Association between genetic risk scores and risk of narcolepsy: a case-control study
Introduction
Narcolepsy is a life-long neurological disorder characterized by excessive daytime sleepiness, cataplexy, sleep paralysis, hallucinations, and disrupted nocturnal sleep (1). While narcolepsy can manifest at any age, symptoms typically start between 15 and 30 years of age, and the prevalence of narcolepsy ranges from 0.002% to 0.167% worldwide. In order to identify patients at risk for narcolepsy and to facilitate the development of enhanced screening or preventive measures, research has focused on genetic risk loci marked by individual single-nucleotide polymorphisms (SNPs) (2-9). Recently, the genetic risk of narcolepsy has been evaluated based on the HLA-DQB1*06:02 marker, because 10–40% of healthy individuals are also carriers, it represents an important but seemingly imperfect predictor of narcolepsy (10-12). In addition to HLA-DQB1*06:02, many other genes have been implicated in narcolepsy risk in international SNP-based genome-wide association studies (GWASs), including carnitine palmitoyltransferase 1B (CPT1B), choline kinase-β (CHKB), T cell receptor alpha (TCRA), purinergic receptor P2Y, G-protein coupled 11 (P2RY11), tumor necrosis factor superfamily member 4 (TNSF4), cathepsin H (CTSH), T cell receptor beta (TCRB), zinc finger protein 365 (ZNF365), chemokine receptor1 (CCR1), chemokine receptor3 (CCR3), eukaryotic initiation factors 3G (EIF3G), and interleukin 10 receptor, beta (IL10RB) (2-9,13). SNPs conferring narcolepsy susceptibility in the human leukocyte antigen (HLA) and other regions have also been identified (4,14,15).
The studies referenced above have provided a rich compendium of genetic variations potentially underlying narcolepsy and prepared the grounds for building new risk prediction tools. However, the effects of common SNPs identified by GWASs are usually minor and of unclear clinical significance. In order to complement GWASs and uncover potential instances of “missing heritability”, several studies investigated the benefits of conflating genomic risk estimates obtained from SNP genotyping into a genetic risk score (GRS) to predict the risk of diseases such as cancer (16), diabetes (17), schizophrenia (18), coronary artery disease (19), Alzheimer’s disease (20), and leukemia (21). These studies have repeatedly demonstrated significant associations between GRSs and disease risk. As for narcolepsy, although GWASs have been performed, few studies have investigated the aggregate effects of genetic variants on narcolepsy risk in large study samples. We therefore selected SNPs discovered in recently reported narcolepsy susceptibility loci (including variants in European and Asian ancestry reported in previous case-control studies) and investigated the association of these SNPs, along with their related GRSs, with narcolepsy risk in a Chinese population.
Methods
Study site and population
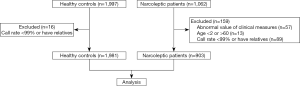
We performed a case-control study comprising 1,062 narcolepsy patients and 1,997 healthy control subjects. The patients were recruited from the sleep laboratory of Peking University People’s Hospital, a unit that receives referrals from all over China. The narcolepsy patients had either hypocretin deficiency (CSF hypocretin-1 ≤110 pg/mL) or clear-cut cataplexy and HLA-DQB1*06:02 (3). Subjects were all Chinese, mostly of Han descent (95%). The majority of study patients were male (68%). The controls were Chinese university employees and students (41% male), and were free of narcolepsy as determined by clinical examination and medical history. Trained interviewers used structured questionnaires to collect information on demographic variables, medical history, and medication. Informed consent (in accordance with governing institutions) was obtained from all subjects. The research protocols were approved by the Institutional Review Board on Medical Human Subjects Research at the Peking University People’s Hospital.
Genotyping and quality control
SNP genotyping and quality control have been described in detail elsewhere (4). In brief, samples were genotyped and analyzed using the Affymetrix Axiom CHB array and the Affymetrix Genotyping Console. All samples used in the current study achieved a call rate of >99%. In total, 903 narcolepsy cases and 1,981 healthy controls were included for further analysis.
