall survival (Figure 6C,D,E, P<0.05), however, no significant prognostic significance was shown in CITED2, ACSL4, MARCKS, or SMO. We further verified the expression levels of 7 hub genes using EdgeR software in TCGA and GTEx data and found that an up-regulation of AURKA, KIF5B, and RHOA in tumor tissues compared with normal tissues (Figure 6F,G,H, P<0.001). In addition, elevated expressions of KIF5B and RHOA correlated with the clinical tumor stage (Figure S3, P<0.05), suggesting LIHCs with high KIF5B and RHOA expression are prone to progress to a more advanced stage.

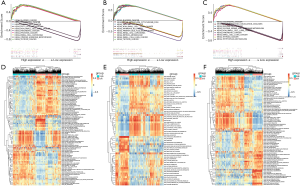
To further study the potential roles of AURKA, KIF5B, and RHOA in HCC, we performed an analysis of GSEA and GSVA on the TCGA-LIHC database. The results of GSEA showed that the hub genes with the highest enrichment score were closely associated with signaling pathways related to cancer (Figure 7A,B,C). Furthermore, GSVA confirmed that many cell cycle-related KEGG pathways, including DNA replication and mismatch repair, were enriched in the groups with high-expression of AURKA, KIF5B, and RHOA, further suggesting that the activation of these hallmark genes might participate in the process of tumor proliferation (Figure 7D,E,F).
Discussion
Over recent decades, studies have emerged, showing the abnormal expression profiles of non-coding RNAs in HCC (19,20). Some investigations have demonstrated circRNA to be closely associated with the occurrence of HCC and its dysfunction to inhibit tumor growth and metastasis (21,22). However, the molecular mechanisms underlying the participation of circRNAs in HCC progression remain elusive.
To explore the molecular regulatory mechanisms of HCC, we studied circRNA microarray profiles and identified 26 DEcircRNAs in HCC tissues and paired normal tissues. Then, qPCR was applied to detect the mRNA expression abundance of the 26 DEcircRNAs in SMMC-7721 cell lines. Finally, hsa_circ_0004913, hsa_circ_0072389, hsa_circ_0000517, hsa_circ_0031027), hsa_circ_0008160, and hsa_circ_0011536 were confirmed by qPCR to be significantly dysregulated in HCC cells. In addition, studies have indicated that circRNAs could act as promising biomarkers for cancer diagnosis (23). Therefore, our findings indicated that circTEX2 (hsa_circ_0004913), circHMGCS1 (hsa_circ_0072389), circRPPH1 (hsa_circ_0000517), circTMCO3 (hsa_circ_0031027), circPIGP (hsa_circ_0008160), and circZMYM4 (hsa_circ_0011536) could represent potentially valuable diagnostic biomarkers for HCC treatment. The previous study demonstrated that circRNAs had been deemed to possess the function of miRNA sponges, which can suppress the binding of miRNAs to target genes, regulating their expression (24). Therefore, we proposed the hypothesis that the circRNA-miRNA-mRNA axis might be the possible molecular regulatory mechanism underlying HCC. In this study, we constructed a ceRNA network based on the above 6 circRNAs and identified 11 DEmiRNAs and 114 mRNAs in this network. Through GO and KEGG pathway analysis, these mRNAs were suggested to participate in many crucial cancer-related biological functions and pathways.
To further identify the crucial circRNAs involving in the network, we screened 7 hub genes, and thereby established the circRNA-miRNA-hub gene network. Among this network, several miRNAs have been identified to play vital roles in the development and progression of HCC, such as miR-577 (25), miR-326 (26), miR-182 (27), miR-892a (28), and miR-581 (29). Moreover, using the TCGA and GTEx databases, we detected the expression of 7 hub genes and found that the expression levels of AURKA, KIF5B, and RHOA were elevated in tumor tissues compared with normal tissues and that higher expression of these genes was dramatically associated with poorer overall survival. Previous studies have shown that circRNAs could regulate host gene expression or work as a miRNA sponge, regulating miRNA-mediated gene expression and thus leading to tumor progression and development (30). In this study, we investigated DEcircRNAs in HCC and then selected the DEcircRNAs that had host genes that were differentially expressed in TCGA-LIHC data and correlated with the prognosis as a candidate circRNAs. Following this, we speculated that these candidate DEcircRNAs (circHMGCS1 and circTMCO3) might participate in the pathogenesis of HCC. In addition, GSEA and GSVA analyses revealed that the functions of AURKA, KIF5B, and RHOA were also closely associated with cancer progression, which indicated that the circRNAs were meaningfully associated with HCC metastasis.