Genetic loci selection
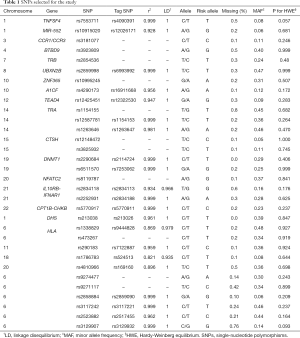
We selected 40 previously discovered narcolepsy risk loci from GWAS studies published before September, 2018 (2,4-6,8,9,14,15,22,23). The narcolepsy susceptibility loci identified in both the Chinese population and European population were selected. The final candidate SNPs were in approximate linkage equilibrium with all pairwise r2<0.25 based on linkage disequilibrium (LD) data from the HapMap Chinese/Japanese combined reference set (CHB/JPT) (24). When a selected variant was not on the Affymetrix Axiom CHB array, an analysis of LD in a 10–20 kb window was performed using Ensembl 1,000 Genomes LD data to identify suitable proxies on the array with an r2≥0.8. SNPs whose call rate was lower than 99% or that violated the Hardy-Weinberg equilibrium (HWE) test were excluded. The final list of candidate SNPs is shown in Table 1. Among the candidate SNPs, 17 included in the GRS were directly genotyped while the remaining SNPs were assessed based on LD with the reported index SNPs with r2≥0.8.
Full table
Statistical analyses
We used the χ2 test to assess whether the SNPs were in HWE and to determine the differences in genotype frequencies between narcolepsy patients and controls. Statistical significance was set to a two-sided P value of less than 0.05. The experiment-wide significance level was derived via Bonferroni correction for the number of lead SNPs.
For the construction of individual GRSs, a genetic score was calculated based on a personal genotype of the SNPs that are associated with narcolepsy, and unweighted GRSs were calculated by adding the number of risk alleles across each SNP. We constructed four different GRSs (the SNPs included in GRSs were shown in Table 2):

Full table
(I) GRS1 excluded the SNPs in the HLA region; (II) GRS2 was limited to the SNPs from the HLA region; (III) GRS3 included all selected SNPs; and (IV) GRS4 only included the SNPs reported in the Chinese population. We considered the GRSs as continuous variables and as tertiles defining low risk, intermediate risk, and high risk classes. Categorical variables were shown in proportions. The associations of the GRSs with narcolepsy risk were evaluated using the logistic regression model adjusted by gender. Odds ratios, 95% CI, and P values were calculated.
We further examined the ability of the four GRSs to discriminate between patients and controls by using receiver-operating characteristics (ROC) curves adjusted for gender. We used Haploview (version 4.2) to calculate the linkage disequilibrium of selected SNPs. The subsequent statistical analyses were performed by using R (version 3.5.0).
Results
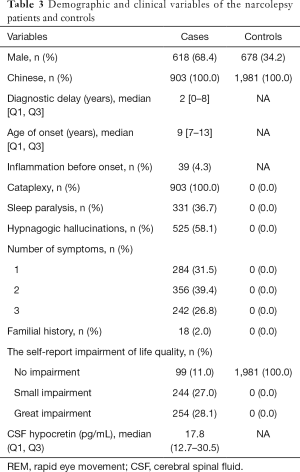
The demographic and clinical data of the narcolepsy patients are presented in Table 3. Among these patients, 860 (95.2%) were of Chinese Han ethnicity, 285 (31.6%) were women, 618 (68.4%) were men, and 18 (2.0%) had a family history of narcolepsy. Among the healthy controls, 678 (34.2%) were men, and 1,303 (65.8%) were women. The flow chart of the inclusion process is shown in Figure 1.
Full table
Association between individual SNP and narcolepsy risk
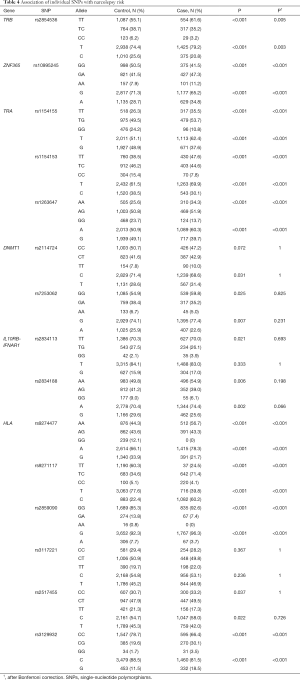
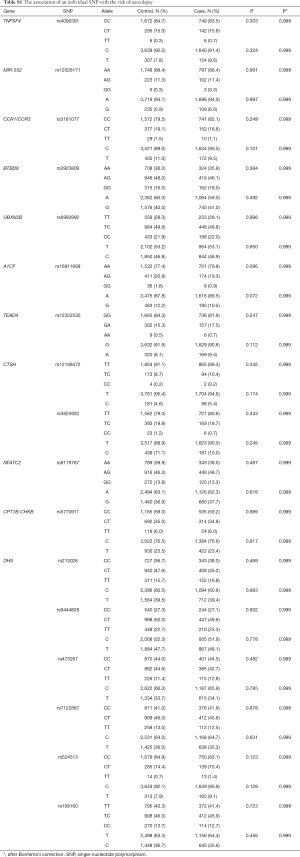
We examined the association between the individual genetic variant and narcolepsy risk. Fourteen loci, including TRB (rs2854536), ZNF365 (rs10995245), DNMT1 (rs2114724, rs7253062), IL10RB-IFNAR1 (rs2834113, rs2834188), TRA (rs1154155, rs1154153, rs1263647), and HLA region (rs9274477, rs9271117, rs2859090, rs3129932, rs2517455) showed significant associations with incident narcolepsy, with P values ranging from <0.001 to 0.005. After the Bonferroni correction, TRB (rs2854536), ZNF365 (rs10995245), TRA (rs1154155, rs1154153, rs1263647), and HLA region (rs9274477, rs9271117, rs2859090, rs3129932) remained statistically significant. The association of each individual SNP with narcolepsy risk is summarized in Tables 4 and S1.
Full table
Full table
Combined genetic effects on narcolepsy risk
We examined the association between the GRSs and narcolepsy risk and observed that the GRSs were significantly associated with narcolepsy risk. Table 5 shows the odds ratios (ORs) for the association between incident narcolepsy and GRSs, comparing the narcolepsy risk of individuals in the low polygenic load categories with the narcolepsy risk of individuals in the intermediate and high polygenic load category. Individuals who carry a higher polygenic load have higher odds of narcolepsy compared with patients carrying a low polygenic load. The OR of narcolepsy risk increased as the genetic load for narcolepsy increased, ranging from an OR of 2.016 (95% CI, 1.657–2.456) to an OR of 4.298 (95% CI, 3.378–5.481). Among the four GRSs, GRS4 (constructed from the narcolepsy susceptibility SNPs identified in the Chinese population) was most closely associated with narcolepsy risk with an OR of 4.298 (95% CI, 3.378–5.481) followed by GRS3 constructed using all narcolepsy-associated SNPs.
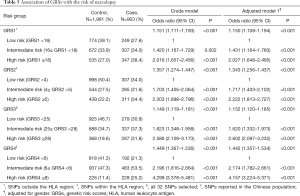
Full table
The discriminatory power of GRS
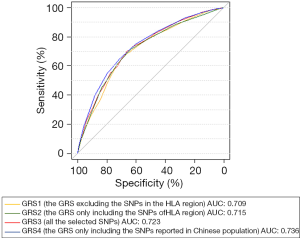
To further refine these results, we examined the discriminatory power of the GRSs for individuals at high risk of narcolepsy by using the ROC curve. The discriminatory ability of the model using GRS1, measured as area under the curve (AUC), was 0.709. The other models using GRS2, GRS3, and GRS4 showed modestly improved discrimination with AUCs of 0.715, 0.723, and 0.736, respectively (Figure 2). These results showed that the GRS constructed from the narcolepsy susceptibility SNPs reported in the Chinese population provided greater discrimination than other GRSs. At a cutoff of 0.335, which was determined according to the maximum of the Youden Index, GRS4 had a sensitivity of 70.9% and a specificity of 65.1% in discriminating narcolepsy patients from healthy controls.