Conclusions
In conclusion, our research revealed circHMGCS1/miR-581/AURKA, circHMGCS1/miR-892a/KIF5B, and circTMCO3/miR-577/RHOA regulatory axes, which could serve as novel biomarkers in the detection of HCC. Finally, bioinformatics analysis indicated that the interaction pairs of the circRNAs mentioned above might be involved in the initiation and development of HCC.
Acknowledgments
Funding: This study was supported by the clinical medical science and technology of Jiangsu Technical Department (BL2014060) and the “Medical Leading Talent and Innovation Team” project (LJ201134) of Jiangsu Province.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2014;20:1829-42. [Crossref] [PubMed]
- Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell 2018;71:428-42. [Crossref] [PubMed]
- Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer 2019;10:5015-21. [Crossref] [PubMed]
- Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol 2018;68:1214-27. [Crossref] [PubMed]
- Huang XY, Huang ZL, Zhang PB, et al. CircRNA-100338 Is Associated With mTOR Signaling Pathway and Poor Prognosis in Hepatocellular Carcinoma. Front Oncol 2019;9:392. [Crossref] [PubMed]
- Li S, Gu H, Huang Y, et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle 2018;17:2349-59. [Crossref] [PubMed]
- Dudekula DB, Panda AC, Grammatikakis I, et al. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 2016;13:34-42. [Crossref] [PubMed]
- Xia S, Feng J, Chen K, et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res 2018;46:D925-9. [Crossref] [PubMed]
- Tan LP, Seinen E, Duns G, et al. A high throughput experimental approach to identify miRNA targets in human cells. Nucleic Acids Res 2009;37:e137. [Crossref] [PubMed]
- Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 2018;46:D296-302. [Crossref] [PubMed]
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015;43:D146-52. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-61. [Crossref] [PubMed]
- Reimand J, Isserlin R, Voisin V, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc 2019;14:482-517. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Klingenberg M, Matsuda A, Diederichs S, et al. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J Hepatol 2017;67:603-18. [Crossref] [PubMed]
- O'Brien A, Zhou T, Tan C, Alpini G, Glaser S. Role of Non-Coding RNAs in the Progression of Liver Cancer: Evidence from Experimental Models. Cancers (Basel) 2019. [Crossref] [PubMed]
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017;66:1151-64. [Crossref] [PubMed]
- Chen Q, Chen Z, Cao S, et al. Role of CircRNAs_100395 in Proliferation and Metastases of Liver Cancer. Med Sci Monit 2019;25:6181-92. [Crossref] [PubMed]
- Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids 2019;16:118-29. [Crossref] [PubMed]
- Xiong DD, Dang YW, Lin P, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med 2018;16:220. [Crossref] [PubMed]
- Wang LY, Li B, Jiang HH, et al. Inhibition effect of miR-577 on hepatocellular carcinoma cell growth via targeting β-catenin. Asian Pac J Trop Med 2015;8:923-9. [Crossref] [PubMed]
- Hu S, Ran Y, Chen W, et al. MicroRNA-326 inhibits cell proliferation and invasion, activating apoptosis in hepatocellular carcinoma by directly targeting LIM and SH3 protein 1. Oncol Rep 2017;38:1569-78. [Crossref] [PubMed]
- Cao MQ, You AB, Zhu XD, et al. miR-182-5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J Hematol Oncol 2018;11:12. [Crossref] [PubMed]
- Jia B, Tan L, Jin Z, et al. MiR-892a Promotes Hepatocellular Carcinoma Cells Proliferation and Invasion Through Targeting CD226. J Cell Biochem 2017;118:1489-96. [Crossref] [PubMed]
- Wang YQ, Ren YF, Song YJ, et al. MicroRNA-581 promotes hepatitis B virus surface antigen expression by targeting Dicer and EDEM1. Carcinogenesis 2014;35:2127-33. [Crossref] [PubMed]
- Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 2018;37:555-65. [Crossref] [PubMed]