Discussion
The present study reports two main findings: first, we confirmed the significant associations between reported individual SNPs and narcolepsy risk. Second, we found that the GRSs constructed from SNPs that were located in different regions showed significant associations with narcolepsy. Together, these findings buttress the feasibility of discovering genetic biomarkers for disease risk prediction or prognosis.
To the best of our knowledge, the present study is the first detailed and large-scale genetic study of narcolepsy in the Chinese population and the largest study applying the GRS approach to evaluate the polygenic risk of narcolepsy. Major strengths of the present study include the following: (I) we constructed several GRSs from SNPs from different regions and compared their discriminate power; (II) we created GRSs using common narcolepsy susceptibility variants from both Asian and European populations. It is thus likely that our findings apply to multiple ethnicities.
We assessed the association of the reported narcolepsy-related variants at 32 loci in the Chinese ethnic group. The trends observed with 14 narcolepsy-associated loci were consistent with previous studies (2-4,25-27). In particular, nine loci, including TRB, ZNF365, TRA, and SNPs within the HLA region, showed significant association with narcolepsy risk after Bonferroni correction. While these variants were mostly identified in European populations, they each showed association with broader global populations, including groups of Asian ancestry (4,9). On the other hand, we were unable to confirm, in our Chinese sample, the individual associations for several genetic variants previously reported to be associated with narcolepsy risk. This could be due to insufficient statistical power to identify variants with small individual effects or to the diverse genetic backgrounds of a variety of ethnicities.
GRSs have been previously employed to increase statistical power when investigating the association with disease-related phenotypes. Our results confirmed the slight increase in narcolepsy risk carried by genetic variants identified by GWAS and justify the use of GRSs to increase the statistical power to identify these associations. The results were consistent with previous studies indicating that the combined effects of these genetic loci had a more sizable influence on narcolepsy risk (28,29). The significant association between GRSs with narcolepsy indicated that disease risk is not only associated with the variants in the HLA region but that SNPs outside the HLA region also play an important role. Our results did not entirely replicate previous studies that emphasized the importance of HLA genes in narcolepsy (10,12,30). This indicates contribution of genetic factors besides those implicated in the dysfunction of the immune system (9,31-33). GRSs constructed without HLA-DQB1*06:02 could be a promising tool for the risk stratification of HLA-DQB1*06:02-negative populations. Among the four GRSs assembled in our study, GRS4 had the best predictive power for narcolepsy risk, followed by GRS3. Since all SNPs included in GRS4 were identified in the Chinese population, the genetic variation underlying narcolepsy may differ across different ethnicities.
The high predictive power of GRS3 constructed from all narcolepsy-associated SNPs indicates that narcolepsy is a polygenic disease involving many common genetic variants of small effect rather than being driven by rare monogenic mutations (34-36). This finding adds further credence to the concept that GRSs are superior to individual variants in identifying those subgroups at a higher risk of narcolepsy that might require earlier intervention and monitoring.
Our study encountered several limitations. First, larger sample sizes will be needed to identify additional narcoleptic genetic factors that remain unknown. As a second limitation, GRSs were validated only in the Chinese population, although narcolepsy-associated loci are deemed similar across ethnicities, informative SNPs for these loci could vary across ethnicities. In future studies, we will focus on increasing sample sizes to identify additional genetic factors of narcolepsy and on validating GRSs across populations. Moreover, non-genetic factors may contribute to the onset of narcolepsy, and we will thus also explore gene-environment interactions in future studies.
Acknowledgments
We would like to thank all the individuals who provided materials and consent to participate in this study. We thank Prof. Yiqun Wu for advice, Shiying Wang for help with statistics, and all the colleagues who assisted with sample collection.
Funding: National Science Foundation of China (Grant No. 81870996), Research project of central health care special fund (Grant No. W2017BJ52).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Informed consent (in accordance with governing institutions) was obtained from all subjects. The research protocols were approved by the Institutional Review Board on Medical Human Subjects Research at the Peking University People’s Hospital.
References
- Dauvilliers Y, Arnulf I, Mignot E. Narcolepsy with cataplexy. Lancet 2007;369:499-511. [Crossref] [PubMed]
- Faraco J, Lin L, Kornum BR, et al. ImmunoChip study implicates antigen presentation to T cells in narcolepsy. PLoS Genet 2013;9:e1003270. [Crossref] [PubMed]
- Hallmayer J, Faraco J, Lin L, et al. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat Genet 2009;41:708-11. [Crossref] [PubMed]
- Han F, Faraco J, Dong XS, et al. Genome wide analysis of narcolepsy in China implicates novel immune loci and reveals changes in association prior to versus after the 2009 H1N1 influenza pandemic. PLoS Genet 2013;9:e1003880. [Crossref] [PubMed]
- Poujois A, Woimant F. Challenges in the diagnosis of Wilson disease. Ann Transl Med 2019;7S67.
- Toyoda H, Miyagawa T, Koike A, et al. A polymorphism in CCR1/CCR3 is associated with narcolepsy. Brain Behav Immun 2015;49:148-55. [Crossref] [PubMed]
- Luca G, Haba-Rubio J, Dauvilliers Y, et al. Clinical, polysomnographic and genome-wide association analyses of narcolepsy with cataplexy: a European Narcolepsy Network study. J Sleep Res 2013;22:482-95. [Crossref] [PubMed]
- Holm A, Lin L, Faraco J, et al. EIF3G is associated with narcolepsy across ethnicities. Eur J Hum Genet 2015;23:1573-80. [Crossref] [PubMed]
- Miyagawa T, Kawashima M, Nishida N, et al. Variant between CPT1B and CHKB associated with susceptibility to narcolepsy. Nat Genet 2008;40:1324-8. [Crossref] [PubMed]
- Mignot E, Lin L, Rogers W, et al. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am J Hum Genet 2001;68:686-99. [Crossref] [PubMed]
- Han F, Lin L, Schormair B, et al. HLA DQB1*06:02 negative narcolepsy with hypocretin/orexin deficiency. Sleep 2014;37:1601-8. [Crossref] [PubMed]
- Yamasaki M, Miyagawa T, Toyoda H, et al. Evaluation of polygenic risks for narcolepsy and essential hypersomnia. J Hum Genet 2016;61:873-8. [Crossref] [PubMed]
- Bernardini C, Lattanzi W, Bosco P, et al. Genome-wide gene expression profiling of human narcolepsy. Gene Expr 2012;15:171-81. [Crossref] [PubMed]
- Tafti M, Hor H, Dauvilliers Y, et al. DQB1 locus alone explains most of the risk and protection in narcolepsy with cataplexy in Europe. Sleep 2014;37:19-25. [Crossref] [PubMed]
- Hor H, Kutalik Z, Dauvilliers Y, et al. Genome-wide association study identifies new HLA class II haplotypes strongly protective against narcolepsy. Nat Genet 2010;42:786-9. [Crossref] [PubMed]
- Allman R, Dite GS, Hopper JL, et al. SNPs and breast cancer risk prediction for African American and Hispanic women. Breast Cancer Res Treat 2015;154:583-9. [Crossref] [PubMed]
- Dauriz M, Porneala BC, Guo X, et al. Association of a 62 Variants Type 2 Diabetes Genetic Risk Score With Markers of Subclinical Atherosclerosis: A Transethnic, Multicenter Study. Circ Cardiovasc Genet 2015;8:507-15. [Crossref] [PubMed]
- French L, Gray C, Leonard G, et al. Early Cannabis Use, Polygenic Risk Score for Schizophrenia and Brain Maturation in Adolescence. JAMA Psychiatry 2015;72:1002-11. [Crossref] [PubMed]
- Gui L, Wu F, Han X, et al. A multilocus genetic risk score predicts coronary heart disease risk in a Chinese Han population. Atherosclerosis 2014;237:480-5. [Crossref] [PubMed]
- Escott-Price V, Shoai M, Pither R, et al. Polygenic score prediction captures nearly all common genetic risk for Alzheimer's disease. Neurobiol Aging 2017;49:214.e7-11. [Crossref] [PubMed]
- Abelson S, Collord G, Ng SWK, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 2018;559:400-4. [Crossref] [PubMed]
- Han F, Lin L, Li J, et al. TCRA, P2RY11, and CPT1B/CHKB associations in Chinese narcolepsy. Sleep Med 2012;13:269-72. [Crossref] [PubMed]
- Shimada M, Miyagawa T, Kawashima M, et al. An approach based on a genome-wide association study reveals candidate loci for narcolepsy. Hum Genet 2010;128:433-41. [Crossref] [PubMed]
- Johnson AD, Handsaker RE, Pulit SL, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008;24:2938-9. [Crossref] [PubMed]
- Thebault S, Vincent A, Gringras P. Narcolepsy and H1N1 vaccination: a link? Curr Opin Pulm Med 2013;19:587-93. [Crossref] [PubMed]
- Liblau RS, Vassalli A, Seifinejad A, et al. Hypocretin (orexin) biology and the pathophysiology of narcolepsy with cataplexy. Lancet Neurol 2015;14:318-28. [Crossref] [PubMed]
- Lecendreux M. Pharmacological management of narcolepsy and cataplexy in pediatric patients. Paediatr Drugs 2014;16:363-72. [Crossref] [PubMed]
- Kraft P, Hunter DJ. Genetic risk prediction--are we there yet? N Engl J Med 2009;360:1701-3. [Crossref] [PubMed]
- Qi L, Parast L, Cai T, et al. Genetic susceptibility to coronary heart disease in type 2 diabetes: 3 independent studies. J Am Coll Cardiol 2011;58:2675-82. [Crossref] [PubMed]
- Capittini C, De Silvestri A, Terzaghi M, et al. Correlation between HLA-DQB1*06:02 and narcolepsy with and without cataplexy: approving a safe and sensitive genetic test in four major ethnic groups. A systematic meta-analysis. Sleep Med 2018;52:150-7. [Crossref] [PubMed]
- Dauvilliers Y, Neidhart E, Lecendreux M, et al. MAO-A and COMT polymorphisms and gene effects in narcolepsy. Mol Psychiatry 2001;6:367-72. [Crossref] [PubMed]
- Kernohan KD, Cigana Schenkel L, Huang L, et al. Identification of a methylation profile for DNMT1-associated autosomal dominant cerebellar ataxia, deafness, and narcolepsy. Clin Epigenetics 2016;8:91. [Crossref] [PubMed]
- Moghadam KK, Pizza F, Tonon C, et al. Polysomnographic and neurometabolic features may mark preclinical autosomal dominant cerebellar ataxia, deafness, and narcolepsy due to a mutation in the DNA (cytosine-5-)-methyltransferase gene, DNMT1. Sleep Med 2014;15:582-5. [Crossref] [PubMed]
- Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet 2012;13:135-45. [Crossref] [PubMed]
- Golan D, Lander ES, Rosset S. Measuring missing heritability: inferring the contribution of common variants. Proc Natl Acad Sci U S A 2014;111:E5272-81. [Crossref] [PubMed]
- Fuchsberger C, Flannick J, Teslovich TM, et al. The genetic architecture of type 2 diabetes. Nature 2016;536:41-7. [Crossref] [PubMed]